Abstract
To explore the correlation between platelet endothelial aggregation receptor-1 (PEAR1) genetic polymorphism and pulmonary thromboembolism (PTE).
Variant loci of the PEAR1 gene were screened in a PTE pedigree, followed by verification using Sanger sequencing. These polymorphic loci were validated in 101 PTE patients and 132 matched normal patients using MassARRAY single nucleotide polymorphism (SNP) genotyping methods. The frequency differences between the allele and genotypes were compared using the Hardy–Weinberg equilibrium test and Chi-square test. The correlation between the PEAR1 gene SNP and PTE was analyzed by comparing the between-group variance differences using the χ2 test.
Three SNPs were identified in the PTE pedigree. There was a heterozygous transition of T>C in rs1952294, and a transition of C>T in rs778026543 in 2 members in the pedigree; however, the rs778026543 was not identified in the 101 PTE patients and 132 healthy controls. The genotype and allele frequencies of rs822442 did not differ significantly between PTE patients and healthy controls (P > 0.05). The variance difference at rs778026543 between pedigree members and healthy controls was significant (P < 0.001), supporting its potential heredity.
The PEAR1 polymorphism, rs778026543, but not rs1952294 and rs822442, may be a susceptibility SNP for PTE.
Keywords: PEAR1, pedigree, pulmonary thromboembolism, SNP, whole exome sequencing
1. Introduction
Pulmonary thromboembolism (PTE), a disease that is determined by interactions between genetic susceptibility and the surrounding environment, is the third most common cause of death in hospitalized patients.[1] PTE usually develops during pathological processes of hemostasis, coagulation, and anticoagulation disorders, and platelets are the basic factors underlying these functions and exerting very important roles in thrombosis.[2–5] As a platelet aggregation receptor, platelet endothelial aggregation receptor-1 (PEAR1) is mainly distributed in the platelet membrane and expressed in unactivated platelets.[6] PEAR1 becomes activated in response to the interactions of platelets, causing the aggregation and adhesion of platelets and participating in the formation and reformulation and stabilization of the thrombus.[7] Many studies have reported an association between the PEAR1 gene and the coagulation system.[8–10] For example, variants of PEAR1 have been reported to be associated with increased platelet aggregation, playing important roles in agonist-induced platelet aggregation.[11,12]
The strong associations between platelet aggregation and a common intronic variant of the PEAR1 gene have been identified in both African Americans and European Americans using sequencing approaches.[10,13] Genetic variation of PEAR1 is also believed to contribute to the functional variability of platelets.[14] The G allele in the rs12041331 A>G single nucleotide polymorphism (SNP) of PEAR1 was associated with increased aggregation in response to all agonists in 2076 healthy persons before and after aspirin treatments.[15] The C allele in the rs2768759 A>C SNP of PEAR1 was generally associated with increased platelet aggregation in response to all agonists at baseline.[11]
However, whether SNPs of the PEAR1 gene increase the susceptibility of PTE patients remains unclear. In this study, variants of the PEAR1 gene in one PTE pedigree were screened, and the correlations between PEAR1 SNPs and PTE were explored in a larger population.
2. Materials and methods
2.1. Ethical statement
This study was approved by the ethics committee of the Second Clinical Medical College (Shenzhen People's Hospital), Jinan University.
2.2. Study population
A total of 3 PTE members and 1 non-PTE member in a Chinese pedigree were recruited for this study. Moreover, 101 PTE patients (exclusion criteria: family history of PTE or deep vein thrombosis (DVT)) who underwent PTE treatment in our hospital from December 2013 to August 2015 along with 132 healthy controls (exclusion criteria: DVT, PTE, endocrine system disease, heart, brain, lung, kidney, blood disease) were also enrolled in the subsequent verification study. The PTE patients were diagnosed according to the PTE diagnosis and treatment guidelines (draft) released by the Chinese Respiratory Diseases Association in 2001. Informed consent was obtained from all participants before inclusion in the study in accordance with the Declaration of Helsinki.
2.3. Whole exome sequencing in a Chinese pedigree with PTE
One milliliter peripheral venous blood was obtained from each 4 family members (Ia, IIb, IId, IIIb) and collected into the EDTA-K3 anticoagulant tubes. Genomic DNA was extracted from the peripheral blood with using the QIAamp DNA BLood Mini kit (QIAGEN GmbH, Hilden, Germany). The OD260/280 and OD260/230 of genomic DNA were examined using a NanoDrop spectrophotometer (Technologies, Rockland, DE, USA), and the DNA integrity was analyzed by agarose gel electrophoresis. After quantification by agarose gel electrophoresis, genomic DNAs were sheared to an average length of 150 to 200 bp using a Covaris S2 sonicator (Covaris, Woburn, Massachusetts, USA), the fragment ends were repaired and adaptors were ligated to the fragments and an extra nucleotide was added to the 3′ end of the end-repaired DNA fragments, then remove the DNA fragment which with a length more than 200 bp. An initial library with 300 bp was generated after ligation-mediated (LM-PCR) procedure was performed for amplification of the above DNA segments. The DNA samples were first subjected to library construction and whole exome sequencing on an Illumina HiSeq2000 platform (Illumina, San Diego, CA). Low quality variants were filtered out according to the following criteria: quality sequencing score <20; sequencing depth coverage <20×; variation detected on a single DNA strand. After the exclusion of all the low quality reads, the variants among the exon region of PEAR1 in the pedigree were identified by comparing the sequences with those in human genome UCSC hg19 version (software version: SOAP aligner v2.21). Then SNP identification was performed using the SOAPsnp program (http://soap.genomics.org.cn, BGI-Shenzhen, Shenzhen, China). All single nucleotide variations (SNVs) or indels annotations were estimated by using dbSNP version 137 (http://www.ncbi.nlm.nih.gov/snp/),[16] 1000 Genome project dataset (1000G, http://www.1000genomes.org/), ESP6500 and dbSNP version 144.[17,18]
2.4. Sanger sequencing
After the exome sequencing, the Sanger sequencing was repeated. Screened SNPs were then validated using the Sanger sequencing method. The utilized primers were designed (Table 1) with Primer Premier 3.0 (Premier Biosoft International, Palo Alto, CA, USA). The DNA samples were first subjected to amplification under the following conditions: 95°C for 5 minutes, followed by 30 cycles at 96°C for 20 seconds, 62°C for 20 seconds, and 72°C for 60 seconds, and a final extension at 72°C for 5 minutes. The PCR products were then purified using ddH2O. Finally, the sequences of the purified PCR product were tested according to the following procedures: 95°C for 15 seconds, followed by 35 cycles at 95°C for 15 seconds, 50°C for 5 seconds, and 60°C for 90 seconds.
Table 1.
Primes sequence for Sanger sequencing (5′–3′).

2.5. MassARRAY SNP genotyping
The identified SNPs were then genotyped in 101 PTE patients and 132 healthy controls using the MassARRAY SNP genotyping system (Sequenom, San Diego, CA). Sequenom's MassARRAY Designer software was used to automatically design the PCR and extension primers for each SNP (Table 2). The following PCR cycling program was used: 94°C for 4 minutes, followed by 45 cycles at 94°C for 20 seconds, 56°C for 30 seconds, and 72°C for 60 seconds, and a final extension at 72°C for 3 minutes. The DNA sample was then maintained at 4°C before the next step. Following transcription, the PCR products were treated with shrimp alkaline phosphatase (SAP) to remove the remaining and nonincorporated dNTPs under the following conditions: 37°C for 40 minutes, 85°C for 5 minutes, and 72°C for 3 minutes. Next, primer extension was performed to detect single base polymorphisms in the amplified DNA. After the extension, the extended reaction products were cleaned up by resin purification. Finally, microarray DNA spotting for genotyping was performed. Gene annotation of the identified SNPs was conducted based on the NCBI database.
Table 2.
Primers sequence for SNP genotyping (5′–3′).

2.6. Statistical analysis
Genotype and gene allele frequencies were calculated for each locus. The observed frequencies in the controls were compared with those predicted by the Hardy–Weinberg equilibrium equation using the Chi-square test. P < 0.05 was considered statistically significant. The variance differences in the identified loci between healthy controls and 101 PTE patients, and the members in the pedigree were determined using the χ2 test.
3. Results
3.1. Baseline characteristics
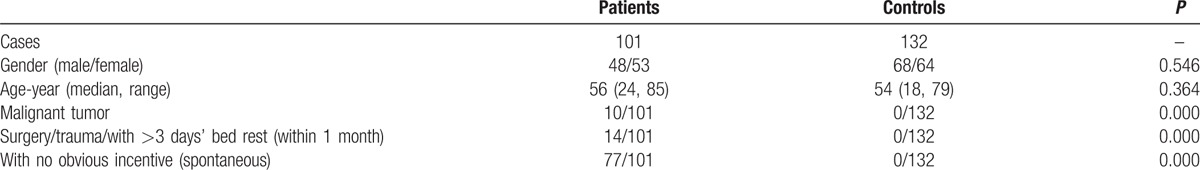
The 4 members (3 affected: IIb, IId, and IIIb; 1 unaffected individuals: Ia) that participated in this study were recruited from a 3-generation Chinese pedigree (Fig. 1). The baseline characteristics of the additional enrolled 101 PTE patients and 132 healthy controls are shown in Table 3. The PTE patients had a median age of 56 years and consisted of 48 males and 53 females. Among all patients, there were 23 traumas and 63 DVT events; the controls had a median age of 54 years and consisted of 68 males and 64 females.
Figure 1.
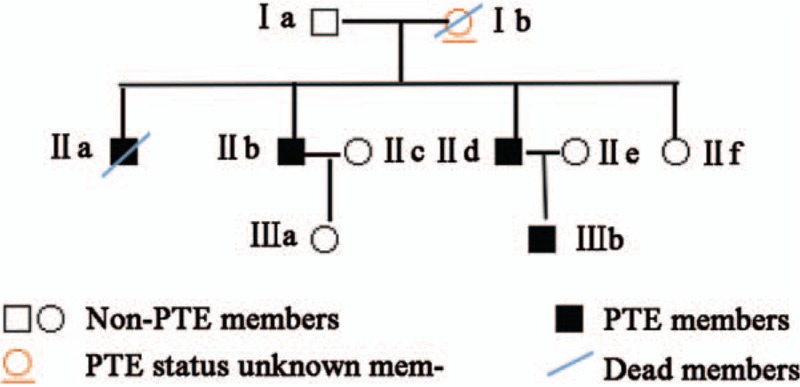
Pedigree of the 3-generation Chinese family with pulmonary thromboembolism (PTE). Squares and circles indicate males and females, respectively. Black symbols indicate affected members, and open symbols indicate unaffected individuals.
Table 3.
Baseline characteristics and clinical features of the 101 PTE (pulmonary thromboembolism) patients and 132 healthy controls.
3.2. Quality control of whole exome sequencing data and identified SNPs
A total of 14,761,440 reads uniquely mapped to the genome, and 75.22% of the effective sequences aligned to a 9580.55 Mb target. The average sequencing depth on the target was 187.16, and the coverage was 99.8%. Moreover, 99.55%, 99.15%, and 98.38% had a target sequence coverage fraction ≥4×, ≥10×, and ≥20×. Three SNPs of the PEAR1 gene were screened: rs1952294, rs822442, and an uncommon rs778026543.
3.3. SNP validation by Sanger sequencing
Sanger sequencing analysis verified the 3 SNPs in the family members from the PTE pedigree (Fig. 2). Both in whole exome sequencing and Sanger sequencing, all detected pedigree members carried the rs1952294 SNP, in addition, a T>C transition was detected in both IIb, IId, IIIb, and Ia. The result of whole exome sequencing showed that IId and IIIb had the rs822442 SNP, and a C>A transition was found. However, Sanger sequencing result showed that IId, IIIb, and Ia had the rs822442 SNP; The results of 2 sequencing method showed that rs778026543 SNP was detected in both IIb and Ia, and a C>T transition was occurred.
Figure 2.
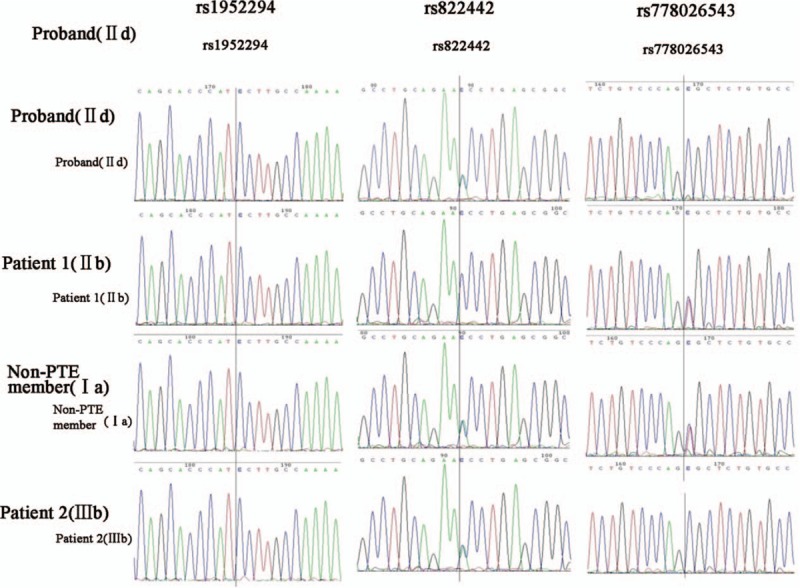
Sequence analysis of the Chinese pedigree with pulmonary thromboembolism (PTE).
3.4. Correlations between the identified SNPs and PTE
In the rs1952294 SNP, a T>C transition was also detected in the 101 PTE patients and 132 healthy controls. The genotype and gene allele frequencies of the rs822442 SNP, which is a A>C transition, are shown in Table 4. The genotype and gene allele frequencies of the rs1952294 SNP and rs778026543 SNP are shown in Tables 5 and 6, respectively. The genotype of the rs822442 SNP followed Hardy–Weinberg equilibrium in the 101 PTE patients (χ2 = 0.053, P = 0.817318) and 132 healthy controls (χ2 = 0.694, P = 0.404863). There were no significant differences in the genotype (P = 0.702) and allele distributions between these 2 groups (P = 0.660).
Table 4.
Genotype and allele gene frequencies of rs822442 SNP.
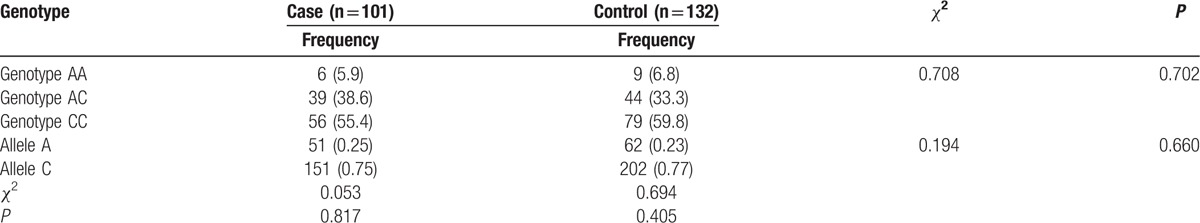
Table 5.
Genotype and allele gene frequencies of rs1952294 SNP.

Table 6.
Genotype and allele gene frequencies of rs778026543 SNP.

The rs778026543 SNP, a C>T transition, was found in 1 affected (IIb) and 1 unaffected (Ia) individual. This SNP was not identified in the 101 PTE patients and 132 healthy controls.
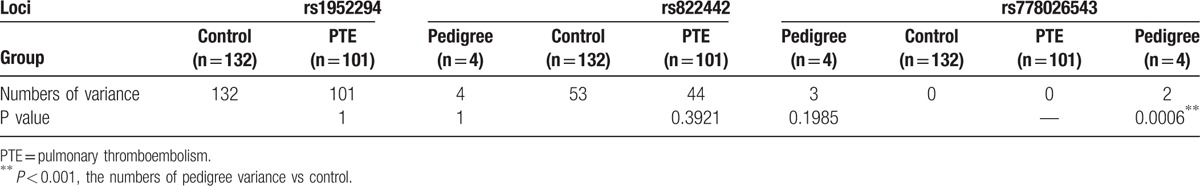
The variance differences in the identified locus between the healthy controls and 101 PTE patients, and the members of the pedigree are displayed in Table 7. The between-group difference was only detected in the rs778026543 SNP: there was a significant difference between the pedigree members and healthy controls (P = 0.0006).
Table 7.
Variance differences at identified SNP locus of healthy controls and PTE subjects.
4. Discussion
PTE is a blockage of an artery in the lungs by a substance that has traveled from elsewhere in the body through the bloodstream. Symptoms of PTE include dysfunction of the pulmonary circulation and respiration.[19,20] PTE is associated with high morbidity and mortality, with a poor prognosis.[21] PTE usually results from the activation, aggregation, and adhesion of platelets, which are regulated by certain genes. PEAR1 is one of the genes that regulates the functions of platelets, and it is highly expressed in platelets and endotheliocytes.[7,10] The PEAR1 gene comprises 23 exons and 22 introns, and its encoded protein participates in extracellular protein–protein interactions. Both intracellular and extracellular zones of the PEAR1 protein bind to phosphotyrosine.[10] The PEAR1 gene has been reported to regulate the function of platelets by controlling the phosphatidylinositol 3 kinase (PI3K)/pentaerythritol tetranitrate (PTEN) pathway.[22,23] The contributions of the PEAR1 gene to the activation of platelets, inhibition of platelet progenitor cell proliferation, and interruption of platelet aggregation have been revealed.[9,11,12,22]
The vital roles of the PEAR1 SNP have been revealed in many diseases: Sokol et al[24] found that patients with hyperlipidemia abortion had a higher incidence of the PEAR1 rs12041331 and rs12566888 SNPs than healthy people, and the T allele in the PEAR1 c. -9-4663G>T gene polymorphism had a protective effect on fetus abortion. Olivi et al[25] reported that there were no correlations between the selected 9 SNP loci of PEAR1 and the changes in diastolic and systolic blood pressure (P > 0.059), or in the incidence in patients with hypertension (P > 0.09). Herreragaleano et al[11] showed that in patients with coronary heart disease, the new variant of the PEAR1 gene (rs2768759), which is located in the promoter region, can induce enhanced agonist-induced platelet aggregation and is a susceptibility factor for platelet aggregation after aspirin therapy. Faraday et al[10] found that in patients with coronary heart disease, the reaction of ADP and epinephrine-induced platelet aggregation was closely associated with the intron 1 variant (rs12041331) of PEAR1. In the study conducted by Würtz et al,[26] AA and GA identified in the PEAR1 rs12041331 SNP were found to attenuate platelet aggregation during aspirin therapy.
Most primary risk factors for PTE are gene variance and SNP-related factors, including blood coagulation factor V, Leiden gene mutation, prothrombin gene G20210A mutation, antithrombin III gene mutation, protein C and protein S gene mutation, hyperhomocysteinemia, and defects in the fibrinolytic system, which influence the formation of thrombosis.[27–30] SNPs of the PEAR1 gene influence these functions and activate platelets and megakaryocytes. Promoted by different agonists, the SNPs also participate in changes in platelet aggregation. The above activities of platelets are the etiologies of PTE. Therefore, it can be assumed that there may be a correlation between PTE and the PEAR1 gene. By conducting whole exome sequencing, a total of 3 SNPs (rs1952294, rs822442, and rs778026543) were found in the PEAR1 gene, and these SNPs were subsequently verified using Sanger sequencing methods. Moreover, the genotype and allele gene distributions were investigated in a subsequent case–control study using samples from the 101 PTE patients and 132 matched healthy controls.
Among all 3 screened SNPs, the rs1952294 locus was identified in all 4 detected pedigree members. This homozygote A>C SNP was then identified in the large sample size (101 patients and 132 controls). However, this SNP was not associated with susceptibility to PTE.
Regarding the rs822442 locus, in a previous study exploring the efficacy of prasugrel, Xiang et al[31] showed that the PEAR1 SNP in a 4-kb area between rs3737224 and rs822442 was associated with ADP-induced platelet aggregation. In this study, there were no significant differences in the representative population (HWE test P > 0.05) between PTE patients and controls in terms of genotype and allele distributions. In addition, there were no differences in SNP loci between pedigree members and healthy controls in terms of variance numbers. The rs822442 SNP was not associated with susceptibility to PTE.
The rs778026543 SNP of PEAR1 is an uncommon variant with a MAF < 0.05 (MAF/minor allele count: T = 0.0000/3) that was first recorded in dbSNP144; however, this variant was observed in two members of the pedigree: 1 PTE patient (IIb) and 1 non-PTE member (Ia). Moreover, the variance numbers at this SNP locus were significantly different between pedigree members and healthy controls. Although the rs778026543 SNP was not identified in the 101 patients and 132 healthy controls, this variant is still a SNP of interest because of its rarity. This is the first report to describe the rs778026543 SNP in patients with PTE, and the associated potential mechanism is described below: the non-PTE member in fact suffered from cerebral thrombosis, which was related to platelet functions and led to a diagnosis of PTE.[32,33] Based on this finding, platelet dysfunction might be the causal factor for PTE and for this variant. Additional research is needed to explore the roles of this variant in PTE in this pedigree to confirm its correlation with the disease. However, if this SNP is verified in a much larger population, it may represent a true susceptibility factor for PTE.
Footnotes
Abbreviations: PEAR1 = platelet endothelial aggregation receptor-1, PTE = pulmonary thromboembolism, SNPs = single nucleotide polymorphisms, SNVs = single nucleotide variations.
Funding: This work was supported by the Knowledge Innovation Program of Shenzhen, China (No: JCYJ20130402111858730). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
YF and SS contributed equally to this study.
The authors have no conflicts of interest to disclose.
References
- [1].Deng C, Yang M, Lin Q, et al. Beneficial effects of inhaled NO on apoptotic pneumocytes in pulmonary thromboembolism model. Theor Biol Med Model 2014;11:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kline DL. Blood coagulation—reactions leading to prothrombin activation. Annu Rev Physiol 1965;27:285–306. [DOI] [PubMed] [Google Scholar]
- [3].Holcomb GR. Hemostasis and Thrombosis: Basic Principles and Clinical Practice. Wolters Kluwer:Lippincott Williams & Wilkins; 2006. [Google Scholar]
- [4].Billon MP, Mckirgan CJ, Mcclure PJ, et al. Prospective validation of the International Society of Thrombosis and Haemostasis scoring system for disseminated intravascular coagulation. Crit Care Med 2004;32:2416–21. [DOI] [PubMed] [Google Scholar]
- [5].Walsh PN. Platelet coagulant activities and hemostasis: a hypothesis. Blood 1974;43:597–605. [PubMed] [Google Scholar]
- [6].Nanda N, Bao M, Lin H, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J Biol Chem 2005;280:24680–9. [DOI] [PubMed] [Google Scholar]
- [7].Nanda N, Phillips DR. Novel targets for antithrombotic drug discovery. Blood Cells Mol Dis 2006;36:228–31. [DOI] [PubMed] [Google Scholar]
- [8].Wu HH, Bellmunt E, Scheib JL, et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat Neurosci 2009;12:1534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pasala T, Sattayaprasert P, Bhat PK, et al. Clinical and economic studies of eptifibatide in coronary stenting [Corrigendum]. Ther Clin Risk Manag 2014;10:603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Faraday N, Yanek LR, Yang XP, et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood 2011;118:3367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herreragaleano JE, Becker DM, Wilson AF, et al. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregabili. Arterioscler Thromb Vasc Biol 2008;28:1484–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lewis JP, Ryan K, O’Connell JR, et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ Cardiovasc Genet 2013;6:184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet 2010;42:608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yao Y, Tang XF, Zhang JH, et al. Association of PEAR1 genetic variants with platelet reactivity in response to dual antiplatelet therapy with aspirin and clopidogrel in the Chinese patient population after percutaneous coronary intervention. Thromb Res 2016;141:28–34. [DOI] [PubMed] [Google Scholar]
- [15].Kim Y, Suktitipat B, Yanek LR, et al. Targeted deep resequencing identifies coding variants in the PEAR1 gene That play a role in platelet aggregation. PLoS ONE 2013;8:e64179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vandamme T, Peeters M, Dogan F, et al. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. J Mol Endocrinol 2015;54:137–47. [DOI] [PubMed] [Google Scholar]
- [17].Yu X, Wang F, Zhang JP. Meta analysis of the association of rs7702187 SNP in SEMA5A gene with risk of Parkinson's disease. Eur Rev Med Pharmacol Sci 2014;18:900–4. [PubMed] [Google Scholar]
- [18].Gable D, Stephens J, Humphries S, et al. The +276G > T adiponectin gene SNP is associated with cardiovascular disease in patients with type 2 diabetes mellitus. Diabetologia 2005. [Google Scholar]
- [19].Tavil B, Kuskonmaz B, Kiper N, et al. Pulmonary thromboembolism in childhood: a single-center experience from Turkey. Heart Lung 2009;38:56–65. [DOI] [PubMed] [Google Scholar]
- [20].Haythe J. Chronic thromboembolic pulmonary hypertension: a review of current practice. Prog Cardiovasc Dis 2012;55:134–43. [DOI] [PubMed] [Google Scholar]
- [21].Gong X, Duan Z, Yuan Y. Long-term prognosis and related factors towards patients with acute pulmonary thromboembolism. Int J Clin Exp Med 2014;8:7906–13. [PMC free article] [PubMed] [Google Scholar]
- [22].Kauskot A, Vandenbriele C, Louwette S, et al. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood 2013;121:5208–17. [DOI] [PubMed] [Google Scholar]
- [23].Fisch AS, Yergesarmstrong LM, Backman JD, et al. Genetic variation in the platelet endothelial aggregation receptor 1 gene results in endothelial dysfunction. PLoS ONE 2015;10:e0138795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sokol J, Biringer K, Skerenova M, et al. Different models of inheritance in selected genes in patients with sticky platelet syndrome and fetal loss. Semin Thromb Hemost 2015;41:330–5. [DOI] [PubMed] [Google Scholar]
- [25].Olivi L, Vandenbriele C, Gu YM, et al. PEAR1 is not a human hypertension-susceptibility gene. Blood Press 2015;24:1–4. [DOI] [PubMed] [Google Scholar]
- [26].Würtz M, Nissen PH, Grove EL, et al. Genetic determinants of on-aspirin platelet reactivity: focus on the influence of PEAR1. PLoS ONE 2014;9:e111816–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Peng F, Triplett D, Barna L, et al. Pulmonary embolism and premature labor in a patient with both factor V Leiden mutation and methylenetetrahydrofolate reductase gene C677T mutation. Thromb Res 1996;83:243–51. [DOI] [PubMed] [Google Scholar]
- [28].Soylu A, Tokaç M, Çora T, et al. Platelet glycoprotein Ibalpha gene polymorphism and massive or submassive pulmonary embolism. J Thromb Thrombolysis 2008;27:259–66. [DOI] [PubMed] [Google Scholar]
- [29].Charafeddine KM, Mahfouz RA, Ibrahim GY, et al. Massive pulmonary embolism associated with Factor V Leiden, prothrombin, and methylenetetrahydrofolate reductase gene mutations in a young patient on oral contraceptive pills: a case report. Clin Appl Thromb Hemost 2010;16:594–8. [DOI] [PubMed] [Google Scholar]
- [30].Kotuličová D, Chudý P, Ivanková MŠ, et al. Variability of GP6 gene in patients with sticky platelet syndrome and deep venous thrombosis and/or pulmonary embolism. Blood Coagul Fibrinolysis 2012;23:543–7. [DOI] [PubMed] [Google Scholar]
- [31].Xiang Q, Cui Y, Zhao X, et al. Identification of PEAR1 SNPs and their influences on the variation in prasugrel pharmacodynamics. Pharmacogenomics 2015;14:1179–89. [DOI] [PubMed] [Google Scholar]
- [32].Keatinge WR, Coleshaw SRK, Easton JC, et al. Increased platelet and red cell counts, blood viscosity, and plasma cholesterol levels during heat stress, and mortality from coronary and cerebral thrombosis. Am J Med 1986;81:795–800. [DOI] [PubMed] [Google Scholar]
- [33].Choudhri TF, Hoh BL, Zerwes HG, et al. Reduced microvascular thrombosis and improved outcome in acute murine stroke by inhibiting GP IIb/IIIa receptor-mediated platelet aggregation. J Clin Invest 1998;102:1301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]


