Abstract
Frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) is an important cause of dementia in individuals under age 65. Common variants in the TMEM106B gene were previously discovered by genome-wide association to confer genetic risk for FTLD-TDP (p = 1 × 10− 11, OR = 1.6). Furthermore, TMEM106B may act as a genetic modifier affecting age at onset and age at death in the Mendelian subgoup of FTLD-TDP due to expansions of the C9orf72 gene. Evidence suggests that TMEM106B variants increase risk for developing FTLD-TDP by increasing expression of Transmembrane Protein 106B (TMEM106B), a lysosomal protein. To further understand the functional role of TMEM106B in disease pathogenesis, we investigated the cell biological effects of increased TMEM106B expression. Here, we report that increased TMEM106B expression results in the appearance of a vacuolar phenotype in multiple cell types, including neurons. Concomitant with the development of this vacuolar phenotype, cells over-expressing TMEM106B exhibit impaired lysosomal acidification and degradative function, as well as increased cytotoxicity. We further identify a potential lysosomal sorting motif for TMEM106B and demonstrate that abrogation of sorting to lysosomes rescues TMEM106B-induced defects. Finally, we show that TMEM106B-induced defects are dependent on the presence of C9orf72, as knockdown of C9orf72 also rescues these defects. In sum, our results suggest that TMEM106B exerts its effects on FTLD-TDP disease risk through alterations in lysosomal pathways. Furthermore, TMEM106B and C9orf72 may interact in FTLD-TDP pathophysiology.
Introduction
Frontotemporal lobar degeneration (FTLD) is a leading cause of presenile dementia (1,2). The most common neuropathological subtype of disease, FTLD-TDP, is characterized by inclusions of TAR DNA-binding protein of 43 kDa (TDP-43) (3). Two of the major Mendelian causes of FTLD-TDP have been identified as (i) non-coding hexanucleotide repeat expansions in C9orf72 (4,5) and (ii) haploinsufficiency mutations in GRN, which encodes the growth factor progranulin (6–8). In addition, a recent genome-wide association study (9) revealed multiple common variants in the largely uncharacterized gene TMEM106B that significantly associated with FTLD-TDP (p = 1 × 10−11, OR = 1.6).
In addition to acting as a genetic risk factor for FTLD-TDP, TMEM106B has also been shown to act as a genetic modifier in both GRN mutation-associated FTLD-TDP (10) and C9orf72 expansion-associated FTLD-TDP (11,12), affecting age at onset of disease and age at death. Moreover, while genotypes at TMEM106B do not appear to confer risk for development of amyotrophic lateral sclerosis (ALS), another disease defined by TDP-43 proteinopathy (13), TMEM106B variants associated with increased FTLD-TDP risk correlate with development of dementia in ALS (14). Most recently, TMEM106B variants associated with increased FTLD-TDP risk have been reported to correlate with increased burden of TDP-43 proteinopathy in aged individuals without overt clinical FTLD (15).
Since its initial discovery as an FTLD-TDP risk factor, TMEM106B has been characterized as a Type II transmembrane protein localized to late endosomes/lysosomes (16–18), with widespread expression in human brain (19). Recent data suggests that TMEM106B is involved in lysosomal transport in neurons, with knockdown of TMEM106B resulting in increased retrograde lysosomal transport in one report (17) and increased bidirectional transport in another report (16). TMEM106B has also been demonstrated to affect lysosomal size, acidification and degradative capacity in immortalized cell lines (16,20,21), and lysosomal size and number in neurons (16).
While functional characterization of FTLD-TDP-associated genetic variants at TMEM106B remains incomplete, we and others have demonstrated that TMEM106B genotypes associated with disease also correlate with increased TMEM106B expression (9,15,21,22). To further understand the contribution of TMEM106B to FTLD-TDP disease pathogenesis, we investigated the cell biological effects of disease-associated increases in TMEM106B expression.
Results
Increased expression of TMEM106B results in altered endolysosomal morphology
We (21) and others (20) have previously demonstrated that increased expression of TMEM106B results in enlargement of organelles positive for the late endosomal/lysosomal marker LAMP1 (Lysosomal-Associated Membrane Protein 1) in immortalized cells. By live cell imaging, we confirmed this effect of increased TMEM106B expression in HeLa cells, using a previously described GFP-tagged TMEM106B construct (20). These enlarged organelles were readily visible by brightfield imaging and did not occur with increased expression of GFP-tagged LAMP1, another transmembrane late endosomal/lysosomal protein (Fig. 1a).
Figure 1.
Increased expression of TMEM106B results in a vacuolar phenotype. (a) Live image of HeLa cells transfected with GFP-TMEM106B or GFP-LAMP1. Expression of TMEM106B resulted in the appearance of enlarged vacuolar structures (left) visible by fluorescence or brightfield microscopy. This phenotype was not observed upon expression of GFP-LAMP1, another transmembrane lysosomal protein (right). (b) In primary mouse hippocampal neurons nucleofected with GFP-TMEM106B, the enlarged vacuolar structures demonstrate co-localization of TMEM106B (green) and the lysosomal marker LAMP1 (red). The two right panels show the merged images, which are shown in monochrome in the first and second panels. Scale bar for images excluding magnified right panel = 10 µm. (c) Primary mouse hippocampal neurons nucleofected with GFP-TMEM106B (left) exhibit enlarged >2–3 µm vacuolar structures, whereas neurons nucleofected with GFP-LAMP1 (right) do not. Scale bar = 10 µm. (d) The diameter of LAMP1+ organelles in TMEM106B over-expressing neurons is significantly larger than that of neighboring neurons not over-expressing TMEM106B. p < 0.001 for four replicate experiments.
We next extended our investigations of TMEM106B over-expression to neurons. As shown in Figure 1b, primary mouse hippocampal neurons over-expressing TMEM106B also exhibited multiple, enlarged vacuolar structures >2–3 μm in size concentrated in the cell body and dispersed along the cell processes as well. In contrast, over-expression of LAMP1 in neurons did not result in the appearance of these enlarged vacuolar structures (Fig. 1c). These enlarged vacuoles appeared within a day post-nucleofection with TMEM106B and were observed in both hippocampal and cortical neurons. Mirroring our prior results in immortalized cell lines (21), these vacuoles were positive for both TMEM106B itself and for LAMP1, which co-localized with TMEM106B at the limiting membrane of these vacuoles (Fig. 1b). Indeed, the average size of LAMP1+ organelles in neurons over-expressing TMEM106B was 50% larger than a control non-over-expressing condition (Fig. 1d). Moreover, vacuoles were negative for EEA-1, a marker of early endosomes, and SV2, a marker of synaptic vesicles; neither of these markers demonstrated strong co-localization with TMEM106B (Supplementary Material, Fig. 1).
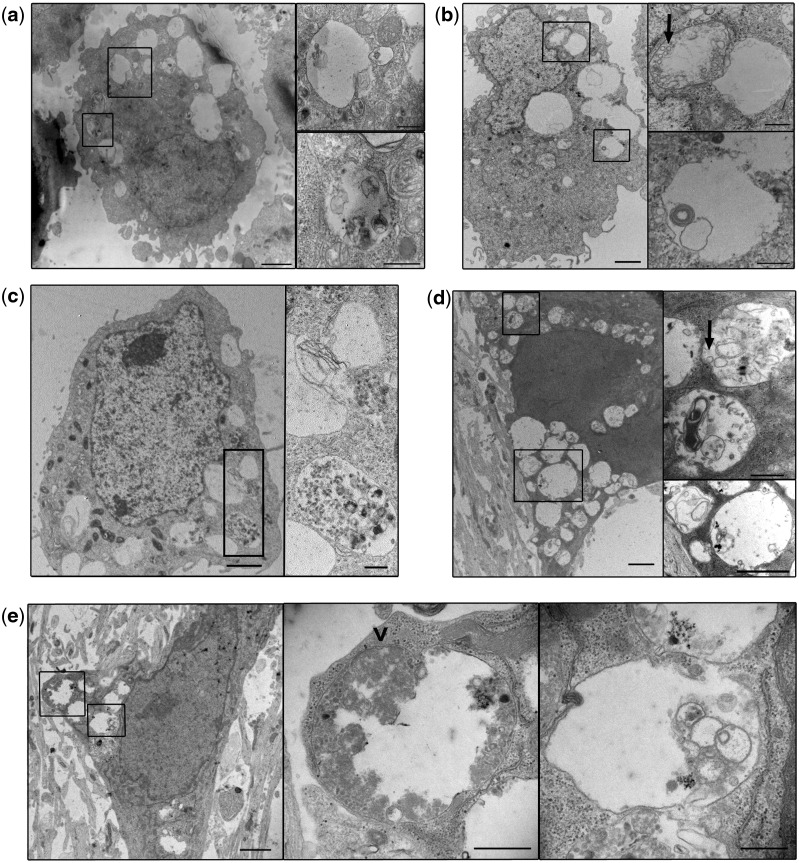
To better characterize these enlarged LAMP1+ TMEM106B+ organelles, we performed ultrastructural analyses by electron microscopy. Exogenous expression of empty vector, GFP or LAMP1 resulted in no notable changes in ultrastructure (Supplementary Material, Fig. 2a). However, in HeLa cells (Fig. 2a and Supplementary Material, Fig. 2b), COS-7 cells (Fig. 2b), HEK293 cells (Fig. 2c) and DIV7 mouse hippocampal neurons (Fig. 2d and e), expression of TMEM106B resulted in the appearance of a striking vacuolar phenotype. Specifically, TMEM106B over-expressing cells contained multiple enlarged, electron-lucent, single-membrane-delimited cytoplasmic organelles >2 μm in diameter. These enlarged organelles often appeared largely empty, although at times they contained multilamellar structures and cytoplasmic material or organelles in varying states of degradation. Because of the “empty” appearance of some of these vacuoles, we investigated the possibility that they could be lipid droplets. However, immunofluorescence microscopy demonstrated that these enlarged vacuoles were negative for BODIPY 493/503, which labels neutral lipids (Supplementary Material, Fig. 2c).
Figure 2.
Ultrastructural characterization of TMEM106B-induced vacuolar phenotype. (a) HeLa cells transfected with TMEM106B exhibit multiple large electron-lucent, single-membraned organelles. These organelles often contain cytosolic components in varying states of degradation and multilamellar structures (see insets), an ultrastructural phenotype consistent with late autophagic vacuoles (autolysosomes or amphisomes). (b) COS-7 cells transfected with TMEM106B also display a similar phenotype, with intraluminal vesicles (ILVs, arrow) and multilamellar structures within the enlarged vacuoles (see insets). (c) HEK293 cells transfected with TMEM106B also display the enlarged vacuoles, some containing components in varying states of degradation (see inset). (d, e) Primary mouse hippocampal neurons nucleofected with TMEM106B display the same ultrastructural phenotype. The organelle depicted in the middle panel of (e) demonstrates a small area of still visible double membrane (arrowhead), consistent with identification as a late autophagic vacuole (double membrane-autophagosome fusing with a lysosome, resulting in degradation of the inner membrane). Occasional internal ILVs are similarly noted as well (arrow in top inset for (d)). In all panels, right panels show insets of the lower-power view in each set of images. Scale bars = 2 µm for the lower-power view, and 0.5 µm for the insets.
We next quantified this ultrastructural phenotype in both HeLas and primary neurons. Because TMEM106B was over-expressed in cells by transient transfection or nucleofection, not all cells over-expressed TMEM106B. Thus, we enriched for TMEM106B-expressing cells by expressing GFP-TMEM106B and sorting into GFP-positive and GFP-negative populations using flow cytometry (Supplemementary Material, Fig. 3). In GFP-positive TMEM106B-transfected HeLa cells, 26% (26/100) exhibited enlarged vacuoles >1 µm in diameter. In the corresponding GFP-negative population (cells which presumably express little or no TMEM106B), only 4% (4/100) displayed this phenotype (p < 0.001, Fisher exact test). As GFP sorting of nucleofected neurons was complicated by cell death, we could not similarly confine our quantitative analyses to neurons with confirmed GFP-TMEM106B expression. However, 22% (11/50) of the total primary hippocampal neurons nucleofected with TMEM106B exhibited the enlarged organelles, whereas only 2% (1/50) of GFP-LAMP1-nucleofected neurons did (p = 0.004, Fisher exact test).
Increased expression of TMEM106B results in multiple lysosomal abnormalities
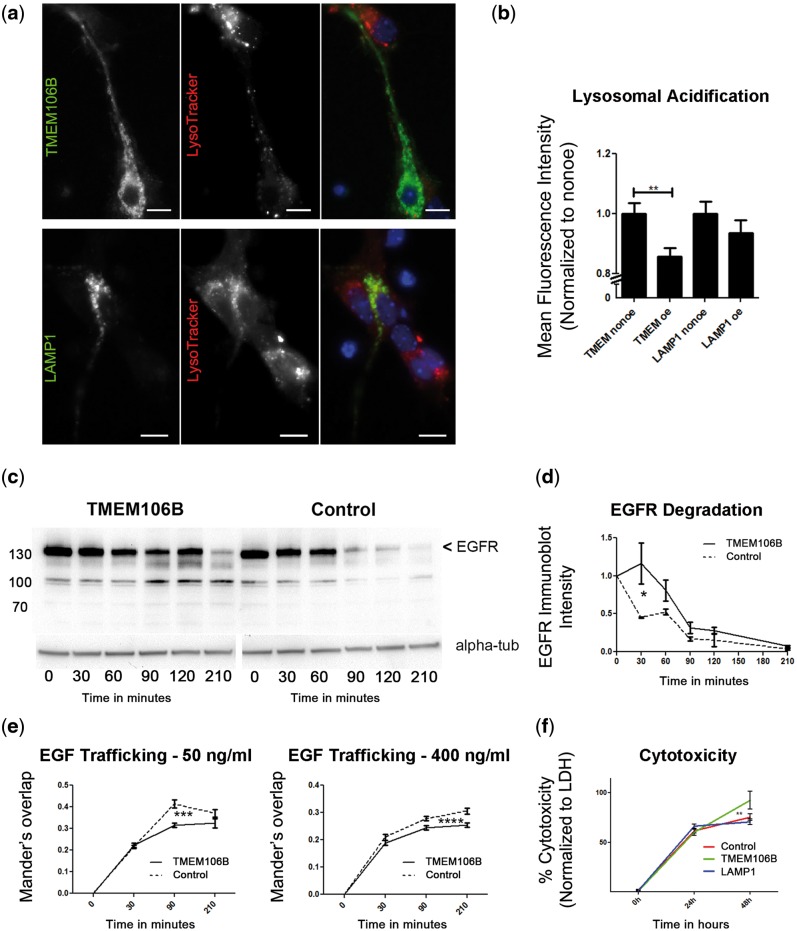
Having observed that TMEM106B over-expression results in a striking vacuolar phenotype, we next asked what the functional consequences of this phenotype might be. Previous data in immortalized cell lines has demonstrated that TMEM106B over-expression results in impaired acidification of the lysosomal compartment as well as compromised lysosomal degradative capability (20,21). To expand this to more disease-relevant cell types, we over-expressed TMEM106B in primary mouse hippocampal neurons and assessed lysosomal acidification with the pH-sensitive dye, LysoTracker. As shown in Figure 3a, TMEM106B over-expressing neurons showed visibly decreased mean fluorescence intensity (MFI) of LysoTracker compared with non-over-expressing neurons, suggesting that elevated levels of TMEM106B impaired acidification. Indeed, quantification of the LysoTracker MFI for TMEM106B over-expressing neurons revealed a significant decrease compared with neighboring non-over-expressing neurons (14.23 ± 3.45% decrease, p = 0.002). In contrast, LAMP1 over-expression did not result in significant changes in Lysotracker MFI comparing LAMP1 over-expressing cells with neighboring non-over-expressers (Fig. 3b).
Figure 3.
Increased expression of TMEM106B results in lysosomal dysfunction. (a) Expression of GFP-TMEM106B (top three panels) in DIV4 primary mouse hippocampal neurons results in an apparent decrease in intensity of LysoTracker, a pH-sensitive dye that fluoresces intensely at low pH and weakly at higher pH. Expression of GFP-LAMP1 (bottom three panels) does not affect LysoTracker intensity. In each set of three panels, the right-most panel shows the merged images, with LysoTracker in red, and TMEM106B or LAMP1 in green. Cells over-expressing TMEM106B or LAMP1 fluoresce brightly in green compared to non-over-expressing neighbors. (b) Quantification of lysosomal acidification data combined from seven replicates performed on 4 days in primary neurons. GFP-TMEM106B over-expressing neurons demonstrate a significant decrease in MFI of LysoTracker, compared with neighboring non-over-expressers (Mann–Whitney p=0.002). Over-expression of GFP-LAMP1 does not significantly affect LysoTracker MFI. (c, d) Addition of EGF in the presence of cycloheximide results in rapid EGFR lysosomal degradation in vector-transfected cells (right immunoblot). EGFR degradation in TMEM106B-transfected cells, however, was impaired (left immunoblot). EGFR is indicated by the arrowhead; other bands are non-specific. Shown are representative immunoblots of EGFR and an alpha-tubulin loading control (alpha-tub) (c) and quantification of four replicates (d) (two-way ANOVA p = 0.011). (e) The endolysosomal trafficking of EGF to LAMP1+ organelles is delayed in TMEM106B over-expressing cells, as demonstrated by decreased co-localization between EGF and LAMP1 under TMEM106B over-expressing conditions, compared with neighboring control cells. Two different concentrations of EGF were tested, as the lower concentration (left) is internalized via EGFR-mediated endocytosis, while the higher concentration (right) may also utilize other internalization mechanisms. Co-localization was quantified by Mander’s overlap (two-way ANOVA p = 0.001 for 50 ng/ml EGF, p < 0.0001 for 400 ng/ml EGF). (f) Increased expression of TMEM106B results in cytotoxicity. Cytotoxicity was quantified in HeLa cells over-expressing TMEM106B or controls (LAMP1, 5TO vector) using measurements of LDH release (which accompanies loss of cell membrane integrity); data are combined for 18 replicates performed on 3 days. Over a 48-h time-course, TMEM106B over-expression (green) resulted in significantly greater cytotoxicity than over-expression of LAMP1 (blue, two way ANOVA p < 0.001) or 5TO vector control (5TO in red, two-way ANOVA p = 0.026).
The loss of lysosomal acidification observed here in neurons and previously described in other cell types has consequences for lysosomal degradative function. Corroborating the reports of others (20), we found that over-expression of TMEM106B results in impaired lysosomal degradation of the epidermal growth factor receptor (EGFR) (Fig. 3c). Under normal conditions, EGFR rapidly targets to the lysosome for degradation upon internalization via endocytosis of its ligand, epidermal growth factor (EGF) (23,24). Quantification of EGFR degradation demonstrated a significant delay in EGFR degradation in the context of increased TMEM106B expression versus vector-expressing control (two-way ANOVA p = 0.011, Fig. 3d).
The delay in EGFR degradation upon conditions of increased TMEM106B expression could be solely because of inefficient lysosomal degradation. Alternatively, trafficking defects induced by increasing TMEM106B expression could also be playing a role. To assess these possibilities, we evaluated the trafficking of fluorescently-labeled EGF from introduction into the cell culture medium to internalization in a LAMP1+ vesicle. As shown in Figure 3e, TMEM106B over-expression resulted in delayed delivery of EGF to LAMP1+ vesicles at both higher and lower concentrations of EGF (two-way ANOVA p = 0.001 for 50 ng/ml EGF, p < 0.001 for 400 ng/ml EGF).
These endolysosomal disturbances appear to be associated with decreased cell survival, since we observed that HeLa cells expressing TMEM106B “disappeared” from culture ∼48 h post-transfection, whereas LAMP1-expressing cells remained readily apparent (Supplementary Material, Fig. 4a). To further quantify these observations, we assessed cytotoxicity in several ways. First, we measured lactate dehydrogenase (LDH) release from cultured cells, which occurs when cell membranes are compromised. After transient transfection with TMEM106B or control constructs, we assessed cytotoxicity at multiple timepoints. Compared with both vector control and LAMP1 over-expressing cells, cells over-expressing TMEM106B exhibited significantly higher cytotoxicity at 48 h post-transfection (two-way ANOVA p = 0.026 compared with vector control, p = 0.010 compared with LAMP1, Fig. 3f), suggesting that TMEM106B over-expression is toxic to cells. Second, because we were concerned that LDH release could also result from our transfection methods, we performed Trypan Blue cell counts to quantitate non-viable cells in each condition. These studies corroborated the cytotoxic effect of increased TMEM106B expression (Supplementary Material, Fig. 4b). Finally, in order to investigate the contribution of apoptotic cell death mechanisms to the cytotoxicity observed with TMEM106B expression, we performed TUNEL staining. Corroborating the results of others (20), we did not see a difference in numbers of cells undergoing apoptosis in conditions with versus without TMEM106B over-expression, with very few TUNEL+ cells in both conditions. (Supplementary Material, Fig. 4c).
Taken together, our data demonstrate that increased expression of TMEM106B in multiple cell types, including neurons, results in prominent morphological alterations in LAMP1+ organelles such as late endosomes, lysosomes, or autolysosomes. Moroever, these alterations affect trafficking to lysosomes, lysosomal acidification and degradative function, ultimately resulting in cytotoxicity.
The cell biological effects of increased TMEM106B expression are dependent on localization of TMEM106B to lysosomes
The preceding experiments demonstrate that increased expression of TMEM106B results in changes in LAMP1+ organelles such as late endosomes, lysosomes, amphisomes or autolysosomes, with concomitant cytotoxic effects. Furthermore, these effects are relatively specific to TMEM106B, since they are not seen upon over-expression of another lysosomal protein, LAMP1, or a control protein, GFP. We next asked whether the localization of TMEM106B to lysosomes is necessary for these changes to occur.
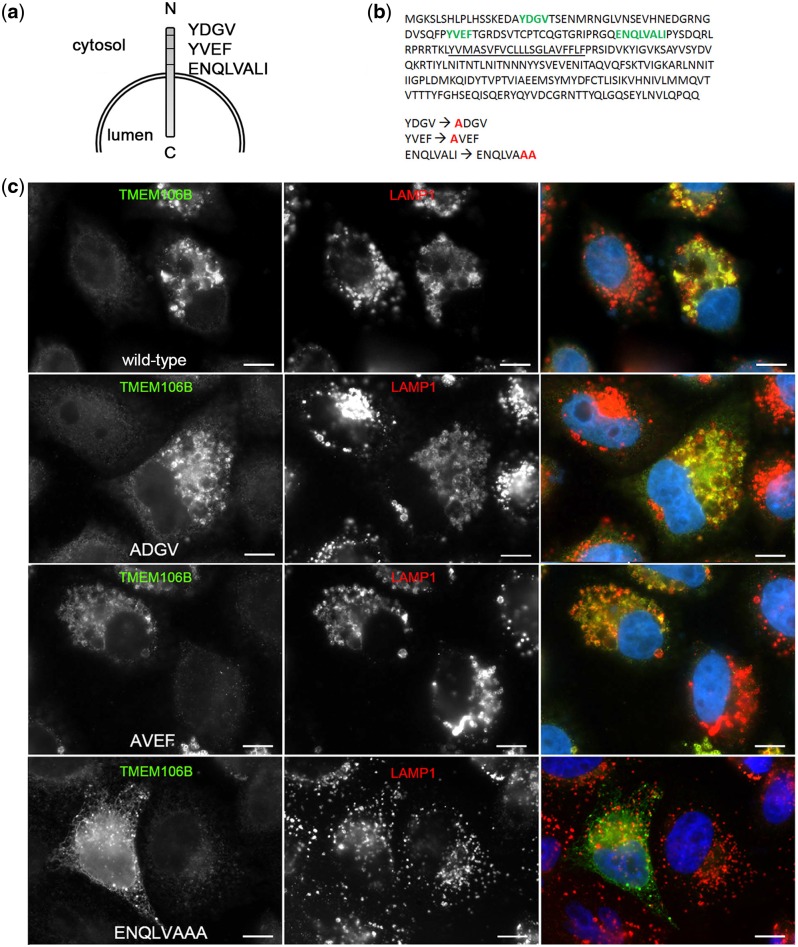
To answer this question, we first had to determine, and then abrogate, potential lysosomal sorting motifs for TMEM106B. Inspection of the cytosolic, N-terminal domain of TMEM106B revealed three potential canonical lysosomal sorting motifs (Fig. 4a)—two tyrosine-based (YDGV, YVEF) and one extended dileucine-based motif (ENQLVALI), fitting the tyrosine consensus sequence YXX(ϕ) and dileucine motif [DE]XXXL[LI], respectively (25). We therefore used site-directed mutagenesis to individually mutate each potential motif. The potential lysosomal sorting motifs are identified in Figure 4b along with the mutations introduced: YDGV → ADGV, YVEF → AVEF and ENQLVALI → ENQLVAAA. We assessed the subcellular localization of each of these motif mutants by immunofluorescence microscopy.
Figure 4.
ENQLVALI is a potential lysosomal sorting motif for TMEM106B. (a) Three potential classical lysosomal targeting motifs—two tyrosine motifs and one isoleucine/dileucine motif—were identified in the N-terminal domain of TMEM106B. (b) The primary amino acid sequence of TMEM106B, with potential lysosomal targeting motifs depicted in green, is shown. Specific residues were individually mutated to alanine residues, with mutated residues indicated in red. (c) Double-label immunofluorescence microscopy demonstrates that wild-type TMEM106B (top row), ADGV-TMEM106B (second row) and AVEF-TMEM106B (third row) continue to co-localize strongly with LAMP1. In contrast, ENQLVAAA-TMEM106B (bottom row) appears diffusely throughout the cytoplasm of HeLa cells and exhibits decreased co-localization with LAMP1. For all constructs, the right-most panel shows merged channels for TMEM106B (green) and LAMP1 (red), with individual channels shown in monochrome in the left and middle panels. Scale bars = 10 µm. TMEM106B is detected by N2077 antibody.
While the constructs containing mutated tyrosine-based motifs (YDGV → ADGV, YVEF → AVEF) continued to localize to lysosomes, mutation of the leucine and isoleucine residues within the extended dileucine-based motif abrogated lysosomal localization of TMEM106B, as demonstrated by decreased co-localization with LAMP1 (Fig. 4c). Instead, in the ENQLVAAA-TMEM106B mutant, TMEM106B expression was more diffusely cytoplasmic. In addition, we verified that the ENQLVAAA-TMEM106B mutant was not retained in the endoplasmic reticulum (ER) by confirming cell surface expression under non-permeabilized immunofluorescence microscopy conditions (Supplementary Material, Fig. 5a) and by Endoglycosidase H (EndoH) and Peptide-N-Glycosidase F (PNGaseF) deglycosylation experiments (Supplementary Material, Fig. 5b). Specifically, both wild-type TMEM106B and ENQLVAAA-TMEM106B showed similar EndoH-resistant bands ∼35 kDa by immunoblot, demonstrating that ENQLVAAA-TMEM106B achieves complex glycosylation in the Golgi and is not retained in the ER. This 35 kDa band fully collapsed down to TMEM106B’s predicted 31 kDa size when deglycosylated with PNGaseF for both ENQLVAAA-TMEM106B and wild-type TMEM106B.
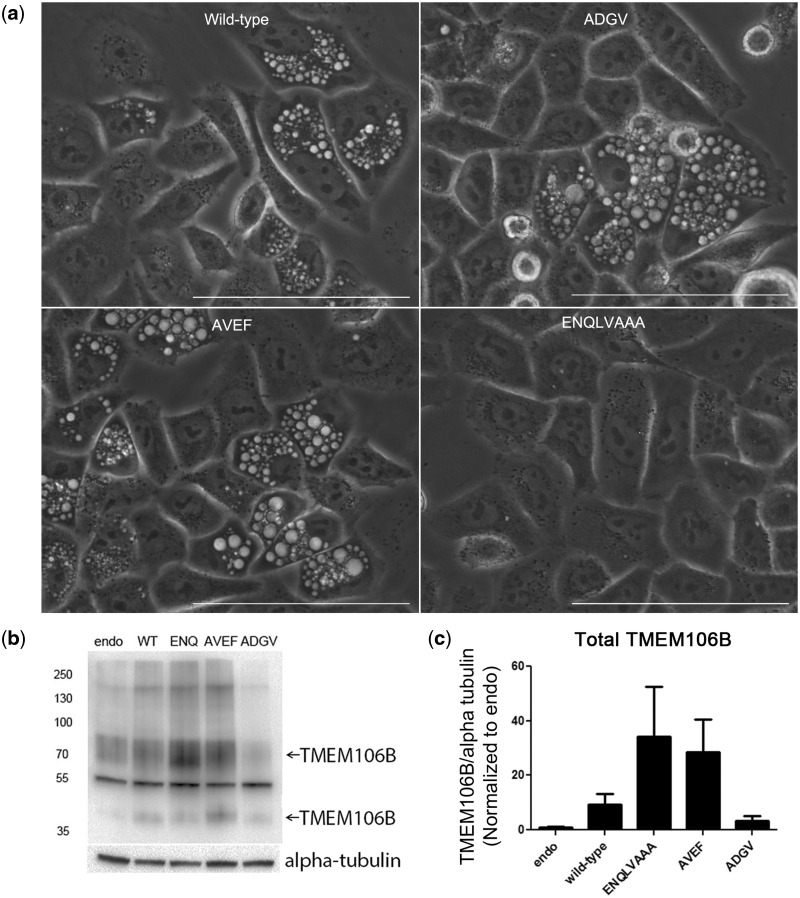
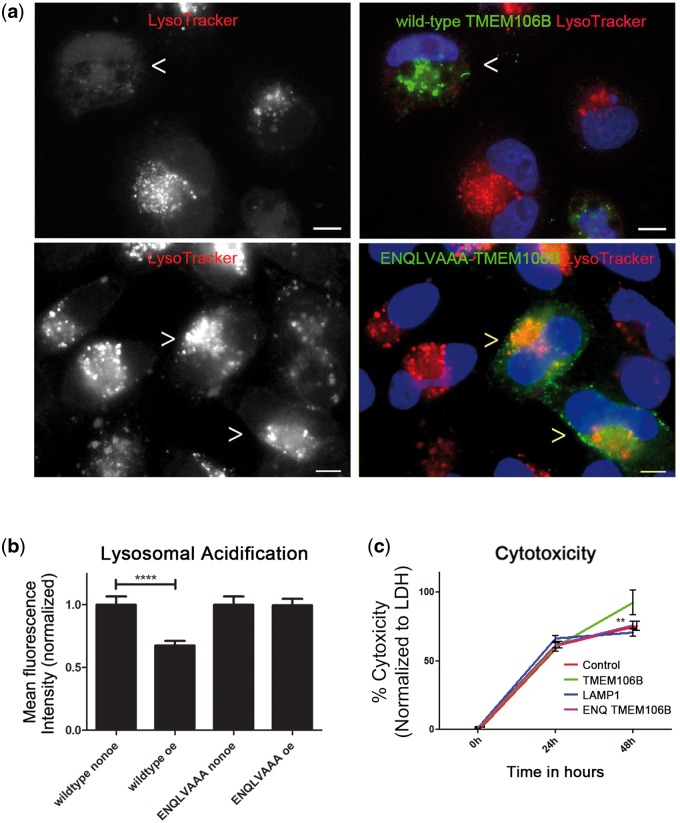
Accompanying the abrogation of lysosomal localization, ENQLVAAA-TMEM106B also demonstrated a striking loss of the vacuolar phenotype (Figs. 4c and 5a). In fact, on brightfield microscopy, cells expressing ENQLVAAA-TMEM106B showed virtually no vacuoles (Fig. 5a). We considered the possibility that the loss of phenotype might be because of lower expression levels of this construct. We found, however, that, whereas our wild-type TMEM106B construct demonstrated over-expression levels 2-10X over baseline, the ENQLVAAA-TMEM106B mutant construct showed much higher expression (Fig. 5b and c).
Figure 5.
The vacuolar phenotype induced by TMEM106B expression depends on proper localization of TMEM106B to lysosomes. (a) Live imaging of HeLas over-expressing either wild-type TMEM106B or lysosomal motif mutants by bright-field microscopy demonstrates loss of the vacuolar phenotype in cells expressing the ENQLVAAA-TMEM106B mutant. In contrast, cytoplasmic vacuoles ranging in size were readily seen in cells expressing wild-type, ADGV-, or AVEF-TMEM106B. Scale bar = 100 µm. (b, c) Lysosomal motif mutants all exhibit expression levels that are comparable to, or higher than, wild-type TMEM106B, with the ENQLVAAA-TMEM106B construct expressing at the highest levels in HeLa cells. Endo=endogenous TMEM106B, WT = wild-type TMEM106B, ENQ = ENQLVAAA-TMEM106B, AVEF = AVEF-TMEM106B, and ADGV = ADGV-TMEM106B. In the example immunoblot (b), two bands (arrows) for TMEM106B at 70 kDa (dimer) and 40 kDa (monomer) are detected (N2077 antibody), and alpha-tubulin is shown as a loading control. Quantification of TMEM106B protein expression levels for four replicate experiments is shown in (c), normalized in each blot to the endogenous condition (mean ± SEM).
Given the loss of the vacuolar phenotype in cells over-expressing ENQLVAAA-TMEM106B, we then asked if the other cell biological effects of TMEM106B over-expression were also abrogated upon mutation of this motif. Indeed, ENQLVAAA-TMEM106B expressing cells did not exhibit decreased lysosomal acidification as seen with wild-type TMEM106B (Fig. 6a and b). In addition, the cytotoxicity effects exerted by wild-type TMEM106B were also rescued in this point mutant, since expression of ENQLVAAA-TMEM106B caused no more toxicity than controls (Fig. 6c, and Supplementary Material, Fig. 5c).
Figure 6.
The effects of increased TMEM106B expression depend on proper localization of TMEM106B to lysosomes. (a, b) While expression of wild-type TMEM106B (FLAG-TMEM106B construct) in HeLa cells impairs lysosomal acidification (as demonstrated by decreased LysoTracker MFI, top row, arrowhead indicates TMEM106B over-expressing cell), expression of ENQLVAAA-TMEM106B does not significantly alter organelle acidification as compared with neighboring non-over-expressers (bottom row, arrowheads indicate ENQLVAAA-TMEM106B over-expressing cells). Representative images are shown in (a), and means ± SEM for six replicates performed on 3 days are shown in (b). Scale bars = 10 µm. Wild-type TMEM106B and ENQLVAAA-TMEM106B are detected by their FLAG tags in (a). (c) Cytotoxicity is rescued by mutation of critical residues within the potential dileucine lysosomal sorting motif (ENQLVALI to ENQLVAAA) in TMEM106B. While wild-type TMEM106B expression in HeLa cells induces cytotoxicity by 48 h (TMEM106B, green line), the loss of lysosomal localization (ENQ TMEM106B, pink) rescues this cytotoxicity to levels seen with transfection of vector only (control, red line, almost entirely overlapped by pink line). Cytotoxicity seen with expression LAMP1 (blue line) is also shown for comparison purposes; the LAMP1 data is repeated from Figure 3f. Beyond the 48-h time point, wild-type TMEM106B-expressing cells are largely lost from the culture medium due to cell death. % cytotoxicity is calculated in comparison to the maximal LDH release induced by treatment with Triton X as described in Materials and Methods section. Data are combined for six replicates performed on three different days. **p < 0.01. ****p < 0.0001.
Lysosomal acidification and cytotoxicity effects of increased TMEM106B expression are dependent on C9orf72
We have previously demonstrated that TMEM106B is a genetic modifier affecting age at death in C9orf72-associated FTLD-TDP (12), suggesting that TMEM106B and C9orf72 may interact in the pathophysiology of FTLD-TDP (11,26,27). The protein product of the C9orf72 gene, in which hexanucleotide repeat expansions are the most common Mendelian cause of both FTLD-TDP and ALS (4,5), is predicted to be a DENN protein by structural analysis (28). As DENN proteins are guanine exchange factors (GEFs) regulating Rab GTPases, which in turn regulate cell biological events such as vesicular trafficking and fusion (29), we asked whether TMEM106B's cell biological effects might be dependent on C9orf72.
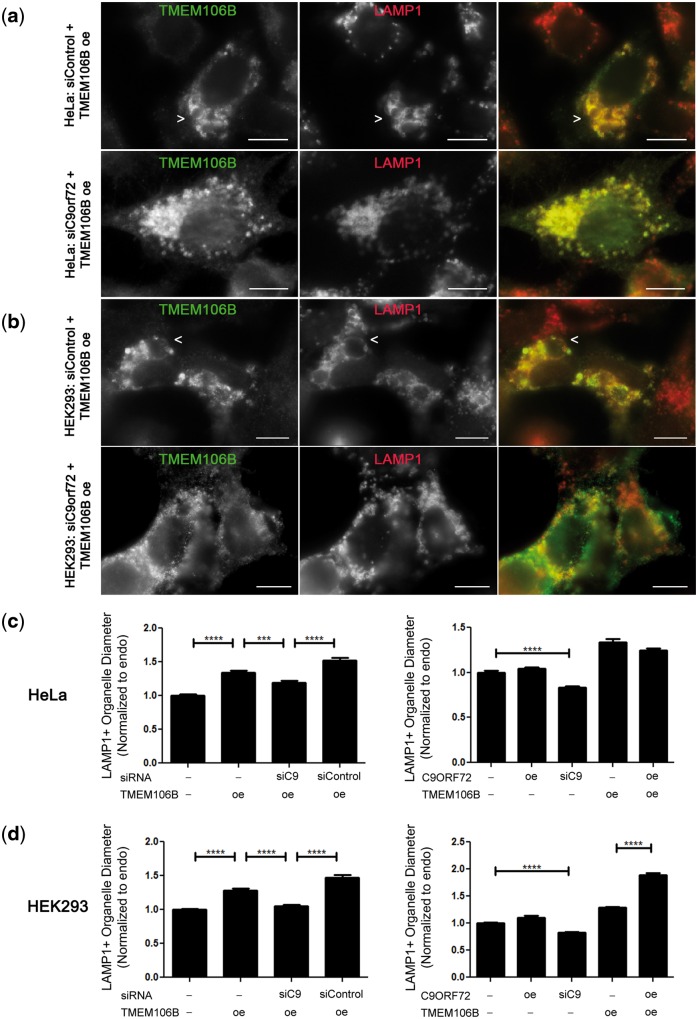
Strikingly, siRNA-mediated knockdown of C9orf72 expression abrogated the vacuolar phenotype seen upon over-expression of TMEM106B. This effect was found in multiple cell types (Fig. 7a and b), and did not occur with control siRNA knockdown. Moreover, quantification of LAMP1+ organelle size demonstrated that siRNA knockdown of C9orf72, but not control siRNA knockdown, rescued the LAMP1+ organelle diameter size in TMEM106B over-expressing cells to levels near that of cells with endogenous levels of C9orf72 and TMEM106B (Fig. 7c and d). Intriguingly, in HEK293 cells but not in HeLa cells, concomitant over-expression of both C9orf72 and TMEM106B resulted in the most dramatic increases in vacuolar size (Fig. 7d). In addition, in both cell types, knockdown of C9orf72 without over-expression of TMEM106B resulted in a small, but significant, decrease in the LAMP1+ organelle diameter size, compared with cells without knockdown of C9orf72 (Fig. 7c and d, Supplementary Material, Fig. 6a).
Figure 7.
Knockdown of C9orf72 rescues TMEM106B-induced vacuolar phenotype. (a, b) In HeLa cells (a) and in HEK293 cells (b), treatment with control siRNA does not affect the LAMP1+ vacuolar phenotype seen in TMEM106B over-expressing cells (arrows, top row), but siRNA knockdown of C9orf72 mitigates the phenotype (bottom row). For all panels, the right panel shows the merged channels for TMEM106B (green) and LAMP1 (red); individual channels are shown in monochrome in the left and middle panels. Scale bars = 10 µm. TMEM106B detected by N2077 antibody. (c, d) The diameter of LAMP1+ organelles was quantified in HeLa (c) and HEK293 (d) cells. In the left graph for both cell types, LAMP1+ diameter was assessed under endogenous conditions (first column), TMEM106B over-expression (second column), TMEM106B over-expression with C9orf72 knockdown (third column), and TMEM106B over-expression with control siRNA knockdown (fourth column). In the right graph, LAMP1+ diameter is quantified under endogenous conditions (first column), C9orf72 over-expression (second column), C9orf72 knockdown alone (third column), TMEM106B over-expression alone (fourth column), and TMEM106B and C9orf72 concomitant over-expression (fifth column). All values are normalized to the endogenous condition (first column) and means ± SEM from nine replicates performed on three separate days are shown. In both HeLa (c) and HEK293 (d) cells, TMEM106B over-expression significantly increased LAMP1+ organelle size (p < 0.0001, left graph, compare first and second columns). In both cell types, however, knockdown of C9orf72 abrogated the effects of TMEM106B over-expression, resulting in a return of LAMP1+ organelle size toward that of the endogenous baseline (p < 0.001, left graph, comparing third column with second and fourth columns). ****p < 0.0001, ***p < 0.001.
Finally, to ensure that the reversal of TMEM106B over-expression effects by C9orf72 knockdown were not due to off-target effects of the siRNA used, we repeated our experiments with a different knockdown construct: specifically, an shRNA targeting C9orf72. As shown in Supplementary Material, Figure 6a, knockdown of C9orf72 by a second method in TMEM106B over-expressing cells again rescued LAMP1+ organelle diameter sizes to near-baseline.
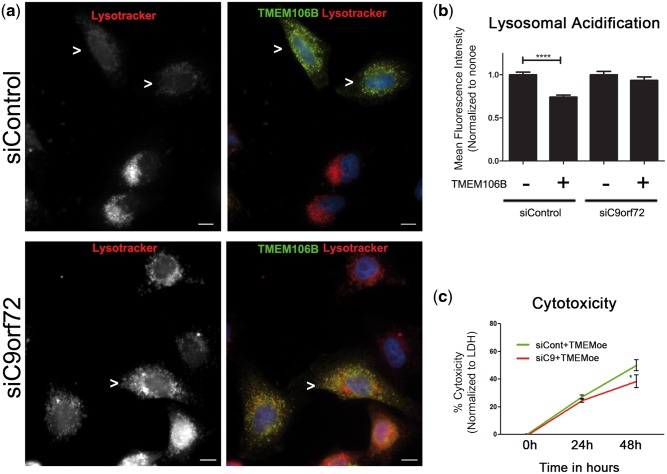
We then asked if other cell biological phenotypes observed with TMEM106B over-expression were similarly dependent on the presence of C9orf72 protein. Specifically, we evaluated cytotoxicity as well as the lysosomal acidification defect induced by TMEM106B over-expression. As shown in Figure 8a and b, the difference in LysoTracker MFI typically seen in TMEM106B over-expressing HeLa cells as compared with non-over-expressing cells is abrogated in the context of C9orf72 knockdown, but maintained in the context of control siRNA knockdown. In addition, knockdown of C9orf72, by siRNA or by shRNA, abrogated the cytotoxic effects of TMEM106B over-expression (Fig. 8c and Supplementary Material, Fig. 6b).
Figure 8.
Knockdown of C9orf72 mitigates TMEM106B-induced acidification defects and cytotoxicity. (a, b) Expression of wild-type TMEM106B in HeLa cells impairs lysosomal acidification, as demonstrated by decreased LysoTracker MFI. Concomitant siRNA knockdown of C9orf72 rescues this defect (bottom row). In contrast, treatment with control siRNA does not rescue this defect (top row). Representative images are shown in (a), and means ± SEM for eight replicates performed on 3 days are shown in (b). Myc-TMEM106B construct is expressed and detected by its myc tag. Cells over-expressing TMEM106B are highlighted by arrowheads. ****p < 0.0001. Scale bars = 10 μm. (c) TMEM106B over-expression induces cytotoxicity in HeLa cells, as quantified by LDH release over a 48-h time-course. Concomitant siRNA knockdown of C9orf72 abrogates cytotoxicity, whereas treatment with control siRNA does not. Means ± SEM for >12 replicates performed on 3 days shown, and cytotoxicity compared by two-way ANOVA. *p < 0.05.
Of note, immunoblots were run in parallel to confirm C9orf72 knockdown for all experiments and demonstrated nearly complete disappearance of the endogenous C9orf72 band; validation of the specificity for the C9orf72 antibody used was demonstrated with C9orf72 knockdown and over-expression in immortalized cells (Supplementary Material, Fig. 6c). In addition, quantitative PCR experiments demonstrated efficient knockdown and over-expression of C9orf72 in our siRNA experiments (Supplementary Material, Fig. 6d). Finally, we considered the possibility that C9orf72 knockdown and over-expression might secondarily affect TMEM106B expression levels, thus rescuing TMEM106B over-expression phenotypes. However, manipulation of C9orf72 levels minimally affected TMEM106B expression (Supplementary Material, Fig. 6e and f).
Discussion
Genotypes at TMEM106B linked to increased risk for FTLD-TDP are associated with increased expression of TMEM106B (21). Moreover, in human FTLD-TDP brain, TMEM106B expression is also increased (21), with an altered subcellular distribution (19). Here, we explore the cell biological consequences of this disease-associated increase in TMEM106B expression, demonstrating that it results in multiple perturbations to endolysosomal pathways, and that these changes are dependent on the presence of C9orf72.
We and others have previously shown that TMEM106B over-expression results in enlargement of LAMP1+ organelles (16,21,30). We corroborate and expand this result here with ultrastructural characterization in neurons, as well as demonstration that the lysosomal acidification defect extends to neurons. We note that the ultrastructural characteristics of these organelles—single-membrane-bound, electron-lucent, containing possible degradation products—suggest that they may be late autophagic vacuoles (amphisomes and autolysosomes). As such, investigation of the role of TMEM106B in autophagy pathways may be a valuable addition to the data presented here. Indeed, it has been recently reported that increased expression of TMEM106B results in increased nuclear translocation of Transcription Factor EB (TFEB) (16). Because TFEB has been proposed as a master regulator of lysosomal and autophagosomal gene expression (31,32), our current findings may also support a potential interaction between TMEM106B and TFEB, although how direct this interaction might be remains to be seen.
We identify for the first time here a potential lysosomal sorting motif for TMEM106B, demonstrating that point mutations to an extended dileucine motif (33,34) abrogate lysosomal localization. The elucidation of this lysosomal sorting motif is important for two reasons. First, it provides domain-level detail for a protein that was virtually uncharacterized prior to 2010. Second, it provides a valuable tool for mechanistic exploration of TMEM106B function. We exploit the latter use in this study as an important control, demonstrating that the effects seen with over-expression of TMEM106B depend on both the identity of the over-expressed protein (since they are not seen with LAMP1 or GFP over-expression) and on the proper subcellular localization of TMEM106B.
The abrogation of TMEM106B-induced effects by concomitant knockdown of C9orf72 also supports the assumption that the vacuolar phenotype, lysosomal acidification defect and cytotoxicity described here are disease-relevant effects of TMEM106B over-expression. Indeed, these findings provide the first mechanistic evidence for a link between the two FTLD-TDP-associated genes TMEM106B and C9orf72. Hexanucleotide repeat expansions in C9orf72 are the most common Mendelian cause of both FTD and ALS (4,5,35). C9orf72-associated FTD and ALS cases demonstrate large expansions (800–4400 repeats), while healthy controls have <33 repeats (35,36). The effects of C9orf72 repeat expansion are an area of active research, with evidence for multiple mechanisms playing a role in disease. In particular, proposed pathophysiological mechanisms for C9orf72 repeat expansions include toxic repeat RNA species (37–40), toxic dipeptide repeats resulting from repeat-associated, non-ATG (RAN) translation (41–43), and reduced levels/loss of function of the normal C9orf72 protein (4,5,44,45).
We and others have previously demonstrated a genetic interaction between TMEM106B and C9orf72 (11,12), with TMEM106B genotypes associated with increased risk for FTLD-TDP in non-C9orf72-expansion cases paradoxically correlating with later age at death and age at onset in C9orf72-expansion-associated FTLD. This sign epistatic effect has been described in other contexts before, and in these other contexts, the protein products of the two genes involved have been shown to mechanistically interact (46,47). Our present study implicates TMEM106B and C9orf72 in the same mechanistic pathways as well, since knockdown of C9orf72 mitigates most lysosomal effects of TMEM106B over-expression.
While the normal function of C9orf72 protein is yet unknown, it is predicted by structural analysis to be a DENN protein (28,48). DENN proteins function as GDP–GTP exchange factors (GEFs) for Rab GTPases and, as such, they play a role in regulating autophagy, vesicular trafficking and fusion, and membrane trafficking events. Proper regulation of Rab GTPase activity is known to be important for neuronal function; furthermore, dysregulation of Rabs by DENN proteins or other protein families with similar GEF activity has previously been linked to neurodegeneration (49–51). Thus, it is possible that many of the effects of increased TMEM106B expression are mediated through alterations to one or more Rab GTPases, which are in turn regulated by C9orf72. Such a model of pathogenesis implies that C9orf72 loss-of-function mechanisms may play a role in development of disease.
We note that C9orf72 knockdown in the context of TMEM106B over-expression here has a phenotype rescue effect. While this result may be counter-intuitive to straightforward assumptions about loss-of-function mechanisms in disease, it is congruent with the sign epistatic genetic modifier effect we previously reported for TMEM106B and C9orf72 in a 30-site international cohort of C9orf72 expansion-associated FTLD-TDP cases (12). That is, in human FTLD-TDP and in cell culture, increased TMEM106B expression (proxied by TMEM106B risk genotype or manipulated by transient transfection) appears deleterious in most cases, but is rescued in the specific context of reduced C9orf72 (proxied by C9orf72 expansion carrier state or manipulated by C9orf72 knockdown). We further note that C9orf72 knockdown, without concomitant TMEM106B over-expression, results in a small, but significant, decrease in the size of LAMP1+ organelles. These data also support the notion that C9orf72 and TMEM106B may function in the same cell biological pathways regulating the biogenesis, size, trafficking, or turnover of these organelles.
Several limitations of our study deserve consideration. First, because many common variant correlates of disease risk may act through cis-regulatory effects on expression of specific genes (52), we modeled these effects in our specific FTLD-TDP case by over-expression of TMEM106B. Such a strategy is not without its dangers, since over-expression may cause cellular artifacts. We guarded against such a possibility in several important ways. First, we evaluated levels of TMEM106B over-expression by immunoblot in all experiments, allowing us to verify that the observed cellular phenotypes occurred reliably at TMEM106B over-expression levels ranging from ∼2 to 10× (Fig. 5b and c). In addition, we employed as controls both over-expression of a different lysosomal protein (LAMP1) and over-expression of a version of TMEM106B that did not target efficiently to lysosomes, in each case confirming that expression levels of control were equivalent to or greater than wild-type TMEM106B over-expression levels. Finally, we demonstrated that effects of TMEM106B over-expression on organelle morphology, acidification, and toxicity could be rescued by manipulation of a second disease-relevant protein, C9orf72. Taken together, these control measures greatly decrease the possibility that the observed effects of increased TMEM106B expression are non-specific over-expression artifacts.
A second limitation of this study is that we have modeled effects of TMEM106B over-expression by transient transfection. While the advantage of such an approach is that it allows for comparisons of cells over-expressing versus not over-expressing TMEM106B within the same field, longer-term effects of alterations in expression might not be captured.
Third, this study focused on the effects of disease-relevant increases in TMEM106B expression on lysosomes, finding, in the process, that observed phenomena are dependent on the presence of C9orf72. We note here that previous work by our group (21) and others (10,18,20) also suggests a potential connection between TMEM106B and progranulin based on genetic and cell-based analyses. Since progranulin localizes to lysosomes (20), and humans without functional progranulin manifest with a lysosomal storage disorder (53), the potential for TMEM106B-induced effects on lysosomes to affect progranulin pathways as well, either in neurons themselves or in other support cells in which progranulin expression is very high (e.g. microglia), is also an area worthy of further investigation.
In summary, since the discovery of the TMEM106B locus as a genetic risk factor for FTLD-TDP in 2010 (9), multiple groups have worked to understand the significance of this gene and protein for disease pathophysiology (16,18,20–22,30). Here, we dissect at a cell biological level the effects of disease-associated increases in TMEM106B expression, demonstrating significant effects on lysosomal morphology and impairment in lysosonal acidification, with associated cytotoxicity. We moreover demonstrate that these effects depend on the presence of a second FTLD-TDP-associated protein, C9orf72. This study thus establishes the importance of lysosomal pathways to the development of FTLD-TDP and suggests a previously unsuspected mechanistic interaction between TMEM106B and C9orf72.
Materials and Methods
Constructs
C9orf72 constructs—FLAG-C9orf72: open reading frame of human C9orf72 (transcript variant 2 NM_018325.3) in pCMV6 vector (Origene); this construct is dual-tagged with myc-FLAG inserted after the C-terminus; however, because only the FLAG was used for antibody detection, we hereinafter refer to this construct as FLAG-C9orf72.
Control constructs—5TO: pcDNA/5TO backbone vector (Invitrogen). GFP: GFP was cloned into the pcDNA/5TO vector (Invitrogen). GFP-LAMP1: gift from L. Volpicelli-Daley (University of Alabama, Birmingham, AL, USA). LAMP1: human LAMP1 in pCMV6-XL5 (Origene).
TMEM106B constructs – (i) FLAG-TMEM106B: the 5′ untranslated region and open reading frame of TMEM106B and a C-terminal FLAG tag were cloned into pcDNA 3.0 vector (Life Technologies). (ii) GFP-TMEM106B: cloned into pEGFP-C3 vector (Clontech) by F. Hu (Cornell University, Ithaca, NY, USA) as previously described (20). (iii) myc-TMEM106B: the 5′ untranslated region and open reading frame of TMEM106B and an N-terminal myc tag were cloned into pcDNA 3.0 (Life Technologies).
Lysosomal motif mutants
Point mutations were introduced to putative lysosomal sorting motifs in the parent FLAG-TMEM106B construct (see previous section). Mutations to the tyrosine and dileucine-based motifs were created using the QuikChange II Site-Directed Mutagenesis Kit and primer design software (Agilent Technologies). The primer used to mutate YVEF → AVEF was tcctgtaaattccacagctggaaactgagagacatctccatttcttcc, YDGV → ADGV was gatgtgactccatcagcagcatcttctttgcttgaatgcaaagg, and ENQLVALI → ENQLVAAA was tctctgatcactatatggagccgctgccaccagttggttttcttgccccc. For precise locations of these motifs, please see Figure 4b.
Cell culture, transfection and nucleofection
Primary cortical and hippocampal mouse neurons were prepared from embryonic day 18 (E18) to E20 C57BL/6 mice as previously described (54). Exogenous plasmids were introduced by nucleofection (Lonza Amaxa Nucleofector 2b). For each nucleofection, 5 million neurons in suspension were spun down at 80×g and resuspended in 100 µl Mirus BioIngenio Electroporation Solution (Mirus #50111). Details of nucleofection protocols can be found in the Supplementary Materials and Methods.
COS-7, HEK293 and HeLa cells were also used for experiments. Exogenous plasmids were introduced by Lipofectamine-mediated transfection. Details of their maintenance and transfection are provided in Supplementary Materials and Methods.
C9orf72 knockdown
Two rounds of knockdown with 120 pg of siRNA against C9orf72 (GE Healthcare ON-TARGETplus SMARTpool #L-013341-01 Human C9orf72) or 120 pg of siControl (GE Healthcare ON-TARGETplus Non-targeting Pool #D-001810-10-05) were performed at 24 and 48 h after cell plating. For experiments that required concomitant over-expression of another construct, the over-expression construct was transfected in parallel with the siRNA during the second round of knockdown.
As a second method to confirm specificity of C9orf72 knockdown effects, we used 4 μg of shRNA against C9Orf72 (Origene pRS construct # TR305711 construct C) or 4 μg of an shRNA to GFP as a negative control (Origene pRS construct # TR30003). Transfections were performed 24 and 48 h after plating, with overexpression of TMEM106B occurring at the same time as the 48-h knockdown transfection.
Live and immunofluorescence microscopy
Live images were taken with the EVOS FL Cell Imaging System (Life Technologies) at 40× or 60× magnification.
Double- and triple-label immunofluorescence labeling experiments were performed as previously described (21). The following primary antibodies and conditions were used: C9orf72: anti-human rabbit C9orf72 (Sigma #HPA023873) at 0.8 µg/ml (1:250); EEA-1: rat anti-mouse EEA-1 (BD #610456) at 1:100; GFP: goat polyclonal anti-GFP (abcam #6673) at 0.98 µg/ml (1:1000); LAMP1 1D4B: rat anti-mouse 1D4B (DSHB) at 1 µg/ml; LAMP1 H4A3: mouse anti-human H4A3 (DSHB) at 1 µg/ml; SV-2 mouse anti-mouse SV-2 (DSHB #SV2) at 1:3000. TMEM106B: custom rabbit anti-human, anti-mouse N2077 polyclonal antibody raised against amino acids 4–19 of TMEM106B at 1 µg/ml; custom chicken anti-human N2077 serum raised against the same peptide at 1:2000. TMEM106B antibody characterization has been previously reported (21). Secondary antibodies: goat anti-rabbit Alexa Fluor 488 (Life Technologies #A-11008); goat anti-rabbit Alexa Fluor 594 (Life Technologies #A-11037); goat anti-chicken Alexa Fluor 488 (Life Technologies #A-11039) all used at 1:1000. AMCA anti-mouse (Vector Laboratories #CI-2000) at 1:100. LysoTracker DND-99 (Life Technologies #L-7528) was used as previously described (21). Images were taken on a Nikon 80i upright fluorescence microscope and analyzed with Nikon NIS-Elements AR Imaging Software.
BODIPY 493/503 (Life #D-3922) was used per manufacturer’s instructions. In brief, cells grown on coverslips were washed with PBS and fixed with 2% paraformaldehyde. BODIPY stock (1 mg/ml in 100% EtOH) was freshly diluted at 1:500 in DPBS and incubated on the coverslips in the dark at room temperature for 15 min. Coverslips were then rinsed with DPBS three times, then counterstained with DAPI and mounted with ProLong Gold mounting medium (Life #P36930).
Immunoblotting
Immunoblots were performed as previously described (55). The following antibodies and conditions were used: N2077 rabbit TMEM106B antibody at 1 µg/ml (21); EGFR (Cell Signaling #2232) at 1:1000; LAMP1 H4A3 at 0.5 µg/ml; alpha tubulin: mouse monoclonal DM1A (abcam #ab7291) at 0.1333 µg/ml (1:7500); C9orf72: rabbit anti-human at 1:500 (Sigma #023873).
EGF-R degradation assay
HeLa cells were transfected with FLAG-TMEM106B, GFP or 5TO (vector) constructs. The next day media was replaced with serum-free DMEM media. After 24-h starvation, 25 mg/ml cycloheximide (Sigma #C1988) was added to the wells; cells were then stimulated with 100 ng/ml recombinant EGF (Invitrogen # PHG0311). At each timepoint, cells were lysed and harvested into 1× RIPA on ice and immunoblots were performed with anti-EGFR antibody at 1:1000. Densitometry was used to quantify the amount of EGFR as normalized to alpha tubulin for four replicate experiments.
EGF trafficking assay
A previously published protocol was adapted (56). In brief, HeLa cells were transfected with TMEM106B; 24-h later, media was changed to serum-free media containing 1 µM leupeptin in DMSO (Sigma #L2023) and 1 µM pepstatin in DMSO (Sigma #P4265). EGF-488 (Invitrogen #E-13345) was then added at 50 or 400 ng/ml. The 0-h timepoint was immediately fixed and immunofluorescently labeled with anti-LAMP1 H4A3 and N2077. For subsequent timepoints, after 20 min of pulsing with EGF-488, media was removed, cells were washed and media containing unlabeled recombinant EGF, leupeptin and pepstatin was added to the cells. Coverslips were fixed at each timepoint and immunofluorescently labeled. Z-stack images were taken at 100× and deconvolved using Nikon NIS-Elements AR Imaging Software. For each timepoint, approximately eight fields were taken; non-over-expressing and over-expressing cells (>12 each) were outlined using the region-of-interest tool and Mander’s overlap was determined between the LAMP1- and EGF channels with the Nikon NIS-Elements AR Imaging Software, comparing neighboring cells with versus without TMEM106B over-expression.
Cytotoxicity assays
The Clontech LDH assay (Clontech #630117), an LDH-based colorimetric kit, was used. HeLa cells were transfected with FLAG-TMEM106B, LAMP1, ENQLVAAA-TMEM106B or 5TO vector backbone constructs. For experiments requiring C9orf72 knockdown, two rounds of knockdown were performed, as described above. At 0, 24 and 48 h after transfection, 150 μl media was removed from each condition and processed according to manufacturer instructions for three independent experiments. Additional details can be found in Supplementary Materials and Methods.
Trypan Blue assays were also used to assess cytotoxicity. Forty-eight hours after transfection, media was collected, adherent cells were trypsinized and added to the collected media, and cells were spun down at 800×g for 5 min. Media was then removed and a 50/50 mix of DMEM and 0.4% Trypan Blue were used to resuspend the cells. The number of blue and total cells were counted using a hemacytometer.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were used to assess for cells with apoptosis-induced DNA damage. HeLa cells were transfected with FLAG-TMEM106B or control empty vector. At 24 h after transfection, staurosporine (Sigma #S6942-200) was added to a final concentration of 1 µM to positive control wells and cells were returned to the incubator for an additional 4 h. Coverslips were then removed from the wells for immunofluorescent labeling, with TMEM106B over-expressing cells identified by staining against the FLAG tag. The TUNEL assay was then performed according to manufacturer’s protocol (Roche #11 684 795 910), with the TUNEL fluorescein reaction mixture incorporated into the secondary antibody step of immunofluorescent labeling. Images were taken on a Nikon 80i upright fluorescence microscope and analyzed with Nikon NIS-Elements AR Imaging Software.
Electron microscopy
Tissues for electron microscopic examination were fixed, washed and dehydrated prior to embedding in EMbed-812 (Electron Microscopy Sciences #14120). Images were acquired with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software. For analysis, grids with cells or neurons expressing either control or TMEM106B constructs were then scanned systematically; non-dividing, non-apoptotic cells were imaged in an unbiased manner. In some cases, as indicated in the text, cells expressing GFP-TMEM106B constructs were first captured by flow cytometric cell sorting, selecting for cells expressing GFP versus cells not expressing GFP. To quantify the percentage of sorted cells exhibiting the vacuolar phenotype, two independent investigators scored GFP-positive and GFP-negative flow-sorted populations of GFP-TMEM106B transfected HeLa cells for the presence or absence of the vacuolar phenotype as captured by electron microscopy; disagreements were reviewed by both investigators so that a consensus decision could be reached. The vacuolar phenotype was defined as the presence of one or more single membrane-delimited vacuoles with long-width diameter >1 μm. Additional details of tissue, coverslip and grid preparation can be found in the Supplementary Material.
Quantification of LysoTracker MFI
Detailed quantification procedures are provided in the Supplementary Material. In brief, LysoTracker MFI was determined as previously described (21), with >180 neurons per condition imaged at 60× and quantified for MFI. In all cases, quantification was performed blinded to the identity of the over-expressed construct.
Quantification of LAMP1+ organelle diameter
The maximum diameter of the 10 largest LAMP1+ organelles (by visual inspection) within each cell analyzed was measured using the Nikon NIS-Elements AR Imaging Software, with >150 LAMP1+ organelles (>15 cells) measured for each condition from images captured at 40×. Additional details regarding quantification and normalization are found within the Supplementary Material.
Statistical tests
Two-tailed nonparametric (Mann–Whitney) tests were used except for time-course experiments, for which two-way ANOVAs were used instead. Fisher exact tests were used for electron microscopy analysis given low event counts in some control categories. Calculations were performed using GraphPad Prism 5.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Margie Maronski and the “Neurons R Us” core facility for providing primary murine neurons, the Electron Microscopy Resource Laboratory for processing and assistance with electron microscopy, Dr. Fenghua Hu for providing the GFP-TMEM106B construct, Emily Bill and Jordan Mak for technical assistance, and Dr. Mickey Marks for his input and advice regarding lysosomal motif mutations.
Conflict of Interest statement. The authors declare no competing financial interests.
Funding
This study was supported by the National Institutes of Health [R01 NS082265]; the Burroughs Wellcome Fund; and the Benaroya Fund. J.I.B. was additionally supported by training grants from the National Institutes of Health [F31 NS086428-01, T32 AG 255-16].
References
- 1.Neary D., Snowden J., Mann D. (2005) Frontotemporal dementia. Lancet Neurol., 4, 771–780. [DOI] [PubMed] [Google Scholar]
- 2.Ratnavalli E., Brayne C., Dawson K., Hodges J.R. (2002) The prevalence of frontotemporal dementia. Neurology, 58, 1615–1621. [DOI] [PubMed] [Google Scholar]
- 3.Baborie A., Griffiths T.D., Jaros E., McKeith I.G., Burn D.J., Richardson A., Ferrari R., Moreno J., Momeni P., Duplessis D, et al. (2011) Pathological correlates of frontotemporal lobar degeneration in the elderly. Acta Neuropathol., 121, 365–371. [DOI] [PubMed] [Google Scholar]
- 4.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J, et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron, 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L, et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S, et al. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature, 442, 916–919. [DOI] [PubMed] [Google Scholar]
- 7.Gass J., Cannon A., Mackenzie I.R., Boeve B., Baker M., Adamson J., Crook R., Melquist S., Kuntz K., Petersen R, et al. (2006) Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet., 15, 2988–3001. [DOI] [PubMed] [Google Scholar]
- 8.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J, et al. (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature, 442, 920–924. [DOI] [PubMed] [Google Scholar]
- 9.Van Deerlin V.M., Sleiman P.M., Martinez-Lage M., Chen-Plotkin A., Wang L.S., Graff-Radford N.R., Dickson D.W., Rademakers R., Boeve B.F., Grossman M, et al. (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet., 42, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruchaga C., Graff C., Chiang H.H., Wang J., Hinrichs A.L., Spiegel N., Bertelsen S., Mayo K., Norton J.B., Morris J.C, et al. (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol., 68, 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Blitterswijk M., Mullen B., Nicholson A.M., Bieniek K.F., Heckman M.G., Baker M.C., DeJesus-Hernandez M., Finch N.A., Brown P.H., Murray M.E, et al. (2014) TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol., 127, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher M.D., Suh E., Grossman M., Elman L., McCluskey L., Van Swieten J.C., Al-Sarraj S., Neumann M., Gelpi E., Ghetti B, et al. (2014) TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol., 127, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen-Plotkin A.S., Lee V.M., Trojanowski J.Q. (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol., 6, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vass R., Ashbridge E., Geser F., Hu W.T., Grossman M., Clay-Falcone D., Elman L., McCluskey L., Lee V.M., Van Deerlin V.M, et al. (2011) Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol., 121, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L., De Jager P.L., Yang J., Trojanowski J.Q., Bennett D.A., Schneider J.A. (2015) The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology, 84, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stagi M., Klein Z.A., Gould T.J., Bewersdorf J., Strittmatter S.M. (2014) Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol. Cell. Neurosci., 61, 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwenk J.M., Lindberg J., Sundberg M., Uhlen M., Nilsson P. (2007) Determination of binding specificities in highly multiplexed bead-based assays for antibody proteomics. Mol. Cell. Proteomics, 6, 125–132. [DOI] [PubMed] [Google Scholar]
- 18.Lang C.M., Fellerer K., Schwenk B.M., Kuhn P.H., Kremmer E., Edbauer D., Capell A., Haass C. (2012) Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J. Biol. Chem., 287, 19355–19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busch J.I., Martinez-Lage M., Ashbridge E., Grossman M., Van Deerlin V.M., Hu F., Lee V.M., Trojanowski J.Q., Chen-Plotkin A.S. (2013) Expression of TMEM106B, the frontotemporal lobar degeneration-associated protein, in normal and diseased human brain. Acta Neuropathol. Commun., 1, 36-5960-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brady O.A., Zheng Y., Murphy K., Huang M., Hu F. (2013) The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet., 22, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen-Plotkin A.S., Unger T.L., Gallagher M.D., Bill E., Kwong L.K., Volpicelli-Daley L., Busch J.I., Akle S., Grossman M., Van Deerlin V, et al. (2012) TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci., 32, 11213–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson A.M., Finch N.A., Wojtas A., Baker M.C., Perkerson R.B., Castanedes-Casey M., Rousseau L., Benussi L., Binetti G., Ghidoni R, et al. (2013) TMEM106B p.T185S regulates TMEM106B protein levels: Implications for frontotemporal dementia. J. Neurochem., 126, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eden E.R., White I.J., Futter C.E. (2009) Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem. Soc. Trans., 37, 173–177. [DOI] [PubMed] [Google Scholar]
- 24.Katzmann D.J., Odorizzi G., Emr S.D. (2002) Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol., 3, 893–905. [DOI] [PubMed] [Google Scholar]
- 25.Bonifacino J.S., Traub L.M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem., 72, 395–447. [DOI] [PubMed] [Google Scholar]
- 26.Cooper-Knock J., Shaw P.J., Kirby J. (2014) The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol., 127, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deming Y., Cruchaga C. (2014) TMEM106B: a strong FTLD disease modifier. Acta Neuropathol., 127, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine T.P., Daniels R.D., Gatta A.T., Wong L.H., Hayes M.J. (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN rab-GEFs. Bioinformatics, 29, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D., Iyer L.M., He F., Aravind L. (2012) Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet., 3, 283.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwenk B.M., Lang C.M., Hogl S., Tahirovic S., Orozco D., Rentzsch K., Lichtenthaler S.F., Hoogenraad C.C., Capell A., Haass C, et al. (2014) The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. Embo J., 33, 450–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P, et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science, 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S, et al. (2009) A gene network regulating lysosomal biogenesis and function. Science, 325, 473–477. [DOI] [PubMed] [Google Scholar]
- 33.Letourneur F., Klausner R.D. (1992) A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell, 69, 1143–1157. [DOI] [PubMed] [Google Scholar]
- 34.Pond L., Kuhn L.A., Teyton L., Schutze M.P., Tainer J.A., Jackson M.R., Peterson P.A. (1995) A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J. Biol. Chem., 270, 19989–19997. [DOI] [PubMed] [Google Scholar]
- 35.Beck J., Poulter M., Hensman D., Rohrer J.D., Mahoney C.J., Adamson G., Campbell T., Uphill J., Borg A., Fratta P, et al. (2013) Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am. J. Hum. Genet., 92, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Blitterswijk M., DeJesus-Hernandez M., Niemantsverdriet E., Murray M.E., Heckman M.G., Diehl N.N., Brown P.H., Baker M.C., Finch N.A., Bauer P.O, et al. (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (xpansize-72): a cross-sectional cohort study. Lancet Neurol., 12, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fratta P., Mizielinska S., Nicoll A.J., Zloh M., Fisher E.M., Parkinson G., Isaacs A.M. (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep., 2, 1016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gendron T.F., Bieniek K.F., Zhang Y.J., Jansen-West K., Ash P.E., Caulfield T., Daughrity L., Dunmore J.H., Castanedes-Casey M., Chew J, et al. (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol., 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C, et al. (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell. Rep., 5, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizielinska S., Lashley T., Norona F.E., Clayton E.L., Ridler C.E., Fratta P., Isaacs A.M. (2013) C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol., 126, 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ash P.E., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., 3rd, Rademakers R, et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron, 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizielinska S., Gronke S., Niccoli T., Ridler C.E., Clayton E.L., Devoy A., Moens T., Norona F.E., Woollacott I.O., Pietrzyk J, et al. (2014) C9orf72 repeat expansions cause neurodegeneration in drosophila through arginine-rich proteins. Science, 345, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori K., Arzberger T., Grasser F.A., Gijselinck I., May S., Rentzsch K., Weng S.M., Schludi M.H., van der Zee J., Cruts M, et al. (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol., 126, 881–893. [DOI] [PubMed] [Google Scholar]
- 44.Ciura S., Lattante S., Le Ber I., Latouche M., Tostivint H., Brice A., Kabashi E. (2013) Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol., 74, 180–187. [DOI] [PubMed] [Google Scholar]
- 45.Gijselinck I., Van Langenhove T., van der Zee J., Sleegers K., Philtjens S., Kleinberger G., Janssens J., Bettens K., Van Cauwenberghe C., Pereson S, et al. (2012) A C9orf72 promoter repeat expansion in a flanders-belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol., 11, 54–65. [DOI] [PubMed] [Google Scholar]
- 46.Schenk M.F., Szendro I.G., Salverda M.L., Krug J., de Visser J.A. (2013) Patterns of epistasis between beneficial mutations in an antibiotic resistance gene. Mol. Biol. Evol., 30, 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva R.F., Mendonca S.C., Carvalho L.M., Reis A.M., Gordo I., Trindade S., Dionisio F. (2011) Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genet., 7, e1002181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y.Q., Gamarra S., Garcia-Effron G., Park S., Perlin D.S., Rao R. (2010) Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog., 6, e1000939.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azzedine H., Bolino A., Taieb T., Birouk N., Di Duca M., Bouhouche A., Benamou S., Mrabet A., Hammadouche T., Chkili T, et al. (2003) Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of charcot-marie-tooth disease associated with early-onset glaucoma. Am. J. Hum. Genet., 72, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Del Villar K., Miller C.A. (2004) Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in alzheimer's disease brain and hippocampal neurons. Proc. Natl Acad. Sci. U. S. A., 101, 4210–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadano S., Otomo A., Kunita R., Suzuki-Utsunomiya K., Akatsuka A., Koike M., Aoki M., Uchiyama Y., Itoyama Y., Ikeda J.E. (2010) Loss of ALS2/alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PLoS One, 5, e9805.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilad Y., Rifkin S.A., Pritchard J.K. (2008) Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet., 24, 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith K.R., Damiano J., Franceschetti S., Carpenter S., Canafoglia L., Morbin M., Rossi G., Pareyson D., Mole S.E., Staropoli J.F, et al. (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet., 90, 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng H.C., Ruegg S.J., Maronski M., Messam C.A., Grinspan J.B., Dichter M.A. (2006) Injuring neurons induces neuronal differentiation in a population of hippocampal precursor cells in culture. Neurobiol. Dis., 22, 88–97. [DOI] [PubMed] [Google Scholar]
- 55.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M, et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. [DOI] [PubMed] [Google Scholar]
- 56.Urwin H., Authier A., Nielsen J.E., Metcalf D., Powell C., Froud K., Malcolm D.S., Holm I., Johannsen P., Brown J, et al. (2010) Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet., 19, 2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.