Abstract
Alterations in oxidative metabolism and defects in mitochondrial Ca2+ handling have been implicated in the pathology of Huntington’s disease (HD), but existing data are contradictory. We investigated the effect of human mHtt fragments on oxidative metabolism and Ca2+ handling in isolated brain mitochondria and cultured striatal neurons from the R6/2 mouse model of HD. Non-synaptic and synaptic mitochondria isolated from the brains of R6/2 mice had similar respiratory rates and Ca2+ uptake capacity compared with mitochondria from wild-type (WT) mice. Respiratory activity of cultured striatal neurons measured with Seahorse XF24 flux analyzer revealed unaltered cellular respiration in neurons derived from R6/2 mice compared with neurons from WT animals. Consistent with the lack of respiratory dysfunction, ATP content of cultured striatal neurons from R6/2 and WT mice was similar. Mitochondrial Ca2+ accumulation was also evaluated in cultured striatal neurons from R6/2 and WT animals. Our data obtained with striatal neurons derived from R6/2 and WT mice show that both glutamate-induced increases in cytosolic Ca2+ and subsequent carbonilcyanide p-triflouromethoxyphenylhydrazone-induced increases in cytosolic Ca2+ were similar between WT and R6/2, suggesting that mitochondria in neurons derived from both types of animals accumulated comparable amounts of Ca2+. Overall, our data argue against respiratory deficiency and impaired Ca2+ handling induced by human mHtt fragments in both isolated brain mitochondria and cultured striatal neurons from transgenic R6/2 mice.
Huntington’s disease (HD) is an incurable neurodegenerative disorder characterized by progressively worsening motor, psychiatric, and cognitive maladies (1). In HD, the exon 1 CAG repeat stretch of the gene that encodes the huntingtin protein (Htt) is mutated, resulting in elongation of this domain (2). Mutant huntingtin (mHtt) possesses an extended polyglutamine (polyQ) tract that, when expanded in humans beyond 35 glutamines, leads to striatal and cortical degenerations and, subsequently, to development of HD symptoms (2). The exact mechanism by which mHtt exerts its deleterious effects in neurons is not clear, but bioenergetic defects and aberrant mitochondrial Ca2+ handling have been implicated as possible factors contributing to neuronal dysfunction in HD (3,4).
In our previous studies, we investigated the effect of human full-length mHtt on respiratory activity and Ca2+ uptake capacity in brain synaptic and non-synaptic mitochondria as well as striatal and cortical neurons from transgenic YAC128 mice (5,6). Despite significant effort, we found no evidence for mHtt-induced alterations in respiration and Ca2+ uptake capacity of mitochondria from wild-type (WT) and YAC128 mice. Whether HD pathogenesis is mediated by full-length mHtt or fragments of mHtt remains not completely understood. Previous studies suggested that mHtt fragments might be more toxic than full-length mHtt (7,8) and it was shown that reduction in mHtt fragment generation improved the phenotype of HD mice (9–11). Here, we hypothesize that fragments of human mHtt, contrary to full-length human mHtt, are more deleterious and exert a detrimental effect on mitochondrial respiration and Ca2+ handling. Consequently, in the present study, we assessed the effect of mHtt fragments on mitochondrial respiratory activity and Ca2+ handling in synaptic and non-synaptic brain mitochondria and striatal neurons from the R6/2 mouse model of HD.
The R6/2 mouse model is one of the first developed and most well-studied transgenic mouse models of HD (12). These mice express the N-terminal fragment of human mHtt with a 144-glutamine stretch and display overt behavioral abnormalities by 6 weeks. In this study, symptomatic 6–8-week old R6/2 mice were utilized to probe the effect of mHtt fragments on mitochondrial respiratory function and Ca2+ uptake capacity. The major findings of the present study are that (i) there is no difference in respiratory rates and Ca2+ uptake capacities between brain mitochondria isolated from R6/2 and WT mice; and (ii) that primary striatal neurons from R6/2 and WT mice showed no difference in oxygen consumption rates (OCRs), cellular ATP levels, and mitochondrial Ca2+ accumulation.
Results
In our experiments, we used the R6/2 mouse model of HD, which expresses exon 1 of human mHtt (12). These mice exhibit a behavioral phenotype that manifests by 6 weeks of age as limb clasping when suspended by the tail (12,13). The presence of this phenotype is consistent with previous reports describing this and other mouse models of HD (5,12,14).
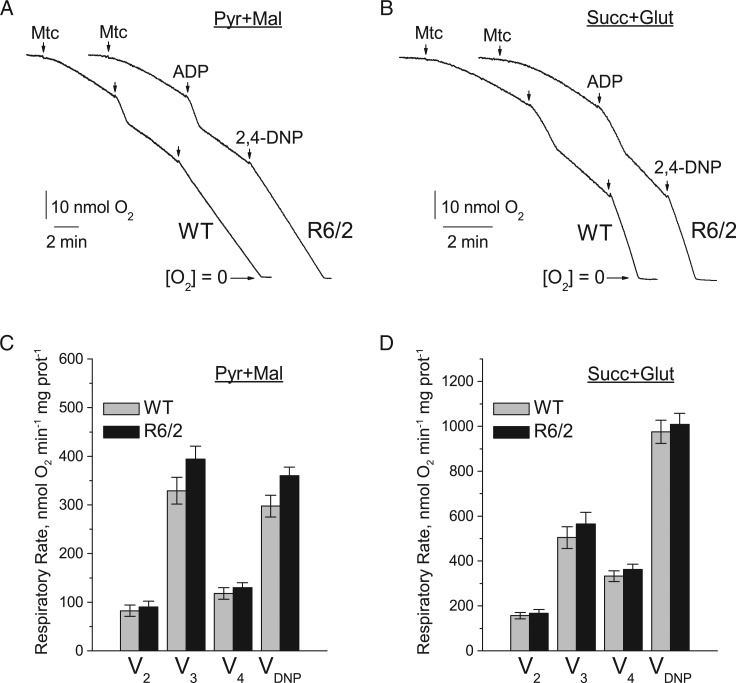
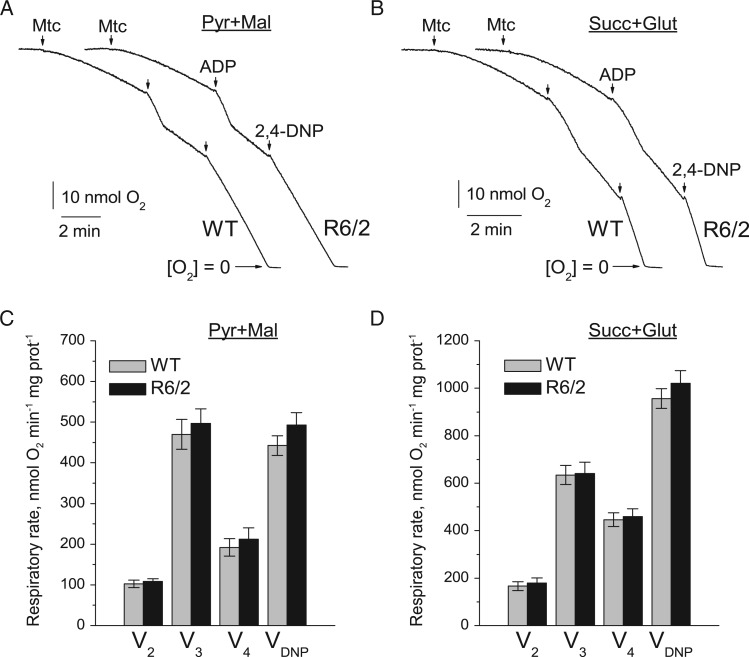
To assess the effect of mHtt fragments on mitochondrial respiration, we used Percoll gradient-purified brain non-synaptic (neuronal plus glial) and synaptic (pure neuronal) mitochondria isolated from 6- to 8-week-old R6/2 and background B6CBA (WT) mice. Each R6/2 mouse demonstrated clasping behavior and each animal was genotyped to confirm the presence of the mutation in the Htt gene (Fig. 1). Previously, it was hypothesized that bovine serum albumin (BSA) may displace mHtt from the outer mitochondrial membrane and hence preclude it from exerting deleterious effects on mitochondrial functions (15). Therefore, although BSA is commonly used during mitochondrial isolation and purification to maintain mitochondrial integrity (16), we omitted BSA from all of our experiments involving isolated mitochondria unless stated otherwise. We assessed mitochondrial respiratory activity in both non-synaptic (Fig. 2) and synaptic (Fig. 3) mitochondria using either a combination of the complex I substrates pyruvate (3 mM) and malate (1 mM) or the complex II substrate succinate (3 mM). In experiments with succinate, the incubation medium was supplemented with glutamate (3 mM) to remove oxaloacetate by transaminase reaction and prevent oxaloacetate-mediated inhibition of succinate dehydrogenase (17,18). For all experiments, basal respiration of mitochondria was measured in the presence of substrates only (V2), followed by ADP-stimulated respiration (V3), controlled respiration after ADP-depletion (V4), and, finally, maximal, uncoupled respiration stimulated by 2,4-dinitrophenol (VDNP). In these experiments, we found that under all tested conditions non-synaptic and synaptic mitochondria from R6/2 animals had similar respiratory rates compared with mitochondria from WT animals (Figs. 2 and 3).
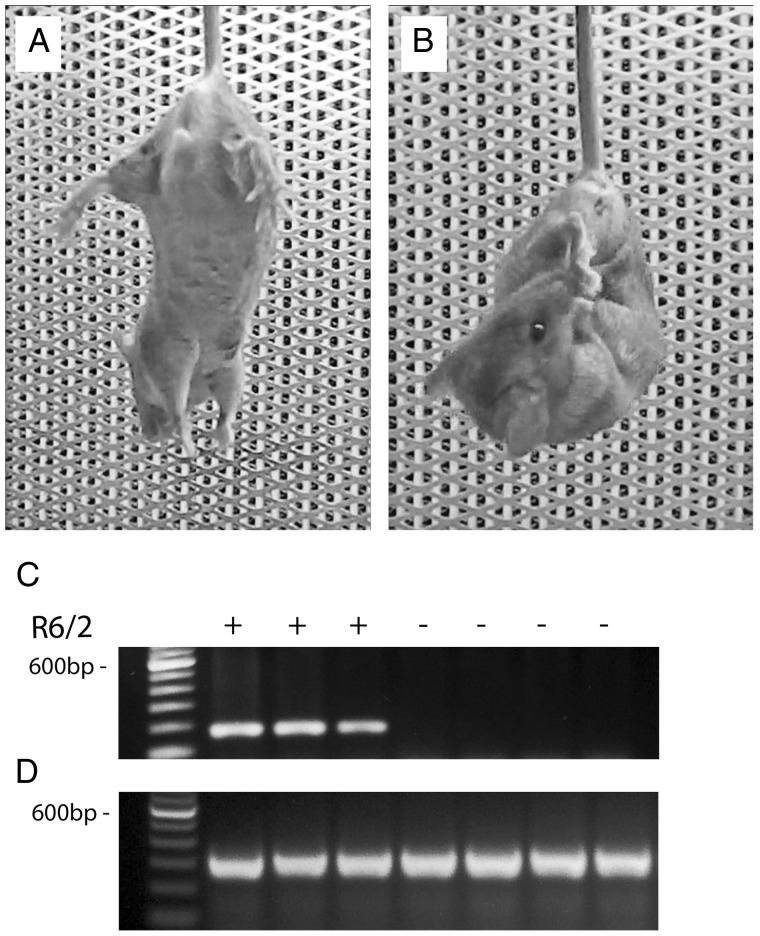
Figure 1.
Comparison of motor phenotype of 6-week old R6/2 and WT mice and representative genotyping. (A) typical posture of a WT mouse following suspension by the tail with fore- and hind-limbs extended away from the body (B) the usual feet-clasping posture adopted by an R6/2 mouse within 10 s of suspension by the tail. (C) representative genotyping data of tail tissue from WT and R6/2 mice. (D) internal positive control.
Figure 2.
Respiratory activity of brain non-synaptic mitochondria isolated from 6- to 8-week-old WT (thin traces) and R6/2 (thick traces) mice. In A and B are representative traces of mitochondrial O2 consumption for mitochondria at 37 °C in incubation medium supplemented with pyruvate (3 mM) plus malate (1 mM) or succinate (3 mM) plus glutamate (3 mM), respectively. Arrows indicate the addition of either non-synaptic mitochondria (Mtc), 200 µM ADP, or 60 µM 2,4-DNP. In (C) and (D) are statistical analyses of respiratory rates. Data are presented as mean ± SEM from 5 separate experiments. Here and in all other experiments with isolated mitochondria, three WT and three transgenic R6/2 mice were used in each experiment.
Figure 3.
Respiratory activity of brain synaptic mitochondria isolated from 6- to 8-week-old WT (thin traces) and R6/2 (thick traces) mice. In (A) and (B) are representative traces of mitochondrial O2 consumption for mitochondria at 37 °C in incubation medium supplemented with pyruvate (3 mM) plus malate (1 mM) or succinate (3 mM) plus glutamate (3 mM), respectively. Arrows indicate the addition of either synaptic mitochondria (Mtc), 200 µM ADP, or 60 µM 2,4-DNP. In (C) and (D) are statistical analyses of respiratory rates. Data are presented as mean ± SEM from five separate experiments.
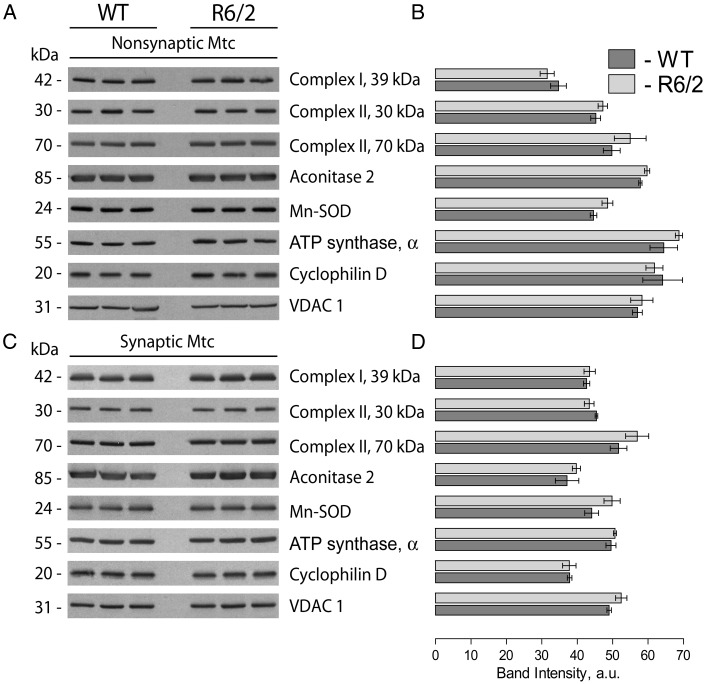
In line with our data, Yano et al. (19) did not find a difference in respiratory activity of synaptic and non-synaptic mitochondria from R6/2 mice compared with mitochondria from WT animals. On the other hand, the authors found evidence for mHtt-induced inhibition of protein import into mitochondria, and proposed that such an inhibition might alter mitochondrial functions at later stages of HD (19). We analyzed expression of several nuclear-encoded mitochondrial proteins, including a 39 kDa subunit of Complex I, 30 and 70 kDa subunits of Complex II, aconitase 2, Mn-dependent superoxide dismutase (MnSOD), cyclophilin D (CyD) and α-subunit of ATP synthase in brain non-synaptic and synaptic mitochondria isolated from 8-week-old R6/2 and WT mice. VDAC1, (voltage-dependent anion channel 1 or mitochondrial porin), a protein in the outer mitochondrial membrane, was used as a loading control. In these experiments, we did not find any evidence for decreased expression of the analyzed proteins in mitochondria from R6/2 compared with mitochondria from WT mice (Fig. 4). These findings argue against mHtt-induced inhibition of mitochondrial protein import and its potential role in inhibition of mitochondrial respiration.
Figure 4.
Expression of nuclear encoded mitochondrial proteins in non-synaptic and synaptic mitochondria isolated from 8-week-old WT and R6/2 mice. In (A) and (C), representative western blots of non-synaptic (A) or synaptic (C) mitochondria generated with antibodies against nuclear encoded mitochondrial proteins including 39 kDa subunit of Complex I, 30 and 70 kDa subunits of Complex II, aconitase 2, MnSOD, CyD, and α-subunit of ATP synthase (ATP synthase, α). VDAC1 was used as a loading control. In B and D, the results of densitometry performed with NIH ImageJ 1.48v software. Data are mean ± SEM from six separate experiments.
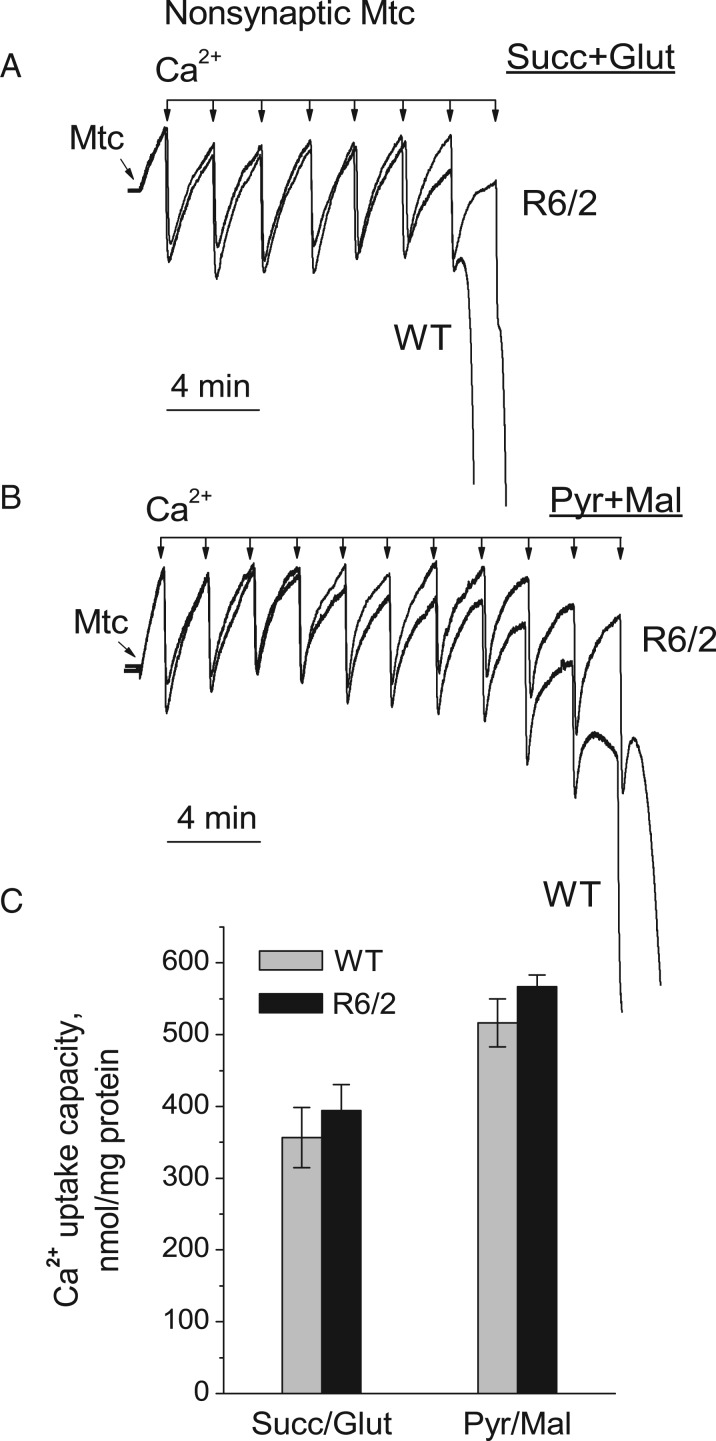
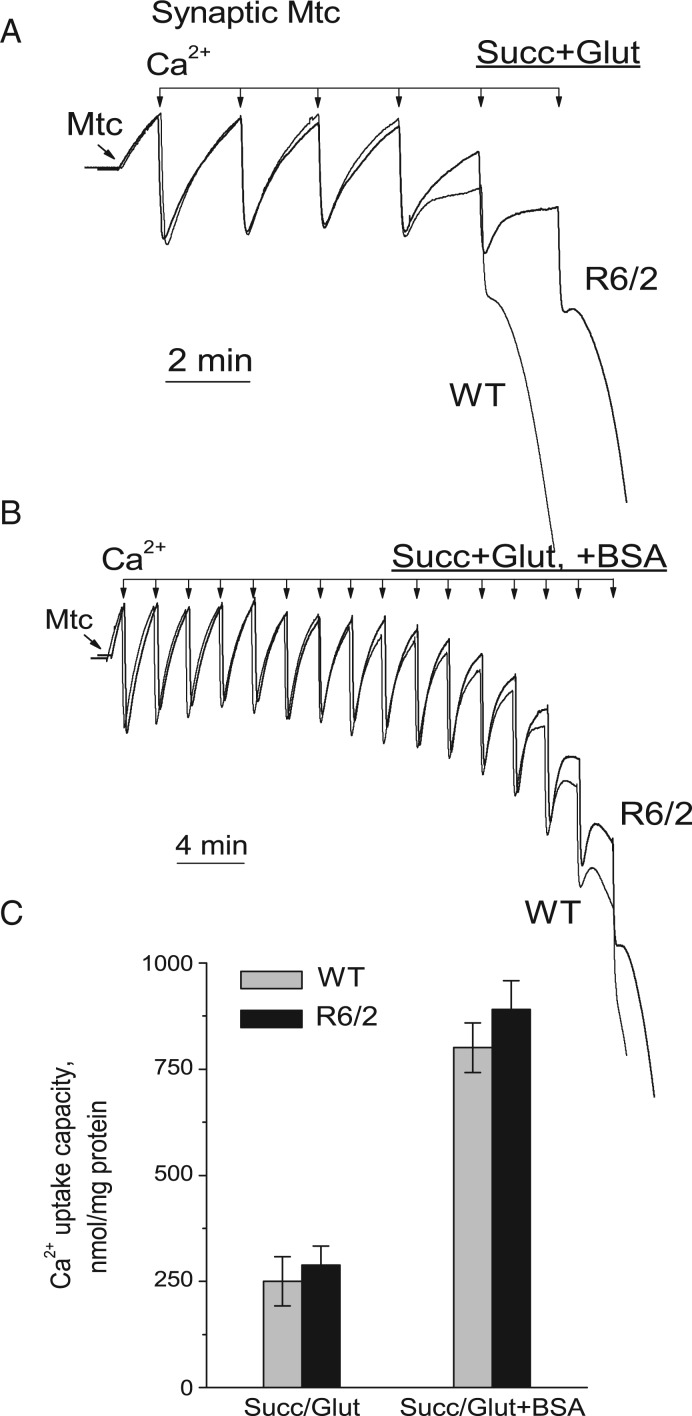
Next, we compared the Ca2+ uptake capacity in synaptic and non-synaptic mitochondria isolated from 6- to 8-week-old R6/2 and WT mice. Mitochondria were subjected to 10 µM Ca2+ pulses every 2 min (Figs. 5 and 6). The Ca2+ uptake capacities in synaptic and non-synaptic mitochondria from R6/2 and WT mice appeared to be similar. In experiments with synaptic mitochondria, Ca2+ uptake capacity was lower than in experiments with non-synaptic mitochondria (Fig. 6). To improve Ca2+ uptake capacity in synaptic mitochondria, we used BSA, which increases Ca2+ uptake capacity, as shown in our previous study (6). In this study, BSA increased Ca2+ uptake capacity of synaptic mitochondria. Importantly, BSA similarly increased Ca2+ uptake capacity in synaptic mitochondria from both R6/2 and WT mice (Fig. 6B and C). Similar results were obtained with non-synaptic mitochondria (not shown).
Figure 5.
Ca2+ uptake capacity of brain non-synaptic mitochondria isolated from 6-to 8-week-old WT (thin traces) and R6/2 (thick traces) mice. Ca2+ uptake was assessed in mitochondria incubated at 37 °C in the standard incubation medium supplemented with either 3 mM succinate plus 3 mM glutamate (A) or 3 mM pyruvate plus 1 mM malate (B). In all Ca2+ uptake experiments, incubation medium was additionally supplemented with 100 µM ADP and 1 µM oligomycin. In (C) is statistical analysis of Ca2+ uptake capacity of mitochondria from R6/2 and WT mice. Data are mean ± SEM, N = 5–6 separate experiments.
Figure 6.
Ca2+ uptake capacity of brain synaptic mitochondria isolated from 6- to 8-week-old WT (thin traces) and R6/2 (thick traces) mice. Ca2+ uptake was assessed in mitochondria incubated at 37 °C in the standard incubation medium supplemented with 3 mM succinate plus 3 mM glutamate either without (A) or with (B) 0.1% BSA in the incubation medium. In all Ca2+ uptake experiments, incubation medium was additionally supplemented with 100 µM ADP and 1 µM oligomycin. In (C) is statistical analysis of Ca2+ uptake capacity of mitochondria from R6/2 and WT mice. Data are mean ± SEM from 5 separate experiments.
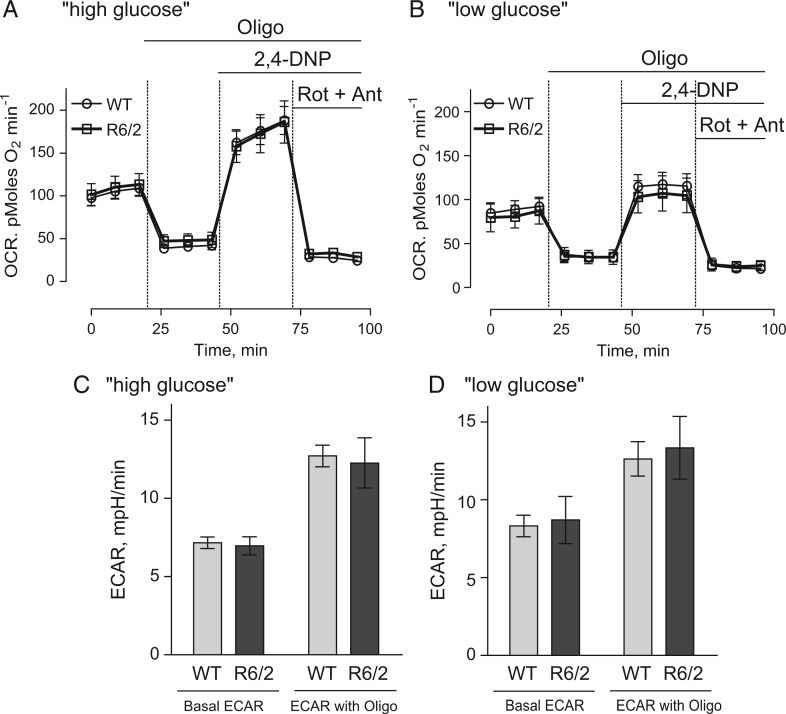
Although mitochondria isolated from brains of R6/2 and WT mice had similar respiratory activities, we could not rule out the effect of mHtt fragments on mitochondrial respiration under more physiological conditions. Striatum is the most vulnerable region of the brain in HD (20). Consequently, we examined the respiratory activities of cultured striatal neurons derived from R6/2 and WT mice using the Seahorse XF24 extracellular flux analyzer. Additionally, in these experiments we evaluated glycolytic activities in striatal neurons. OCR was used to evaluate cellular respiration and extracellular acidification rate (ECAR) was used to assess glycolytic activity (5). Each experiment followed the same protocol with sequential measurements of basal respiratory activity, oligomycin-sensitive respiration coupled to ATP synthesis, and 2,4-DNP-stimulated, maximal respiration. At the end of the experiment, neurons were treated with rotenone and antimycin A to completely inhibit mitochondrial respiration. It was previously suggested that supplementing the bath solution with 15 mM pyruvate in addition to 10 mM glucose would accentuate mitochondrial respiration (21). Under these ‘high glucose plus pyruvate’ conditions, we did not find any difference in respiration and glycolytic activities of striatal neurons from R6/2 and WT mice (Fig. 7A and C).
Figure 7.
OCR and ECAR of cultured striatal neurons from WT and R6/2 mice in ‘high’ and ‘low’ glucose medium conditions. Striatal neurons from post-natal day 1 R6/2 and WT mice were grown in culture for 8 DIV before measurements. Where indicated, the bath solution contained either ‘high glucose’ (10 mM glucose, 15 mM pyruvate) or ‘low glucose’ (2.5 mM glucose). Where indicated, cells were treated with 1 µM oligomycin (Oligo), 60 µM 2,4-DNP and 1 µM rotenone (Rot) plus 1 µM antimycin A (Ant). In (A) and (B), OCR of striatal neurons in ‘high glucose’ or ‘low glucose’ medium conditions, respectively. In (C) and (D), ECAR of striatal neurons in ‘high glucose’ or ‘low glucose’ medium conditions, respectively. A Seahorse XF24 flux analyzer (Seahorse Bioscience, Billerica, MA, USA) was used to measure both OCR and ECAR at 37 °C, seeded at 105 cells per well. Data are mean ± SEM from five to nine separate experiments.
Recently, it was proposed, that impaired respiratory activity may become apparent in mHtt-expressing neurons when the cells are incubated in a ‘low glucose’ medium that contained no pyruvate (22). Under these conditions, neurons from BACHD rats had decreased carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP)-stimulated maximal respiratory rates compared with neurons from WT animals. We assessed OCR and ECAR of striatal neurons from R6/2 and WT animals in ‘low glucose’ medium (2.5 mM glucose, no pyruvate) and did not find any statistically significant difference in respiratory and glycolytic activities of these cells (Fig. 7B and D). Consistent with the lack of impairment in respiratory and glycolytic activities in striatal neurons, ATP levels in neurons from R6/2 mice (17.9 ± 1.1 nmol ATP/mg protein, N = 5) were not statistically different compared with ATP levels in neurons from WT mice (18.7 ± 1.0 nmol ATP/mg protein, N = 5).
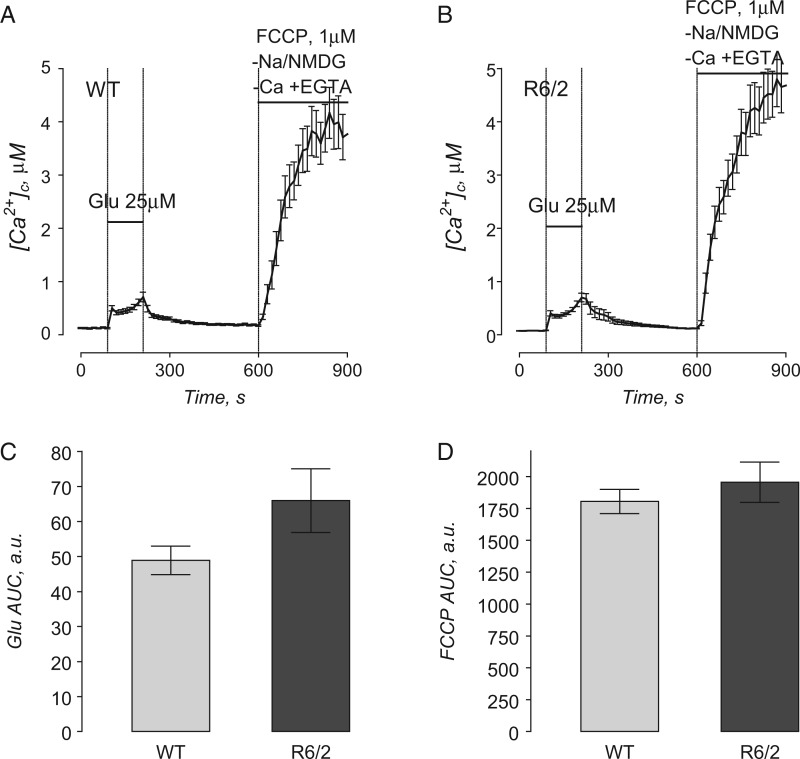
We also assessed mitochondrial Ca2+ accumulation in striatal neurons derived from R6/2 and WT mice. For these measurements, we stimulated Ca2+ influx into neurons and subsequently into mitochondria by briefly exposing cells to glutamate (25 µM glutamate plus 10 µM glycine) (Fig. 8). Changes in cytosolic Ca2+ were monitored by following fluorescence of Fura-2FF. After glutamate removal and return of cytosolic Ca2+ to near resting level, the release of mitochondrial Ca2+ into the cytosol was triggered by mitochondrial depolarization with 1 µM FCCP (Fig. 8). The magnitude of cytosolic Ca2+ elevation following depolarization was taken as a measure of mitochondrial Ca2+ accumulation in situ as described previously (6,23). To avoid Ca2+ influx into cells from the extracellular medium, Ca2+ was omitted from the bath solution. To prevent Ca2+ extrusion from the cell via the Na+/Ca2+ exchanger during FCCP-induced increase in cytosolic Ca2+, external Na+ was replaced with equimolar N-methyl-D-glucamine (NMDG) during FCCP application (24). NMDG is a bulk organic cation that cannot be transported by the Na+/Ca2+ exchanger and, therefore, precludes the extrusion of Ca2+ via this mechanism. In these experiments, neurons from R6/2 and WT mice showed comparable transient increases in cytosolic Ca2+ during glutamate application (Fig. 8C). The increase in cytosolic Ca2+ triggered by FCCP was also not different between neurons from R6/2 and WT mice (Fig. 8D). This result, similar to the results obtained with Ca2+ uptake capacity in isolated mitochondria (Figs. 5 and 6), argues against mHtt fragment-mediated alterations in mitochondrial Ca2+ accumulation in the R6/2 mouse model of HD.
Figure 8.
Mitochondrial Ca2+ accumulation following transient glutamate-induced elevations in cytosolic Ca2+ in striatal neurons derived from WT and R6/2 mice. In A and B, the averaged fluorescence signals (mean ± SEM) from the representative experiments are shown. Cytosolic Ca2+ was followed by monitoring Fura-2FF F340/F380 fluorescence ratio at 37 °C. In these experiments, striatal neurons (10–13 DIV) were exposed to 25 µM glutamate with 10 µM glycine for 2 min as indicated. Then, glutamate and glycine were removed to let cytosolic Ca2+ concentration ([Ca2+]c) recover. After [Ca2+]c reached near resting level (7 min after glutamate removal) neurons were treated with 1 µM FCCP to depolarize mitochondria and release accumulated Ca2+. To avoid ambiguity concerning possible Ca2+ influx from the outside of the cell, external Ca2+ was removed simultaneously with glutamate and glycine. In addition, to prevent Ca2+ extrusion from the cell by Na+/Ca2+ exchanger, the external Na+ was replaced by equimolar NMDG as indicated. In (C) and (D), the areas under the curve (AUC) for the averaged fluorescence signals are shown. The AUC for glutamate-induced increase in [Ca2+]c (Glu AUC) was calculated for the 120-s period beginning with glutamate application. The AUC for FCCP-induced increase in [Ca2+]c (FCCP Glu) was calculated for the 300-s period following FCCP application. Data are mean± SEM from five to six separate experiments with 20–25 individual neurons analyzed in each experiment.
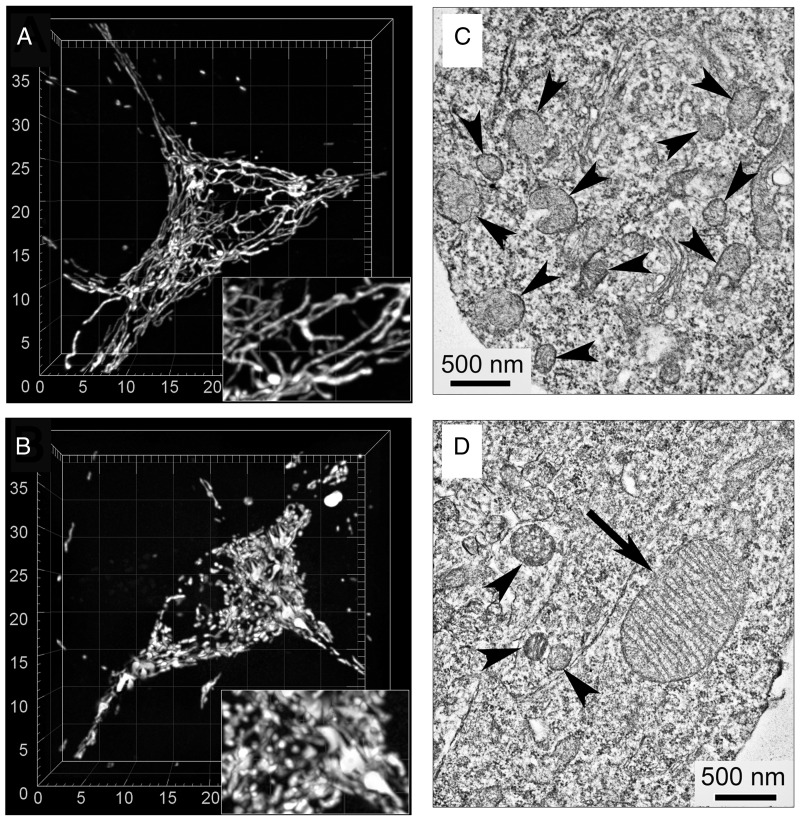
Nevertheless, we believe that the lack of detectable functional defects does not rule out possible alterations in mitochondrial morphology in HD. Previously, morphological imbalance and a shift toward augmented fission was reported in mouse and cell models of HD (25–28). In addition, a dramatic enlargement of mitochondria in neurons (29,30) and peripheral cells (31,32) expressing mHtt was reported. In our experiments, we used live-cell laser spinning-disk confocal microscopy and 3D reconstruction of the mitochondrial network in neurons from WT and R6-2 mice. In parallel experiments, we employed transmission electron microscopy to substantiate data obtained with confocal microscopy. In all analyzed striatal neurons from WT mice (142 neurons), mitochondria had a normal worm-like appearance whereas in about 24% of neurons from R6/2 mice (35 out of 148 analyzed neurons) most of mitochondria were fragmented while some mitochondria were enlarged (Fig. 9). Yet, despite altered mitochondrial morphology in this subpopulation of neurons, the entire neuronal population from R6/2 mice had similar respiratory activities and Ca2+ handling compared with neurons from WT mice. Whether neurons with abnormal mitochondrial morphology observed in our study have alterations in oxidative metabolism and mitochondrial Ca2+ handling is not clear. We will address this question in our future studies.
Figure 9.
Representative 3D reconstructions of mitochondrial networks and electron micrographs of cultured striatal neurons from WT (A, C) and R6/2 (B, D) mice. In A and B, the representative 3D maximal fluorescence intensity projection images of the mitochondrial network in live cultured striatal neurons (9 DIV) from WT and R6/2 mice, respectively (total number of neurons analyzed in these experiments—142 neurons from WT mice and 148 neurons from R6/2 mice). Cells were analyzed in a blind manner. The scales are in µm. In C and D, representative electron micrographs of cultured striatal neurons (9 DIV) from WT and R6/2 mice, respectively. Arrowheads point to small-size, normal-looking mitochondria; the arrow points to enlarged mitochondrion.
Taken together, our results suggest the lack of difference in respiratory activities and Ca2+ handling in isolated brain mitochondria and cultured striatal neurons from R6/2 mice compared with mitochondria and neurons from WT animals.
Discussion
In earlier studies, mitochondrial aberrations were proposed to contribute to HD pathogenesis (for review see (33)). However, despite significant effort, the role of mitochondrial dysfunction in HD still remains controversial. In this study, we assessed the effect of mHtt fragments on mitochondrial respiratory activity and Ca2+ handling using both isolated brain mitochondria and cultured striatal neurons from R6/2 mice. It remains not completely clear whether full-length mHtt or mHtt fragments mediate HD pathogenesis. However, some studies suggest that mHtt fragments might be more prone to aggregation and therefore might be more toxic for neurons (8,9). In our previous study, we demonstrated the lack of mitochondrial respiratory or Ca2+ handling defects in mitochondria and cultured neurons from YAC128 mice, which express human full-length mHtt (5,6). Consequently, in this study, we hypothesized that fragments of mHtt are more harmful to mitochondria than full-length mHtt. If this would be the case, we may expect to detect mitochondrial functional abnormalities induced by mHtt fragments. In this study, we evaluated mitochondrial functions in the R6/2 mouse, a mouse model of HD that expresses fragments of human mHtt with 144 glutamines (12).
In our experiments, we used two model systems: isolated brain mitochondria and primary cultured striatal neurons. It is conceivable that there might be some difference in the results produced with these two model systems. Mitochondria isolated from adult brains may better reflect changes that take place in HD over time. However, isolated mitochondria taken away from their natural intracellular environment may lead to unwanted artifacts (e.g. loss of association with mHtt) and, consequently, to inaccurate interpretations. On the other hand, mitochondria in young post-natal neurons are immersed within their natural intracellular environment, but they might be too young for developing overt functional defects. The use of both, mitochondria isolated from adult brains and mitochondria in post-natal neurons, complement each other and help us to better assess possible effects of mHtt on the organelles.
Earlier studies reported mitochondrial respiratory dysfunction and a decreased ability of mitochondria to accumulate Ca2+ in HD mouse models expressing full-length or fragments of mHtt (34–37). On the other hand, studies from our group (5,6) and from other investigators (23,38,39) suggest the lack of defects in respiration and Ca2+ uptake in mitochondria isolated from brains of HD mice compared with mitochondria from brains of WT animals. Consistent with these previous findings in other mouse models of HD, our data presented here indicate the lack of defects in respiration and Ca2+ uptake in brain mitochondria isolated from R6/2 mice compared with brain mitochondria from WT mice. Similarly, in experiments with cultured striatal neurons, our data demonstrate the lack of difference in cellular respiratory activity and Ca2+ handling between neurons from R6/2 and WT mice.
In this study, we used isolated brain mitochondria from symptomatic 6- to 8-week-old R6/2 mice and age-matched WT mice. The R6/2 mice used in our experiments displayed a clasping phenotype typical for HD animals (12). The presence of clasping behavior assured us that if mitochondrial respiratory abnormalities and aberrant Ca2+ handling by mitochondria were involved in neuronal dysfunction leading to these behavioral aberrations, then we should be able to detect such changes in respiration, Ca2+ handling or both. The lack of difference between mitochondria from WT and R6/2 mice suggests that fragments of mHtt do not impair mitochondrial respiratory and Ca2+ uptake activities. Accordingly, our data argue against defects of mitochondrial functions as a contributing factor to HD-related pathology in R6/2 mice.
Recently, Yano et al. (19) reported the lack of difference in respiration of synaptic and non-synaptic mitochondria from R6/2 mice compared with mitochondria from WT animals. Nevertheless, the authors presented evidence for mHtt-induced inhibition of protein import into mitochondria, and suggested that such an inhibition might affect mitochondrial functions later in the disease progression (19). According to the authors, this inhibition of mitochondrial protein import might result in a decreased expression of nuclear-encoded mitochondrial proteins, but the authors did not provide experimental evidence supporting this notion (19). Our data with mitochondria isolated from R6/2 mice indicate the lack of difference in expression of several randomly selected nuclear-encoded mitochondrial proteins, suggesting the lack of defects in mitochondrial protein import. This is consistent with the study by Orr et al. (2008) that found that expression of nuclear encoded MnSOD as well as 30 and 70 kDa subunits of succinate dehydrogenase (Complex II) is similar in brain mitochondria isolated from 3- and 10-month-old heterozygous knock-in 150Q/7Q mice as well as in 3-month-old WT 7Q/7Q mice (40). Milakovic and Johnson (2005) also failed to demonstrate a difference in expression of 30 and 70 kDa subunits of Complex II in mutant STHdhQ111/Q111 striatal cells compared with WT STHdhQ7/Q7 cells (41).
Previously, Rosenstock et al. (42) reported a significant increase in cytosolic Ca2+ induced by 1 mM glutamate in cortical and striatal slices of 9 month-old R6/1 transgenic mice. Following glutamate removal, FCCP-mediated mitochondrial depolarization led to Ca2+ release from mitochondria and resulted in an increase in cytosolic Ca2+ that was similar in slices from both R6/1 and WT animals. This suggests the lack of defects in mitochondrial Ca2+ accumulation in brain slices from R6/1 mice.
The lack of defects in Ca2+ uptake by non-synaptic mitochondria isolated from brains of R6/2 mice was also found by David Nicholls’ group (21). Contrary to expectations, they found a moderate but statistically significant increase in Ca2+ uptake capacity of mitochondria isolated from brains of R6/2 mice compared with mitochondria from WT animals. Although we did not find a statistically significant increase in Ca2+ uptake capacity in isolated brain mitochondria from R6/2 mice, we confirm the lack of deterioration of mitochondrial Ca2+ uptake capacity in these mitochondria. The authors also found an increase in Ca2+ uptake capacity in non-synaptic mitochondria from YAC128 mice, which express full-length mHtt, and the lack of difference in mitochondria from knock-in Hdh150/+ mice (21). In addition, the authors did not find a significant difference in respiratory activity of striatal neurons from Hdh150/+ and WT mice.
The reason for the increased Ca2+ uptake capacity in mitochondria from HD mice is not clear but might be related to compensatory changes in mitochondria and alterations in susceptibility to induction of the permeability transition pore (PTP), which determines mitochondrial Ca2+ uptake capacity (43). Interestingly, Choo et al. (44) reported a significantly increased level of glutathione in cortical and striatal mitochondria from R6/2 mice. The authors interpreted this finding as an indication of compensatory reaction to protect cells against an increase in mitochondrial oxidative stress. An increase in mitochondrial glutathione may also suggest increased resistance to Ca2+-induced damage that manifests in induction of the PTP. Indeed, glutathione was found to be protective against Ca2+-induced PTP in liver (45), heart (46) and brain mitochondria (47). Consequently, the same mechanism might contribute to the lack of difference in Ca2+ uptake capacity in our study of mitochondria from R6/2 and WT mice.
Mitochondrial CyD is a component of the mitochondrial PTP that sensitizes it to Ca2+ (48). Genetic ablation of CyD inhibits PTP induction (49) and significantly increases Ca2+ uptake capacity in brain mitochondria (50). Considering this, it was logical to propose that if mitochondria from HD mice have increased propensity to PTP induction and, concomitant decrease in Ca2+ uptake capacity and if this defect in Ca2+ handling plays an important role in HD pathology, then CyD deletion should protect mitochondria, increase Ca2+ uptake capacity, and rescue behavioral deficits in R6/2 mice. Perry et al. reported elegant experiments in which the authors crossed R6/2 with CyD-knockout mice (CyD−/−) and evaluated mitochondrial Ca2+ uptake capacity in mitochondria from cerebral cortex of crosses of CyD−/− and R6/2 mice (51). The authors found a significant increase in Ca2+ uptake capacity in mitochondria from R6/2:CyD−/− mice but no indication of behavioral improvement or alleviation of HD neuropathological features. From these results, the authors concluded that increasing neuronal mitochondrial Ca2+ uptake capacity is not beneficial in the R6/2 model of HD. Unfortunately, in this study the authors failed to compare Ca2+ uptake capacity of brain mitochondria from R6/2 and WT mice. Based on our data and results from David Nicholls’ group (21), Ca2+ uptake capacities of isolated brain mitochondria and mitochondria in cultured neurons from R6/2 mice are not decreased compared with Ca2+ capacity of mitochondria from WT animals. Consequently, alterations in Ca2+ uptake capacity most likely do not play a role in HD pathology and one should not expect significant improvement in HD pathology due to an increase in mitochondrial Ca2+ uptake capacity.
Overall, the data presented in this article, recent findings by our group (5,6), and by other investigators (23,38,39) show no significant differences in respiratory activity and Ca2+ handling in mitochondria exposed to human full-length or truncated mHtt. Consequently, based on our results and data from other groups, the dearth of defects in bioenergetics and Ca2+ handling makes their contribution to HD pathology unlikely. However, this does not rule out the potential contribution of mitochondria to HD pathogenesis. The neurotoxic effect of mHtt or its fragments may be due to mechanisms that are not directly related to mitochondrial bioenergetics or Ca2+ transport but may indirectly affect these functions. These mechanisms may include oxidative stress associated with elevated NAD(P)H oxidase activity (52), alterations in cholesterol metabolism (53), aberrant vesicular trafficking (27). In addition, numerous reports indicate alterations in mitochondrial morphology and traffic in cell and mouse models of HD (28,54,55). Our observations of alterations in mitochondrial morphology in neurons from R6/2 mice made with spinning-disk confocal and electron microscopy are consistent with previously reported mitochondrial morphological abnormalities found in cells expressing mHtt (25–32). It is conceivable that these alterations in mitochondrial morphology as well as in non-mitochondrial mechanisms may play a significant role in neuronal dysfunction and over time may lead to the neuronal loss and behavioral deficits seen in HD.
Materials and Methods
Materials
Pyruvate, malate, succinate, glutamate, Ethylene glycol tetraacetic acid (EGTA), ADP, oligomycin, rotenone, antimycin A, 2,4-DNP, NMDG and carbonylcyanide-p-trifluoromethoxyphenylhydra-zone (FCCP) were purchased from Sigma (St. Louis, MO, USA). BSA, free from free fatty acids, was from MP Biomedicals (Irvine, CA, USA).
Animals
All procedures with animals were performed in accordance with the Institutional Animal Care and Use Committee approved protocol. Mice were purchased from Jackson Laboratories (Bar Harbor, ME) and breeding colonies were established in Laboratory Animal Resource Center at Indiana University School of Medicine, Indianapolis, IN. R6/2 mice and their WT littermates were generated by crossing WT CBAxC57Bl/6 F1 males with CBAxC57Bl/6 females that had ovary transplants from R6/2 mice. All offspring were genotyped with a PCR assay on tail DNA (12). The mice were housed under standard conditions with free access to water and food. In experiments with isolated brain mitochondria, we used symptomatic 6- to 8-week-old R6/2 mice and their WT littermates (background: B6CBA). For preparing cultured neurons, we used post-natal day 1 (PN1) mouse pups. R6/2 mice express the N-terminus fragment of the human mHtt gene that encodes a pathologically expanded polyglutamine stretch with 144 glutamines (12). In our experiments, we used animals of both sexes. However, in every individual experiment we only used animals of the same sex. We did not find significant difference in the results obtained with either sex and, therefore, the data were pooled together for statistical analysis.
Genotyping
All offspring were genotyped using a PCR assay on tail DNA (12). Briefly, PCR of tail DNA was carried out following the protocol provided by Jackson Laboratories with oligonucleotide primers oIMR1594 (CCGCTCAGGTTCTGCTTTTA) and oIMR1596 (TTGAAGGACTTGAGGGACTC) obtained from Invitrogen. Oligonucleotide primers oIMR7338 (CTAGGCCACAGAATTGAAAGATCT) and oIMR7339 (GTAGGTGGAAA-TTCTAGCATCATCC) (both from Invitrogen) were used as internal positive control. The PCR reaction mixture consisted of 1 µl DNA template and 23 µl Platinum PCR SuperMix (Invitrogen) supplemented with 0.5 µM of each primer (Invitrogen), total volume 25 µl. Cycling conditions were 2 min at 94°C, 10 cycles for 20 s at 94°C followed by 15 s at 65°C with −0.5°C per cycle temperature decrease and 10 s at 68°C. Then, 28 cycles for 15 s at 94 °C followed by 15 s at 60 °C and 10 s at 72 °C, concluded by 1 min at 72°C (Jackson Laboratories, Bar Harbor, ME). Reaction products were analyzed on 1.2% agarose gel run at 100 V for 60 min with Tris-acetate-EDTA running buffer containing 1× GelRed Nucleic Acid Gel Stain (Biotium, CA).
Isolation of non-synaptic and synaptic brain mitochondria
Percoll gradient-purified brain non-synaptic and synaptic mitochondria from 6- to 8-week old R6/2 and WT mice were isolated as we described previously (56,57). Previously, it was proposed that BSA may displace mHtt from its binding sites on mitochondria (15). For this reason, we omitted BSA from all solutions used in our experiments with isolated mitochondria unless otherwise mentioned.
Mitochondrial respiration
Mitochondrial respiration was assessed in a 0.4 ml, continuously stirred, chamber containing the standard incubation medium which was composed of 125 mM KCl, 0.5 mM MgCl2, 3 mM KH2PO4, 10 mM Hepes, pH 7.4, 10 µM EGTA, supplemented either with 3 mM pyruvate plus 1 mM malate or 3 mM succinate plus 3 mM glutamate. Experiments with succinate were supplemented with glutamate to prevent inhibition of succinate dehydrogenase by oxaloacetate (17,18). The chamber was maintained at 37°C and was equipped with a Clark-type oxygen electrode and a tightly sealed lid. The slope of the oxygen electrode trace corresponded to the respiratory rate.
Mitochondrial Ca2+ uptake capacity
Mitochondrial Ca2+ uptake was measured using a 0.3 ml chamber at 37°C under continuous stirring that was outfitted with a miniature Ca2+-selective electrode. Mitochondrial Ca2+ uptake was indicated by a decrease in the external Ca2+ concentration. The standard incubation medium contained 125 mM KCl, 0.5 mM MgCl2, 3 mM KH2PO4, 10 mM Hepes, pH 7.4, 10 µM EGTA, and was supplemented either with 3 mM pyruvate plus 1 mM malate or 3 mM succinate plus 3 mM glutamate. Succinate was used in combination with glutamate to prevent oxaloacetate inhibition of succinate dehydrogenase (17,18). The incubation medium was further supplemented with 0.1 mM ADP and 1 µM oligomycin as described previously (43). Ca2+ was delivered to mitochondria as 10 µM CaCl2 pulses.
Cell culturing
Primary neuronal cultures were prepared from the striatum of post-natal day 1 R6/2 pups and WT littermates as described previously (58), but without pooling cells from different pups together. For respirometry experiments, neurons were plated as described for Ca2+ imaging experiments (58), but neurons were seeded onto 24-well Seahorse plates. To inhibit proliferation of non-neuronal cells, 35 mg·ml−1 uridine plus 15 mg·ml−1 5-fluoro-2′-deoxyuridine were added 24 h after plating. Neurons were maintained in a 5% CO2 atmosphere at 37 °C in Neurobasal Medium with B27 supplement (Life Technologies).
Cell respirometry
A Seahorse XF24 flux analyzer (Seahorse Bioscience, Billerica, MA, USA) was used to measure OCRs of cultured striatal neurons (8 DIV) following the manufacturer’s instructions. Neuronal cultures were seeded in the 24-well assay plates at 105 cells per well. Prior to the experiment, the growth medium was exchanged for the standard bath solution supplemented with 10 mM glucose and 15 mM pyruvate or 2.5 mM glucose alone as stated. The standard bath solution contained 139 mM NaCl, 3 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 10 mM HEPES, pH 7.4, 37 °C. For experiments with bath solution containing 2.5 mM glucose, sucrose was added to maintain osmolarity similar to that of the growth medium (280 mosm).
ATP measurements
ATP content was determined by using a luciferin/luciferase-based ATP bioluminescent somatic cell assay kit (Sigma) and a GloMax 20/20 luminometer (Promega). Cultured striatal neurons (7 DIV) were lysed on ice with 1× Releasing Reagent (Sigma), following manufacturer’s instructions. ATP was measured following precipitation of proteins by perchlorate (4%) and subsequent neutralization of extracts by KOH according to manufacturer’s instructions.
Immunoblotting
Brain isolated mitochondria pretreated with Protease Inhibitor Cocktail (Roche) were solubilized by incubation in NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA, USA) supplemented with a reducing agent at 70°C for 15 min. Bis-Tris Mops gels (12%, Invitrogen) was used for electrophoresis (20 µg protein per lane). After electrophoresis, proteins were transferred to Hybond-ECL nitrocellulose membrane (Amersham Biosciences). Blots were incubated for 1 h at room temperature in blocking solution of 5% BSA, phosphate-buffered saline, pH 7.2, and 0.15% Triton X-100. Then, blots were incubated with one of the following primary antibodies: mouse monoclonal anti-Complex I 39 kDa subunit (Invitrogen, 1:1000), mouse monoclonal anti-Complex II 30 kDa subunit (Invitrogen, 1:1000), mouse monoclonal anti-Complex II 70 kDa subunit (Invitrogen, 1:1000), mouse monoclonal anti-aconitase 2 (Abcam, 1:1000), rabbit polyclonal anti-manganese superoxide dismutase (MnSOD, Millipore, 1:2000), mouse monoclonal anti-ATP synthase α subunit (Abcam, 1:1000), mouse monoclonal anti-aconitase 2 (Abcam, 1:1000), mouse monoclonal anti-CyD antibody (Calbiochem, 1:500), rabbit polyclonal anti-VDAC1 (Calbiochem, 1:1000). Blots were incubated with goat anti-mouse or goat anti-rabbit IgG (1:20 000) coupled with horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) and developed with Supersignal West Pico chemiluminescent reagents (Pierce, Rockford, IL, USA). Molecular mass markers See Blue Plus 2 Standards (5 µl) and HiMark Pre-stained High Molecular Weight Protein Standards (10 µl) (Invitrogen) were used to determine molecular masses of the bands. NIH ImageJ 1.48v software (http://rsb.info.nih.gov//ij, last accessed: April 1, 2016) was used to quantify band densities.
Calcium imaging
Cytosolic Ca2+ was assessed in striatal neurons (10–12 days in vitro, DIV) by loading neurons at 37 °C with 2.6 µM Fura-2FF-AM (Molecular Probes, Eugene, OR) in a bath solution containing 139 mM NaCl, 3 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 10 mM NaHEPES, pH 7.4 and 5 mM glucose. Osmolarity of the bath solutions was measured with an Osmette II osmometer (Precision Systems Inc., Natick, MA) to ensure that it was close to the osmolarity of the growth medium (280 mosm). A Nikon Eclipse TE2000-U inverted microscope using a Nikon CFI Plan Fluor 20× 0.45 NA objective and a back-thinned EM-CCD camera, Hamamatsu C9100-12 (Hamamatsu Photonic Systems, Bridgewater, NJ) controlled by Simple PCI software 6.1 (Compix Inc., Sewickley, PA) was used to take fluorescent images. A Lambda-LS system (Sutter Instruments, Novato, CA) was used to deliver excitation light. A Lambda 10-2 optical filter changer (Sutter Instruments, Novato, CA) was used to control excitation filters (340 ± 5 and 380 ± 7 nm). Fluorescence was recorded through a 505 nm dichoroic mirror at 535 ± 25 nm. Images were taken every 15 s for the duration of the experiment. The exposure time was chosen that provided satisfactory image quality but minimal exposure. The ratio of F340/F380, calculated following background subtraction in both channels, was used to monitor changes in cytosolic Ca2+ concentration ([Ca2+]c). After 90 s of baseline fluorescence recording in standard bath solution, glutamate (25 µM) plus glycine (10 µM) was applied to the neuronal culture. At the end of the experiment, the glutamate-containing solution was replaced with a glutamate-, Ca2+- and Na+-free solution. The Na+ was replaced by equimolar NMDG. Then 1 µM FCCP was applied to the neurons, causing neuronal mitochondria to depolarize and leading to release of accumulated mitochondrial Ca2+ into the cytosol. The Grynkiewicz method (59) was used for calculation of [Ca2+]c from Fura-2FF signals, using an assumed Kd for Fura-2FF of 5.5µM. Fluorescence background was subtracted in every experiment. As has been suggested previously (60,61), it should be noted that the Ca2+ binding and spectroscopic properties of fluorescent dyes can differ depending on the intracellular environment. Therefore calculations of [Ca2+]c should be considered estimates.
Neuronal transfection
To visualize mitochondria within live cells, cultured striatal neurons were transfected in suspension during plating using an electroporator BTX 630 ECM (Harvard Apparatus, Holliston, MA) with a plasmid encoding mitochondrially-targeted enhanced yellow fluorescent protein (mito-eYFP, generously provided by Dr. Roger Tsien, UCSD) as we described before for hippocampal neurons (62). This procedure usually provided a 10–15% transfection rate in primary cultures of rat hippocampal neurons compared with <1% efficacy with commercial cationic lipid liposomes. The transfected neurons were imaged 8–10 days after transfection.
Live-cell laser spinning-disk confocal microscopy
Mitochondrial morphology in live striatal neurons was analyzed as described previously (62). Briefly, serial images of neuronal mitochondria, visualized with mito-eYFP, were collected using spinning-disk confocal microscopy. A Nikon Eclipse TE2000-U microscope equipped with a Yokogawa spinning-disk confocal unit CSU-10, a back-thinned EM-CCD camera Andor iXonEM+ DU-897 (Andor Technology, South Windsor, CT), and a motorized stage Prior H-117 (Prior Scientific, Rockland, MA) was employed. This setup was controlled by Andor iQ 1.4 software (Andor Technology, South Windsor, CT). To image mitochondria, neurons were exposed to excitation light at 488 nm using an air-cooled Kr/Ar laser T643-RYB-A02 (Melles Griot, Carlsbad, CA). The laser power was set to the minimum (<5%) to prevent photobleaching. Fluorescence was collected through a 505 nm dichroic mirror and a 535 ± 25 nm emission filter using a Nikon CFI Plan Apo 100× 1.4 NA objective. Serial images (z-stacks) were collected using the piezoelectric positioning device PIFOC P-721 (Physik Instrumente, Auburn, MA) with a z-step of 0.1 µm. The spatial resolution of the Andor iXonEM+ DU-897 camera was increased by installing a 2× extender lens in front of the camera. Three-dimensional blind deconvolution of z-stacks and 3D rendering were performed using AutoDeblur Gold CF 1.4.1 software (MediaCybernetics, Silver Spring, MD). Three-dimensional maximal projection reconstructions of the mitochondrial network in live neurons were performed using Imaris 6.4.0 (Bitplane Inc., St. Paul, MN).
Transmission electron microscopy
Electron microscopy of cultured striatal neurons was performed similarly to the procedure described earlier (63). Briefly, neurons were fixed in 2% glutaraldehyde in the standard bath solution at room temperature for 15 min. The cells were pelleted by centrifugation and the supernatant was discarded. The pellet was layered with a fresh solution of 2% glutaraldehyde in the standard bath solution and left overnight at 4 °C. The samples were postfixed in 1% osmium tetroxide for 1 h and dehydrated through a series of graded ethyl alcohols before embedding in the resin Embed 812 (Electron Microscopy Sciences, Fort Washington, PA). Thick sections were cut on an Ultracut UCT microtome (Leica, Bannockburn, IL), and then thin sections were cut using a diamond knife (Diatome, Electron Microscopy Sciences) at 70–90 nm and stained with uranyl acetate and lead citrate. Digital electron micrographs were taken using a Tecnai G12 BioTwin electron microscope (FEI, Hillsboro, OR) equipped with an AMT2.6 × 2.6K digital CCD camera. Mitochondrial morphology in striatal neurons was analyzed in a blind manner.
Statistics
Power analysis was performed using G*Force software version 3.1.9.2 (by Franz Faul, Universitat Kiel, Germany) to establish the sample size necessary to detect a 20% difference between mitochondria from WT and YAC128 mice. Based on this power analysis, the number of experiments that gives an 80% likelihood of detecting 20% difference between two means at the significance level of α = 0.05 is 5–6. Data are shown as mean ± SEM of indicated number of separate experiments. Statistical analysis of the experimental results consisted of unpaired t-test or one-way ANOVA followed by Bonferroni’s post hoc test if applicable (GraphPad Prism 4.0, GraphPad Software Inc., San Diego, CA, USA).
Acknowledgements
The authors are very grateful to Caroline Miller (Electron Microscopy Center, Indiana University School of Medicine) for help with electron microscopy.
Conflict of Interest Statement. None declared.
Funding
This work was supported by National Institutes of Health grant (R01 NS078008 to N.B.). N.B. is a Showalter Scholar.
References
- 1.Roze E., Bonnet C., Betuing S., Caboche J. (2010) Huntington’s disease. Adv. Exp. Med. Biol., 685, 45–63. [PubMed] [Google Scholar]
- 2.MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N. et al. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 3.Beal M.F., Hyman B.T., Koroshetz W. (1993) Do defects in mitochondrial energy metabolism underlie the pathology of neurodegenerative diseases? Trends Neurosci., 16, 125–131. [DOI] [PubMed] [Google Scholar]
- 4.Giacomello M., Hudec R., Lopreiato R. (2011) Huntington’s disease, calcium, and mitochondria. Biofactors, 37, 206–218. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton J., Pellman J.J., Brustovetsky T., Harris R.A., Brustovetsky N. (2015) Oxidative metabolism in YAC128 mouse model of Huntington’s disease. Hum. Mol. Genet., 24, 4862–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellman J.J., Hamilton J., Brustovetsky T., Brustovetsky N. (2015) Ca(2+) handling in isolated brain mitochondria and cultured neurons derived from the YAC128 mouse model of Huntington’s disease. J. Neurochem., 134, 652–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Li S.H., Johnston H., Shelbourne P.F., Li X.J. (2000) Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet., 25, 385–389. [DOI] [PubMed] [Google Scholar]
- 8.Kim M., Lee H.S., LaForet G., McIntyre C., Martin E.J., Chang P., Kim T.W., Williams M., Reddy P.H., Tagle D. et al. (1999) Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J. Neurosci., 19, 964–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wellington C.L., Singaraja R., Ellerby L., Savill J., Roy S., Leavitt B., Cattaneo E., Hackam A., Sharp A., Thornberry N. et al. (2000) Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J. Biol. Chem., 275, 19831–19838. [DOI] [PubMed] [Google Scholar]
- 10.Tian J., Yan Y.P., Zhou R., Lou H.F., Rong Y., Zhang B.R. (2014) Soluble N-terminal fragment of mutant Huntingtin protein impairs mitochondrial axonal transport in cultured hippocampal neurons. Neurosci. Bull., 30, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong B.K., Ehrnhoefer D.E., Graham R.K., Martin D.D., Ladha S., Uribe V., Stanek L.M., Franciosi S., Qiu X., Deng Y. et al. (2015) Partial rescue of some features of Huntington disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol. Dis., 76, 24–36. [DOI] [PubMed] [Google Scholar]
- 12.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell, 87, 493–506. [DOI] [PubMed] [Google Scholar]
- 13.Stack E.C., Kubilus J.K., Smith K., Cormier K., Del Signore S.J., Guelin E., Ryu H., Hersch S.M., Ferrante R.J. (2005) Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington’s disease transgenic mice. J. Comp Neurol., 490, 354–370. [DOI] [PubMed] [Google Scholar]
- 14.Reddy P.H., Charles V., Williams M., Miller G., Whetsell W.O, Jr, Tagle D.A. (1999) Transgenic mice expressing mutated full-length HD cDNA: a paradigm for locomotor changes and selective neuronal loss in Huntington’s disease. Philos. Trans. R. Soc. Lond B Biol. Sci., 354, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panov A.V., Burke J.R., Strittmatter W.J., Greenamyre J.T. (2003) In vitro effects of polyglutamine tracts on Ca2+-dependent depolarization of rat and human mitochondria: relevance to Huntington’s disease. Arch. Biochem. Biophys., 410, 1–6. [DOI] [PubMed] [Google Scholar]
- 16.Lai, J.C.K. and Clark, J.B. (1989) In A.A. Boulton, G.B. Baker, R.F. Butterworth (eds.), Neuromethods. pp. 43–98.
- 17.Wojtczak L., Wojtczak A.B., Ernster L. (1969) The inhibition of succinate dehydrogenase by oxalacetate. Biochim. Biophys. Acta, 191, 10–21. [DOI] [PubMed] [Google Scholar]
- 18.Oestreicher A.B., van den Bergh S.G., Slater E.C. (1969) The inhibition by 2,4-dinitrophenol of the removal of oxaloacetate formed by the oxidation of succinate by rat-liver and -heart mitochondria. Biochim. Biophys. Acta, 180, 45–55. [DOI] [PubMed] [Google Scholar]
- 19.Yano H., Baranov S.V., Baranova O.V., Kim J., Pan Y., Yablonska S., Carlisle D.L., Ferrante R.J., Kim A.H., Friedlander R.M. (2014) Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci., 17, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuccato C., Valenza M., Cattaneo E. (2010) Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev., 90, 905–981. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira J.M., Jekabsons M.B., Chen S., Lin A., Rego A.C., Goncalves J., Ellerby L.M., Nicholls D.G. (2007) Mitochondrial dysfunction in Huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J. Neurochem., 101, 241–249. [DOI] [PubMed] [Google Scholar]
- 22.Gouarne C., Tardif G., Tracz J., Latyszenok V., Michaud M., Clemens L.E., Yu-Taeger L., Nguyen H.P., Bordet T., Pruss R.M. (2013) Early deficits in glycolysis are specific to striatal neurons from a rat model of Huntington disease. PLoS. One, 8, e81528.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang D.T., Rintoul G.L., Pandipati S., Reynolds I.J. (2006) Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis., 22, 388–400. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Van B.C. (1995) Na(+)-Ca2+ exchange in intact endothelium of rabbit cardiac valve. Circ. Res., 76, 396–404. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Lim P.J., Karbowski M., Monteiro M.J. (2009) Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum. Mol. Genet., 18, 737–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., Malorni W., Davies K.J., Carafoli E., Scorrano L. (2010) Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol. Med., 2, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., Mao P., Reddy P.H. (2012) Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet., 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R, et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tellez-Nagel I., Johnson A.B., Terry R.D. (1974) Studies on brain biopsies of patients with Huntington’s chorea. J. Neuropathol. Exp. Neurol., 33, 308–332. [DOI] [PubMed] [Google Scholar]
- 30.Bayram-Weston Z., Jones L., Dunnett S.B., Brooks S.P. (2012) Light and electron microscopic characterization of the evolution of cellular pathology in HdhQ92 Huntington’s disease knock-in mice. Brain Res. Bull., 88, 171–181. [DOI] [PubMed] [Google Scholar]
- 31.Squitieri F., Cannella M., Sgarbi G., Maglione V., Falleni A., Lenzi P., Baracca A., Cislaghi G., Saft C., Ragona G, et al. (2006) Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech. Ageing Dev., 127, 217–220. [DOI] [PubMed] [Google Scholar]
- 32.Squitieri F., Falleni A., Cannella M., Orobello S., Fulceri F., Lenzi P., Fornai F. (2010) Abnormal morphology of peripheral cell tissues from patients with Huntington disease. J. Neural Transm., 117, 77–83. [DOI] [PubMed] [Google Scholar]
- 33.Brustovetsky N. (2015) Mutant huntingtin and elusive defects in oxidative metabolism and mitochondrial calcium handling. Mol. Neurobiol., 53, 2944–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panov A.V., Gutekunst C.A., Leavitt B.R., Hayden M.R., Burke J.R., Strittmatter W.J., Greenamyre J.T. (2002) Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci., 5, 731–736. [DOI] [PubMed] [Google Scholar]
- 35.Lim D., Fedrizzi L., Tartari M., Zuccato C., Cattaneo E., Brini M., Carafoli E. (2008) Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J. Biol. Chem., 283, 5780–5789. [DOI] [PubMed] [Google Scholar]
- 36.Kim S.H., Thomas C.A., Andre V.M., Cummings D.M., Cepeda C., Levine M.S., Ehrlich M.E. (2011) Forebrain striatal-specific expression of mutant huntingtin protein in vivo induces cell-autonomous age-dependent alterations in sensitivity to excitotoxicity and mitochondrial function. ASN. Neuro, 3, e00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damiano M., Diguet E., Malgorn C., D'Aurelio M., Galvan L., Petit F., Benhaim L., Guillermier M., Houitte D., Dufour N, et al. (2013) A role of mitochondrial complex II defects in genetic models of Huntington’s disease expressing N-terminal fragments of mutant huntingtin. Hum. Mol. Genet., 22, 3869–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guidetti P., Charles V., Chen E.Y., Reddy P.H., Kordower J.H., Whetsell W.O., Jr, Schwarcz R., Tagle D.A. (2001) Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp. Neurol., 169, 340–350. [DOI] [PubMed] [Google Scholar]
- 39.Olah J., Klivenyi P., Gardian G., Vecsei L., Orosz F., Kovacs G.G., Westerhoff H.V., Ovadi J. (2008) Increased glucose metabolism and ATP level in brain tissue of Huntington’s disease transgenic mice. Febs J., 275, 4740–4755. [DOI] [PubMed] [Google Scholar]
- 40.Orr A.L., Li S., Wang C.E., Li H., Wang J., Rong J., Xu X., Mastroberardino P.G., Greenamyre J.T., Li X.J. (2008) N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci., 28, 2783–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milakovic T., Johnson G.V. (2005) Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J. Biol. Chem., 280, 30773–30782. [DOI] [PubMed] [Google Scholar]
- 42.Rosenstock T.R., Bertoncini C.R., Teles A.V., Hirata H., Fernandes M.J., Smaili S.S. (2010) Glutamate-induced alterations in Ca2+ signaling are modulated by mitochondrial Ca2+ handling capacity in brain slices of R6/1 transgenic mice. Eur. J. Neurosci., 32, 60–70. [DOI] [PubMed] [Google Scholar]
- 43.Chalmers S., Nicholls D.G. (2003) The relationship between free and total calcium concentrations in the matrix of liver and brain mitochondria. J. Biol. Chem., 278, 19062–19070. [DOI] [PubMed] [Google Scholar]
- 44.Choo Y.S., Mao Z., Johnson G.V., Lesort M. (2005) Increased glutathione levels in cortical and striatal mitochondria of the R6/2 Huntington’s disease mouse model. Neurosci. Lett., 386, 63–68. [DOI] [PubMed] [Google Scholar]
- 45.Chernyak B.V., Bernardi P. (1996) The mitochondrial permeability transition pore is modulated by oxidative agents through both pyridine nucleotides and glutathione at two separate sites. Eur. J. Biochem., 238, 623–630. [DOI] [PubMed] [Google Scholar]
- 46.Aon M.A., Cortassa S., Maack C., O’Rourke B. (2007) Sequential opening of mitochondrial ion channels as a function of glutathione redox thiol status. J. Biol. Chem., 282, 21889–21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heales S.J., Bolanos J.P. (2002) Impairment of brain mitochondrial function by reactive nitrogen species: the role of glutathione in dictating susceptibility. Neurochem. Int., 40, 469–474. [DOI] [PubMed] [Google Scholar]
- 48.Rasola A., Bernardi P. (2007) The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis., 12, 815–833. [DOI] [PubMed] [Google Scholar]
- 49.Baines C.P., Kaiser R.A., Purcell N.H., Blair N.S., Osinska H., Hambleton M.A., Brunskill E.W., Sayen M.R., Gottlieb R.A., Dorn G.W, et al. (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature, 434, 658–662. [DOI] [PubMed] [Google Scholar]
- 50.Li V., Brustovetsky T., Brustovetsky N. (2009) Role of cyclophilin D-dependent mitochondrial permeability transition in glutamate-induced calcium deregulation and excitotoxic neuronal death. Exp. Neurol., 218, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perry G.M., Tallaksen-Greene S., Kumar A., Heng M.Y., Kneynsberg A., van G.T., Detloff P.J., Albin R.L., Lesort M. (2010) Mitochondrial calcium uptake capacity as a therapeutic target in the R6/2 mouse model of Huntington’s disease. Hum. Mol. Genet., 19, 3354–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valencia A., Sapp E., Kimm J.S., McClory H., Reeves P.B., Alexander J., Ansong K.A., Masso N., Frosch M.P., Kegel K.B. et al. (2013) Elevated NADPH oxidase activity contributes to oxidative stress and cell death in Huntington’s disease. Hum. Mol. Genet., 22, 1112–1131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Trushina E., Canaria C.A., Lee D.Y., McMurray C.T. (2014) Loss of caveolin-1 expression in knock-in mouse model of Huntington’s disease suppresses pathophysiology in vivo. Hum. Mol. Genet., 23, 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trushina E., Dyer R.B., Badger J.D., Ure D., Eide L., Tran D.D., Vrieze B.T., Legendre-Guillemin V., McPherson P.S., Mandavilli B.S. et al. (2004) Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol., 24, 8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brustovetsky N., Brustovetsky T., Jemmerson R., Dubinsky J.M. (2002) Calcium-induced cytochrome c release from CNS mitochondria is associated with the permeability transition and rupture of the outer membrane. J. Neurochem., 80, 207–218. [DOI] [PubMed] [Google Scholar]
- 57.Shalbuyeva N., Brustovetsky T., Brustovetsky N. (2007) Lithium desensitizes brain mitochondria to calcium, antagonizes permeability transition, and diminishes cytochrome C release. J. Biol. Chem., 282, 18057–18068. [DOI] [PubMed] [Google Scholar]
- 58.Dubinsky J.M. (1993) Intracellular calcium levels during the period of delayed excitotoxicity. J. Neurosci., 13, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grynkiewicz G., Poenie M., Tsien R.Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem., 260, 3440–3450. [PubMed] [Google Scholar]
- 60.Dietz R.M., Kiedrowski L., Shuttleworth C.W. (2007) Contribution of Na(+)/Ca(2+) exchange to excessive Ca(2+) loading in dendrites and somata of CA1 neurons in acute slice. Hippocampus, 17, 1049–1059. [DOI] [PubMed] [Google Scholar]
- 61.Stanika R.I., Pivovarova N.B., Brantner C.A., Watts C.A., Winters C.A., Andrews S.B. (2009) Coupling diverse routes of calcium entry to mitochondrial dysfunction and glutamate excitotoxicity. Proc. Natl. Acad. Sci. U S A, 106, 9854–9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brustovetsky T., Li V., Brustovetsky N. (2009) Stimulation of glutamate receptors in cultured hippocampal neurons causes Ca2+-dependent mitochondrial contraction. Cell Calcium, 46, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shalbuyeva N., Brustovetsky T., Bolshakov A., Brustovetsky N. (2006) Calcium-dependent spontaneously reversible remodeling of brain mitochondria. J. Biol. Chem., 281, 37547–37558. [DOI] [PubMed] [Google Scholar]