Abstract
In vivo evidence for brain mitochondrial dysfunction in animal models of Huntington disease (HD) is scarce. We applied the novel 17O magnetic resonance spectroscopy (MRS) technique on R6/2 mice to directly determine rates of oxygen consumption (CMRO2) and assess mitochondrial function in vivo. Basal respiration and maximal CMRO2 in the presence of the mitochondrial uncoupler dinitrophenol (DNP) were compared using 16.4 T in isoflurane anesthetized wild type (WT) and HD mice at 9 weeks. At rest, striatal CMRO2 of R6/2 mice was equivalent to that of WT, indicating comparable mitochondrial output despite onset of motor symptoms in R6/2. After DNP injection, the maximal CMRO2 in both striatum and cortex of R6/2 mice was significantly lower than that of WT, indicating less spare energy generating capacity. In a separate set of mice, oligomycin injection to block ATP generation decreased CMRO2 equally in brains of R6/2 and WT mice, suggesting oxidative phosphorylation capacity and respiratory coupling were equivalent at rest. Expression levels of representative mitochondrial proteins were compared from harvested tissue samples. Significant differences between R6/2 and WT included: in striatum, lower VDAC and the mitochondrially encoded cytochrome oxidase subunit I relative to actin; in cortex, lower tricarboxylic acid cycle enzyme aconitase and higher protein carbonyls; in both, lower glycolytic enzyme enolase. Therefore in R6/2 striatum, lowered CMRO2 may be attributed to a decrease in mitochondria while the cortical CMRO2 decrease may result from constraints upstream in energetic pathways, suggesting regionally specific changes and possibly rates of metabolic impairment.
Introduction
In Huntington’s disease (HD, the expanded CAG repeat expansion in the ubiquitously expressed huntingtin protein produces energetic dysfunction along with other physiological problems. Much effort has been expended trying to pinpoint whether the metabolic compromise in HD brains arises from deficits in glycolysis or mitochondrial function. Different experimental models and strategies have uncovered abnormalities in glucose uptake (1,2), glycolytic flux (3), metabolite and ATP levels (4–7), mitochondrial biogenesis (8), dynamics (9), protein import (10), transport (11) and enzyme function (12). Enzymatically, a long standing debate exists over whether deficits in energy generation are attributable to glycolytic, tricarboxylic acid cycle (TCA) (3) or electron transport chain (ETC) abnormalities and whether these are causal or consequence of disease progression (12,13). Abnormalities in glycolysis and TCA cycle have been reported (3,14,15) as well as downstream constrictions in high energy phosphate storage capacity (16). In the rapidly progressive R6/2 mouse model of HD, respiration has been reported to be unchanged in isolated brain mitochondria at 3–4 weeks and impaired in 12-week-old striatal homogenates (10,17). Normal electron transport function has been reported in brain mitochondria from several HD mice (13,14,18) and several cellular preparations (19,20), while abnormal fluxes have only been reported for end stage R6/2 brains (17). In vivo in humans at early disease stages, glycolytic dysfunction has been implicated (21) while postmortem enzymatic assessments indicate electron transport defects (22). These conflicting results may be attributable to varying onset of regionally specific enzymatic deficits at different stages of disease that are not well aligned among multiple models of the human disease. Methodologically, the majority of these studies have been conducted in a variety of reduced preparations necessary for measuring the particular biochemistry of interest.
High field strength, magnet-based techniques offer ways to make the same measurements in both humans and mouse models of HD. 17O magnetic resonance spectroscopy (MRS) provides a simple, noninvasive way to assess flux at one down-stream step of the energy generating pathway, complex IV (cytochrome c oxidase, CIV) of the ETC. In vivo, the O produced by CIV from inhaled 17O2 can be monitored by 17O MRS at high magnetic field (23–26). 17O MRS has been used successfully to assess regional differences in the cerebral metabolic rate of oxygen utilization (CMRO2) and to detect areas of infarct following middle cerebral artery occlusion (24,25).
Recently we demonstrated the feasibility of measuring regional CMRO2 using 17O MRS in mice with high within-session test–retest reproducibility (27). In the experiments reported here, we sought to measure CMRO2 in both basal and metabolically challenged states in the well described R6/2 mouse model of HD. To maximally stimulate electron transport flux in vivo and to obtain the maximal velocity of the CIV reaction, di-nitrophenol (DNP) was injected to uncouple CIV from oxidative phosphorylation (28). In an isolated mitochondrial preparation, the maximal enzymatic flux through CIV would be considered to be proportional to the amount of enzyme present. This would determine whether striatum and cortex in mouse HD had similar abilities to respond to increases in metabolic demand as in control mouse brains. Similarly, examining 17O MRS in the presence of oligomycin (oligo) to block oxidative phosphorylation would determine the coupling between complex IV and V in mouse HD. We used mice at 9 wk of age when disease is manifest but before its overt mitochondrial defects have been reported in reduced preparations (17,29).
Results
Physiological studies
At 9 weeks of age, R6/2 mice show symptoms of manifest disease (4,29,30). All R6/2 mice used in this study demonstrated clasping behavior; wild type mice did not.
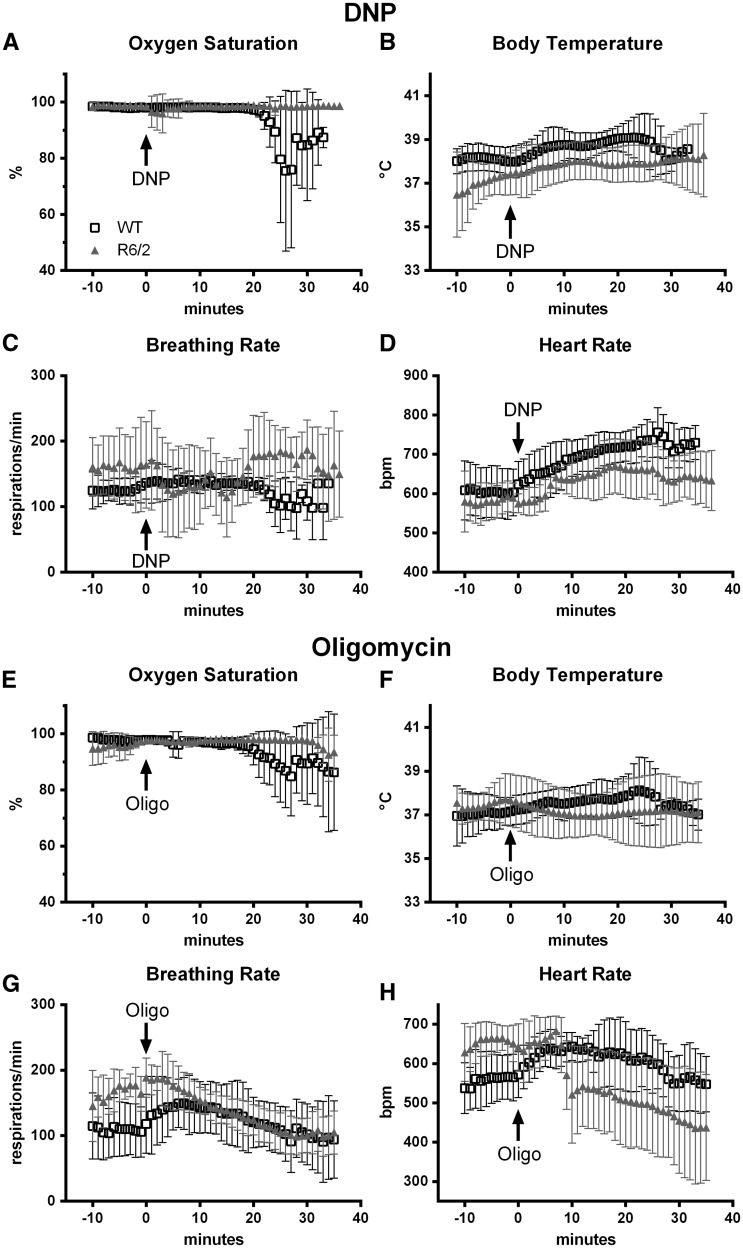
Previous work established the reliability of repeated 17O MRS measurements of CMRO2 in mouse striatum under basal conditions (27). To achieve reliable CMRO2 measurements after mitochondrial inhibition, 17O MRS scans would have to be obtained at a time when drug was active in the brain but prior to physiological deterioration. Primarily, the blood oxygen saturation could not drop and secondarily, the respiration rate should not change during the two 17O MRS scans for each mouse (the first one at baseline, prior to drug injection and the second one following drug injection). Since the pharmacokinetic properties of DNP and oligomycin distribution, absorption and bioavailability in mice were uncertain, we performed bench studies to determine how long various physiological parameters remained stable following injection. The dose of each drug to be used in the MRS studies was determined during these initial bench studies to provide equivalent survival times between genotypes, a surrogate measure of bioavailability. For a 21 mg/kg dose of DNP, survival times were 31 ± 6 min, N = 5 for wt and 44 ± 18 min, N = 5 for R6/2 (p = 0.15, 2 tailed t test). For a 2.25 mg/kg dose of oligomycin, survival times were 35 ± 7 min, N = 6 for wt and 41 ± 6 min, N = 4 for R6/2 (p = 0.15, two-tailed t test). These chosen dosages provided a period of relative physiological stability in which to measure drug effects upon CMRO2 (Fig. 1).
Figure 1.
Mouse physiology changes in response to the mitochondrial uncoupler dinitrophenol (DNP, A–D) or Complex V inhibitor, oligomycin (E–H) during bench studies. Drug was injected via a cannula in the femoral vein at time 0. Data are mean ± SD for (A–D) N = 5 WT (black squares) and N = 5 R6/2 (grey triangles); (E-H) WT N = 6, R6/2 N = 4.
In the bench studies, R6/2 breathing rate and body temperature were more variable than those of the wild type mice (Fig. 1B, C, F and G). Changes in the heart rate signaled onset of DNP or oligomycin effects in the periphery (Fig. 1D and H). For wild type mice, these occurred almost immediately after drug injection. For R6/2 mice, changes in heart rate were more delayed, by up to 10 min. Most importantly, blood oxygen saturation was generally very stable and high (>95%) for both R6/2 and wild type mice, both initially and for at least 20 min following injection of either drug (Fig. 1A and E). Decreases in the oxygen concentration in the blood signaled deterioration leading to death. This demonstrates that despite variation in breathing rates controlling oxygen intake and heart rate controlling blood flow to the brain, the concentration of oxygen in the blood delivered to the brain was virtually constant for 20–25 min following injection. Given these physiological profiles following drug injection, the second 17O MRS scan was acquired beginning 10 min after drug injection. The second 17O2 inhalation was completed 15.5 min post-injection.
Only breathing rate and body temperature could be monitored during the magnet studies without compromising the 17O MRS signal. These remained stable for both genotypes of mice (Table 1, examples in Fig. 2C, D, G and H). Minor differences in body temperature did reach statistical significance across genotypes and drug delivery in a few cases. However, these were of no practical significance because the values were all within normal physiological ranges. Since variations in breathing rate, body temperature and anesthesia level could influence CMRO2, these physiological parameters were all considered as covariates in subsequent statistical evaluation of the CMRO2 values.
Table 1.
Physiological parameters during MRS scanning
| Breathing Rate | Body Temperature | Isoflurane Level | ||||
|---|---|---|---|---|---|---|
| wild type | R6/2 | wild type | R6/2 | wild type | R6/2 | |
| Experiment | mean (sd) | mean (sd) | mean (sd) | mean (sd) | mean (sd) | mean (sd) |
| Basal | 109.8 (11.8) | 120.3 (12.5) | 37.9 (0.3) | 36.9 (0.4)** | 1.4 (0.1) | 1.5 (0.2) |
| After saline | 104.0 (20.6) | 119.0 (13.2) | 37.9 (0.2) | 37.4 (0.3)*ˆ | 1.3 (0.3) | 1.4 (0.1) |
| N | 5 | 3 | 5 | 3 | 5 | 3 |
| Basal | 104.1 (16.4) | 121.6 (14.8)* | 37.6 (1.1) | 37.5 (0.5) | 1.3 (0.2) | 1.2 (0.4) |
| After DNP | 105.0 (16.7) | 108.6 (19.9) | 37.9 (0.9) | 37.8 (0.5) | 1.2 (0.3) | 1.2 (0.3) |
| N | 10 | 11 | 10 | 11 | 10 | 11 |
| Basal | 107.9 (10.3) | 120.6 (23.3) | 37.6 (0.5) | 38.2 (0.6)* | 1.1 (0.3) | 1.1 (0.3) |
| After oligo | 108.3 (19.8) | 111.5 (23.1) | 37.8 (0.4)ˆˆˆ | 38.1 (0.4) | 1.1 (0.3) | 1.0 (0.2) |
| N | 10 | 11 | 10 | 11 | 10 | 11 |
p < 0.05, p < 0.01 compared to wild type, unpaired t test.
p < 0.05, p = 0.001 compared to basal condition, paired t test.
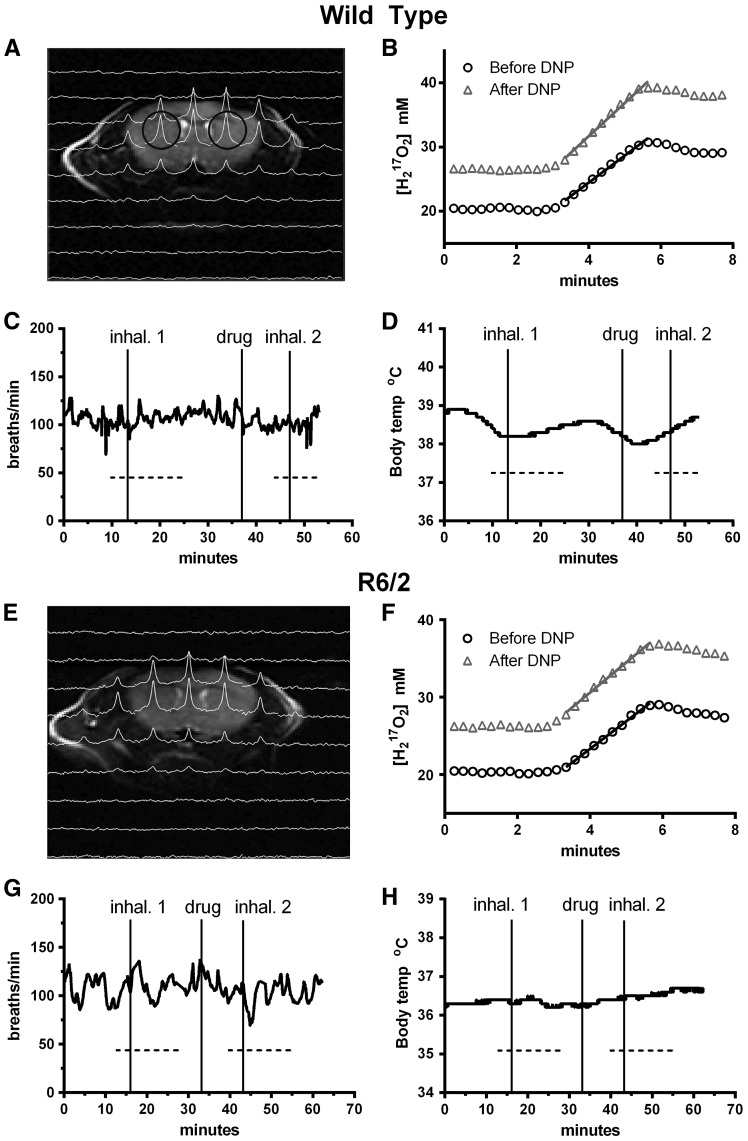
Figure 2.
Example 17O2 MRS data and mouse physiology used for CMRO2 calculations for a wild type (A–D) and an R6/2 (E–H) mouse. (A, E) 1H T2 weighted coronal images overlaid with the corresponding slice of 17O 3D CSI image, 1.5 min into the first 2.5 min inhalation. Voxels are indicated by black circles. (B, F) 17O signal from the right striatum during first (circles) baseline inhalation and second (triangles) inhalation after injection of DNP. Signal slopes during the inhalation indicated rate of oxygen consumption. During MRS scanning (dotted lines), breathing rate (C, G) and body temperature (D, H) remained stable.
17O MRS
During the 2.5min 17O2 inhalation, the peak 17O MRS signal increased (Fig. 2B and F) as the 17O became incorporated into water by complex IV of the ETC (see Fig. 7, (25,27)). The rate of increase of the 17O signal represents the CMRO2 (fitted slopes in Fig. 2B and F). Under anesthesia, the basal rates of CMRO2 in R6/2 striatum and cortex were not different from each other or those in the corresponding regions of wild type mice (cortex: 2.74 ± 0.45 µmole/g/min, N = 25 WT versus 2.57 ± 0.47 µmole/g/min N = 25, R6/2, p = 0.20, two-tailed t test; striatum: 2.72 ± 0.46 µmole/g/min WT versus 2.59± 0.39 µmole/g/min R6/2, p = 0.29; Fig. 3, all basal bars).
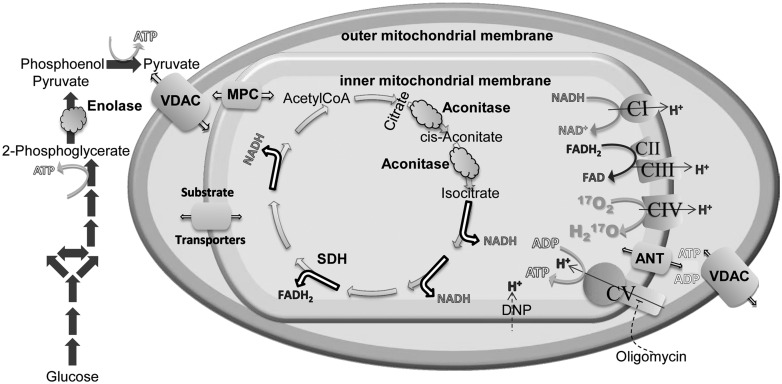
Figure 7.
Metabolic pathways and mitochondrial proteins altered or implicated in the altered function observed in this study. Changes in protein expression in enolase (round cloud) may slow glycolytic ATP production (glycolysis, solid grey arrows). Pyruvate, as well as other mitochondrial substrates, crosses the mitochondrial outer membrane when the voltage dependent anion channel (VDAC, large barrel) is open. Pyruvate is transported across the inner mitochondrial membrane through the mitochondrial pyruvate carrier (MPC). Other substrates utilize a variety of transporters (small barrels) to enter the mitochondrial matrix. Decreases in striatal VDAC expression may limit substrate availability. Inside the matrix, pyruvate enters the TCA cycle (open single headed arrows). Decreases in expression of cortical aconitase (oval cloud) may limit flux through TCA. These upstream alterations may limit flux through the electron transport chain complexes and oxidative phosphorylation, whose overall expression levels were largely unchanged. In vivo CMRO2 was measured from the conversion of 17O2 to O by complex IV (curved arrow). ATP produced by complex V and ADP cross the inner mitochondrial membrane through the adenine nucleotide transporter (ANT) and the outer mitochondrial membrane through VDAC. Decreases in striatal VDAC could also limit ATP and ADP movement into and out of mitochondria. The site of action of DNP and oligomycin are also indicated.
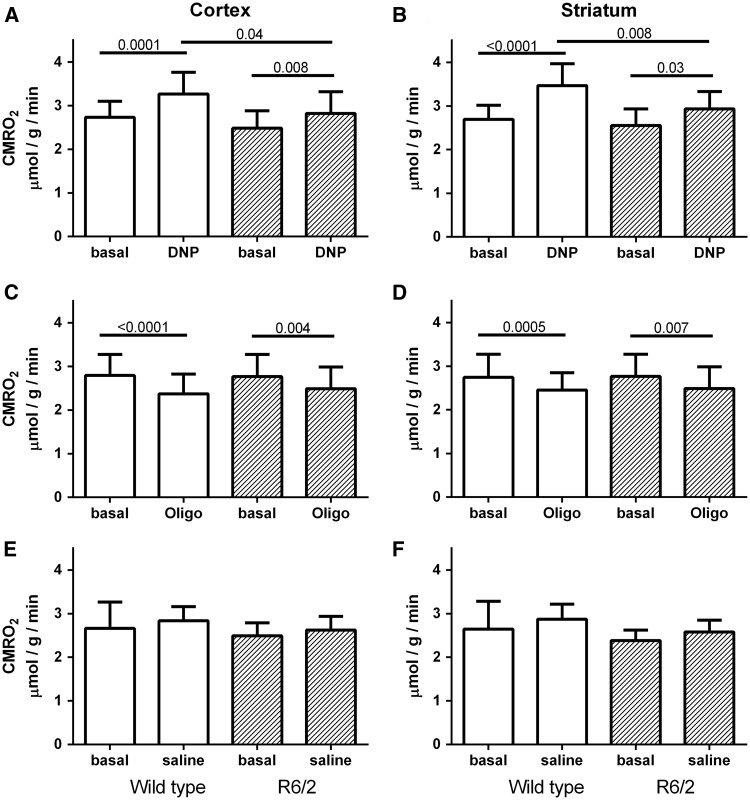
Figure 3.
Summary of CMRO2 data from wild type (white) and R6/2 (hatched) cortex (left column) and striatum (right column). (A, B) DNP injection significantly increased CMRO2 in both regions of all mice, but the increase in R6/2 (hatched) was less than in wild type (white) mice. (C, D) Oligomycin injection decreased the CMRO2 in striatum and cortex of both R6/2 (hatched) and wild type (white) mice. No differences were observed between R6/2 and WT mice in either region. (E, F) No differences in CMRO2 were observed between regions or genotype when saline was injected prior to the second 17O2 inhalation. Data are mean + SD. N = 10–11 for A, B. N = 3–5 for E, F. P values appear above lines indicating significant comparisons.
As a mitochondrial uncoupler, DNP was expected to deplete the hydrogen ion gradient across the mitochondrial inner membrane, stimulating the ETC to operate maximally. After DNP injection, the slopes of the 17O signal over time increased in both wild type and R6/2 mouse striata, indicating an increase in metabolic flux (Fig. 2B and F). In both striatum and cortex, the stimulation of CMRO2 induced by DNP was significantly greater in wild type mice than in the R6/2 (Fig. 3A and B), indicating that the mutant huntingtin mice had a reduced ability to produce oxidative energy in response to metabolic stress. Spare energy capacity was calculated as the difference between CMRO2 in the presence of DNP minus basal CMRO2. Significant differences were observed in spare capacity between WT and R6/2 mice in striatum (0.78 ± 0.48 µmole/g/min versus 0.38 ± 0.33 µmole/g/min, p = 0.02) but not in cortex (0.54 ± 0.34 µmole/g/min versus 0.34 ± 0.26 µmole/g/min, p = 0.22, 2 tailed t test).
By blocking the oxidative phosphorylation synthesizing ATP in complex V, oligomycin stabilizes a high proton gradient across the inner mitochondrial membrane, slowing the ETC and oxygen utilization. When fully active, oligomycin should decrease the CMRO2 by an amount representing the oxidative metabolism being used under basal conditions. Within 10 min of injection, oxidative metabolism was expected to decrease proportional to oligomycin bioavailability to that point. Oligomycin did indeed decrease the CMRO2 in both striatum and cortex of both wild type and R6/2 mice (Fig. 3C and D). However, no regional nor genotype specific differences were observed, suggesting that oxidative phosphorylation contributed to basal metabolism equally in both regions of diseased and non-diseased mice. The coupling efficiency between ATP production and ETC function was calculated by dividing the fraction of basal CMRO2 used for ATP synthesis (basal rate minus CMRO2 in the presence of oligomycin) by the basal CMRO2. No differences were observed in coupling efficiencies between R6/2 and WT in either region (striatum 0.089 ± 0.049 WT, 0.089 ± 0.134 R6/2, p = 1.0; cortex 0.100 ± 0.083 WT, 0.147 ± 0.097 R6/2, p = 0.26, two-tailed t test).
When saline was delivered for the drug injection, the second CMRO2 measurements were equivalent to the first, as in the previous study (27). This was true for R6/2 mice showing manifest disease, as well as the wild type mice (Fig. 3E and F).
Ex vivo biochemistry
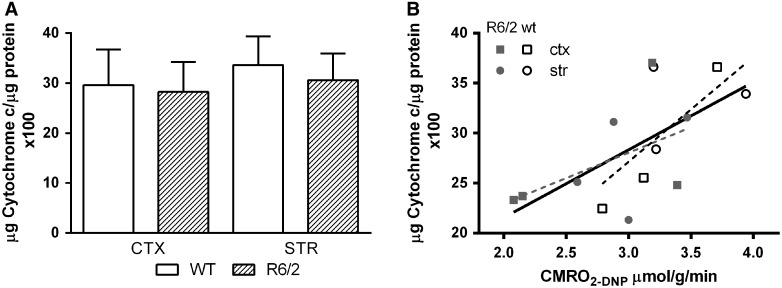
The lower maximal CMRO2 in R6/2 mice suggested that the amount of CIV enzyme and possibly the number of mitochondria could be lower in these brains. To examine the first possibility, cortical and striatal tissue from 9-week-old wild type and R/2 mice were assayed ex vivo for CIV activity. Surprisingly, no differences were observed between diseased and normal mice in either brain region (Fig. 4A), indicating a reduction in enzyme levels or mitochondrial mass was unlikely. The ex vivo CIV activities and DNP-stimulated CMRO2 values from the same mice were correlated (Fig. 4B, r2 = 0.40, p = 0.016), indicating that the 17O measurements indeed reflected oxygen consumption at complex IV. However, when compared by genotype, the correlation coefficient for R6/2 mice (r2 = 0.25, p = 0.21) was lower than that for wild type mice (r2 = 0.54, p = 0.10). This discrepancy suggests the possibility that in R6/2 brain, substrate availability may limit maximal energetic flux.
Figure 4.
Cytochrome c oxidase enzyme activity in homogenates of cortex and striatum of wild type and R6/2 mice at 9 weeks. (A) ex vivo cytochrome oxidase activity for cortex and striatum of wild type and R6/2 mice, mean + SD, N = 10 each. (B) Correlation of in vitro CIV activity with in vivo CMRO2 values in the presence of DNP. Data are from cortex and striatum of three wild type and four R6/2 mice (r2 = 0.40 for all data points, solid black line; r2 = 0.25 for R6/2, dotted grey line; r2 = 0.54 for wt, dotted black line).
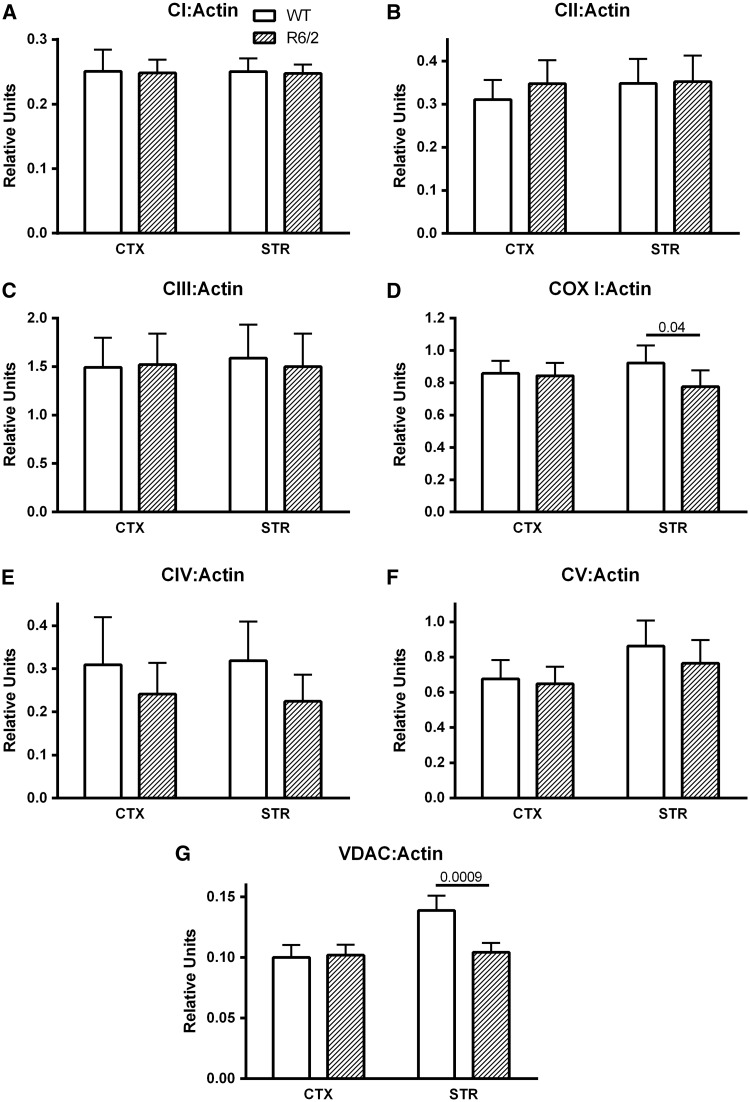
To determine if mitochondrial protein was decreased in R6/2 striatum and cortex, expression levels for representative proteins from each electron transport complex and the voltage-dependent anion channel (VDAC) were examined and ratioed to actin as a marker of cell volume (Fig. 5). No significant differences were observed in the amount of actin detected among all gel columns (data not shown). For complex IV, antibodies against the entire holoenzyme (CIV) and a mitochondrially encoded subunit (COX I) were both probed. Consistent with the activity measurements, no differences in cellular expression levels were observed for complexes I, II, III and holoenzyme IV in either region between R6/2 and wild type mice (Fig. 5A–C and E). The large variation in CIV:Actin precluded the trend towards lower values in R6/2 from approaching significance (Fig. 5E, p = 0.55, 0.19 for cortex and striatum, respectively). However, the mitochondrially encoded COX I expression in striatum was significantly lower in R6/2 striatum (Fig. 5D). Similarly, expression of another mitochondrial marker, the outer mitochondrial membrane protein VDAC was lower in R6/2 striatum (Fig. 5G).
Figure 5.
Relative expression levels of representative electron transport chain proteins (complexes I, II, III, IV, V and mitochondria encoded subunit 1 of complex IV (COX I)) and mitochondrial voltage dependent anion channel (VDAC) in cortical and striatal tissue from wild type and R6/2 mice. Expression levels are ratioed to actin (A–G). Data are mean + sem, N = 10 per genotype in A–F; N = 11–12 in G.
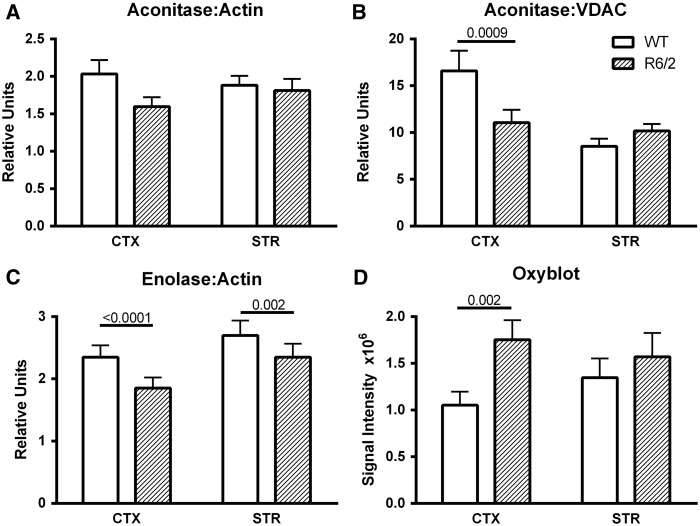
While the in vivo decrease in striatal R6/2 CMRO2 could possibly be easily explained by mitochondrial loss, this explanation or the lack of changes in the ETC would not account for the cortical decrease in DNP-stimulated CMRO2. Another explanation might be that metabolic flux may have been compromised due to restrictions in upstream metabolic pathways. To investigate this possibility, we examined the expression of aconitase and enolase as representative proteins from the TCA cycle and glycolysis, respectively. Each has previously been implicated in HD pathophysiology (31–34). When ratioed to actin, cortical aconitase trended lower in the R6/2 brains (Fig. 6A). When ratioed to VDAC as a mitochondrial marker, cortical aconitase expression was twice that of the striatum in WT brains (Fig. 6B). Aconitase/VDAC was markedly reduced in the R6/2 cortex, but did not appear so in the striatum because of the lower VDAC expression (Fig. 6B).
Figure 6.
Analysis of the TCA cycle enzyme, aconitase (A, B) and the glycolytic enzyme, enolase (C) in cortical and striatal tissue from wild type and R6/2 mice. (A, B) Western blots of regional brain homogenates of aconitase ratioed to actin (A) or VDAC (B). Data are mean + sem, N = 13–14 per group. (C) Western blots of regional brain homogenates of enolase ratioed to actin, N = 8 per group. (D) Protein oxidation measured by Oxyblot signal intensity of all proteins in the regional brain homogenates of WT and R6/2. Data are mean ± sem, N = 9 per group.
Neuron-specific enolase expression ratioed to actin was greatly reduced in both cortex and striatum of R6/2 brains (Fig. 6C). Aconitase, enolase and VDAC become oxidized in R6/2 whole brain at 10 weeks, providing another possible explanation for restricted metabolic flux (31). In keeping with this, when we probed all proteins for the presence of protein carbonyl groups, cortical proteins from R6/2 mice were significantly more oxidized than those in wild type cortex (Fig. 6D). In this analysis, a main effect was found for genotype (p = 0.004), but the R6/2 striatal carbonyl levels were not significantly different from those of wild type (p = 0.33).
Discussion
These in vivo cortical and striatal CMRO2 measurements indicate that at rest, the ETC flux appears normal in the R6/2 mouse. Oxidative phosphorylation in the basal state also seems normal in this mouse HD model, as seen from the oligomycin experiments. However, when maximally stressed in the DNP experiments, both cortex and striatum of R6/2 brain have less spare energy capacity than non-diseased mice. Naively, one would expect this lower maximal CMRO2 to be paralleled by both lowered maximal CIV activity and protein expression levels ex vivo. Surprisingly, this was not universally the case. In striatum, VDAC and COX I decreases suggested some loss of mitochondria, but this was not reflected in expression levels of other ETC proteins. The lower maximal spare capacity in R6/2 may be attributable to additional in vivo restrictions on metabolic flux from upstream processes. While we did not perform exhaustive metabolomics or proteomic analyses, our survey of key enzymes in the ETC, TCA and glycolysis indicated that metabolic flux was likely restricted through glycolysis and TCA in cortex and through glycolysis and possibly TCA in striatum. Most interestingly, these data raise the possibility that substrate influx into mitochondria via VDAC might also be restricted in the striatum in this mouse model.
The variability among mice for the current basal CMRO2 measurements was comparable to that reported in a previous test-retest study (27). Since the CMRO2 values are influenced by oxygen intake and physiological status, keeping breathing rate and body temperature within normal limits was a priority during the 17O MRS scans. To determine that the CMRO2 comparisons reflected disease and not anesthetic status, a general linear model that incorporated the physiological variables as covariates was used in all statistical evaluations.
Other measures of CMRO2 in HD
This is the first report of CMRO2 in an animal model of HD. The unaltered brain CMRO2 in the anesthetized state indicated that basal mitochondrial output was adequate to support quiescent brain function in both wild type and diseased mice. This does not however rule out the possibility that differences might be found in the awake state where neural activity generated increases in intracellular calcium might differentially stimulate metabolism in diseased and normal mice. The absence of a difference in the CMRO2 in the presence of oligomycin between R6/2 and WT brains suggests that oxidative phosphorylation in R6/2 brain mitochondria was not impaired. This similarity also rules out an increase in mitochondrial uncoupling in the R6/2 brain.
In mouse, sensory stimulation-induced changes in cortical CMRO2 measured using photoacoustical microscopy were reported to peak at ∼15% (35). The DNP-stimulated CMRO2 measured here represented a 20% increase for cortex and 30% increase for striatum in WT mice. As expected, DNP produced a greater effect than natural stimulation. In the R6/2 brain, the DNP-induced maximal stimulation was limited to 14-15%, a level that would be sufficient to respond to natural stimulation but not excessive stress. The true maximal effects of the injected drugs likely occurred after our measurements were taken when mouse physiology became unstable. Therefore the values reported here for DNP-stimulated CMRO2 are likely underestimates of maximum and should not be compared directly to those obtained from reduced preparations.
Never the less, in striatal neurons cultured from embryonic HD rat, oxygen consumption rate, respiratory control ratio (RCR) and spare capacity were normal in abundant glucose but decreased when glucose was restricted to 2.5 mM (14). Alternate energy substrates (lactate and pyruvate) supplement this respiratory function but do not fully restore it (14). Substrate availability in the current in vivo studies was not measured, but all animals had ad lib access to food until they were anesthetized. We can estimate the RCR by dividing the CMRO2 in the presence of DNP by the CMRO2 in the presence of oligomycin. For wild type animals, the RCR estimates were 1.41 and 1.38 in striatum and cortex, respectively. For R6/2, the estimated RCRs were 1.19 and 1.13 for striatum and cortex, respectively, clearly less than normal.
In humans, basal measures of occipital ATP generation, Pi/ATP and Pi/PCR measured with 31P MRS, were not different between early stage HD patients and controls but after visual stimulation, these measures increased in the controls but not in the HD brains (6). This functional stress parallels the uncoupling stress applied in the current study. Both suggest the HD brain is unable to meet the energy generating needs of functional demands. Indeed, HPLC measurements of ATP concentration in R6/2 striatum and cortex at 8 wk is decreased by 6–9% (7), but this deficit was too small to observe by noninvasive MRS methods (5). Phosphocreatine levels, representing the stored cellular energy, progressively increase in R6/2 brains over their lifetime, consistent with a compensatory attempt to replenish energy or a restriction in PCr breakdown and use (4,5,16).
Glycolysis
The evidence is mixed for abnormalities in glycolytic metabolism in HD. While striatal glucose uptake exceeds that of the cortex in normal mice (36), cerebral glucose uptake has been reported to be unchanged (37) or decreased early and progressively in R6/2 (1,38). Decreased spare capacity and higher glycolytic flux characterize a striatal cell line expressing mutant huntingtin with a 111 repeat expansion compared to a sister line with only seven repeats (39). An early PET study in asymptomatic at risk individuals identified a subgroup with markedly decreased caudate glucose metabolism (40). Moreover, PET measurements of occipital CMRO2, cerebral blood flow and oxygen extraction fraction were not different between early stage HD and matched controls at rest (21). However, the cerebral metabolic rate of glucose consumption (CMRglc) was decreased in human striatum, suggesting glycolytic compromise. As a consequence, the ratio CMRO2/CMRglc rose from a normal value of 5.1 to the maximum theoretical value of 6.0, indicating an increased efficiency for energy generation. Essentially everything processed through glycolysis was utilized to produce ATP, a result inconsistent with dysfunctional electron transport (21). In addition, in vitro ETC function in platelets taken from these patients prior to scanning, was not correlated with the in vivo CMRO2/CMRglc measures (41). Our CMRO2 measurements in R6/2 mice suggest they possess a similar phenotype.
Protein expression
Cortical aconitase activity may be reduced by both the lower level of aconitase expression, and the possibility that this protein is oxidized (31), suggesting that the decrease in DNP-stimulated cortical CMRO2 observed in the R6/2 brain may be attributable to reductions in TCA flux. Previously, aconitase was reported decreased in R6/2 striatum but not cortex at 12 wk, while CIV activity was decreased in both regions (32). At 9 weeks, we saw no difference between R6/2 vs WT in ex vivo CIV activity and aconitase expression was lower in R6/2 cortex. In R6/2, respiration has been reported to be unchanged in isolated brain mitochondria at 3-4 weeks or impaired in striatal homogenates at 12 wk (10,17). If we assume that our R6/2 data add a snapshot of metabolism at an additional 9-week time point, together these data suggest that expression levels and activity of various metabolic enzymes are dynamically changing over disease progression. Protein carbonyls previously had been shown to increase at 13 weeks but not earlier in R6/2 whole brain (34). We now show significant oxidized proteins in cortex at 9 weeks. Although we did not identify individual oxidized proteins, VDAC1, aconitase and neuron-specific enolase are each oxidized in R6/2 striatum at 10 weeks (31) and would be expected to contribute to both the Oxyblot signal reported here and to decreased flux through glycolysis and the TCA cycle. VDAC is also oxidized in Alzheimer’s disease brains and after traumatic injury to rat brain (42,43).
In addition to mitochondrial loss, some other critical function of VDAC may be absent in the R6/2 striatum at 9 weeks. In the outer mitochondrial membrane, VDAC forms the pathway for substrates of electron transport to cross this membrane (Fig. 7, (44)). Specific transport proteins shuttle substrates across the inner mitochondrial membrane. Thus, VDAC is in a position to limit substrate availability for energy generation. In cancer cells, VDAC opening and closing limits mitochondrial substrate availability and ATP production. One consequence of VDAC closure is glycolytic ATP production in aerobic conditions, termed the Warburg effect (44,45). We speculate that a similar process could be occurring in R6/2 striatum. VDAC1 is also responsible for Ca2+ influx across the outer mitochondrial membrane. Reduced VDAC1 levels might homeostatically reduce mitochondrial calcium accumulations to apoptotic stimuli (46).
Limitations
A central problem in understanding the progressive changes in protein expression in HD models is what to use as a reference for comparison. Some studies compare expression levels to a marker protein while others ratio to one region or to wild type levels (31,47). VDAC1 expression was previously examined by ratioing 2D gels among paired R6/2 and control mice (31,47). Results were contradictory, with one study reporting slight whole brain increases at 8 weeks (47) and another reporting no change in 10 weeks striatum (31). VDAC and actin are typically used as markers for mitochondrial and cellular mass. Both may be changing in HD as neuronal cells shrink (or fail to grow) and glial cells proliferate (4,47,48). Nevertheless, by ratioing to actin, Figure 5 reports the relative amount of energy generating protein machinery available to cells and mitochondria. Relative VDAC and COX I decreases suggest mitochondrial mass has shrunk but the majority of ETC protein levels remained normal in the 9-week R6/2 cortex and striatum. The acknowledged, significant variability among gels precluded resolving additional changes in other ETC proteins. Decreased VDAC/actin levels have been reported in StHdHQ111/Q7 cells in the absence of decreases in other electron transport subunits (49). In these cells, CIV activity increased when Cox17, another CIV subunit, expression and CI activity decreased, demonstrating that compensatory responses occur even within the ETC (49). In R6/2 mice, loss of body temperature indicative of PGC1a, the master regulator of mitochondrial biogenesis (50), stimulated abnormalities in mitochondrial biogenesis beginning at 10 weeks (8), so a deficit in mitochondrial biogenesis would not be expected to be a major contributor to measurements in our study. Given that relative ETC complex levels vary by tissue type and that mitochondria are regulated by transcriptional, translational and functional factors (51), VDAC or COX I expression may be regulated differently than the other ETC complexes.
These data do not generally distinguish neuronal and glial contributions. Only the enolase expression was specific to neurons. We cannot rule out compensatory changes in glial mitochondrial protein expression that might counteract or complement neuronal energy deficits (52). Similarly, changes in electron transport protein expression in a subset of mitochondria might be masked by the assay variability. Synaptosomal, but not non-synaptosomal, mitochondria have less ATP in CAG150 knock in HD mouse brains compared to controls (53,54). Mutant htt binds to a transport protein in the inner mitochondrial membrane responsible for import of nuclear encoded proteins and impairs uptake into synaptosomal but not non-synaptosomal mitochondria (10). The deficit in maximal CMRO2 reported here was pharmacologically produced and should reflect activation of the ETC in all cerebral mitochondria combined, not just those from a specific compartment. Similarly, the protein evaluations were performed on regional tissue homogenates, not purified mitochondria, to obtain overall expression changes that might reflect the in vivo CMRO2 measurements.
Conclusion
Our current data, from a single time point in an overtly diseased mouse, illustrate how a single measure of mitochondrial function, CMRO2, might actually reflect multiple dynamic changes in upstream metabolic pathways. Metabolic dysfunction, at least in the R6/2 mouse, and possibly more generally in HD, develops in a longitudinally progressive manner that may be regionally specific, or desynchronized among brain regions (4). Since the mutant gene is present from gestation, compensatory homeostatic mechanisms must be invoked initially and throughout a mouse or human lifetime to explain the superficially normal phenotype prior to onset of debilitating function. Given the strong homeostatic regulation of ATP levels, compensatory shifts in flux through energy pathways almost certainly occur. Thus a documented deficit at one point in a pathway may not result in an overall downstream loss of energy production until the system is stressed. Such stresses shift the entire energy production system out of its normal operating space. The current data illustrated this. Despite deficits in glycolytic and TCA enzymes and a possible loss of striatal mitochondria, CMRO2 and probably energy production remained normal in R6/2 under basal conditions but exhibited limited spare capacity to respond to increased demands. We do not know if maximal CMRO2 is compromised at earlier ages in the R6/2 mouse but this might be expected. Correspondingly, as disease progresses, the currently documented changes may expand across more regions and additional deficits could accrue, as occurs in humans (55). Excessive mitochondrial dysfunction, however, may be limited to end stage disease.
Materials and Methods
Animals
All animal procedures were approved by the University of Minnesota Institutional Animal Care and Use Committee. Male R6/2 mutant huntingtin mice and C57B6/CBA wild type controls were obtained from Jackson Laboratories. CAG repeat length ranged from 120 to 128 glutamines. Animals were housed and cared for in accordance with the Guide for the Care and Use of Laboratory Animals. In pilot studies female mice were also tested. Survival rates and physiological parameters (described below) were more variable for female mice than male mice of both genotypes. Therefore, only male mice were used in these experiments. Across all studies, a total of 68 mice were used.
At nine weeks of age, mice were weighed, checked for a clasping response and anesthetized with isoflurane (3.0% for induction, ∼1.5% for maintenance) in a 1:1 mixture of nitrous oxide and oxygen. Isoflurane levels were adjusted manually to maintain breathing rates stable during all experiments. The right femoral vein was cannulated for drug delivery. To prevent occlusion 10 µl saline with 20U heparin was administered. For the physiological bench studies and during cannulation, a MouseOx Plus system (Starr Life Sciences, Oakmont, PA, USA) was used to monitor heart rate, oxygen saturation, breathing rate, and body temperature of mice. For the magnet studies, a Model 1025 Monitoring & Gating System (Small Animal Instruments, Inc, Stony Brook, NY) was used to monitor breathing rate and body temperature. Body temperature (maintained between 36 and 39 °C) was regulated by a thermostat and experimenter oversight of a lamp on the bench and both a circulating warm water system and a heating fan system in the magnet. Spontaneously breathing mice were placed in a custom holder with a tightly fitting nose cone connected to a custom built pneumatic switching system to deliver the anesthetic mix in normal O2 or 17O2 (70% enriched, Cambridge Isotope Laboratories, Tewksbury, MA, USA) during scanning (27). After physiological parameters were stable for a minimum of 10 min, drug (saline, 21mg/kg dinitrophenol or 2.25 mg/kg oligomycin) was injected intravenously at a rate of 200µl/min.
Magnetic resonance protocol
All MRI studies were performed using a 16.4 T Varian/Magnex (26 cm bore) system. A home built linear surface 17O coil combined with a quadrature 1H coil designed for minimal cross-talk was used to acquire axial anatomic 1H images and three dimensional 17O MRSI (magnetic resonance spectroscopy imaging) data with 15.4 seconds temporal resolution using previously described parameters (27). The coil was positioned with the most sensitive area over the striatum and anterior cortex. After initial 17O MRSI data were acquired for 3 min during non-labeled O2 inhalation, the respiration gas was switched to the 17O2 mixture while 17O MRSI data were continuously acquired. After 2.5 min of 17O2 inhalation, the gas was switched back to the unlabeled O2/N2O gas mixture, and the 17O MRSI acquisition was continued for up to 15 minutes (27). Drug was injected intravenously 15–20 min following the first inhalation. Ten minutes later, a second MRSI acquisition sequence with 2.5 min 17O2 inhalation ensued.
Cerebral metabolic rate of oxygen (CMRO2) calculation
The cylindrical voxel size was nominally 9.3 mm3 (diameter 2.2 mm, height 2.4 mm). For striatal CMRO2, single voxels were placed over each striatum, lateral to the ventricle (Fig. 2A and E). For ‘cortical’ CMRO2, a bilateral voxel was placed over the midline prefrontal and motor cortex extending ventrally into the septal area. The O resonance intensities from each voxel were converted to absolute O concentrations using the natural abundance signal (20.35 μmol/g) from the same voxel as an internal reference. Linear regression of the brain O concentration time courses during the 17O2 inhalation period was applied and the slopes were used for calculating the CMRO2 values. CMRO2 values are presented as μmoles O2/min/g brain water. We previously determined that the water content of R6/2 brains did not differ from littermates (4). This simplified model has been previously validated in rat brains with short 17O2 inhalation where fast exchange of the oxygen gas is expected (25,26). To verify the appropriateness of the linear model, a subset of the data were also analyzed by a polynomial model (25,26). The variability was increased by the more complex model as it overfit the limited data points during the inhalation. Thus the CMRO2 fits from the linear model were reported.
Tissue harvesting
Following completion of the magnet protocol, mice were returned to 3% isoflurane and rapidly sacrificed. Brains were removed on ice and cortex and striatum were dissected into homogenization buffer (50 mM KCl, 5 mM EGTA, 5 mM MgCl2, 6 mM PMSF, 1% Protease Inhibitor Cocktail (Sigma-Aldrich P8340), 9% Lauryl Maltoside Solution (Abcam), pH 7.4). Tissue was flash frozen in liquid nitrogen and stored in −80 °C. Additional tissue was harvested from R6/2 and wild type mice not used in the magnet study.
Complex IV activity assay
Frozen tissue was thawed on ice, and homogenized in ice cold homogenization buffer using a Minilys Tissue Homogenizer (Bertin Technolgies, Villeurbanne, France). The tissue lysate was incubated on ice for 30 min and subsequently centrifuged at 14 000 r.p.m. for 15 min at 4 °C. The supernatant was collected and protein concentration was determined using the Bio-Rad DC Protein Assay (Bio-Rad, Hercules, CA). Enzyme activity was determined using a Cytochrome Oxidase Activity Colorimetric Assay Kit (Biovision, Milpitas, CA) according to the manufacturer’s protocol, following the oxidation of reduced cytochrome C at an absorption of 550nm. Absorption slopes were then compared to a standard activity curve of pure Cytochrome C Oxidase from Bovine Heart (Sigma-Aldrich, St. Louis, MO) (56).
Western blotting
Homogenized protein (30 μg) was mixed with Laemmlli Buffer (5% ß-mercaptoethanol) and run on a 10–20% tris-glycine gel (Life Technologies, Carlsbad, CA), transferred to Invitrolon™ PVDF membrane (Life Technologies, Carlsbad, CA), and immunoblotted according to standard procedures. Primary antibodies were 1:500 Mitoprofile total OXPHOS Rodent WB cocktail including mouse anti-CI, CII, CIII, Cox I (CIV mitochondria encoded subunit 1), and CV (Abcam 110413 Cambridge, MA), and 1:1000 mouse anti-CIV antibody (native holoenzyme, Molecular Probes, Thermo Fischer Scientific, Waltham MA) diluted in Odessey™ TBS Blocking Buffer and 0.2% Tween-20 (Bio-Rad, Hercules, CA). Secondary antibodies were IR Dye 800CW goat anti-mouse (1:10,000, LICOR, Lincoln, NE). Band intensity was analyzed using Image Studio Lite 3.1 (LICOR, Lincoln NE). After visualization using the LI-COR Odyssey CLx imaging system, membranes were stripped of antibody in Restore PLUS Western Blot Stripping Buffer (Life Technologies Carlsbad, CA). The immunoblotting process was then repeated from the blocking step with the substitution of primary antibodies: 1:1000 rabbit anti-beta actin antibody (Abcam, Cambridge, MA) and 1:1000 rabbit anti-VDAC1 antibody (Voltage Dependent Anion Channel, Cell Signaling, Danvers, MA) and secondary antibody IR Dye 680RD goat anti-rabbit (LICOR, Lincoln, NE). For aconitase (1:1000 mouse anti-aconitase 2, Abcam, Cambridge MA; 1:10000 IR Dye 800CW goat anti-rabbit, LICOR, Lincoln NE) and NSE (1:2000 mouse anti-NSE, Abcam, Cambridge, MA; 1:10,000 IR Dye 680RD goat anti-mouse, LICOR, Lincoln, NE), actin and VDAC immunoreactivity were visualized simultaneously.
Protein oxidation
Previously homogenized aliquots of tissue were thawed on ice. The OxyBlot Protein Oxidation Detection Kit (Millipore, S7150) was used to determine protein oxidation following manufacturer’s protocol with the exception of secondary fluorescent antibody detection (IR Dye 700CW goat anti-rabbit antibody (LICOR) at 1:10,000) instead of chemiluminescence. Immunoblotted membranes were visualized and quantified as in the Westerns.
Statistics
CMRO2 and protein markers were each analyzed in a linear mixed model with a random effect for mouse to control for repeated measures within mouse across regions. Predictors in the CMRO2 model to test genotype differences and pre- vs. post-treatment differences included genotype (WT, R6/2), experiment (pre, post), and their interaction, with adjustment for physiological variables (isoflurane level, body temperature, and breathing rate); these were run separately for each region (cortex and striatum). Genotype differences within experiment, and pre/post differences within genotype, were compared using F-tests of the least square means. Predictors in the CMRO2 model to test region differences included genotype, regions, experiment and all interactions, with adjustment for physiological variables. Regions were compared using an F test of the least square means within each combination of genotype and experiment. All these CMRO2 analyses were carried out separately for each of the three treatments used: oligo, DNP and saline. Predictors in the protein marker models were genotype, regions, and their interaction, adjusting for individual gels. This acknowledged the significant, practical variability arising from running tissues from only four animals per gel. Genotypes were compared within region using an F-test of the least square means. All analyses were run in SAS version 9.4. Two-way ANOVAs in Table 1 and the correlations in Figure 4B were run in GraphPad Prism version 6.01.
Acknowledgements
We would like to acknowledge prior support from the Winston and Maxine Wallin Neuroscience Discovery Fund for validation of the 17O MRS method, without which the current project would not have been feasible.
Conflict of Interest statement. None declared.
Funding
This work was supported by CHDI, Inc. [A-7107 to JMD] and National Institutes of Health [2P41EB015894 to LEE]. The Center for MR Research is supported by the National Institute of Biomedical Imaging and Bioengineering [P41 EB015894], the Institutional Center Cores for Advanced Neuroimaging [P30 NS076408] and National Center for Research Resources (NCRR) [S10 RR025031].
References
- 1.Cepeda-Prado E., Popp S., Khan U., Stefanov D., Rodriguez J., Menalled L.B., Dow-Edwards D., Small S.A., Moreno H. (2012) R6/2 Huntington's disease mice develop early and progressive abnormal brain metabolism and seizures. J. Neurosci., 32, 6456–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClory H., Williams D., Sapp E., Gatune L.W., Wang P., Difiglia M., Li X. (2014) Glucose transporter 3 is a rab11-dependent trafficking cargo and its transport to the cell surface is reduced in neurons of CAG140 Huntington's disease mice. Acta Neuropathol. Commun., 2, 179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira I.L., Cunha-Oliveira T., Nascimento M.V., Ribeiro M., Proenca M.T., Januario C., Oliveira C.R., Rego A.C. (2011) Bioenergetic dysfunction in Huntington's disease human cybrids. Exp. Neurol., 231, 127–134. [DOI] [PubMed] [Google Scholar]
- 4.Zacharoff L., Tkac I., Song Q., Tang C., Bolan P., Mangia S., Henry P.G., Li T., Dubinsky J.M. (2012) Cortical Metabolites as Biomarkers in the R6/2 Model of Huntington's Disease. J. Cereb. Blood Flow Metab., 32, 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tkac I., Henry P.G., Zacharoff L., Wedel M., Gong W., Deelchand D., Li T., Dubinsky J.M. (2012) Homeostatic adaptations in brain energy metabolism in mouse models of Huntington disease. J. Cereb. Blood Flow Metab., 32, 1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mochel F., N'guyen T.M., Deelchand D., Rinaldi D., Valabregue R., Wary C., Carlier P.G., Durr A., Henry P.G. (2012) Abnormal response to cortical activation in early stages of Huntington disease. Mov. Disord., 27, 907–10. [DOI] [PubMed] [Google Scholar]
- 7.Mochel F., Durant B., Meng X., O'Callaghan J., Yu H., Brouillet E., Wheeler V.C., Humbert S., Schiffmann R., Durr A. (2012) Early alterations of brain cellular energy homeostasis in huntington disease models. J. Biol. Chem., 287, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D, et al. (2006) Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab., 4, 349–362. [DOI] [PubMed] [Google Scholar]
- 9.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R, et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yano H., Baranov S.V., Baranova O.V., Kim J., Pan Y., Yablonska S., Carlisle D.L., Ferrante R.J., Kim A.H., Friedlander R.M. (2014) Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci., 17, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trushina E., Dyer R.B., Badger J.D., Ure D., Eide L., Tran D.D., Vrieze B.T., Legendre-Guillemin V., McPherson P.S., Mandavilli B.S, et al. (2004) Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol., 24, 8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brustovetsky N. (2015) Mutant huntingtin and elusive defects in oxidative metabolism and mitochondrial calcium handling. Mol. Neurobiol., 53, 2944–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton J., Pellman J.J., Brustovetsky T., Harris R.A., Brustovetsky N. (2015) Oxidative metabolism in YAC128 mouse model of Huntington's disease. Hum. Mol. Genet., 24, 4862–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouarne C., Tardif G., Tracz J., Latyszenok V., Michaud M., Clemens L.E., Yu-Taeger L., Nguyen H.P., Bordet T., Pruss R.M. (2013) Early deficits in glycolysis are specific to striatal neurons from a rat model of huntington disease. PLoS One, 8, e81528.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naseri N.N., Xu H., Bonica J., Vonsattel J.P., Cortes E.P., Park L.C., Arjomand J., Gibson G.E. (2015) Abnormalities in the tricarboxylic Acid cycle in Huntington disease and in a Huntington disease mouse model. J Neuropathol. Exp. Neurol., 74, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J., Amante D.J., Moody J.P., Edgerly C.K., Bordiuk O.L., Smith K., Matson S.A., Matson W.R., Scherzer C.R., Rosas H.D, et al. (2010) Reduced creatine kinase as a central and peripheral biomarker in Huntington's disease. Biochim. Biophys. Acta, 1802, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aidt F.H., Nielsen S.M., Kanters J., Pesta D., Nielsen T.T., Norremolle A., Hasholt L., Christiansen M., Hagen C.M. (2013) Dysfunctional mitochondrial respiration in the striatum of the Huntington's disease transgenic R6/2 mouse model. PLoS Curr., 5, doi: 10.1371/currents.hd.d8917b4862929772c5a2f2a34ef1c201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guidetti P., Charles V., Chen E.Y., Reddy P.H., Kordower J.H., Whetsell W.O.J., Schwarcz R., Tagle D.A. (2001) Early degenerative changes in transgenic mice expressing mutant huntingtin involve dendritic abnormalities but no impairment of mitochondrial energy production. Exp. Neurol, 169, 340–350. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira J.M., Jekabsons M.B., Chen S., Lin A., Rego A.C., Goncalves J., Ellerby L.M., Nicholls D.G. (2007) Mitochondrial dysfunction in Huntington's disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J. Neurochem., 101, 241–249. [DOI] [PubMed] [Google Scholar]
- 20.Milakovic T., Johnson G.V. (2005) Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J. Biol. Chem., 280, 30773–30782. [DOI] [PubMed] [Google Scholar]
- 21.Powers W.J., Videen T.O., Markham J., McGee-Minnich L., Antenor-Dorsey J.V., Hershey T., Perlmutter J.S. (2007) Selective defect of in vivo glycolysis in early Huntington's disease striatum. Proc. Natl. Acad. Sci. USA, 104, 2945–2949., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. (1997) Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann. Neurol., 41, 646–653. [DOI] [PubMed] [Google Scholar]
- 23.Lu M., Zhang Y., Ugurbil K., Chen W., Zhu X.H. (2013) In vitro and in vivo studies of (17) O NMR sensitivity at 9.4 and 16.4 T. Magn Reson. Med., 69, 1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X.H., Chen J.M., Tu T.W., Chen W., Song S.K. (2013) Simultaneous and noninvasive imaging of cerebral oxygen metabolic rate, blood flow and oxygen extraction fraction in stroke mice. Neuroimage, 64, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X.H., Zhang Y., Zhang N., Ugurbil K., Chen W. (2007) Noninvasive and three-dimensional imaging of CMRO(2) in rats at 9.4 T: reproducibility test and normothermia/hypothermia comparison study. J Cereb. Blood Flow Metab., 27, 1225–1234. [DOI] [PubMed] [Google Scholar]
- 26.Zhang N., Zhu X.H., Lei H., Ugurbil K., Chen W. (2004) Simplified methods for calculating cerebral metabolic rate of oxygen based on 17O magnetic resonance spectroscopic imaging measurement during a short 17O2 inhalation. J. Cereb. Blood Flow Metab., 24, 840–848. [DOI] [PubMed] [Google Scholar]
- 27.Cui W., Zhu X.H., Vollmers M.L., Colonna E.T., Adriany G., Tramm B., Dubinsky J.M., Oz G. (2013) Non-invasive measurement of cerebral oxygen metabolism in the mouse brain by ultra-high field (17)O MR spectroscopy. J. Cereb. Blood Flow Metab., 33, 1846–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellon E.A., Beesam R.S., Elliott M.A., Reddy R. (2010) Mapping of cerebral oxidative metabolism with MRI. Proc. Natl. Acad. Sci. USA, 107, 11787–11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell, 87, 493–506. [DOI] [PubMed] [Google Scholar]
- 30.Menalled L., El-Khodor B.F., Patry M., Suarez-Farinas M., Orenstein S.J., Zahasky B., Leahy C., Wheeler V., Yang X.W., MacDonald M, et al. (2009) Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol. Dis., 35, 319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perluigi M., Poon H.F., Maragos W., Pierce W.M., Klein J.B., Calabrese V., Cini C., De M.C., Butterfield D.A. (2005) Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol. Cell Proteomics., 4, 1849–1861. [DOI] [PubMed] [Google Scholar]
- 32.Tabrizi S.J., Workman J., Hart P.E., Mangiarini L., Mahal A., Bates G., Cooper J.M., Schapira A.H.V. (2000) Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann. Neurol., 47, 80–86. [DOI] [PubMed] [Google Scholar]
- 33.Tabrizi S.J., Cleeter M.W., Xuereb J., Taanman J.W., Cooper J.M., Schapira A.H. (1999) Biochemical abnormalities and excitotoxicity in Huntington's disease brain. Ann. Neurol., 45, 25–32. [DOI] [PubMed] [Google Scholar]
- 34.Zourlidou A., Gidalevitz T., Kristiansen M., Landles C., Woodman B., Wells D.J., Latchman D.S., de B.J., Tabrizi S.J., Morimoto R.I., Bates G.P. (2007) Hsp27 overexpression in the R6/2 mouse model of Huntington's disease: chronic neurodegeneration does not induce Hsp27 activation. Hum. Mol. Genet., 16, 1078–1090. [DOI] [PubMed] [Google Scholar]
- 35.Yao J., Wang L., Yang J.M., Maslov K.I., Wong T.T., Li L., Huang C.H., Zou J., Wang L.V. (2015) High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat. Methods, 12, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casteels C., Vunckx K., Aelvoet S.A., Baekelandt V., Bormans G., Van L.K., Koole M. (2013) Construction and evaluation of quantitative small-animal PET probabilistic atlases for [(1)(8)F]FDG and [(1)(8)F]FECT functional mapping of the mouse brain. PLoS One, 8, e65286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooms M., Rietjens R., Rangarajan J.R., Vunckx K., Valdeolivas S., Maes F., Himmelreich U., Fernandez-Ruiz J., Bormans G., Van L.K., Casteels C. (2014) Early decrease of type 1 cannabinoid receptor binding and phosphodiesterase 10A activity in vivo in R6/2 Huntington mice. Neurobiol. Aging, 35, 2858–2869. [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Sarkar A., Cicchetti F., Yu M., Zhu A., Jokivarsi K., Saint-Pierre M., Brownell A.L. (2005) Cerebral PET imaging and histological evidence of transglutaminase inhibitor cystamine induced neuroprotection in transgenic R6/2 mouse model of Huntington's disease. J. Neurol. Sci., 231, 57–66. [DOI] [PubMed] [Google Scholar]
- 39.Siddiqui A., Rivera-Sanchez S., Castro M.R., Acevedo-Torres K., Rane A., Torres-Ramos C.A., Nicholls D.G., Andersen J.K., Ayala-Torres S. (2012) Mitochondrial DNA damage is associated with reduced mitochondrial bioenergetics in Huntington's disease. Free Radic. Biol Med., 53, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazziotta J.C., Phelps M.E., Pahl J.J., Huang S.C., Baxter L.R., Riege W.H., Hoffman J.M., Kuhl D.E., Lanto A.B., Wapenski J.A. and (1987) Reduced cerebral glucose metabolism in asymptomatic subjects at risk for Huntington's disease. N. Engl. J. Med., 316, 357–362. [DOI] [PubMed] [Google Scholar]
- 41.Powers W.J., Haas R.H., Le T., Videen T.O., Markham J., Perlmutter J.S. (2011) Platelet mitochondrial complex I and I+III activities do not correlate with cerebral mitochondrial oxidative metabolism. J. Cereb. Blood Flow Metab., 31, e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sultana R., Poon H.F., Cai J., Pierce W.M., Merchant M., Klein J.B., Markesbery W.R., Butterfield D.A. (2006) Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol. Dis., 22, 76–87. [DOI] [PubMed] [Google Scholar]
- 43.Opii W.O., Nukala V.N., Sultana R., Pandya J.D., Day K.M., Merchant M.L., Klein J.B., Sullivan P.G., Butterfield D.A. (2007) Proteomic identification of oxidized mitochondrial proteins following experimental traumatic brain injury. J. Neurotrauma, 24, 772–789. [DOI] [PubMed] [Google Scholar]
- 44.Maldonado E.N., Lemasters J.J. (2014) ATP/ADP ratio, the missed connection between mitochondria and the Warburg effect. Mitochondrion., 19 Pt A, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemasters J.J., Holmuhamedov E. (2006) Voltage-dependent anion channel (VDAC) as mitochondrial governator–thinking outside the box. Biochim. Biophys. Acta, 1762, 181–190. [DOI] [PubMed] [Google Scholar]
- 46.De Stefani D., Bononi A., Romagnoli A., Messina A., De, P V, Pinton P., Rizzuto R., (2012) VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death. Differ., 19, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zabel C., Mao L., Woodman B., Rohe M., Wacker M.A., Klare Y., Koppelstatter A., Nebrich G., Klein O., Grams S, et al. (2009) A large number of protein expression changes occur early in life and precede phenotype onset in a mouse model for huntington disease. Mol. Cell Proteomics., 8, 720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stack E.C., Kubilus J.K., Smith K., Cormier K., Del Signore S.J., Guelin E., Ryu H., Hersch S.M., Ferrante R.J. (2005) Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. J. Comp. Neurol., 490, 354–370. [DOI] [PubMed] [Google Scholar]
- 49.Napoli E., Wong S., Hung C., Ross-Inta C., Bomdica P., Giulivi C. (2013) Defective mitochondrial disulfide relay system, altered mitochondrial morphology and function in Huntington's disease. Hum. Mol. Genet., 22, 989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scarpulla R.C., Vega R.B., Kelly D.P. (2012) Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab., 23, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koopman W.J., Distelmaier F., Smeitink J.A., Willems P.H. (2013) OXPHOS mutations and neurodegeneration. Embo J., 32, 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez-Fernandez S., Almeida A., Bolanos J.P. (2012) Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem. J., 443, 3–11. [DOI] [PubMed] [Google Scholar]
- 53.Orr A.L., Li S., Wang C.E., Li H., Wang J., Rong J., Xu X., Mastroberardino P.G., Greenamyre J.T., Li X.J. (2008) N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci., 28, 2783–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Z.X., Li S.H., Evans J., Pillarisetti A., Li H., Li X.J. (2003) Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci., 23, 2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., Ferrante R.J. (2010) Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum. Mol. Genet., 19, 3919–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melendez-Ferro M., Rice M.W., Roberts R.C., Perez-Costas E. (2013) An accurate method for the quantification of cytochrome C oxidase in tissue sections. J. Neurosci. Methods, 214, 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]