Abstract
In the course of influenza A virus (IAV) infections, a secondary bacterial infection frequently leads to serious respiratory conditions provoking high hospitalization and death tolls. Although abundant pro-inflammatory responses have been reported as key contributing factors for these severe dual infections, the relative contributions of cytokines remain largely unclear. In the current study, mathematical modelling based on murine experimental data dissects IFN-γ as a cytokine candidate responsible for impaired bacterial clearance, thereby promoting bacterial growth and systemic dissemination during acute IAV infection. We also found a time-dependent detrimental role of IL-6 in curtailing bacterial outgrowth which was not as distinct as for IFN-γ. Our numerical simulations suggested a detrimental effect of IFN-γ alone and in synergism with IL-6 but no conclusive pathogenic effect of IL-6 and TNF-α alone. This work provides a rationale to understand the potential impact of how to manipulate temporal immune components, facilitating the formulation of hypotheses about potential therapeutic strategies to treat coinfections.
Retrospective studies performed on victims of the 1918/1919 influenza A virus (IAV) pandemic and also the recent H1N1 IAV pandemic revealed a high incidence of coinfections with unrelated bacterial pathogens1,2,3,4,5. In fact, 71% of the high death toll during the 1918/1919 outbreak was attributed to coinfection with Streptococcus pneumoniae (S. pneumoniae)5. This copathogen is a human adapted gram-positive colonizer of the nasopharynx in asymptomatic children, adults and individuals over 65 years of age but at the same time remains to be the most common cause of community-acquired pneumonia6. Animal and human studies have shown that preceding IAV infection enhances all aspects of S. pneumoniae pathogenesis from nasopharyngeal colonization to invasive pneumococcal disease7, leading to the strong predisposition to lethal secondary pneumococcal infection in IAV infected patients.
Coinfections can be either concurrent or sequential and can involve both acute and chronic infections8. Synergistic interactions between pathogens have been well documented for chronic viral infections, for example the influence of prior HIV infection on the development of chronic hepatitis B infection9. Regarding IAV-S. pneumoniae coinfections, several mechanisms have been implicated in the viral-bacterial synergism which together demonstrated a multifactorial and complex nature of copathogenesis. However, holistic understanding of the effects between IAV, bacteria and immune modulation remain largely unknown.
One central dogma in the viral-bacterial synergism is the disruption of the protective alveolar epithelial cell barrier due to the cytolytic mode of influenza A replication which exposes otherwise cryptic bacterial adherence factors on the basal membrane and thereby promotes invasive pneumococcal disease. More debatable mechanisms are the IAV-mediated immune modulations such as immune cell dysfunction and apoptosis causing an aberrant production of inflammatory mediators in the case of a secondary bacterial encounter. Experimental reports indicate dampened innate inflammatory responses to the bacteria in IAV pre-infected hosts due to an enhanced activation threshold of lung innate immune cells that renders them hypo-responsive10. In contrast, a number of studies describe a massive and overshooting inflammatory cell influx due to the hyper-production of pro-inflammatory cytokines such as type I Interferons (IFN-I), Interferon-γ (IFN-γ), Interleukin-6 (IL-6) and Tumor Necrosis Factor-α (TNF-α) during secondary bacterial infection. These are often linked to pulmonary edema due to irreparable damage to the alveoli and immunopathology leading to mortality during coinfections7,11,12,13,14. Taken together, these studies strongly reflect an exacerbated cytokine and chemokine production that may significantly contribute to the detrimental changes in the lung microenvironment that favor secondary bacterial infections. However it is still largely unknown what the relative contribution of these cytokines to bacterial outgrowth, morbidity and mortality in secondary infection is and whether they work alone or in synergism as ‘friend or foe’ to the coinfected host.
Dissecting the detailed contributions of the identified players in enhancing susceptibility to severe secondary bacterial disease following IAV as well as their respective interactions is crucial to develop prophylactic and therapeutic strategies. In the past, mathematical modelling has made valuable contributions to our understanding of IAV infection, focusing either on IAV replication15,16,17,18,19,20,21 and/or host immune responses to IAV22,23,24,25,26,27,28,29,30,31,32. Regarding the interactions between IAV and S. pneumoniae, the only modelling approach proposed so far has been through the pioneering work of Smith et al.33. However, to the best of the authors knowledge, untangling the contributions of the different mechanisms by which changes in the immune response affect bacterial clearance in a temporal manner has not been attempted until now. Therefore, by combining the results of murine in vivo experiments and mathematical modelling approaches, we aimed at clarifying the relative contributions of different underlying mechanisms of the IAV-S. pneumoniae synergism.
Results
Study Design
The dynamics of IAV and S. pneumoniae coinfection were investigated by establishing a murine model displaying disease upon subsequent infection with sub-lethal infection doses of both copathogens. Secondary infection with 1 × 106 colony forming units (CFU) of Streptococcus pneumoniae strain TIGR4 (T4) was performed on day 7 after IAV infection based on previous experimental observations that indicated peak susceptibility to pneumococcal disease at this time point during acute IAV infection3,34. Bacterial burden, viral titers, cytokine concentrations and alveolar macrophage (AM) counts were determined in the respiratory tract for three experimental groups: coinfected (IAV + T4), single IAV and single T4 infected animals. A schematic representation of the experiments is provided in Fig. 1.
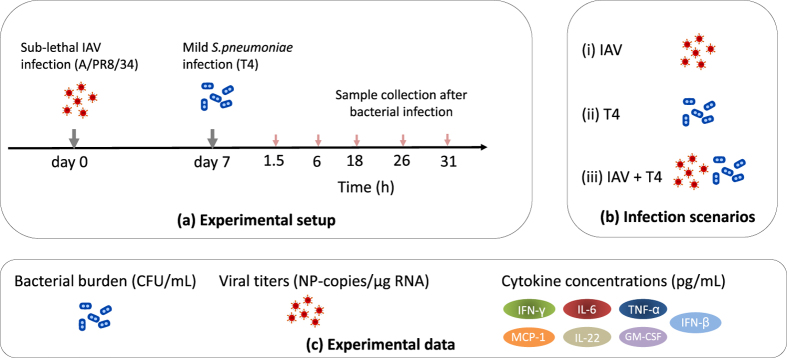
Figure 1. Experimental scheme.
(a) C57BL/6 J wildtype mice were intranasally infected with a sub-lethal dose of IAV (A/PR8/34) followed by bacterial infection with the S. pneumoniae strain T4 on day 7. Bronchoalveolar lavage (BAL), post-lavage lung and blood were collected at the indicated time points post secondary bacterial infection (hpi). (b) The infection groups were single viral infection (IAV), single bacterial infection (T4) and coinfection (IAV+T4). (c) The bacterial burden, viral titers and cytokine concentrations were determined as the experimental readouts.
The complexity and at times redundancy of immune responses to infections often render to arduous and expensive experimental settings when attempting to identify the key components and their temporal contributions during coinfections. Thus, merging mathematical modelling with the relevant in vivo data is a promising tool to unravel complex interactions25,31,33,35. In order to dissect the dynamics observed in our experiments, mathematical modelling was employed not as a quantitative recapitulation of experimental data but as a tool to evaluate various hypotheses on the basis of mathematical models and the corrected Akaike information criterion (AICc) for the model selection process.
Bacterial growth kinetics during IAV-pneumococcal coinfection
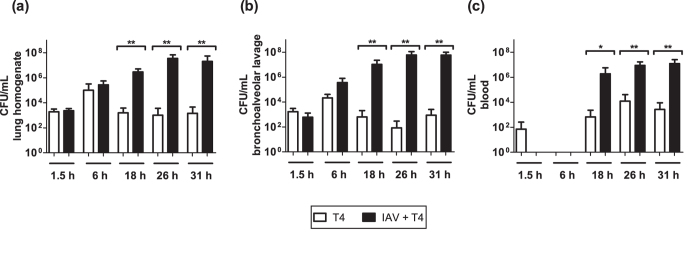
Mouse coinfection experiments revealed that the bacterial load in the post-lavage lung tissue and bronchoalveolar lavage (BAL) was comparable between the coinfected (IAV+T4) and single T4 infected groups until 6 hours post infection (hpi) (Fig. 2a and b). At 18 hpi, a significantly higher bacterial load was observed in both the lung tissue and BAL of the coinfected compared to the single T4 infected group. The bacterial load further increased until 31 hpi in the coinfected group. At the later time points, the high bacterial numbers in the respiratory tract were accompanied by the systemic spread via the blood (bacteremia) in all the coinfected animals (Fig. 2c). Significantly lower grade bacteremia was observed in the single T4 infected mice starting 18 hpi (Fig. 2c).
Figure 2. Organ-wide bacterial burden in the single and coinfected animals.
Bacterial titers in single T4 infected and IAV+T4 coinfected mice were determined in (a) post-lavage lung, (b) BAL and (c) blood. All experiments were performed in groups of 4–7 WT C57BL/6 J mice, raw data can be found in the Supplementary Fig. S1. Statistical analysis was performed using the Mann-Whitney test. Asterisks indicate significant differences between single and coinfected mice: *p < 0.05; **p < 0.01.
Taken together, assessing the kinetics of bacterial growth and clearance in the respiratory tract and blood following IAV-S. pneumoniae coinfection revealed a “turning-point” between 6 and 18 hpi. At 18 hpi bacterial outgrowth became clearly evident in the coinfected group and was in strong contrast to the onset of bacterial clearance in the T4 only infected group (also see CFU data for individual mice in Supplementary Fig. S1).
Absolute AM numbers in the lung homogenate of the coinfected animals may not determine bacterial outgrowth
Previous results in ref. 36 revealed that 90% of resident AMs were depleted in the first week after influenza infection forming a favourable niche for a secondary pneumococcal infection. In contrast, we did not observe IAV-induced AM depletion in our mouse studies but detected significant AM depletion 18 hours post coinfection37. To further evaluate dynamic AM numbers in our experimental setting and mathematical modelling approach, we determined the absolute numbers of AMs in the lung tissue at several time-points early after secondary S. pneumoniae infection. Experimental results in Fig. 3 show high numbers of AMs in the lung following coinfection that start decreasing at 18 hpi. At this time point, a significant reduction of AMs was detectable in the coinfected animals compared to the bacterial infection alone, which coincided with the established bacterial outgrowth in these mice.
Figure 3. Absolute numbers of alveolar macrophages (AM) in post-lavage lungs of single and coinfected animals.
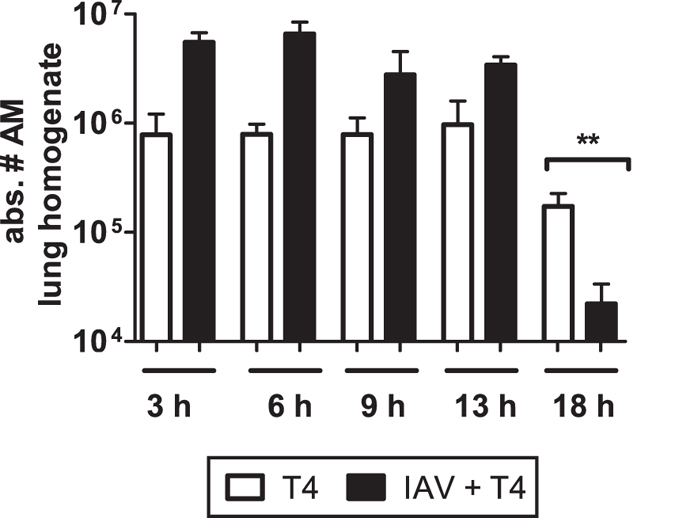
Statistical analysis was performed using the Mann-Whitney test. Asterisks indicate significant differences between single and coinfected mice: **p < 0.01. Experiments were performed in groups of 3–4 WT C57BL/6 J mice.
Thus, we evaluated the hypothesis of AM dynamics as a determinant to represent the bacterial kinetics in the single S. pneumoniae infected or in the coinfected group. To this end, by using AM count dynamics for the different groups, we proposed several mathematical models to represent the dynamics observed in the single S. pneumoniae infected (Supplementary 2) and coinfected animals (Supplementary 3). A model selection process by AIC revealed that assuming bacterial clearance with a constant number of functional AMs (model D2 or D3 at Supplementary 2) provided the best fitting to the data in the single T4 group. In contrast, considering dynamic AM numbers resulted in the worst fitting among the different models for both the single T4 group and the coinfection group. Thereby our model selection study may not support the hypothesis that bacterial clearance is mainly driven by the absolute AM numbers. More likely, only a fraction of functional AMs is required to clear the bacterial infection. Nevertheless, we assume constant AM numbers after 18 hpi for the model M1, while other models in Table 1 are based on experimental data till 31 hpi. Thus, numerical results of the model M1 may need to be taken with caution. Further experiments in mice dissecting functional and non-functional AMs will help to fully determine the effects on the bacterial clearance. To include the experimental data variability in the mathematical model M1, bootstrapping procedures are presented in Supplementary 4.
Table 1. Selected list of the coinfection models to test different hypotheses that facilitate S. pneumoniae colonization.
| No | Hypothesis | fx | Parameters | RSS | AICc |
|---|---|---|---|---|---|
| M1 | AM number dynamics MA(t) are sufficient to facilitate bacterial outgrowth36. | MA(t) | 47.74 | 12.27 | cbB |
| M2 | Single change in bacterial clearance rate is enough to explain bacterial outgrowth. | 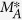 |
cbB | 21.84 | −15.87 |
| M3 | IFN-γ responses alone can impair bacterial clearance facilitating bacterial colonization12. | 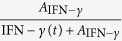 |
AIFN-γ(t) | 16.13 | −26.78 |
| M4 | TNF-α responses alone can impair bacterial clearance facilitating bacterial colonization63. | 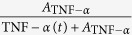 |
ATNF-α | 39.82 | 5.74 |
| M5 | IL-6 responses alone can impair bacterial clearance facilitating bacterial colonization63. | 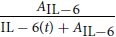 |
AIL-6 | 27.22 | −7.94 |
| M6 | A synergistic effect of the IFN-γ (X1), IL-6 (X2), and TNF-α (X3) cytokine responses in facilitation of bacterial outgrowth. | 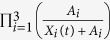 |
AIFN-γ, AIL-6 ATNF–α | 14.57 | −25.82 |
| M7 | A synergistic effect of the IFN-γ (X1) and IL-6 (X2) cytokine responses in facilitation of bacterial outgrowth. | 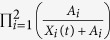 |
AIFN-γ, AIL-6 | 14.57 | −28.21 |
Best models based on AICc difference lower than 2 units are in bold. A complete model list with estimated parameters can be found in Supplementary 3.
Coinfection leads to a significant increase of IFN-γ, IL-6 and TNF-α airway concentrations
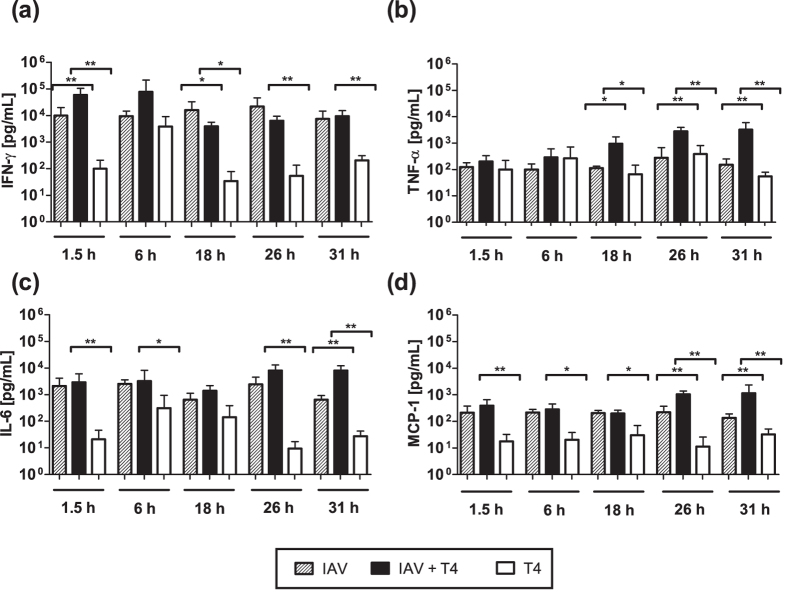
In contrast to previous animal studies that often focused on a single time point post secondary infection11,12, we assessed the early kinetics of pro-inflammatory cytokines in the respiratory tract. For this purpose, we determined IFN-γ, TNF-α, IL-6, Monocyte Chemoattractant Protein-1 (MCP-1), Interferon-β (IFN-β), Interleukin 22 (IL-22) and Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) protein concentrations in the BAL fluid of single and coinfected mice. A time-dependent significant increase in the protein concentrations of IFN-γ, TNF-α, and IL-6 was observed in the coinfected animals when compared to the single T4 infected animals (Fig. 4).
Figure 4. Pro-inflammatory cytokine profiles of the BAL of coinfected and single T4 infected mice.
Protein concentrations of (a) IFN-γ, (b) TNF-α, (c) IL-6 and (d) MCP-1 were determined in the BAL fluid at the indicated time points after secondary T4 infection on day 7 post IAV or single T4 infection on day 7 post PBS treatment. Raw data can be found in the Supplementary Fig. S2. Statistical analysis was performed using the Mann-Whitney test. Asterisks indicate significant differences between single and coinfected mice: *p < 0.05; **p < 0.01.
Bacterial infection alone led to a transient increase in IFN-γ at 6 hpi and IL-6 at 6 hpi and 18 hpi, which hardly reached the elevated IFN-γ and IL-6 levels detected in the IAV-infected group at all time points analyzed (Fig. 4). For IFN-γ, coinfection led to a further increase early at 1.5 hpi and 6 hpi compared to the single IAV infection whereas the levels remained constant compared to the underlying IAV infection for the later time points and a significant increase was only observed when compared to the single T4 infection. Overshooting IL-6 responses in the coinfected mice were detected at 26 hpi and 31 hpi compared to the single T4 infection. At the same time, TNF-α levels hardly changed between mice infected with IAV alone and T4 alone but were increasingly and significantly elevated in coinfected mice from 18 hpi on. This late excess in IL-6 and TNF-α compared to both the single IAV and T4 infected groups indicated a steady induction of these two pro-inflammatory cytokines in the coinfected animals (Fig. 4b and c). Of note, the chemokine MCP-1 was also significantly increased in the IAV+T4 group compared to the single T4 infected group and marginally increased to the IAV only group at 26 hpi and 31 hpi (Fig. 4d). The protein concentrations of the other inflammatory mediators did not show significant changes between the groups at all time points (see Supplementary Fig. S3). Note that different studies using C57BL/6 J mice reported the baseline levels of the mediators studied here to be significantly lower than what is detected following IAV infection12,37,38.
Decisive role of IFN-γ in bacterial outgrowth
To dissect the temporal contribution of the measured pro-inflammatory cytokines in preventing bacterial clearance, mathematical models fitted from single S. pneumoniae infection were challenged in order to assess the effects of pro-inflammatory responses on bacterial lung titers. Hence, we adopted the best model for single S. pneumoniae infection (model D2 from the Supplementary 2) and challenged the mathematical term representing bacterial clearance (cbB) with different functions (cbfxB) to evaluate which of the pro-inflammatory cytokines or their combinations provided the best fit to the bacterial burden detected in the coinfected mice. A brief list of mathematical models tested is presented in Table 1 and a complete version with estimated parameters and parameter uncertainty analysis is shown in Supplementary 3 and 4, respectively.
Considering the AICc scores, criterion of small differences (less than 2 units) were not significant (see Materials and Methods), the best group of models was M3 and M7 (Table 1). The common component of these two models is the IFN-γ kinetic and, remarkably, a mechanism only based on the IFN-γ response (M3) provided a better fit than mechanisms based on only TNF-α (M4) or IL-6 (M5) even though a more conservative AICc criterion is considered (e.g ≤10). Additionally, different models presented in the Supplementary 3 (M6, M7, M8 and M9) that also had IFN-γ dynamics involved in the impairment function fx scored closely to M3. In agreement with previous work12, our model selection process dissected IFN-γ as a key and sufficient modulator in the impairment of bacterial clearance.
The IL-6 response contributes to the impairment of bacterial clearance in a temporal manner
Both the models M6 and M7 suggested that IL-6 may contribute to impairing bacterial clearance during coinfection. However, the AICc for the model M5 pointed out that it may not be as crucial as IFN-γ for impairing bacterial clearance. However, it could be deduced that IL-6 is involved in the majority of the best models (e.g M6, M7, M8 and M9), indicating a time-dependent detrimental role in curtailing bacterial outgrowth.
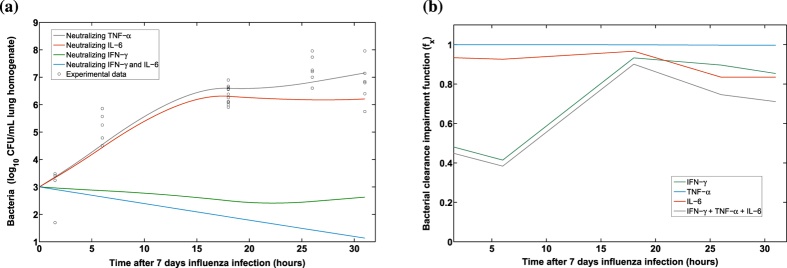
To disentangle the time-dependent roles of significant pro-inflammatory cytokines, we selected the model M6 to perform in silico experiments. Simulation results suggested that a single neutralization of IFN-γ directly modulates the bacterial clearance. In contrast, neutralization of IL-6 or TNF-α did not present a clear role in impairing bacterial clearance (Fig. 5a). Interestingly, when the IFN-γ response was neutralized in the model, simulations suggested that the IL-6 response may increase the duration of bacterial colonization in the lungs (Fig. 5a). This possibly explains the experimental observation that coinfected mice still presented a marginal increase in the bacterial burden and degree of lethality even after in vivo IFN-γ blockade12.
Figure 5. Simulations for the coinfection model M6.
In silico neutralizations of the different pro-inflammatory responses in model M6 are presented in the panel (a). The time-dependent contributions of pro-inflammatory cytokines to the impairment of bacterial clearance by the function fx are depicted in the panel (b). When the mathematical function fx is 1 means that there is no impairment to the bacterial clearance.
To weigh the inhibitory effects of different pro-inflammatory cytokines on bacterial clearance, we simulated the time evolution of the bacterial clearance impairment function (fx) for the different pro-inflammatory cytokines using the model M6. Indeed, the main modulator early after S. pneumoniae coinfection was IFN-γ. However, after 18 hours the detrimental effects of IL-6 on the bacterial clearance became more apparent (Fig. 5b). In contrast, model selection procedures and numerical simulations did not support that TNF-α kinetics contributed to the impairment of bacterial clearance, yielding one of the worst models (Table 1). These in silico results were supported by other model structures (M7, M8 and M9 at Supplementary 3) and parameter uncertainty studies (Supplementary 4).
Impairment of bacterial clearance by direct IAV kinetics is not supported
Viral titers in the single IAV infected group remained stable at day 7 post the viral infection, i.e 1.5 and 6 hpi post secondary challenge, and the viral load then declined from 26 hpi (see Supplementary Fig. S4b). In accordance with previous experimental observations by Smith et al.33, viral titers of the coinfected group showed a marginal, however not statistically significant viral rebound at 31 hpi, as the coinfected mice yielded higher viral loads than the single infected mice.
The modelling work by Smith et al.33 tested the hypothesis that influenza infection modulates the bacterial clearance by AM phagocytosis. However, their model did not include aspects of host defense33. As a comparison, we considered the inhibitory function proposed by Smith et al.33 using the kinetics of the viral load detected in our experiments (M10 at Supplementary 3). Model selection procedures however did not support that a time-dependent modulation by IAV kinetics contributes to the impairment of bacterial clearance.
Discussion
The threat of newly emerging pandemic IAV strains together with the increasing prevalence of antibiotic-resistant bacterial pathogens underline the need for a complete understanding of the mechanisms for secondary pneumonia. Recently, a substantial number of studies have uncovered several mechanisms through which IAV compromises efficient anti-bacterial defense. These different contributing factors for the viral-bacterial synergism highlight the multifactorial nature of copathogenesis.
At times however, the identified mechanisms are contradictory, e.g both de-sensitization and hyper-sensitization have been reported to favor bacterial infection post influenza infection10,13,39. Nevertheless, a common observation is an altered pro-inflammatory cytokine response to the bacterial infection if preceded by influenza and many studies have attributed the altered lung cytokine milieu to be sufficient to skew host susceptibility to severe secondary pneumococcal infection11,12,40.
The relative contribution of the different identified players however has not been addressed comprehensively to date. Although murine experiments are crucial to advance our knowledge of mechanisms which underlie virus-bacteria interactions, examining all different factors in detail is extremely difficult work. In this study, we combined an in vivo experimental approach and mathematical modelling to dissect the contributions of some of the circulating hypotheses proposed in driving bacterial outgrowth in IAV pre-infected hosts with the main focus on pro-inflammatory cytokines.
Unlike described by previous reports10,11,41, in our experiments the inflammatory responses to secondary pneumococcal infection were not dampened but instead heightened when compared to the single bacterial infection. Through mathematical modelling we identified a definite role for IFN-γ in impairing anti-pneumococcal clearance leading to outgrowth and systemic spread. This was in line with the finding that IFN-γ released during IAV-infection suppresses alveolar macrophage phagocytosis and increases oxidative radicals by downregulating their expression of the scavenger receptor MARCO in a state of coinfection, favoring bacterial outgrowth10,12,39. As in vivo neutralization of IFN-γ failed to fully alleviate bacterial outgrowth following coinfection with IAV and S. pneumoniae, other mediators are most likely to also contribute to the strongly impaired bacterial clearance12.
In accordance with other possible key players during coinfection, we found in our murine experiments significantly higher concentrations of IL-6 and TNF-α during coinfection compared to the single infections13,40. The exact role of these cytokines in the pathogenesis of coinfections still remains debatable. Here, our in silico studies suggest that the inhibitory effect of IL-6 is both concentration and time-dependent but not as conclusive for bacterial outgrowth as IFN-γ. Early after pneumococcal infection, IL-6 is likely to play a predominantly protective role due to its immune-regulatory function in the feedback circuit of cytokines. Indeed, it was reported that IL-6 KO mice displayed a significantly enhanced susceptibility to S. pneumoniae infection with elevated concentrations of TNF-α, IL1-β and IFN-γ in the lungs in comparison to wild-type mice42. On the other hand, due to its potent pro-inflammatory functions, IL-6 is also known to be a marker for disease severity in pneumococcal infections43. Importantly, our finding that increased amounts of IL-6 predominantly impair bacterial clearance in synergism with the IAV-dependent IFN-γ present in the coinfected lung reflects how alterations in the inflammatory status affect host susceptibility in a dynamic temporal manner.
Interestingly, we observed that TNF-α showed the least contribution in impairing bacterial clearance following influenza. This was well in line with previous findings describing that TNF-α neutralization elevated mortality during single S. pneumoniae infection and IAV coinfections suggesting more of a protective role for TNF-α44,45. However, TNF-α is in contrast known to induce apoptosis of various pulmonary cells and to disrupt the epithelial barrier integrity and therefore TNF-α blockade ameliorates pulmonary immunopathology in single IAV infected animals46,47,48. Nevertheless, in our study, effects such as the detrimental contribution of immunopathology on S. pneumoniae outgrowth were not modelled. Therefore it is likely that an excessive production of TNF-α during coinfections may potentiate host tissue damage and thereby may still exert an indirect negative influence on bacterial clearance. Regarding the protein MCP-1, as this chemokine regulates migration and infiltration of AMs, we assumed that the downstream effects of MCP-1 were reflected in the AM dynamics (Fig. 3 and the model M1). However, in order to analyse the hypothesis that MCP-1 may directly play a role during coinfections, the model M12 in Supplementary 3 was tested. The AICc criterion did not rank MCP-1 kinetics as a key mechanism to describe the bacterial burden in coinfected animals.
Under homeostatic conditions, tightly regulated immune responses are co-ordinated by the action of pro- and anti-inflammatory cytokines and chemokines with the goal to clear pathogens and at the same time curtail immunopathology. By our experimental and mathematical modelling approach, we found that during coinfections this balance is lost due to the exaggerated amounts of pro-inflammatory cytokines causing a pathogenic effect on bacterial clearance. Notably, this finding was further supported by the results of challenging our mathematical models for the IAV-S. pneumoniae coinfection with the cytokine data from the single S. pneumoniae infection group, in silico results predicted bacterial clearance as in the single bacterial infection without further parameter fitting (Fig. S5).
Previous studies showed that the abundance of IFN-γ driven mainly by T cells enhances the susceptibility to secondary pneumococcal coinfection12. Nevertheless, IFN-γ plays a central role in host defence and immunity against single infections to IAV49 and S. pneumoniae50,51, pointing out an essential role for the recruited neutrophils in the production of IFN-γ in response to S. pneumoniae50,51. Furthermore, experiments in mice showed that deletion of IFN-γ resulted in a more severe impairment of lung function49. Therefore, further modelling and experimental efforts are needed to uncover the bifurcation point that shifts IFN-γ from benign to detrimental in coinfections.
In conclusion, by combining tailored experimental data and mathematical modelling our study suggested a strong detrimental effect of IFN-γ alone and in synergism with IL-6 but no conclusive pathogenic effect of IL-6 or TNF-α alone. In silico predictions pave the way for further murine experiments to prove or reject the advantage of a double IFN-γ and IL-6 neutralization approach. Ultimately, these findings correlate well with our previous results31 suggesting that the increased levels of pro-inflammatory cytokines (the “inflammaging” state), in particular IFN-γ, contribute to the reported impaired responses in people over 65 years of age. Thereby, IFN-γ plays a pivotal role in driving severe disease during primary IAV infection in the elderly as well as bacterial outgrowth during coinfections. Ultimately, such full understanding of the downstream effects of the altered inflammatory response in bacterial coinfection following IAV will be crucial when attempting to design future prophylactic and therapeutic interventions.
Materials and Methods
Mice
7–8 weeks old wildtype C57BL/6J female mice were purchased from Harlan Winkelmann (Borchen, Germany) and housed in specific pathogen-free conditions.
Ethics Statement
All animal experiments were approved by the local ethical body “Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit” (file number 33.9-42502-04-11/0443) and were conducted in conformity with the German animal welfare act and the European Communities Council Directive 2010/63/EU.
Viral and bacterial pathogens
For viral infection experiments, the influenza A virus strain A/Puerto Rico/8/1934 (A/PR8/34) subtype H1N1 was used34. The virus was grown and stored as described previously in ref. 34. The tissue culture infectious dose (TCID50) was calculated using the Reed and Muench endpoint calculations. For bacterial infection experiments S. pneumoniae serotype 4, TIGR4 (ATCC BAA-334TM) was used. Bacteria were grown to mid-logarithmic phase in Todd-Hewitt yeast medium (THY; THB Sigma-Aldrich, Germany and Yeast extract, Roth, Germany) at 37 °C for the preparation of frozen stocks. Bacterial counts were determined by plating 10-fold serial dilutions on blood agar plates (BD Diagnostic Systems, Columbia Agar with 5% sheep blood) overnight for 16–18 h at 37 °C with 5% CO2. Before each infection, aliquots were thawed, centrifuged at 8000 rpm and resuspended in the desired amount of phosphate buffered saline (PBS, Gibco, UK) followed by plating of serial dilutions to confirm the infection dose.
Infection experiments
Mice were sedated via intraperitoneal injection of ketamine/rompun (working concentration of 0.1 ml/10 g per mouse). For viral challenges, a sub-lethal dose of 0.31 TCID50 was administered intranasally in 25 μl PBS. For bacterial challenges, mice were placed with their backs on an intubation slope, and an infectious dose of 1 × 106 CFU in 25 μl PBS was instilled at the end of their nasopharynx using a long flexible pipet tip. All the mice were monitored and scored for the following clinical symptoms: weight loss, mobility, posture, pilo-erection, respiration, response to stimuli and eye infection. Animals with a severe score of any one of the parameters or moderate scores for two to three parameters were euthanized.
Determination of bacterial colony forming units in blood, bronchoalveolar lavage and lung tissue
Bacterial CFU were determined post-mortem by plating 10-fold serial dilutions of the blood, BAL and lung tissue homogenate. For the blood, 5 μl of heart blood were collected and diluted in 45 μl PBS. BAL samples were obtained by flushing the lung once with 1 mL PBS. Next, the lungs were perfused with PBS and excised. Whole lungs were homogenized in 1 mL PBS though a 100 μm cell stainer (Corning Inc, USA). Serial dilutions of BAL and lung homogenates were prepared and 10 μl were plated onto blood agar plates. Plates were incubated overnight at 37 °C and 5% CO2. For determination of CFU the colonies of each dilution were counted manually and calculated as CFU/mL.
Quantification of viral load with qRT-PCR
Lungs were perfused with PBS and the RNA was extracted from lung tissue homogenates with the RNeasy mini kit (Qiagen, Germany) according to the manufacturer’s protocol. After extraction and purification, cDNA synthesis was performed with 100 ng RNA using the SuperScript III first-strand synthesis System (Invitrogen, USA). For cDNA quality control, a PCR of the housekeeping gene RPS9 (Eurofins MWG, Germany) was performed. Viral nucleoprotein copy numbers were quantified by RT-PCR on a Light cycler (Roche) using a plasmid standard with defined NP copy numbers. The following primers were used: rps9 5′CTGGACGAGGGCAAGATGAAGC and 3′TGACGTTGGCGGATGAGCACA; np 5′GAGGGGTGAGAATGGACGAAAAAC and 3′CAGGCAGGCAGGCAGGACTT.
Flow cytometry for alveolar macrophages
Single cell suspensions of perfused excised lungs were prepared by enzymatic digestion for 45 minutes at 37 °C with Iscove’s Modified Dulbecco’s Medium (IMDM) containing GlutaMax-1 (Gibco, USA) supplemented with 5% fetal bovine serum (FBS; PanBiotech, Germany), 0.2 mg/ml collagenase D (Roche, Germany) and 1 mg/ml DNAse (Sigma-Aldrich, Germany). The enzymatic reaction was stopped with 5 mM EDTA and cell suspensions were filtered through a 100 μm strainer.
Cells were pelleted by centrifugation and resuspended in LIVE/DEAD® fixable blue stain (ThermoFisher, USA) and anti-mouse CD16/CD32 antibody (purified; BioLegend, USA) as Fc-block following erythrocyte lysis. Cell surface marker staining was performed using anti-mouse CD11b (BioLegend, USA) and anti-mouse F4/80 (BioLegend, USA). All samples were acquired on a BD LSRII Fortessa instrument with FACS DIVA software (BD) and analyzed using FlowJo software (Tree Star). Following debris and dead cell exclusion, AMs were identified as SSChigh, FSChigh, CD11b- and F4/80+ autofluorescent cells. In separate analyses the AM phenotype of the cells identified by this strategy was confirmed by CD11c staining. Absolute cell counts were calculated from the manually counted overall cell number in a sample and the population frequency assessed by flow cytometry.
Cytokine measurement
The protein concentration of the cytokines IFN-γ, TNF-α, IL-6, IFN-β, IL-22 and the chemokines MCP-1 and GM-CSF was measured with a customized Mouse LegendPlex™ kit (BioLegend, USA) according to the manufacturers protocol. All samples were acquired on a BD LSRII Fortessa with FACS DIVA software (BD) and analyzed with the provided LegendPlex™ Data Analysis Software (BioLegend, USA).
Mathematical Models
This work aimed to disentangle the mechanisms modulated by IAV that can contribute to the colonization of S. pneumoniae. Although, we recognized that bacterial infection may enhance viral release from infected cells33, these interactions were outside of the aim of the work proposed here. Therefore, we modelled the S. pneumoniae infection (B) with the following ordinary differential equation (ODE):
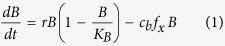 |
where r is the bacterial proliferation rate with a maximum carrying capacity KB. Phagocytosis of the bacteria is considered by the multiplicative term cbfx, where cb is the constant clearance rate and the term fx is the mathematical function which served to evaluate different hypotheses.
The previous work by Smith et al.33 proposed and tested that IAV induces phenotypic changes in AM. However, the work in ref. 33 assumed the restrictive condition that the phagocytosis rate cb is impaired by the viral load (V(t)). This hypothesis was written with the term 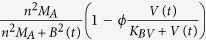 where the values for the uptake of bacteria by AM were expressed by n and the number of AM(MA) was chosen to be equal to the quasi steady state value
where the values for the uptake of bacteria by AM were expressed by n and the number of AM(MA) was chosen to be equal to the quasi steady state value 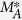 33. The reduction of the bacterial clearance due to the viral effects was included by the saturation function ϕV(t)/(KBV + V(t)), where ϕ is the maximal reduction of the phagocytosis rate and KBV is the half-saturation constant.
33. The reduction of the bacterial clearance due to the viral effects was included by the saturation function ϕV(t)/(KBV + V(t)), where ϕ is the maximal reduction of the phagocytosis rate and KBV is the half-saturation constant.
In order to evaluate the AM dynamics, our model uses the AM experimental data to build a piecewise linear function to determine the model term MA(t). Additionally, we examined the hypothesis that AM depletion by IAV is a sufficient mechanism to facilitate bacterial outgrowth. This is represented by a direct input of AMs data in equation (1).
Beyond the work of ref. 33, different functions (fx) were tested to dissect the contributions of pro-inflammatory cytokines (IFN-γ, TNF-α, and IL-6) to modulate the bacterial clearance. To this end, we used the experimental data for the different cytokines to build piecewise linear functions to dynamically determine the equation (1). We opted for this approach instead of mechanistic modelling with an ODE for each cytokine for different reasons. First, our main aim was not to quantify dynamics but to determine the contribution of important pro-inflammatory cytokines in promoting bacterial colonization. Second, modelling with an ODE for each cytokine may not improve the model selection process but only increase the complexity of mathematical models and parameter fitting procedures. Finally, our experimental data were measured frequently enough allowing us to use the data as an input in the equation (1).
To evaluate if IAV infection modulates a specific pro-inflammatory cytokine X(t), which subsequently facilitates bacterial colonization, we considered the saturating term 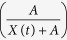 with A as the “half-saturation constant”. Different mathematical terms and other term combinations to dissect the pro-inflammatory effects responsible to impair bacterial clearance are summarized in Table 1 and Supplementary 3.
with A as the “half-saturation constant”. Different mathematical terms and other term combinations to dissect the pro-inflammatory effects responsible to impair bacterial clearance are summarized in Table 1 and Supplementary 3.
Parameter Fitting
Note that the purpose of our in silico work was not biological quantification of the experiments but using a model selection process to provide mechanistic insights into coinfection. Thus, in this case, finding a working set of parameter values deemed sufficient. Nevertheless, for good practice procedures, we checked identifiability and parameter uncertainty expanded in the Supplementary 4.
To focus on the mechanisms that promote S. pneumoniae colonization, we fixed the growth rate (r = 1.13 h−1) and the carrying capacity (KB = 2.3 × 108 CFU/ml) corresponding to single S. pneumoniae infection from previous works20. Using similar reasoning as Smith et al.20, we assumed that only a proportion of the bacterial inoculum may reach the lung since some bacteria can be removed by mucocilliary mechanisms. Thus, we considered 1000 CFU/mL S. pneumoniae as inoculum to fit the model parameters in model (1). We would like to remark different assumptions of parameters and inoculation will only rescale the fitted parameters, but not the mechanistic insights from the model selection procedures.
Other model parameters were obtained minimizing the residual sum of squares (RSS) between the model output and the experimental measurement, both on log scales. ODEs were solved in Matlab software using the ode45 solver. The minimization of RSS was performed using different optimization solvers, including both deterministic and stochastic methods. However, the Differential Evolution algorithm (DE), a global optimization algorithm, was selected to avoid relying on any initial parameter guesses and producing more robust results than other tested methods52,53. In separate form, model fitting was performed to all the models summarized in Table 1 for the single infection (T4) and the IAV+T4 coinfection starting at day 7 post IAV infection respectively.
Model Selection
The Akaike information criterion (AIC) was used to compare the goodness-of-fit of models that represent different hypotheses54. A lower AIC value means that a given model describes the data better than other models with higher values. Absolute difference in AIC greater than two units indicates stronger evidence for one model over the other54. For a small number of data points, the corrected Akaike information criterion (AICc) has the form:
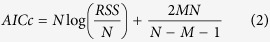 |
where N is the number of data points, M is the number of unknown parameters and RSS is the residual sum of squares obtained from the fitting routine.
Parameter uncertainty
Bootstrapping is a statistical method for assigning measures of accuracy to estimates55,56. Due to the large variability of viral and bacterial infections, bootstrapping methods have been applied to different viral infectious diseases53,57. Here, the non-parametric bootstrap was considered. Of note, instead of using all replications at each time point, three replications at each time point were randomly selected and used to generate a new data set. Note that this procedure was based exclusively on the observed measurement data. For each repetition, the model parameters were refitted to obtain the corresponding parameter distribution. The 95% Confidence Interval (CI) of parameter estimates was computed using the outcome of the bootstrap method56. For each parameter, the 2.5% and 97.5% quantiles of the estimates were used to form the 95% CI.
Parameter identifiability
A relevant aspect to verify in quantifying mechanisms is whether model parameters are identifiable based on the measurements of output variables58,59,60,61. A system that is algebraically identifiable may still be practically non-identifiable if the amount and quality of the measurements is insufficient and the data shows large deviations. The computational approach proposed in ref. 62 exploits the profile likelihood to determine identifiability and was considered here. This method is able to detect both structurally and practically non-identifiable parameters. Briefly, the idea behind this approach is to explore the parameter space for each parameter θi by re-optimizing the RSS with respect to all other parameters θj≠i. The main task is to detect directions where the likelihood flattens out62. The resulting profiles are plotted versus each parameter range with the respective 95% CI to assess the parameter identifiability.
Statistical Analysis
All statistical analyses were performed by the Mann-Whitney test using the Graph Pad Prism software (Graph Pad Software, La Jolla/USA).
Additional Information
How to cite this article: Duvigneau, S. et al. Hierarchical effects of pro-inflammatory cytokines on the post-influenza susceptibility to pneumococcal coinfection. Sci. Rep. 6, 37045; doi: 10.1038/srep37045 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by iMed - the Helmholtz Initiative on Personalized Medicine. Dunja Bruder was supported by the President’s Initiative and Networking Fund of the Helmholtz Association of German Research Centers (HGF) under contract number W2/W3-029 and by a grant from the German Research Foundation (DFG) in frame of the SFB854 (subproject A23N). Niharika Sharma-Chawla received support from the International Research Training Group (IRTG) 1273. In addition, Esteban Hernandez-Vargas thanks for the support provided by the Department of Systems Immunology (HZI) through the Measures for the Establishment of Systems Medicine (e:Med) projects in Systems Immunology and Image Mining in Translational Bio-marker Research (SYSIMIT), contract number 01ZX1308B and in identification of predictive response and resistance factors to targeted therapy in gastric cancer using a systems medicine approach (SYS-Stomach), contract number 01ZX1310 C by the Federal Ministry of Education and Research (BMBF), and by the Human Frontier Science Program (HFSP), RGP0033/2015.
Footnotes
Author Contributions D.B. and E.A.H.V. designed the experiments, modelling and the whole work. S.D. and N.S. developed the experimental work. S.D., V.K.N., A.B. and E.A.H.V. analysed the data and performed the simulations. S.D., N.S., A.B., S.S., D.B. and E.A.H.V. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Ballinger M. N. & Standiford T. J. Postinfluenza bacterial pneumonia: host defenses gone awry. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research 30, 643–652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J. K. & Morens D. M. The pathology of influenza virus infections. Annual review of Pathology 3, 499–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A. & Rehg J. E. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. The Journal of Infectious Diseases 186, 341–350 (2002). [DOI] [PubMed] [Google Scholar]
- McCullers J. A. Insights into the interaction between influenza virus and pneumococcus. Clinical Microbiology Reviews 19, 571–582 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nature Reviews Microbiology 12, 252–262 (2014). [DOI] [PubMed] [Google Scholar]
- Ortqvist A., Hedlund J. & Kalin M. Streptococcus pneumoniae: Epidemiology, Risk Factors, and Clinical Features. Seminars in Respiratory and Critical Care Medicine 1, 563–574 (2005). [DOI] [PubMed] [Google Scholar]
- Mina M. J., McCullers J. A. & Klugman K. P. Live attenuated influenza vaccine enhances colonization of Streptococcus pneumoniae and Staphylococcus aureus in mice. mBio 5, e01040-13- (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman L. The complication of coinfection. Yale Journal of Biology and Medicine 85, 127–132 (2012). [PMC free article] [PubMed] [Google Scholar]
- Gatanaga H., Yasuoka A., Kikuchi Y., Tachikawa N. & Oka S. Influence of Prior HIV-1 Infection on the Development of Chronic Hepatitis B Infection. European Journal of Clinical Microbiology & Infectious Diseases 19, 237–239 (2000). [DOI] [PubMed] [Google Scholar]
- Goulding J. et al. Lowering the threshold of lung innate immune cell activation alters susceptibility to secondary bacterial superinfection. Journal of Infectious Diseases 204, 1086–1094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahangian A. et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. Journal of Clinical Investigation 119, 1910–1920 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K. & Metzger D. W. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nature Medicine 14, 558–564 (2008). [DOI] [PubMed] [Google Scholar]
- Damjanovic D., Lai R., Jeyanathan M., Hogaboam C. M. & Xing Z. Marked improvement of severe lung immunopathology by influenza-associated pneumococcal superinfection requires the control of both bacterial replication and host immune responses. American Journal of Pathology 183, 868–880 (2013). [DOI] [PubMed] [Google Scholar]
- Cauley L. S. & Vella A. T. Why is coinfection with influenza virus and bacteria so difficult to control? Discovery Medicine 19, 33–40 (2015). [PMC free article] [PubMed] [Google Scholar]
- Marchuk G. I. Mathematical modelling of immune response in infectious diseases, vol. 395 (Springer Science & Business Media, 1997).
- Möhler L., Flockerzi D., Sann H. & Reichl U. Mathematical model of influenza A virus production in large-scale microcarrier culture. Biotechnology and Bioengineering 90, 46–58 (2005). [DOI] [PubMed] [Google Scholar]
- Baccam P., Beauchemin C., Macken C. a., Hayden F. G. & Perelson A. S. Kinetics of influenza A virus infection in humans. Journal of Virology 80, 7590–7599 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C. A. & Handel A. A review of mathematical models of influenza A infections within a host or cell culture: lessons learned and challenges ahead. BMC Public Health 11 Suppl 1, S7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H. et al. Higher level of replication efficiency of 2009 (H1N1) pandemic influenza virus than those of seasonal and avian strains: kinetics from epithelial cell culture and computational modeling. Journal of Virology 85, 1125–1135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., McCullers J. A. & Adler F. R. Mathematical model of a three-stage innate immune response to a pneumococcal lung infection. Journal of Theoretical Biology 276, 106–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boianelli A. et al. Modeling Influenza Virus Infection: A Roadmap for Influenza Research Viruses, 7(10), 5274–5304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancioglu B., Swigon D. & Clermont G. A dynamical model of human immune response to influenza A virus infection. Journal of Theoretical Biology 246, 70–86 (2007). [DOI] [PubMed] [Google Scholar]
- Lee H. Y. et al. Simulation and prediction of the adaptive immune response to influenza A virus infection. Journal of Virology 83, 7151–7165 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz R. A. et al. Dynamics of Influenza Virus Infection and Pathology. Journal of Virology 84, 3974–3983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H. et al. Quantifying the Early Immune Response and Adaptive Immune Response Kinetics in Mice Infected with Influenza A Virus. Journal of Virology 84, 6687–6698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M. et al. Effect of 1918 PB1-F2 expression on influenza A virus infection kinetics. PLoS Computational Biology 7, e1001081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canini L. & Carrat F. Population modeling of influenza A/H1N1 virus kinetics and symptom dynamics. Journal of Virology 85, 2764–2770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek K. A. et al. Modeling Within-Host Dynamics of Influenza Virus Infection Including Immune Responses. PLoS Comput Biol 8, e1002588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas E. A. & Meyer-Hermann M. Innate Immune System Dynamics to Influenza Virus. In 8th IFAC Symposium on Biological and Medical Systems 45, 260–265 (2012). [Google Scholar]
- Dobrovolny H. M., Reddy M. B., Kamal M. a., Rayner C. R. & Beauchemin C. a. a. Assessing Mathematical Models of Influenza Infections Using Features of the Immune Response. PLoS ONE 8, e57088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas E. A. et al. Effects of aging on influenza virus infection dynamics. Journal of Virology 88, 4123–4131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boianelli A., Sharma-Chawla N., Bruder D. & Hernandez-Vargas E. A. Oseltamivir PK/PD modeling and simulation to evaluate treatment strategies against influenza-pneumococcus coinfection. Frontiers in Cellular and Infection Microbiology 6, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M. et al. Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathogens 9, e1003238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S. et al. Increased Susceptibility for Superinfection with Streptococcus pneumoniae during Influenza Virus Infection Is Not Caused by TLR7-Mediated Lymphopenia. PLoS ONE 4, e4840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R., Lewin S. R., Elliott J. H. & Perelson A. S. Modeling the Effects of Vorinostat In Vivo Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells. PLOS Pathogens 11, e1005237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim H. E., Thomas P. G. & McCullers J. A. Depletion of Alveolar Macrophages during Influenza Infection Facilitates Bacterial Superinfections. The Journal of Immunology 191, 1250–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma-Chawla N. et al. Influenza A virus infection predisposes hosts to secondary infection with different Streptococcus pneumoniae serotypes with similar outcome but serotype-specific manifestation. Infection and Immunity 00422-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szade A. et al. Effect of crossing C57BL/6 and FVB mouse strains on basal cytokine expression. Mediators of Inflammation 2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T. & Cavanagh M. M. The innate immune rheostat: influence on lung inflammatory disease and secondary bacterial pneumonia. Biochemical Society transactions 37, 811–813 (2009). [DOI] [PubMed] [Google Scholar]
- McNamee L. A. & Harmsen A. G. Both influenza-induced neutrophil dysfunction and neutrophil-independent mechanisms contribute to increased susceptibility to a secondary Streptococcus pneumoniae infection. Infection and Immunity 74, 6707–6721 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didierlaurent A. et al. Sustained desensitization to bacterial Toll-like receptor ligands after resolutionof respiratory influenza infection. The Journal of Experimental Medicine 205, 323–329 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Poll T. et al. Interleukin-6 gene-deficient mice show impaired defense against pneumococcal pneumonia. The Journal of Infectious Diseases 176, 439–444 (1997). [DOI] [PubMed] [Google Scholar]
- Norrby-Teglund A., Pauksens K., Norgren M. & Holm S. E. Correlation between serum TNF alpha and IL-6 levels and severity of group A streptococcal infections. Scandinavian Journal of Infectious Diseases 27, 125–130 (1995). [DOI] [PubMed] [Google Scholar]
- Kirby A. C., Raynes J. G. & Kaye P. M. The role played by tumor necrosis factor during localized and systemic infection with Streptococcus pneumoniae. Journal of Infectious Diseases 191, 1538–1547 (2005). [DOI] [PubMed] [Google Scholar]
- Ellis G. T. et al. TRAIL + monocytes and monocyte-related cells cause lung damage and thereby increase susceptibility to influenza-Streptococcus pneumoniae coinfection. EMBO reports 16, 1203–1218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T., Pennycook A. & Openshaw P. J. M. Inhibition of tumor necrosis factor reduces the severity of virus-specific lung immunopathology. European Journal of Immunology 31, 2566–2573 (2001). [DOI] [PubMed] [Google Scholar]
- La Gruta N. L., Kedzierska K., Stambas J. & Doherty P. C. A question of self-preservation: immunopathology in influenza virus infection. Immunology and Cell Biology 85, 85–92 (2007). [DOI] [PubMed] [Google Scholar]
- Marcos-Ramiro B., Garcia-Weber D. & Millan J. TNF-induced endothelial barrier disruption: Beyond actin and Rho. Thrombosis and Haemostasis 112, 1088–1102 (2014). [DOI] [PubMed] [Google Scholar]
- Wiley J. A., Cerwenka A., Harkema J. R., Dutton R. W. & Harmsen A. G. Production of interferon-gamma by influenza hemagglutinin-specific CD8 effector T cells influences the development of pulmonary immunopathology. The American Journal of Pathology 158, 119–130 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M. et al. Interferon-gamma production by neutrophils during bacterial pneumonia in mice. American Journal of Respiratory and Critical Care Medicine 183, 1391–1401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. C. et al. Mechanisms of interferon-gamma production by neutrophils and its function during streptococcus pneumoniae pneumonia. American Journal of Respiratory Cell and Molecular Biology 52, 349–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storn R. & Price K. Differential evolution–a simple and efficient heuristic for global optimization over continuous spaces. Journal of Global Optimization 341–359 (1997). [Google Scholar]
- Nguyen V. K., Binder S. C., Boianelli A., Meyer-Hermann M. & Hernandez-Vargas E. A. Ebola virus infection modeling and identifiability problems. Frontiers in Microbiology 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K. P. & Anderson D. R. Model selection and multimodel inference: a practical information-theoretic approach (Springer Science & Business Media, 2002). [Google Scholar]
- Davison A. C. & Hinkley D. V. Bootstrap Methods and Their Application (Cambridge Univ, 1997). [Google Scholar]
- Xue H., Miao H. & Wu H. Sieve estimation of constant and time-varying coefficients in nonlinear ordinary differential equation models by considering both numerical error and measurement error. The Annals of Statistics 38, 2351–2387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafoura S. et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4 (+) memory T Cells. Nature Communications 5, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. & Moog C. Identifiability of nonlinear systems with application to HIV/AIDS models. IEEE Transactions on Automatic Control 48, 330–336 (2003). [Google Scholar]
- Wu H., Zhu H., Miao H. & Perelson A. S. Parameter identifiability and estimation of HIV/AIDS dynamic models. Bulletin of Mathematical Biology 70, 785–799 (2008). [DOI] [PubMed] [Google Scholar]
- Miao H., Xia X., Perelson A. S. & Wu H. On Identifiability of Nonlinear ODE Models and Applications in Viral Dynamics. Plus Review 53, 3–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.-K. & Hernandez-Vargas E. A. Identifiability Challenges in Mathematical Models of Viral Infectious Diseases. IFAC-PapersOnLine 48, 2–7 (2015). [Google Scholar]
- Raue A. et al. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 25, 1923–1929 (2009). [DOI] [PubMed] [Google Scholar]
- Scheller J., Chalaris A., Schmidt-Arras D. & Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et Biophysica Acta 1813, 878–888 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.