Abstract
Alcohol consumption is a risk factor for breast cancer. Little is known regarding the mechanism, although it is assumed that acetaldehyde or estrogen mediated pathways play a role. We previously showed that long-term exposure to 2.5 mM ethanol (blood alcohol ~0.012%) of MCF-12A, a human normal epithelial breast cell line, induced epithelial mesenchymal transition (EMT) and oncogenic transformation. In this study, we investigated in the human breast cancer cell line MCF-7, whether a similar exposure to ethanol at concentrations ranging up to peak blood levels in heavy drinkers would increase malignant progression. Short-term (1-week) incubation to ethanol at as low as 1–5 mM (corresponding to blood alcohol concentration of ~0.0048–0.024%) upregulated the stem cell related proteins Oct4 and Nanog, but they were reduced after exposure at 25 mM. Long-term (4-week) exposure to 25 mM ethanol upregulated the Oct4 and Nanog proteins, as well as the malignancy marker Ceacam6. DNA microarray analysis in cells exposed for 1 week showed upregulated expression of metallothionein genes, particularly MT1X. Long-term exposure upregulated expression of some malignancy related genes (STEAP4, SERPINA3, SAMD9, GDF15, KRT15, ITGB6, TP63, and PGR, as well as the CEACAM, interferon related, and HLA gene families). Some of these findings were validated by RT-PCR. A similar treatment also modulated numerous microRNAs (miRs) including one regulator of Oct4 as well as miRs involved in oncogenesis and/or malignancy, with only a few estrogen-induced miRs. Long-term 25 mM ethanol also induced a 5.6-fold upregulation of anchorage-independent growth, an indicator of malignant-like features. Exposure to acetaldehyde resulted in little or no effect comparable to that of ethanol. The previously shown alcohol induction of oncogenic transformation of normal breast cells is now complemented by the current results suggesting alcohol's potential involvement in malignant progression of breast cancer.
Keywords: MCF-7, alcohol, microRNAs, gene expression, stemness
Introduction
Excessive chronic alcohol intake is a widely acknowledged risk factor for breast cancer. In a previous study, we summarized the epidemiology and known pathology pertaining to alcohol consumption and breast cancer (1). Briefly, consumption of three or more drinks per day leads to a 40–50% increase in risk and there are ~50,000 alcohol-attributable breast cancer cases per year, worldwide (2–4). The risk of breast cancer increases with the quantity of alcohol consumed, showing a linear dose-response (5). There is a greater risk for lobular rather than ductal breast cancer (5–7), and tumors are more likely to be ER− and HER2+ in the women with high alcohol consumption (8,9).
One proposed mechanism for the putative alcohol carcinogenicity involves stimulation of estrogen levels and/or estrogen responsiveness but other possibilities include effects unrelated to estrogen (2,3,9–13) such as inhibition of DNA methylation, interaction with retinoid metabolism, or oxidative stress. Such changes could operate either by direct ethanol effects and/or through the first ethanol metabolite, acetaldehyde.
As summarized previously (1), a few studies in mice and rats have shown that ethanol consumption promotes mammary tumors via the estrogen pathway (14). The estrogen dependence was originally shown and partially explained in the widely used MCF-7 cell line, a human breast cancer luminal epithelial cell line which is estrogen and progesterone receptor positive and lacks ERBB2 gene amplification or Her2/neu protein overexpression (15–18). It was also shown that ethanol stimulates the in vitro growth, invasiveness and migration of these cells (17–24). However, the common denominator of the previous studies on MCF-7 cells is that the ethanol exposure was limited to <1 week, concentrations were >50 mM, and the effects were modest. A similar situation occurred with studies conducted on other types of more malignant breast cancer cell lines, such as T47D and erbB2 transformed cells (25–30).
Another potential mechanism of ethanol's carcinogenicity is through enrichment of a subpopulation of cancer stem cells, but there are no reports on the effects of ethanol on this type of stem cells (31–33). Cancer stem cells are postulated to be involved in the generation of primary breast tumors and their progression to undifferentiated tumors and metastasis, and are claimed to be enriched within mammospheres (34,35). Although ethanol affects the proliferation and differentiation of normal embryonic and adult stem cells (36,37), it is not known whether it activates and/or increases the number of cancer stem cells. The latter process, as well as the regulation of breast cancer genes in general, is partially regulated by microRNAs (miR) (34,38–41), particularly with regard to the epithelial mesenchymal transition (EMT) (42,43). Ethanol affects the expression of certain miRs in alcoholic liver injury and other pathologies (44,45), but no reports link this to breast cancer. In contrast, there is a substantial recent literature on miRs in relation to estrogen effects, particularly in MCF-7 cells (46–48), but none has been directly linked to ethanol exposure.
In our previous study on the non-malignant epithelial human breast cell line MCF-12A (1) we found that ethanol, but not acetaldehyde, induced oncogenic features and EMT, and stimulated the expression of a collection of mRNAs and miRs, including those associated with these processes, and also stimulated certain protein markers for stem-related properties.
In this study, the effects of short- and long-term exposures to physiologically relevant concentrations of ethanol, and acetaldehyde up to supraphysiological levels were studied using MCF-7 monolayers and mammospheres. Stem cell markers, global transcriptional gene expression signatures including miRs, and in vitro responses in oncogenic assays were carried out to better understand the mechanism of action of alcohol on malignant progression in breast cancer. The aim was to clarify: a) whether the epidemiological relationship between excessive and long-term alcohol consumption and the malignant progression of breast cancer can be elucidated by defining the effects of ethanol on an accepted epithelial breast cancer cell line such as MCF-7 in vitro; b) whether ethanol intensifies some of the MCF-7 malignant features in a dosage- and/or duration of exposure-dependent way; c) the potential mediation of these effects by stem cell enrichment in both monolayers and mammospheres; d) the possible role of acetaldehyde in mediating those changes; and e) the impact of both alcohol and acetaldehyde on the mRNA and miR global signatures, so as to define the more affected pathways and to evaluate putative estrogen mediation.
Materials and methods
Cell culture
The human adherent epithelial adenocarcinoma MCF-7 cell line was obtained from ATCC (Catalog HT-22™ Manassas, VA, USA), and routinely cultured on monolayers at ≤80% confluence in MEME (minimum essential medium Eagle's, ATCC), 10% FBS and 0.01 mg/ml bovine insulin (Sigma, St. Louis, MO, USA). For most experiments, cells were incubated on 6-well plates with 0-25 mM ethanol (Fisher, molecular grade ethanol) or 0–12.5 mM acetaldehyde (Sigma, ACS), using freshly prepared solutions. Medium was replaced 2–3 times/week, including addition of ethanol or acetaldehyde, and cultures were maintained for 1 week (short-term incubations), or for 4 weeks (long-term incubations). In the latter case, cells were passaged on average once a week, with a 1:3 splitting.
Mammospheres were generated for 1 week experiments by seeding 50,000 cells onto Corning Ultra Low Attachment 6-well-plates with 2 ml/well of MEBM medium (Fisher, mammary epithelial cell growth medium), adding 2% (v/v) of B27 Supplement (B27 serum-free supplement, Invitrogen, Carlsbad, CA, USA) and 0.01 mg/ml bovine insulin, and then adding ethanol or acetaldehyde. For 4-week incubations, monolayers were cultured for this period with ethanol or acetaldehyde and used for mammosphere generation that were maintained in the presence of these agents for an additional week. Their number and total area were determined by applying quantitative image analysis (QIA) to digital photographs taken with a Nikon digital camera of 0.005% crystal violet stained mammospheres contained in individual wells of a 6-well plate, using ImagePro-Plus 5.1 software (Media Cybernetics, Silver Spring, MD, USA). After images were calibrated for background lighting, integrated optical density (IOD = area × average intensity) was calculated. Inverted microscopy images were taken under phase contrast at 40× and 100× using a Nikon Eclipse Ti microscope and a Leica VCC digital camera.
Western blot analyses
Medium was decanted from wells and cells were washed twice with PBS at pH 7.4. Boiling buffer (1% SDS, 1 mM sodium orthovanadate, 10 mM Tris pH 7.4 and protease inhibitors) was added to each well, cells were scraped from each well and passed several times through a 26-gauge needle to reduce viscosity, incubated in a boiling water bath for 5 min, and centrifuged at 16,000 g for 5 min, then 20–40 μg of protein was run on 4–15% gradient polyacrylamide gels, transferred electrophoretically to nitrocellulose, and analyzed by immunodetection using antibodies against: i) Oct-4, (rabbit polyclonal, 1:500, BioVision, Mountain View, CA, USA); ii) CEACAM-6 (rabbit polyclonal, 1:500, Novus Biologicals, Littleton, CO, USA); iii) NANOG (rabbit polyclonal, 1:1,000, AVIVA Systems, San Diego, CA, USA) and iv) GAPDH (mouse monoclonal, 1:3,000, Chemicon). Membranes were incubated with secondary polyclonal horse anti-mouse or anti-rabbit IgG linked to horseradish peroxidase (1:2,000; BD Transduction Laboratories, Franklin Lakes, NJ, USA or 1:5,000; Amersham GE, Pittsburgh, PA, USA), bands were visualized with luminol (SuperSignal West Pico, Chemiluminescent, Pierce, Rockford, IL, USA). For the negative controls the primary antibody was omitted.
Immunocytochemistry
Cultures were grown in 8-well-removable-chamber slides, subjected to immunofluorescence detection by quenching in 0.3% H2O2, blocking with goat (or corresponding) serum, and incubated overnight at 4°C with the primary antibody for Oct-4 or NANOG. This was followed by a secondary anti-mouse IgG biotinylated antibody (goat, 1:200, Vector Laboratories) and this complex was detected with streptavidin-Texas Red. After washing with PBS, the sections were mounted with Prolong antifade/DAPI (Molecular Probes, Carlsbad, CA, USA). Negative controls in all cases omitted the first antibodies or were replaced by IgG isotype.
Flow cytometry
Control and 25 mM ethanol-incubated MCF-7 cells were grown in GM-20, washed twice with Hanks buffered salt solution, disaggregated by repeated pipeting in CellStripper (Mediatech, Manassas VA, USA), pelleted, and resuspended in staining buffer consisting of PBS, 3% FBS (SB). Cells were incubated in the presence of antibodies for 30 min on ice, washed twice with SB, and resuspended in SB for flow cytometry on an LSR II (BD Biosciences). Controls included samples without any antibody as well as samples including all combinations of antibodies so as to determine that the Ceacam6 stained cells and the CD44 stained cells were accurately identified. Data analysis and plotting were done using FACSDiva Version 6.1.1 software. Fluorophore-conjugated antibodies and G12-5841 PE (eBiosciences), performed separately, followed cell permeabilization with BD CytoFix/CytoPerm kit. BD CompBeads were used for compensation.
Global DNA microarray transcriptional profile
RNA was isolated from cells using the RNeasy Plus Micro kit (Qiagen) with quality determined using the Agilent 2100 Bioanalyzer. Assays were performed by the UCLA DNA microarray core, applying the Affymetrix Human Gene 1.0 ST array for >30,000 genes. Up- and downregulated genes (by >2-fold) were considered, except where indicated. DNA microarray results are deposited in the GEO library under accession no. GSE72013.
RT/PCR
The expression of some of the down- and upregulated genes identified by DNA microarray analysis was further examined on triplicate RNA samples. cDNA was synthesized by reverse transcription using the Superscript III First-Strand Synthesis SuperMix for qRT-PCr (Life Technologies), and the resulting samples were amplified using PCR. Primers were designed using the NCBI Primer Blast program applied to mRNA sequences and synthesized by Sigma-Aldrich. All primers were designed to include an exon-exon junction except for GAPDH. Negative controls omitted cDNA. PCR results were analyzed by electrophoresis through 1% agarose gels in Tris-acetate EDTA buffer followed by photography under ultraviolet illumination in a UVP Biodoc transilluminator.
Global miR profiles
RNA was isolated from cells using the mirVana™ miRNA isolation kit (Ambion), and analysis was carried out by LC Sciences (Houston, TX, USA) for all miR transcripts listed in the Sanger miRBase Release 18.0. The miR results are deposited in the GEO library under accession no. GSE72013.
Anchorage-independence
Cells were trypsinized, suspended in 1 ml/well of warm (37°C) 0.3–0.5% agar in MEME-10% FBS-bovine serum albumin (soft agar layer) and 10,000 cells/ml were plated in duplicate or triplicate above a layer of 1 ml of 1% agar in the same medium that had previously been allowed to solidify on 6-well plates at 4°C (hard layer agar). Cultures were allowed to grow for 3–4 weeks and when foci were visible, they were stained with 0.005% crystal violet in Hanks solution for 1 h, wells were photographed as described in 'Cell culture', and colonies were counted, also as described above.
Cell invasiveness
Matrigel™ basement membrane matrix was diluted 1:5 with serum free culture medium containing 0.5% BSA according to the manufacturer's instructions. Trypsinized cells were added at 25,000 cells/well to Transwell® Permeable Support 8.0-μm inserts containing Matrigel and culture medium, and cultured in a 24-well plate using FBS as the chemoattractant for 40 h. Cells were fixed and stained with 0.5% toluidine blue.
Response to tamoxifen
MCF-7 cells (control vs 4 weeks 25 mM ethanol) were incubated in estrogen-depletion medium (EDM): phenol red-free DMEM/F12 (Invitrogen) containing 5% charcoal stripped fetal bovine serum (Gibco) for 3 days, washed with Hanks, trypsinized, resuspended in EDM and plated at 1,000 cells/well into a 96-well plate. 4-OH-tamoxifen was added to a concentration of 0, 1, 2, 5 or 10 μM (6-wells for each concentration). Cells were incubated at 37°C in 5% CO2 for 4 days, washed once with Hanks, followed by the addition of 100 μl of EDM. After 30-min additional incubation, each well received 20 μl of the metabolic activity indicator CellTiter 96® AQueous One Solution Cell Proliferation assay (Promega, Madison, WI, USA), and cells were further incubated (37°C in 5% CO2) for 30 min. The absorbance at 490 nm was measured using an automated plate reader.
Statistical analyses
Statistical values are expressed as the mean (± SEM). The normality distribution of the data was established using the Wilk-Shapiro test. Multiple comparisons were analyzed by a single factor ANOVA, followed by post hoc comparisons with the Newman-Keuls test. Differences among groups were considered statistically significant at p<0.05.
Results
Short-term exposure of MCF-7 monolayers to low concentrations of ethanol
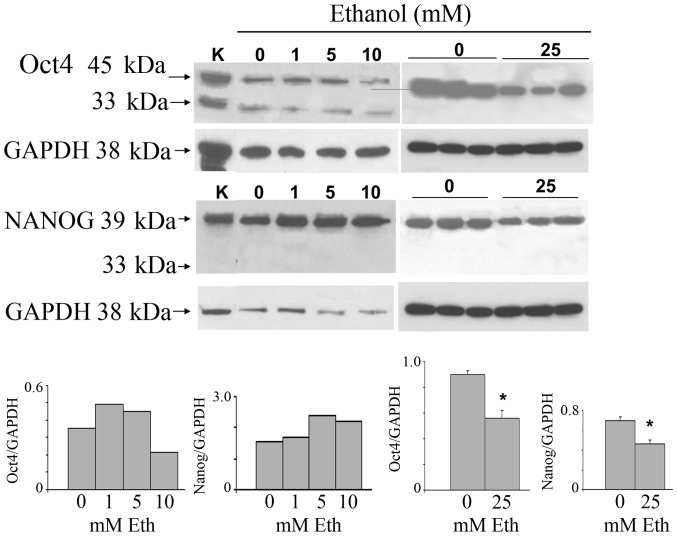
To investigate the possibility that ethanol exerts its oncogenic effects through the stimulation of cancer stem cell proliferation, we carried out experiments to ascertain the effects on the key stem cell marker proteins Oct4 and Nanog. MCF-7 monolayer cultures were incubated at 30–80% confluence for 7 days with 1–25 mM ethanol and subjected to western blot analysis for Oct4 and Nanog (Fig. 1). The highest concentration of ethanol is roughly equivalent to peak serum levels of alcohol in women within 1 h of consumption of 3–4 glasses of wine. The stemness-related nuclear Oct4a isoform (45 kDa) was increased after exposure to 1 and 5 mM ethanol, and was reduced at 10 and 25 mM. Triplicate samples of the 25 mM treatment show downregulation. The cytoplasmic Oct4b (33 kDa), unrelated to stemness, was expressed at low levels and remained unchanged. The main 39 kDa Nanog isoform was increased after exposure to 1–10 mM ethanol, and analogous to the Oct4 result, was also decreased at 25 mM. The nuclear localization of Oct4a, consistent with its known stemness function, and, in addition, the nuclear localization of Nanog, were confirmed by double immunofluorescence with Texas red for the specific antigen and DAPI for nuclei in MCF-7 cells incubated in the absence of ethanol (data not shown).
Figure 1.
Dose response of exposure of MCF-7 cells to ethanol for 1 week. Extracts from the monolayer cultures on 6-well plates, incubated at the indicated ethanol concentrations for 1 week, were analyzed by western blot analysis and corrected for GAPDH expression. Rat kidney protein was used as a positive control and is labeled as K. Top left, dose course to 10 mM for Oct4a (45 kDa) and Nanog (39 kDa), with GAPDH as reference. Top right, 0 and 25 mM only, in triplicate incubations for both control and 25 mM ethanol-treated samples. Bottom, densitometric determinations; *p<0.05.
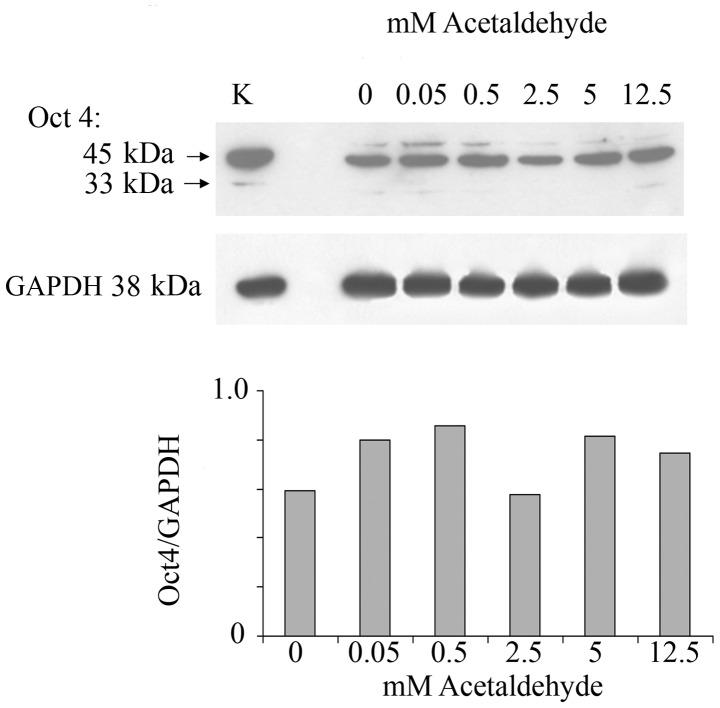
In order to investigate whether ethanol effects are mediated by its first oxidation product, acetaldehyde, a range of acetaldehyde concentrations was applied to MCF-7 monolayers and the effect on Oct4 expression was analyzed. Acetaldehyde over the range of 0.5–12.5 mM and over the same time of incubation did not affect the expression of Oct4 (Fig. 2). These acetaldehyde concentrations, considerably lower than the respective ethanol concentrations, were chosen based on the fact that they are marginally to considerably higher (i.e., approximately one log value) than the acetaldehyde values that would be expected in human serum after substantial drinking. These levels have been measured to be as much as 0.2 mM (49). Even at these levels acetaldehyde had little or no effect on Oct4 levels.
Figure 2.
MCF-7 cells grown in monolayer were subjected to acetaldehyde over the range of 0–12.5 mM for 1 week. Oct4 was analyzed by western blot analysis. The stem cell active upper band Oct4a and the non-stem-active lower band Oct4b are shown. Lower, Oct4a values were normalized according to GAPDH prevalence. K, rat kidney extract as positive marker for Oct4.
Ethanol exposure affects gene expression both in the short-term and long-term
Gene expression analysis of MCF-7 cells grown in monolayer in the presence or absence of ethanol (25 mM) for 1 and 4 weeks was carried out by subjecting RNA samples to DNA microarray analysis using the Affymetrix Human Gene 1.0 ST system. The longer exposure was intended to better model the situation that exists in vivo in chronic drinkers. In order to determine the global transcriptional signature that differentiates the malignant MCF-7 cells from a normal counterpart, we compared the MCF-7 cell line with the spontaneously immortalized but otherwise benign breast epithelial line MCF-12A (1). For each gene sequence, the ratio of MCF-7 expression to MCF-12A expression was determined from duplicate samples. We refer to the collection of MCF-7/MCF-12A gene expression ratios shown in Table I as the MCF-7 oncogenic signature. Some genes related to oncogenic processes were substantially changed, being up- or downregulated by a factor ≥2.0. This transcriptional signature was characterized by 15 genes upregulated by a factor ≥2.4, and 3 oncogenesis-related genes downregulated to a factor of ≤0.27, including some associated with oncogenic transformation and some associated with growth-related hormone receptors.
Table I.
The non-malignant cell line MCF-12A is compared to the malignant cell line MCF-7 in an oncogenic signature (column 3).a
| Gene | Gene description | Oncogenic signature C/C MCF-7/MCF-12A |
1 week
|
4 weeks
|
||
|---|---|---|---|---|---|---|
| C MCF-7 |
Eth/C MCF-7 |
C MCF-7 |
Eth/C MCF-7 |
|||
| CEACAM5 | CEA related cell adhesion molecule | 42.20 | 181 | 1.03 | 223 | 3.14 |
| PGR | Progesterone receptor | 12.80 | 137 | 0.98 | 494 | 1.71 |
| ESR1 | Estrogen receptor 1 | 10.10 | 111 | 0.82 | 0.88 | |
| TET2 | Tet oncogene family member 2 | 9.87 | 392 | 1.08 | 881 | 1.28 |
| BCAS1 | Breast carcinoma amplified sequence 1 | 8.69 | 161 | 1.38 | 528 | 1.08 |
| CEACAM6 | CEA related cell adhesion molecule 6 | 8.03 | 121 | 1.15 | 170 | 2.95 |
| ERBB3 | v-erb-b2 erythro leukemia homolog 3 | 7.95 | 766 | 1.11 | 816 | 1.14 |
| TACSTD1 | Tumor associated Ca signal transducer 1 | 7.91 | 2,715 | 1.02 | 3,386 | 1.06 |
| BCAS2 | Breast ca amplified seq 2 | 7.31 | 3,697 | 1.12 | 4,533 | 0.97 |
| MYB | v-myb oncogene homolog | 6.40 | 474 | 1.33 | 700 | 1.16 |
| TET1 | Tet oncogene 1 | 5.01 | 116 | 1.29 | 147 | 0.78 |
| TOB1 | Transducer of ERBB2 1 | 3.42 | 4,246 | 1.18 | 3,498 | 1.01 |
| BCAS3 | Breast carcinoma amplified seq 3 | 3.34 | 1,426 | 1.07 | 1,359 | 1.11 |
| ERBB2 | v-erb-b2 oncogene homolog 1 | 2.53 | 723 | 1.10 | 437 | 1.17 |
| AR | Androgen receptor | 2.41 | 248 | 1.26 | 71 | 0.98 |
| MTSS1 | Metastasis suppressor 1 | 0.27 | 285 | 1.03 | 171 | 1.17 |
| CD44 | CD44 molecule (Indian blood group) | 0.23 | 591 | 0.99 | 492 | 1.23 |
| CTGF | Connective tissue growth factor | 0.13 | 205 | 0.76 | 86 | 0.96 |
Exposure of MCF-7 to ethanol at 1 week (column 5) and 4 weeks (column 7) are shown. Gene IDs as NCBI Gene data set. C, normalized DNA microarray values for gene expression in untreated cells. Eth, ethanol treated.
We then investigated whether an oncogenic signature reflected the effects of ethanol treatment on MCF-7 by itself. Short-term ethanol incubation for 1 week had little or no effect on the expression of this group of 18 genes (Table I), whereas 4-week exposure increased CEACAM5, CEACAM6, and progesterone receptor (PGR) gene expression (Table I). Neither 1-nor 4-week exposures to ethanol or acetaldehyde affected the transcriptional expression of Oct-4 or Nanog (Table II).
Table II.
Expression of stem-related genes after 1 week of ethanol or acetaldehyde.a
| Gene | Monolayers with ethanol
|
||
|---|---|---|---|
| Control | 25 mM Eth | Eth/cont | |
| OCT-4A | 150 | 144 | 0.96 |
| NANOG | 79 | 70 | 0.89 |
| ALDH2 | 167 | 149 | 0.89 |
| SOX4 | 484 | 475 | 0.98 |
| HEY1 | 109 | 112 | 1.03 |
| JAG1 | 185 | 191 | 1.03 |
| DNER | 124 | 108 | 0.87 |
| DLL1 | 259 | 248 | 0.96 |
|
| |||
| Mammospheres with ethanol
|
|||
| Gene | Control | 25 mM Eth | Eth/cont |
|
| |||
| OCT-4A | 116 | 136 | 1.17 |
| NANOG | 75 | 77 | 1.03 |
| ALDH2 | 124 | 147 | 1.19 |
| SOX4 | 450 | 446 | 0.99 |
| HEY1 | 82 | 95 | 1.16 |
| JAG1 | 178 | 176 | 0.99 |
| DNER | 91 | 99 | 1.09 |
| DLL1 | 248 | 268 | 1.08 |
|
| |||
| Monolayers with acetaldehyde
|
|||
| Gene | Control | 25 mM Acet | Acet/cont |
|
| |||
| OCT-4A | 76 | 85 | 1.12 |
| NANOG | 38 | 42 | 1.11 |
| ALDH2 | 88 | 105 | 1.19 |
| SOX4 | 544 | 558 | 1.03 |
| HEY1 | 69 | 78 | 1.13 |
| JAG1 | 168 | 202 | 1.20 |
| DNER | 79 | 87 | 1.10 |
| DLL1 | 253 | 267 | 1.06 |
Cells (as monolayers or mammospheres) were exposed to 25 mM ethanol or 2.5 mM acetaldehyde, or received no added treatment (control). Upregulation and downregulation induced by ethanol or acetaldehyde is expressed as the ratio of treated expression value divided by control.
Upregulation of stem-related proteins and Ceacam6 protein
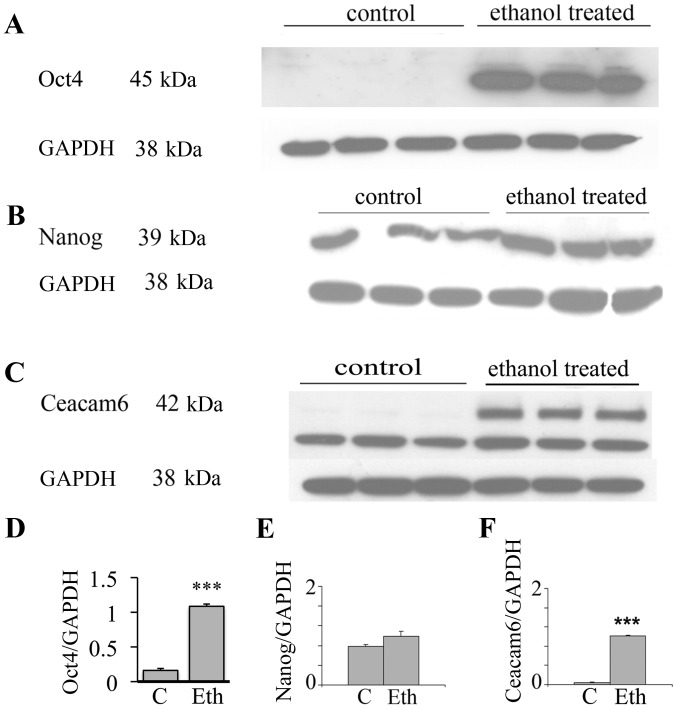
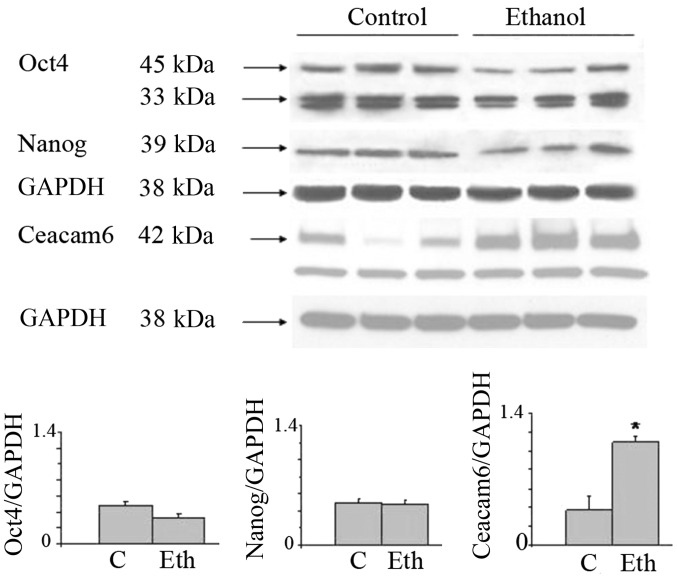
In view of the interesting results on the transcriptional expression of the malignancy related CEACAMs, we evaluated by western blot analysis the effects of long-term ethanol exposure on Ceacam6, an oncogenic protein associated with breast cancer (50), along with the Oct4 and Nanog proteins. Fig. 3 shows that 25 mM ethanol upregulates the expression of these proteins, with particularly strong upregulation in Oct4 and Ceacam6, and a visible but non-significant increase of Nanog. In the case of Oct4, this suggests a post-transcriptional modulation induced by ethanol (see Discussion). These results potentially may also indicate the induction of higher stem cell number or the selection of a subpopulation of cells with some stem-like features.
Figure 3.
Long-term exposure of MCF-7 cells to ethanol at 25 mM increases Oct4a, Nanog, and Ceacam6 protein expression. Triplicate samples (control vs. 25 mM ethanol) grown as monolayer cultures on 6-well plates, incubated for 4 weeks, were analyzed by western blot analysis, and densitometric scans were corrected for GAPDH expression. (A) expression of Oct4a (45 kDa). (B) Nanog (39 kDa). (C) Ceacam6 (42 kDa), all with GAPDH as reference. Densitometric determinations for proteins normalized by Gapdh as (D) Oct4; (E) Nanog; (F) Ceacam6; ***p<0.001.
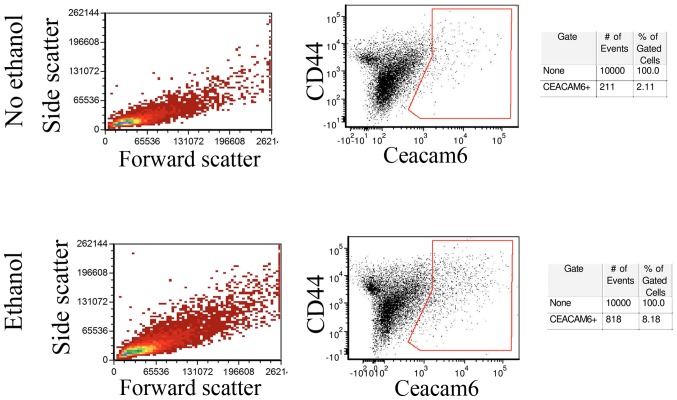
In order to determine whether Ceacam-6 upregulation represented a global increase or was restricted to a subpopulation of cells, untreated control MCF-7 monolayers and monolayer cells exposed to 25 mM ethanol for 4 weeks were analyzed by flow cytometry. Fig. 4 (center) shows that a fraction of the cells did show enrichment of Ceacam-6 following ethanol treatment, from 2.11% in controls to 8.18% in ethanol-treated cells, consistent with levels of upregulation observed in western blot analyses and PCRs. The control forward- and side-scattering (Fig. 4, left) indicate that the ethanol treatment did not appreciably affect cell size or internal complexity, as shown by the lack of observed changes.
Figure 4.
Flow cytometry shows an enrichment of CEACAM6 expressing cells following long-term 25 mM ethanol exposure. MCF-7 monolayers incubated 4 weeks with 25 mM ethanol and their untreated controls were subjected to flow cytometry. Left panels, side scatter vs. forward scatter. Center panels, CD44 vs. Ceacam6 labeling. In the Ceacam6 vs. CD44 panels, the control (no ethanol treatment) sample showed 2.11% of the sample cells in the Ceacam6 positive group (box). The ethanol-treated sample showed a total of 8.18% of cells as positive for Ceacam6 (box). Right panels, counts of total cells in each sample as well as the number of cells showing Ceacam6 positive fluorescence.
Long-term ethanol upregulates Ceacam6 protein in mammospheres, but does not induce an increase in Oct4 or Nanog proteins
In view of the assumption that mammospheres are enriched in stem cells, we investigated whether ethanol affects mammosphere formation and composition. MCF-7 cells were maintained in monolayer culture in the presence or absence of ethanol, following which mammospheres were prepared from each sample. The mammospheres derived from control and ethanol treated cultures were maintained for an additional week in control medium or ethanol medium respectively. The number of mammospheres and their morphology did not visually appear to be modified by ethanol exposure (data not shown).
Despite the observed increase in Oct4a and Nanog proteins induced by long-term 25 mM ethanol treatment in MCF-7 monolayers, it is of interest that these effects were not replicated in mammospheres obtained from these cultures and maintained for an additional week in the presence of ethanol. Fig. 5 shows that the previously observed increase in Oct4a and Nanog in monolayer was not observed in mammospheres where stem cells should be enriched, but Ceacam6 did remain upregulated. More surprisingly, taking into account our original assumption, when mammospheres were quantitated by determining both size and number, long-term exposure to ethanol did not significantly increase their yield, as judged by their relative area (29,352 control vs. 33,889 ethanol-treated), or by their numbers either by counting stained mammospheres under the microscope (1,337 control vs. 1,295 ethanol) or as determined using quantitative image analysis (1,314 control vs. 1,357 ethanol-treated).
Figure 5.
The increased expression of Oct4a, Nanog, and Ceacam6 proteins found in MCF-7 monolayers upon long-term exposure to ethanol is considerably reduced or disappears in mammospheres. Mammospheres were obtained as described and then further incubated for 1 week in the presence or absence of 25 mM ethanol and were analyzed by western blot analysis and corrected for GAPDH expression. Top, expression of Oct4a (45 kDa), Nanog (39 kDa); and Ceacam6 (42 kDa), with GAPDH as reference. Bottom, densitometric determinations; *p<0.05.
RNA was isolated from the mammospheres obtained under 1-week exposure to ethanol or acetaldehyde and subjected to DNA microarray analysis. Table II shows that mammospheres were not enriched in the expression of a collection of stem cell genes. Neither ethanol nor acetaldehyde stimulated the expression of these genes in mammospheres or their original monolayers, which is noteworthy since some are related to breast cancer, such as: ALDH2, SOX4, SOX2, KLF4, LIN28, HEY1, JAG1, DNER or Dll1 (51). No parallel assay was conducted in the mammospheres exposed long-term to ethanol.
Gene expression in MCF-7 after ethanol exposure
As stated above, CEACAM6 protein upregulation induced by long-term ethanol exposure as shown on western blot analyses was in general agreement with mRNA upregulation, so we investigated whether other CEACAM mRNAs or those from other gene families were also upregulated. Table III shows that gene expression of the CEACAMs, cytokines and HLAs were all stimulated by 25 mM ethanol, by factors ranging from 1.7 to 8.1.
Table III.
Some gene families upregulated after long-term 25 mM ethanol.a
| Gene ID | Gene Description | C | Eth/C |
|---|---|---|---|
| Ceacam5 | Carcinoembryonic antigen-related cell adhesion molecule 5 | 223 | 3.14 |
| Ceacam6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 170 | 2.95 |
| Ceacam7 | Carcinoembryonic antigen-related cell adhesion molecule 7 | 70 | 1.9 |
| Ceacam1 | Carcinoembryonic antigen-related cell adhesion molecule 1 | 43 | 1.9 |
| IFI6 | Interferon, α-inducible protein 6 | 333 | 8.1 |
| IFITM1 | Interferon-induced transmembrane protein 1 | 624 | 2.4 |
| IRF9 | Interferon regulatory factor 9 | 119 | 1.8 |
| IL24 | Interleukin 24 | 164 | 2.1 |
| HLA-A | Major histocompatibility complex, Class I,A | 914 | 1.8 |
| HLA-B | Major histocompatibility complex, Class I,B | 937 | 1.9 |
| HLA-C | Major histocompatibility complex, Class I,C | 1,237 | 1.8 |
| HLA-G | Major histocompatibility complex, Class I,G | 518 | 1.7 |
| HLA-H | Major histocompatibility complex, Class I,H | 561 | 1.8 |
Exposure to 25 mM ethanol for 4 weeks reveals families of genes which are upregulated. The results are averaged from 2 sets of DNA microarray assays. C, normalized DNA microarray values for gene expression in untreated cells. Eth/C, ratio of DNA microarray gene expression values for ethanol-treated vs. control cells.
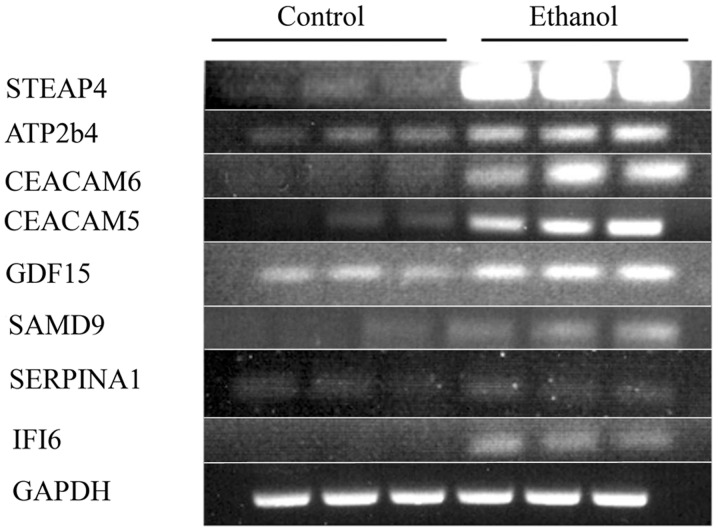
Long-term incubation with ethanol also induces other changes of oncogenic relevance, as confirmed by the substantial upregulation of a series of genes related to breast cancer such as STEAP4, SERPINA3, SAMD9, GDF-15, TP63, PGR, and others, with the transcriptional expression of 13 genes being increased ≥2.0 in two experiments (Table IVA). To confirm the DNA microarray results, selected RNAs from the second of the two experiments were subjected to RT-PCR for some genes in the families mentioned above (Fig. 6). The correspondence between the RT-PCR and DNA microarray values was good to excellent (Table IVB), thus showing that ethanol indeed upregulated these cancer-related genes, and validating in general the DNA microarray data.
Figure 6.
Long-term exposure to 25 mM ethanol increases the transcriptional expression of several oncogenic genes different from the ones in the MCF-7/MCF-12A comparative signature. Confirmation for some selected genes from one of two experiments. PCRs of the selected gene products (triplicate controls vs. triplicate 25 mM ethanol-treated) were run on separate gels.
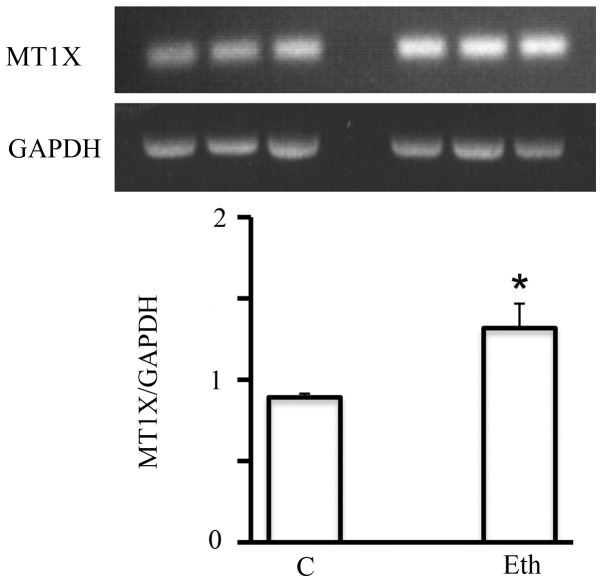
Of particular interest, a family of genes, the metallothioneins (MTs), is known to be induced by ethanol (52,53). Short-term 25 mM ethanol upregulated the mRNA expression of multiple members of this family (MT1F, MT1X, and MT2A, by a factor of ≥2) (Table V). This effect was specific to ethanol, as it was not seen in response to treatment with 2.5 mM acetaldehyde. The upregulation detected by DNA microarray analysis was also confirmed by RT-PCR, as for example MT1X (Fig. 7). However, this significant upregulation seemed to be short-lived, since after the 4-week exposure, it declined to only a marginal increase (not shown) suggesting the adaptation of MCF-7 to ethanol in terms of metallothionein expression.
Table V.
MCF-7 cells treated for 1 week with 25 mM ethanol were analyzed for gene expression using DNA microarray analysis.a
| Gene ID | cont for ETh | Eth/cont | cont for Acet | Acet/cont |
|---|---|---|---|---|
| MT1A | 321 | 1.09 | 188 | 1.15 |
| MT1B | 206 | 1.31 | 66 | 0.99 |
| MT1F | 1,749 | 2.27 | 2,014 | 1.03 |
| MT1G | 748 | 1.71 | 4,768 | 0.78 |
| MT1H | 196 | 1.47 | 133 | 1.29 |
| MT1L | 940 | 1.76 | 501 | 1.00 |
| MT1X | 836 | 2.71 | 812 | 0.85 |
| MT2A | 5,225 | 1.95 | 7,099 | 0.81 |
| MT3 | 395 | 0.89 | 275 | 1.11 |
| MT4 | 155 | 1.08 | 90 | 1.19 |
The results for the metallothionein gene family are shown. Gene ID per NCBI Gene. Eth, values for cells treated with ethanol; Acet, values for cells treated with acetaldehyde.
Figure 7.
Short-term exposure to 25 mM ethanol, but not to 2.5 mM acetaldehyde increases the transcriptional expression of most metallothionein genes. Top, control and ethanol-treated samples analyzed for MT1X by RT-PCR. Bottom, densitometric determination of ethanol-treated vs. control samples. *p<0.05.
Effects of long-term ethanol exposure on the expression of miRs controlling the expression of genes related to stem cells, malignancy, and estrogen effects
MicroRNAs (miRs) function by suppressing the activities of specific target mRNAs. Although it is possible for multiple miRs to affect a specific mRNA, sometimes an inverse relationship is observed between the expression level of a particular miR and the prevalence of the polypeptide coded by its target mRNA. Therefore, identifying the global miR transcriptional signature in response to ethanol exposure is important in clarifying the mechanism(s) of action of ethanol on breast cancer, particularly in evaluating its putative estrogen-like effects. We therefore carried out 4-week incubations of MCF-7 monolayers with and without 25 mM ethanol in duplicate experiments to investigate changes in miR prevalence.
Changes in the global expression signature of miRs induced by ethanol are presented in Table VI, showing that out of 1,904 miRs analyzed, 18 miRs were consistently upregulated in two separate assays by a factor of >2.0 and another 24 were downregulated by at least an equivalent factor (to ≤0.5). Of these, 4 miRs showed substantial upregulation (by >3.0) and 9 miRs were substantially downregulated (to ≤0.33). Within the group of 4 upregulated genes, 3 are linked to cancer, namely miR-3170 which is downregulated in Merkel cell carcinoma (54), miR-335-5p which is linked to fibrosarcoma (55) and colorectal cancer but was negative in at least one study in MCF-7 (56), and miR-424-5p which is increased in breast cancer through an estrogen stimulated pathway (57) but is anti-invasive in another system (58). Within the group of 9 downregulated miRs, 5 are also related to cancer: miR-2861 is upregulated in thyroid carcinoma with lymph node metastases (59) but has been suggested as one element in a circulating miR screen for cervical cancer (see Discussion), miR-3185 for chordoma (60), miR-1915 whose downregulation would predict an antiapoptotic effect (and therefore potential oncogenicity) mediated through Bcl-2 (61), miR-4492 potentially linked to breast cancer (62), and miR-1469 downregulation linked to lymphatic metastasis in gastric cancer (63) but a stimulatory factor for apoptosis in lung cancer cells (64).
Table VI.
Long-term exposure to 25 mM ethanol and expression of miRs involved in cancer and stem cells.a
| miRs upregulated | Cont | Eth/Cont | miRs downregulated | Cont | Eth/Cont |
|---|---|---|---|---|---|
| hsa-miR-3170 | 28 | 3.87 | hsa-miR-4507 | 914 | 0.51 |
| hsa-miR-335-5p | 80 | 3.58 | hsa-miR-4687-3p | 819 | 0.49 |
| hsa-miR-424-5p | 2,046 | 3.18 | hsa-miR-320e | 394 | 0.49 |
| hsa-miR-3607-3p | 411 | 3.01 | hsa-miR-4734 | 1,819 | 0.48 |
| hsa-miR-20b-5p | 226 | 2.92 | hsa-miR-4516 | 6,902 | 0.48 |
| hsa-miR-148a-3p | 548 | 2.70 | hsa-miR-4530 | 6,332 | 0.48 |
| hsa-miR-494 | 2,012 | 2.69 | hsa-miR-638 | 11,664 | 0.46 |
| hsa-miR-4284 | 4,767 | 2.53 | hsa-miR-4508 | 4,645 | 0.44 |
| hsa-miR-23c | 85 | 2.38 | hsa-miR-3656 | 6,310 | 0.43 |
| hsa-miR-126-3p | 168 | 2.31 | hsa-miR-3196 | 6,608 | 0.43 |
| hsa-miR-30a-5p | 1,452 | 2.29 | hsa-miR-5001-5p | 8,936 | 0.42 |
| hsa-miR-3607-5p | 3,251 | 2.28 | hsa-miR-4505 | 1,873 | 0.40 |
| hsa-miR-141-3p | 1,297 | 2.18 | hsa-miR-663a | 9,642 | 0.39 |
| hsa-miR-454-3p | 255 | 2.16 | hsa-miR-762 | 1,375 | 0.37 |
| hsa-miR-29a-3p | 2,275 | 2.16 | hsa-miR-3940-5p | 3,288 | 0.34 |
| hsa-miR-3676-5p | 153 | 2.06 | hsa-miR-1469 | 267 | 0.33 |
| hsa-miR-429 | 369 | 2.04 | hsa-miR-4466 | 5,285 | 0.33 |
| hsa-miR-106b-5p | 1,869 | 2.02 | hsa-miR-4492 | 1,515 | 0.32 |
| hsa-miR-106a-5p | 795 | 1.96 | hsa-miR-4707-5p | 5,583 | 0.30 |
| hsa-miR-1915-3p | 3,695 | 0.29 | |||
| hsa-miR-3185 | 555 | 0.26 | |||
| hsa-miR-4800-3p | 1,374 | 0.24 | |||
| hsa-miR-2861 | 1,437 | 0.22 | |||
| hsa-miR-654-5p | 5,651 | 0.21 |
Upregulated miRs are shown on the left, and downregulated miRs are shown on the right.
In Table VII, we group other miRs affected by long-term ethanol exposure of MCF-7, focusing on whether these miRs may be related to two additional important issues that may underlie the observed intensification of MCF-7 malignant features: a) the finding of upregulation of Oct4 and Ceacam6 protein expression; and b) the putative mediation of alcohol effects by estrogen-like mechanisms. In Table VII, miRs whose expression was affected in general by a factor of >2 are listed based on a relationship with one or more of these issues. In terms of the Oct4 regulation, Let-7 has been reported to repress Oct4 protein expression (65) and after ethanol treatment it is substantially downregulated from high levels seen in the non-exposed control. Considering the observed lack of OCT4 gene transcriptional upregulation in the presence of ethanol, concomitant with the observed stimulation of Oct4 protein, this miR change is very pertinent in this context. In turn, with respect to possible miRs targeting Ceacam6, only one likely miR was downregulated (149-3P) (66) which would be consistent with Ceacam6 upregulation, although three (15a-5P, 16-5P and 195-5P) were upregulated, but their relationship with Ceacam6 is less clear.
Table VII.
Long-term exposure to 25 mM ethanol affects miRs involved in oncogenesis and in estrogen effects.
| miR | C | Eth/C |
|---|---|---|
| Let-7a-5p | 6,095 | 0.6 |
| miR-15A-5p | 310 | 2.57 |
| miR-16-5p | 4,286 | 2.01 |
| miR-195-5p | 910 | 2.32 |
| miR-149-3p | 2,067 | 0.15 |
| Let-7b | 2,737 | 0.66 |
| Let-7c | 4,158 | 0.64 |
| miR-424-5pa | 2,046 | 3.18 |
| miR-494a | 2,012 | 2.69 |
| miR-27a | 3,888 | 2.09 |
| miR-27a | 2,254 | 2.43 |
| miR-429a | 369 | 2.04 |
| miR-16 | 4,286 | 2.01 |
| miR-203 | 2,482 | 2.15 |
| miR-342 | 3,339 | 1.83 |
| miR-200a | 854 | 2.44 |
miRs also listed in Table VI.
In terms of a possible relationship with estrogen pathways, Table VII shows several miRs affected by long-term ethanol exposure that are known to modulate or be modulated by estrogen-mediated processes or estrogen responsiveness, specifically 6 that are upregulated (16, 27a, 27b, 200a, 203, and 342) and 3 downregulated (let7b, 7c and 7d) (see Discussion).
Long-term exposure of MCF-7 cells to ethanol, but not to acetaldehyde, stimulates anchorage-independence
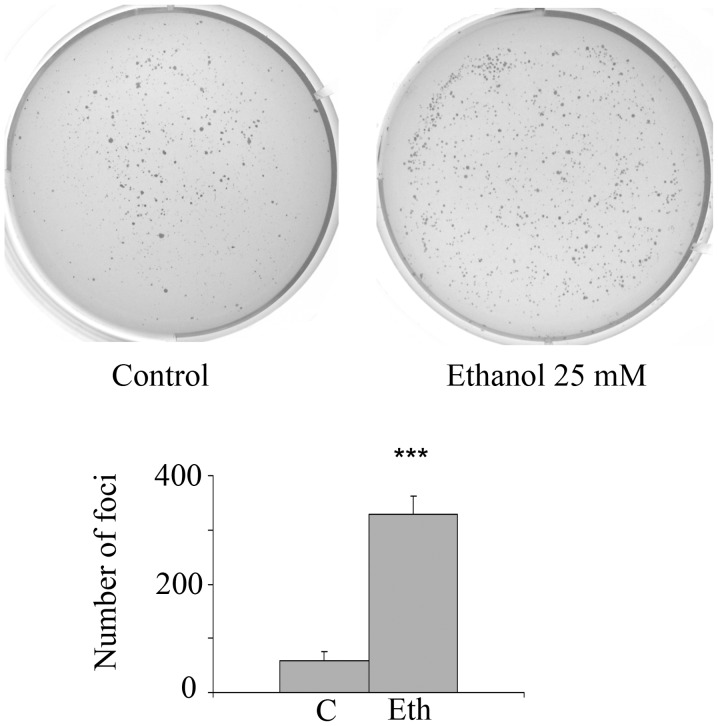
In order to determine whether long-term exposure to ethanol or acetaldehyde stimulates anchorage-independence, an in vitro marker of oncogenic transformation, incubations with 2.5–25 mM ethanol or 2.5 mM acetaldehyde were carried out for 4 weeks followed by 3–4 weeks growth in the clonogenic soft agar assay. Fig. 8 shows that 25 mM ethanol treatment substantially and significantly increases the number of anchorage-independent foci by 5.6-fold (4 separate experiments, each in triplicate), which agrees with the protein expression and transcriptional signature changes related to stem cell and oncogenic features. In addition, after exposure to a lower ethanol concentration (10 mM), there was a substantial and significant increase in anchorage-independence of 3.85-fold (p=0.014) for triplicate ethanol wells and duplicate control wells. However, exposure to further ethanol dilutions, 2.5 or 5 mM, had no significant effect. As in the case of 1-week incubation, long-term treatment with 2.5 mM acetaldehyde did not stimulate foci formation (data not shown).
Figure 8.
Long-term exposure to 25 mM ethanol stimulates MCF-7 anchorage-independence as detected by the soft agar procedure. Top, representative images of the crystal violet staining of typical plates of soft agar growth for 25 mM ethanol. Bottom, foci number for each type of incubation as mean of 4 experiments, each experiment using triplicate wells. ***p<0.001.
In contrast to the clonogenic assay, incubation of 25,000 cells with 25 mM ethanol for 4 weeks was not associated with the stimulation of invasiveness in the Matrigel assay (data not shown). It should be noted that the MCF-7 cell line is not invasive per se. There were only trace numbers of positive cells (mean of 4 samples each: control mean 11.25±2.4 SE; ethanol mean 14.5±2.9 SE) and there was no statistical significance (p=0.68, two-tailed t-test). Moreover, there was no induction of tamoxifen resistance after incubation for 4 weeks with 25 mM ethanol as analyzed by cellular metabolism in the presence of increasing levels of the drug (data not shown).
Discussion
To our knowledge this is the first report on the in vitro long-term effects of ethanol on breast cancer cells at doses comparable to peak exposures in humans, and describing multiple targets of potential cancer-related impact. These effects are not exerted by acetaldehyde, and are largely inconsistent with respect to estrogen-mediated effects. Therefore, our results do not support the prevalent hypothesis that ethanol action in cancer-related processes functions mainly through estrogenic or acetaldehyde mediation, although future, more mechanistic work is needed to test this assumption, particularly in relation to the potential estrogen mediation.
In a previous study (1), we investigated the effects of ethanol exposure on a non-malignant, spontaneously immortalized breast epithelial cell line, MCF-12A. We found that relatively low doses of ethanol stimulated markers for epithelial mesenchymal transition (EMT), as shown by changes in mRNAs and miRs. We also found some upregulation in the stem-related proteins Oct4 and Nanog, as well as in the oncogenic marker Ceacam6. The results with respect to these phenomena are remarkably similar in this study of MCF-7, although the levels of ethanol necessary to achieve the observed changes differed. Ethanol incubation induced a gene expression signature in MCF-12A that includes short-term upregulation of one gene family in particular, the metallothioneins, similar to that observed in MCF-7.
In this study, we have shown that: i) an exposure as short as 1 week to 25 mM ethanol stimulates the transcriptional expression of the metallothionein gene family in MCF-7, showing that even in an established cancer cell line, the effect of ethanol on MT expression occurs; this short-term ethanol treatment does not, however, induce any substantial change in the global transcriptional signature related to the expression of stem cell or oncogenic markers in monolayers or mammospheres; ii) in contrast, longer (4-week) exposure of MCF-7 monolayers to 5–25 mM ethanol leads to upregulation of the key stem cell proteins Oct4 and Nanog, and of a key oncogenic protein, Ceacam6, demonstrating that phenomena previously observed in MCF-12A are maintained in the established cancer line MCF-7; iii) in MCF-7 mammospheres incubated long-term with ethanol, only Ceacam6 remains upregulated, and there is no effect on mammosphere number or size; iv) changes in Ceacam6 protein expression are reflected in RNA expression, but the same phenomenon is not observed for Oct4 or Nanog protein expression, suggesting their regulation at the translational level.
In addition, of even more significant pathological relevance, we showed that: v) in MCF-7 monolayers, long-term effects induced by 25 mM ethanol include substantial changes in the global transcriptional signature, including upregulation of immune-related genes, HLAs, and in particular, a series of genes involved in breast cancer; vi) this long-term oncogenic intensification of the transcriptional signature by ethanol is associated with the stimulation of anchorage-independence, a marker of increased malignancy; vii) this putative transformation exerted by ethanol is not associated with induction of tamoxifen resistance, potentially related to estrogen effects, or of invasiveness; viii) ethanol effects on miR expression are largely inconsistent with the hypothesis that ethanol effects are entirely or even largely mediated through estrogenic pathways; rather, there are conflicting effects such that some miR expression follows this pattern and other miR expression goes in the direction that is opposite to what would be expected under the estrogenic hypothesis; ix) most of the alterations we observed following ethanol treatment do not appear to be mediated by acetaldehyde.
We do not have a conclusive explanation for the temporal pattern showed by oncogenic effects due to long-term exposure to ethanol, but speculate as to two possible mechanisms: i) ethanol effects on gene expression may result from a cascade of regulatory events that requires multiple cell divisions to complete; or ii) longer term exposure to ethanol not only affects gene expression, it selects for a subset of cells within the original population; thus the partially toxic effects of ethanol might be reflected in gene expression changes that accumulate over the course of several cell division cycles.
Before analyzing the mechanistic significance of these alterations, it is important to stress the limitations of this study, in that it is essentially an in vitro proof-of-concept approach, based on the cell line, MCF-7, similar to what we cautiously stated regarding our previous study on the normal MCF-12A cell line (1). The malignant MCF-7 cell line was derived from a metastatic site of a breast cancer tumor and is widely used as a model for breast cancer studies. Our results need to be confirmed and extended in vitro using lower concentrations of ethanol and by examination of the interconnections between observed gene expression alterations. It should be noted that in our previous study on MCF-12A cells, considerably lower levels of ethanol led to substantial effects on protein, mRNA, and miR prevalence, and that their long-term exposure to high 25 mM levels arrested their growth and ultimately was toxic to the cells. In a simplistic comparison, this would mean that the stimulation of malignant features (MCF-7) requires a higher concentration of ethanol than their induction (MCF-12A).
To better extrapolate these MCF-7 results to the human, in vivo confirmation is required in experimental animals. Although the in vitro exposure to ethanol concentrations of 5–25 mM, roughly equivalent in women to peak human serum concentrations after 0.6–3 glasses of wine taken during a 2-h period, was maintained continuously, its duration of only 4 weeks is certainly much shorter than the cumulative years of exposure that a human drinker experiences in a lifetime. There is no easy calculation of the equivalence between cell culture incubations and breast tumor tissue exposures, particularly because MCF-7 cultures may not reflect the complex stromal/epithelial interactions involved in breast cancer progression. Despite the above limitations, this study calls attention to the existence of molecular alterations in breast tumor cells induced by ethanol that may open up directions to further investigations on breast cancer in women.
It is of interest that the metallothioneins are members of a family of genes known to be induced by ethanol (52) and comprising members such as MT-I and MT-II that are antiapoptotic, proliferative, angiogenic, and oncogenic (53). The expression of MT-I and MT-II is increased in breast cancer and other tumors, correlating with higher tumor grade/stage, increased recurrence and poor survival in the highly malignant invasive ductal breast carcinomas, and predicting poor prognosis in estrogen receptor-negative patients. Although the stimulation of metallothionein gene expression is indicative of gene expression effects beginning as soon as a few days after the administration of alcohol, the stimulation of metallothionein gene expression did not persist in our model system, so the long-term effect of these gene changes with respect to oncogenesis remains uncertain. As noted above, the temporal course of action in MT genes are similar for both the malignant MCF-7 cells and the non-malignant MCF-12A cells.
A consistent pattern of changes induced by long-term ethanol, but not acetaldehyde, is the correlation between the upregulation of the key stem cell proteins Oct4a and Nanog (67) and an important cancer marker, Ceacam6 (50), in addition to the induction of anchorage-independence evidenced by soft agar growth. OCT4 is well known as a key gene responsible for the stemness network, and the main factor in the generation of induced pluripotent stem cells (iPS). More recently, OCT4 has emerged as a key oncogenic factor related to the role of cancer stem cells (68). For instance, expression of the Oct4 protein is higher in cancerous tissues than in adjacent-tumor tissues, is related to histological type, lymph node status and molecular type of breast cancer, and together with Her-2, is an independent prognostic factor for breast cancer (69,70), although it seems to occur later than SOX2 activation (71,72). In MCF-7 cells, estrogen stimulates and metformin reduces the size and number of mammospheres and their expression of Oct4 (73).
However, our current results must be viewed as speculative as to whether they connect the observed upregulation of Oct4 to increased malignancy, since in another study, silencing OCT4 promoted invasion and metastasis in MCF-7 cells by inducing EMT (74). The very low expression of Oct4 detected by us in MCF-7 monolayers in the absence of ethanol is consistent with observations from other studies (75,76), and the potentially oncogenic upregulation by ethanol has not been previously reported. As to NANOG, there is extensive evidence linking it to breast cancer directly or through its activation by SOX2, alone or in conjunction with KLF4 (77), and it is known that ethanol induces Nanog in embryonic stem cells and hepatic carcinogenesis (78).
The upregulation of CEACAM6 after long-term 25 mM ethanol exposure is consistent with a more aggressive phenotype of MCF-7 cells in vitro (75). Ceacam6, a membrane-associated cell adhesion protein, is overexpressed in breast cancer and a variety of other tumors, and is considered to be a marker of invasiveness and metastasis, a predictor of breast cancer recurrence, and specifically of invasive breast cancer in women with atypical ductal hyperplastic tissues (79–82). There are no previous reports of ethanol effects on CEACAM6, or on a link between CEACAM6 and OCT4 expression in any other context save our previous study on non-malignant cells (1), and the correlation of this upregulation to the intensification of anchorage-independence has not been reported previously except in our previous report (1), thus showing that this phenomenon is observed in both malignant and non-malignant cells derived from breast epithelium. Similarly, the stimulation of MCF-7 growth in soft agar due to exposure to ethanol at a dose as low as 10 mM has not been described before except in the previous report (1), since previous studies (20–26) used high concentrations of ethanol (90–110 mM), short periods of exposure (48 h), and in some cases, breast cancer cell lines other than MCF-7.
The stimulation by long-term ethanol of the expression of Ceacam6 protein is in agreement with the observed transcriptional upregulation of CEACAM6 and CEACAM5 and with other CEACAMs. The observed stimulation is also paralleled by the increase in mRNA levels for a series of oncogenesis-related genes, STEAP4 (83,84), SERPINA3 (84), SAMD9 (85), GDF-15 (86), KRT15 (87), TP53INPI (88), IF16 or G1P3 (89), and HLA-G (90) as well as other members of the HLA family, all of which have in common their breast cancer-related expression and the fact that none has been reported as being upregulated by ethanol. Some of these genes (SERPINA3, GDF15, IFI6) are also modulated by estrogen, but only one, GDF15, was previously studied in MCF-7. Surprisingly, no reports are available on the relationship of any of these genes with either OCT4 or CEACAM6.
No evidence of preferential stimulation of stem cell accumulation following ethanol treatment in either MCF-7 monolayers or mammospheres could be found by gene expression analysis, as judged by the lack of meaningful changes in the transcriptional expression of a series of stem cell genes, including OCT4 and Nanog. This finding is in agreement with the similarity between the transcriptional signatures of mammospheres and monolayers in the absence of ethanol. This is puzzling within the framework of the significant upregulation of Oct4 and Nanog protein expression induced by long-term ethanol. OCT4 and NANOG are key stem cell genes in a network of other genes involved in stemness, but which are expressed transcriptionally in this study at only a low level, and not changed by ethanol treatment. A possible explanation could involve translational regulation mediated by miRs (see below). It is also conceivable that OCT4 and NANOG act as oncogenic factors per se, unrelated to stem cell activation, or speculatively may be due to the failure of stem cells to form mammospheres, but no supporting data exist in these in respect of other than OCT4 overexpression in breast cancer and other tumors.
The seeming contradiction between Oct4 protein upregulation in the absence of detectable Oct4 mRNA increase suggests a model in which untreated cells exist in a state of translational repression of Oct4 due to some miR or combination of miRs. According to this hypothesis, some miRs such as Let-7A-5P (which is reduced in prevalence by ethanol exposure) allows the induction, by its partial absence, of overexpression of the Oct4 protein. Other Let-7 family members are also substantially expressed in control cells and reduced by ethanol. Let-7A has been shown to function as a tumor suppressor in head and neck cancers and in their associated tumor initiating cells, where it is significantly decreased when OCT4 expression was increased (65). However, it is not clear that this is only translational repression and NANOG is also affected, so it is possible that Let-7 might not be responsible by itself for the selective upregulation of Oct4 protein, and it works in conjunction with the opposite effects of Lin 28b. Another miR, miR-145 functions as a protective miRNA in tumor tissues of lung adenocarcinoma patients and binds to the OCT4 3′-untranslated region (UTR) thus blocking protein expression correlated with anti-oncogenic action (91–93), but we did not detect significant changes in this miR. The same uncertainty applies to the putative interaction between the downregulation of miR-149-3p by long-term ethanol, that would be consistent with the observed upregulation of CEACAM6 mRNA and protein, and the counterintuitive upregulation of miR-15A-5p, 16-5p, and 195-5p that could potentially oppose this effect. However, these are inferences based purely on sequence analogies in the miRBase 18.0 (94) and without mechanistic proof as yet. Therefore, the roles of the changes in miR levels in relation to the upregulation of Oct4, Nanog, and Ceacam6 induced by long-term ethanol in MCF-7 require further study.
The molecular signature of miRs related to estrogen responsiveness is key to understanding whether ethanol may act via estrogenic pathways on luminal-like breast tumor cells, such as the MCF-7, to induce or stimulate their proliferation, survival, and functional status through the estrogen receptor α. Alcohol is assumed to increase sex hormone levels in both premenopausal and postmenopausal women (6) by: a) increased aromatase activity; b) decreased hepatic catabolism of androgens; c) modulation of adrenal steroid production; and d) by increasing the expression or transcriptional activity of ERα. Alcohol may also preferentially enhance cellular proliferation and ERα content in ER-positive cell lines (15–18). Therefore, it might be expected that long-term ethanol effects on miRs in MCF-7 would mimic those exerted by estrogen or those related to estrogen responsiveness, but no reports are available on ethanol effects on any miR profile except in our recently published study on non-malignant cells (1).
One of the miRs we found upregulated in MCF-7, miR-424, was also shown to be induced by 17-β-estradiol in at least two (95,96) of the multiple studies conducted in MCF-7. In some cases, ethanol induced upregulation is also seen after estrogen treatment, such as in miR-27a and b (96). The fact that ethanol affected any particular miR in this study did not necessarily coincide with affects on other miRs that would be expected to change comcomitantly if ethanol action were limited to one specific pathway such as the estrogenic pathway: thus upregulated miRs such as miR-107, miR-424, miR-570, miR-618, and miR-760 (95), miR-21 (28), miR-34b (97), miR-98 (98), miR-19A and miR-24 (96), miR-26a and b (99), miR-17 (100,101), miR-7 (102), or miR-190a (103); or downregulated miRs such as miR-16, miR-143, or miR-203 (104). Interestingly, the latter is the opposite of what we observed with ethanol.
Although there are 4 miRs where both ethanol and estradiol induce the same changes in MCF-7, there are at least 18 miRs known to be affected by estrogen that can target a significant number of transcripts belonging to one or more estrogen-responsive gene clusters, but were not modified by ethanol in this study. In turn, 5 of our ethanol responsive changes have not been reported in global miR transcription studies conducted with estrogen or estrogenic compounds in MCF-7. The scope of this report does not cover the elucidation of how these miRs may act in terms of oncogenic or tumor suppressor effects. However, it is worth mentioning that out of the miRs whose levels have previously been shown to be regulated by estrogen and/or to modulate its receptor, and that were found in our work to be changed by ethanol, only the miR-27a and miR-27b levels were modulated as predicted by their role in estrogen responsiveness. It is also known that miR-27a is oncogenic in the triple-negative MDA-MB-231 breast cancer cell line, and indirectly regulates E2-responsiveness in MCF-7 cells through suppression of ZBTB10, thereby enhancing expression of ERα (47,48). In turn, miR-27a is upregulated in endometrial adenocarcinoma and precancerous lesions in response to estrogen overexposure. However, let-7 behaves similarly in the hyperplastic endometrium (98,105) and most members of this family (b, c and d) are considered to be pro-estrogenic. In this study they were uniformly reduced.
The lack of consistency between the ethanol modulation of miR levels and their relationship with estrogen responsiveness is confirmed by our results with miR-16 (106), miR-200a (47), miR-203 (47), and miR-342 (107), which, as in the case of let-7a, b, and c, are opposite to what could be expected from an estrogen-like effect. However, since miRs exert pleiotropic effects, a mere association with estrogen-mediated pathways is not sufficient to conclude that those may not be involved in the observed ethanol effects on MCF-7 cultures. It should be emphasized that many of these miRs also affect other processes different from estrogenic effects, as described in a large and growing literature. For example, the Let-7 family is highly conserved and is involved in development, stem cell modulation, oncogenesis, and the cardiovascular system among other things (108), while the miR 15/107 group containing miR-15 and miR-16 affects BRCA1 expression (109), and miR-195 and miR-29 have been linked to aortic aneurism (110).
Regarding the ethanol/estradiol comparison of miR signatures, we note the limitation that our study was long-term, whereas the previous estrogen studies varied in duration, but we propose that estradiol effects do not seem to define more than a small proportion of the changes in the global miR transcriptional signature of MCF-7 induced by ethanol. The ethanol induced changes are specific and at least partially different from estrogen-induced changes. Thus this study does not provide strong support for the hypothesis that estrogen mediates ethanol effects. Further studies are needed, vis-à-vis ethanol and estrogen, and with anti-estrogenic agents, to clarify the estrogenic mediation of long-term ethanol effects on MCF-7.
A recent review lists a large number of miRs that are involved in cancer-related processes ranging from tumor suppression to various oncogenic features (111). We note that several miRs listed in that review show ethanol effects in this study, including miRs 15a, 16, the Let-7 family, 27a, 148a, and 149. We should also point out that ethanol does not necessarily promote oncogenesis in every case, for example tumor suppressors miR-15a and miR-16 are upregulated in response to ethanol treatment. However, the same two miRs were found to be upregulated in a biomarker test distinguishing sepsis from systemic inflammatory response (112). In this study, we found that exposure of MCF-7 cells to ethanol results in detectable changes in the expression of miRs such as miR-29a associated with inflammation (113). Interestingly, miR-15a upregulation is found in the serum of humans exposed to particulate air pollution (114). In another study, it was found that miR-29a is potentially able to stimulate the conditions for metastasis by binding to Toll-like receptors (115).
Some miRs that have been proposed for inclusion as circulating cancer markers in the clinical setting show changes in expression levels in response to ethanol. This suggests that such markers should be considered carefully with regard to the ethanol intake status of the patient. For example, miR-424 has been proposed for inclusion in a group of circulating biomarkers for breast cancer (116), and miR-2861 is upregulated in thyroid carcinoma with lymph node metastases (59) but has been suggested as one element in a circulating miR screen for cervical cancer, where it has been shown to be decreased (117).
In conclusion, our overall results suggest that prolonged exposure to ethanol may lead to the intensification of a series of novel oncogenic features in breast cancer by mechanisms that are not directly related to acetaldehyde and which do not strongly indicate estrogen mediation. These features require further clarification, with the caveat that the in vitro incubations using a breast cancer cell line are not directly extrapolatable to alcohol consumption in women. Our results may suggest some kind of stem cell involvement, but one which so far shows contradictory transcriptional and translational aspects. However, in conjunction with our findings on the induction of oncogenic features in a normal breast epithelial cell line (1), the current data may constitute the first comparison of global transcriptional signatures elicited by long-term ethanol exposure on normal and cancer breast cells, and the defining of potentially oncogenic features exacerbated by alcohol. Moreover, miRs in circulation are now being proposed as a diagnostic tool for both carcinomas and metatatic carcinomas (118). Therefore, the observation that ingested ethanol affects miR levels may have clinical significance, particularly with respect to miR values measured in heavy drinkers.
Table IV.
Long-term ethanol exposure, and RT-PCR.
| A, Selected genes with relevance to breast cancer that are upregulated after long-term exposure to ethanol
| |||
|---|---|---|---|
| Gene | Also known as | Control | Eth/Control |
| STEAP4 | Stamp2 | 134 | 8 |
| SERPINA3 | α-1 antitrypsin | 1,235 | 7.7 |
| SAMD9 | Sterile α motif domain 9 | 46 | 5.2 |
| GDF15 | Growth differentiation factor 15 | 2,915 | 3.6 |
| KRT15 | Keratin 15 | 58 | 3.3 |
| ITGB6 | Integrin β | 79 | 3.1 |
| OAS1 | 1–5A synthetase | 91 | 3 |
| FGB | Fibrinogen, β chain | 43 | 5.1 |
| TP53INP1 | p53-dependent damage-inducible nuclear protein | 312 | 2.5 |
| TP63 | Tumor protein p63 | 68 | 2 |
| PGR | Progesterone receptor | 494 | 2 |
| PLAT | Tissue plasminogen activator | 105 | 2 |
| FN1 | Fibronectin 1 | 222 | 1.9 |
| KRT81 | Keratin 81 | 349 | 1.8 |
| ATP2b4 | Plasma membrane calcium-4 transporting ATPase | 516 | 2.4 |
| B, Genes identified by DNA microarray analysis were verified by RT-PCR.a
| ||
|---|---|---|
| Gene ID | DNA microarray ratio (Eth/cont) | PCR ratio (Eth/cont) |
| SAMD9 | 4.3 | 3.1 |
| IFI6 | 4.1 | 3.9 |
| CEACAM6 | 2.2 | 4 |
| STEAP4 | 2.1 | 4.3 |
| ATP2b4 | 1.7 | 2.8 |
| CEACAM5 | 1.6 | 7.1 |
| GDF15 | 1.3 | 1.9 |
| SERPINA1 | 1.1 | 1.8 |
The RNA from one of two experiments was subjected to RT-PCR. The DNA microarray upregulation ratios (Eth/cont) are compared with the PCR upregulation ratios (Eth/cont).
References
- 1.Gelfand R, Vernet D, Bruhn K, Vadgama J, Gonzalez-Cadavid NF. Long-term exposure of MCF-12A normal human breast epithelial cells to ethanol induces epithelial mesenchymal transition and oncogenic features. Int J Oncol. 2016;48:2399–2414. doi: 10.3892/ijo.2016.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz HK, Pelucchi C, Bagnardi V, La Vecchia C. Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012. Alcohol Alcohol. 2012;47:204–212. doi: 10.1093/alcalc/ags011. [DOI] [PubMed] [Google Scholar]
- 3.Coronado GD, Beasley J, Livaudais J. Alcohol consumption and the risk of breast cancer. Salud Publica Mex. 2011;53:440–447. [PubMed] [Google Scholar]
- 4.Pelucchi C, Tramacere I, Boffetta P, Negri E, La Vecchia C. Alcohol consumption and cancer risk. Nutr Cancer. 2011;63:983–990. doi: 10.1080/01635581.2011.596642. [DOI] [PubMed] [Google Scholar]
- 5.Chen WY, Rosner B, Hankinson SE, Colditz GA, Willett WC. Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. JAMA. 2011;306:1884–1890. doi: 10.1001/jama.2011.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narod SA. Alcohol and risk of breast cancer. JAMA. 2011;306:1920–1921. doi: 10.1001/jama.2011.1589. [DOI] [PubMed] [Google Scholar]
- 7.Saxena T, Lee E, Henderson KD, Clarke CA, West D, Marshall SF, Deapen D, Bernstein L, Ursin G. Menopausal hormone therapy and subsequent risk of specific invasive breast cancer subtypes in the California Teachers Study. Cancer Epidemiol Biomarkers Prev. 2010;19:2366–2378. doi: 10.1158/1055-9965.EPI-10-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allemani C, Berrino F, Krogh V, Sieri S, Pupa SM, Tagliabue E, Tagliabue G, Sant M. Do pre-diagnostic drinking habits influence breast cancer survival? Tumori. 2011;97:142–148. doi: 10.1177/030089161109700202. [DOI] [PubMed] [Google Scholar]
- 9.Jelski W, Chrostek L, Szmitkowski M, Markiewicz W. The activity of class I, II, III and IV alcohol dehydrogenase isoenzymes and aldehyde dehydrogenase in breast cancer. Clin Exp Med. 2006;6:89–93. doi: 10.1007/s10238-006-0101-z. [DOI] [PubMed] [Google Scholar]
- 10.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T. Alcohol consumption and oxidative DNA damage. Int J Environ Res Public Health. 2011;8:2895–2906. doi: 10.3390/ijerph8072895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balbo S, Meng L, Bliss RL, Jensen JA, Hatsukami DK, Hecht SS. Time course of DNA adduct formation in peripheral blood granulocytes and lymphocytes after drinking alcohol. Mutagenesis. 2012;27:485–490. doi: 10.1093/mutage/ges008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz HK, Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: Role of genetics in ethanol metabolism. Genes Nutr. 2010;5:121–128. doi: 10.1007/s12263-009-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong AW, Dunlap SM, Holcomb VB, Nunez NP. Alcohol promotes mammary tumor development via the estrogen pathway in estrogen receptor alpha-negative HER2/neu mice. Alcohol Clin Exp Res. 2012;36:577–587. doi: 10.1111/j.1530-0277.2011.01654.x. [DOI] [PubMed] [Google Scholar]
- 15.Singletary KW, Frey RS, Yan W. Effect of ethanol on proliferation and estrogen receptor-alpha expression in human breast cancer cells. Cancer Lett. 2001;165:131–137. doi: 10.1016/S0304-3835(01)00419-0. [DOI] [PubMed] [Google Scholar]
- 16.Etique N, Chardard D, Chesnel A, Merlin JL, Flament S, Grillier-Vuissoz I. Ethanol stimulates proliferation, ERalpha and aromatase expression in MCF-7 human breast cancer cells. Int J Mol Med. 2004;13:149–155. [PubMed] [Google Scholar]
- 17.Etique N, Flament S, Lecomte J, Grillier-Vuissoz I. Ethanol-induced ligand-independent activation of ERalpha mediated by cyclic AMP/PKA signaling pathway: An in vitro study on MCF-7 breast cancer cells. Int J Oncol. 2007;31:1509–1518. [PubMed] [Google Scholar]
- 18.Etique N, Grillier-Vuissoz I, Lecomte J, Flament S. Crosstalk between adenosine receptor (A2A isoform) and ERalpha mediates ethanol action in MCF-7 breast cancer cells. Oncol Rep. 2009;21:977–981. doi: 10.3892/or_00000311. [DOI] [PubMed] [Google Scholar]
- 19.Przylipiak A, Rabe T, Hafner J, Przylipiak M, Runnebaum R. Influence of ethanol on in vitro growth of human mammary carcinoma cell line MCF-7. Arch Gynecol Obstet. 1996;258:137–140. doi: 10.1007/s004040050114. [DOI] [PubMed] [Google Scholar]
- 20.Meng Q, Gao B, Goldberg ID, Rosen EM, Fan S. Stimulation of cell invasion and migration by alcohol in breast cancer cells. Biochem Biophys Res Commun. 2000;273:448–453. doi: 10.1006/bbrc.2000.2942. [DOI] [PubMed] [Google Scholar]
- 21.Luo J, Miller MW. Ethanol enhances erbB-mediated migration of human breast cancer cells in culture. Breast Cancer Res Treat. 2000;63:61–69. doi: 10.1023/A:1006436315284. [DOI] [PubMed] [Google Scholar]
- 22.Izevbigie EB, Ekunwe SI, Jordan J, Howard CB. Ethanol modulates the growth of human breast cancer cells in vitro. Exp Biol Med (Maywood) 2002;227:260–265. doi: 10.1177/153537020222700406. [DOI] [PubMed] [Google Scholar]
- 23.Etique N, Chardard D, Chesnel A, Flament S, Grillier-Vuissoz I. Analysis of the effects of different alcohols on MCF-7 human breast cancer cells. Ann NY Acad Sci. 2004;1030:78–85. doi: 10.1196/annals.1329.010. [DOI] [PubMed] [Google Scholar]
- 24.Etique N, Grillier-Vuissoz I, Flament S. Ethanol stimulates the secretion of matrix metalloproteinases 2 and 9 in MCF-7 human breast cancer cells. Oncol Rep. 2006;15:603–608. [PubMed] [Google Scholar]
- 25.Verma M, Davidson EA. MUC1 upregulation by ethanol. Cancer Biochem Biophys. 1999;17:1–11. [PubMed] [Google Scholar]
- 26.Ma C, Lin H, Leonard SS, Shi X, Ye J, Luo J. Overexpression of ErbB2 enhances ethanol-stimulated intracellular signaling and invasion of human mammary epithelial and breast cancer cells in vitro. Oncogene. 2003;22:5281–5290. doi: 10.1038/sj.onc.1206675. [DOI] [PubMed] [Google Scholar]
- 27.Aye MM, Ma C, Lin H, Bower KA, Wiggins RC, Luo J. Ethanol-induced in vitro invasion of breast cancer cells: The contribution of MMP-2 by fibroblasts. Int J Cancer. 2004;112:738–746. doi: 10.1002/ijc.20497. [DOI] [PubMed] [Google Scholar]
- 28.Ke Z, Lin H, Fan Z, Cai TQ, Kaplan RA, Ma C, Bower KA, Shi X, Luo J. MMP-2 mediates ethanol-induced invasion of mammary epithelial cells over-expressing ErbB2. Int J Cancer. 2006;119:8–16. doi: 10.1002/ijc.21769. [DOI] [PubMed] [Google Scholar]
- 29.Cordes T, Diesing D, Becker S, Diedrich K, Reichrath J, Friedrich M. Modulation of MAPK ERK1 and ERK2 in VDR-positive and -negative breast cancer cell lines. Anticancer Res. 2006;26A:2749–2753. [PubMed] [Google Scholar]
- 30.Xu M, Bower KA, Wang S, Frank JA, Chen G, Ding M, Wang S, Shi X, Ke Z, Luo J. Cyanidin-3-glucoside inhibits ethanol-induced invasion of breast cancer cells overexpressing ErbB2. Mol Cancer. 2010;9:285. doi: 10.1186/1476-4598-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raouf A, Sun Y, Chatterjee S, Basak P. The biology of human breast epithelial progenitors. Semin Cell Dev Biol. 2012;23:606–612. doi: 10.1016/j.semcdb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Bruno RD, Smith GH. Role of epithelial stem/progenitor cells in mammary cancer. Gene Expr. 2011;15:133–140. doi: 10.3727/105221611X13176664479368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feifei N, Mingzhi Z, Yanyun Z, Huanle Z, Fang R, Mingzhu H, Mingzhi C, Yafei S, Fengchun Z. MicroRNA expression analysis of mammospheres cultured from human breast cancers. J Cancer Res Clin Oncol. 2012;138:1937–1944. doi: 10.1007/s00432-012-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie G, Zhan J, Tian Y, Liu Y, Chen Z, Ren C, Sun Q, Lian J, Chen L, Ruan J, et al. Mammosphere cells from high-passage MCF7 cell line show variable loss of tumorigenicity and radioresistance. Cancer Lett. 2012;316:53–61. doi: 10.1016/j.canlet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Nash R, Krishnamoorthy M, Jenkins A, Csete M. Human embryonic stem cell model of ethanol-mediated early developmental toxicity. Exp Neurol. 2012;234:127–135. doi: 10.1016/j.expneurol.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 37.Worley SL, Vaughn BJ, Terry AI, Gardiner CS, DeKrey GK. Time- and dose-dependent effects of ethanol on mouse embryonic stem cells. Reprod Toxicol. 2015;57:157–164. doi: 10.1016/j.reprotox.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krell J, Frampton AE, Jacob J, Castellano L, Stebbing J. miRNAs in breast cancer: Ready for real time? Pharmacogenomics. 2012;13:709–719. doi: 10.2217/pgs.12.15. [DOI] [PubMed] [Google Scholar]
- 40.Shore AN, Herschkowitz JI, Rosen JM. Noncoding RNAs involved in mammary gland development and tumorigenesis: There's a long way to go. J Mammary Gland Biol Neoplasia. 2012;17:43–58. doi: 10.1007/s10911-012-9247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valastyan S. Roles of microRNAs and other non-coding RNAs in breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2012;17:23–32. doi: 10.1007/s10911-012-9241-9. [DOI] [PubMed] [Google Scholar]
- 42.Guttilla IK, Adams BD, White BA. ERα, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol Metab. 2012;23:73–82. doi: 10.1016/j.tem.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Jain P, Alahari SK. Breast cancer stem cells: A new challenge for breast cancer treatment. Front Biosci (Landmark Ed) 2011;16:1824–1832. doi: 10.2741/3824. [DOI] [PubMed] [Google Scholar]
- 44.Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D. MicroRNAs: Master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A, Staloch D, McCarra J, Liu J, Venter J, et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804–817. doi: 10.1016/j.ajpath.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klinge CM. miRNAs and estrogen action. Trends Endocrinol Metab. 2012;23:223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res Treat. 2012;132:75–85. doi: 10.1007/s10549-011-1534-y. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Mertens-Talcott SU, Zhang S, Kim K, Ball J, Safe S. MicroRNA-27a indirectly regulates estrogen receptor {alpha} expression and hormone responsiveness in MCF-7 breast cancer cells. Endocrinology. 2010;151:2462–2473. doi: 10.1210/en.2009-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed TE, Kalant H, Gibbins RJ, Kapur BM, Rankin JG. Alcohol and acetaldehyde metabolism in Caucasians, Chinese and Amerinds. Can Med Assoc J. 1976;115:851–855. [PMC free article] [PubMed] [Google Scholar]
- 50.Tsang JY, Kwok YK, Chan KW, Ni YB, Chow WN, Lau KF, Shao MM, Chan SK, Tan PH, Tse GM. Expression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in breast cancers. Breast Cancer Res Treat. 2013;142:311–322. doi: 10.1007/s10549-013-2756-y. [DOI] [PubMed] [Google Scholar]
- 51.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Ono S, Ishizaki Y, Tokuda E, Tabata K, Asami S, Suzuki T. Different patterns in the induction of metallothionein mRNA synthesis among isoforms after acute ethanol administration. Biol Trace Elem Res. 2007;115:147–156. doi: 10.1007/BF02686026. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen MO, Larsen A, Stoltenberg M, Penkowa M. The role of metallothionein in oncogenesis and cancer prognosis. Prog Histochem Cytochem. 2009;44:29–64. doi: 10.1016/j.proghi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Ning MS, Kim AS, Prasad N, Levy SE, Zhang H, Andl T. Characterization of the Merkel Cell Carcinoma miRNome. J Skin Cancer. 2014;2014:289548. doi: 10.1155/2014/289548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojas F, Hernandez ME, Silva M, Li L, Subramanian S, Wilson MJ, Liu P. The oncogenic response to MiR-335 is associated with cell surface expression of membrane-type 1 matrix metalloproteinase (MT1-MMP) activity. PLoS One. 2015;10:e0132026. doi: 10.1371/journal.pone.0132026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y, Yang H, Yuan L, Liu G, Zhang C, Hong M, Liu Y, Zhou M, Chen F, Li X. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412:235–245. doi: 10.1007/s11010-015-2630-9. [DOI] [PubMed] [Google Scholar]
- 57.Saumet A, Vetter G, Bouttier M, Antoine E, Roubert C, Orsetti B, Theillet C, Lecellier CH. Estrogen and retinoic acid antagonistically regulate several microRNA genes to control aerobic glycolysis in breast cancer cells. Mol Biosyst. 2012;8:3242–3253. doi: 10.1039/c2mb25298h. [DOI] [PubMed] [Google Scholar]
- 58.Ruiz-Llorente L, Ardila-González S, Fanjul LF, Martínez-Iglesias O, Aranda A. microRNAs 424 and 503 are mediators of the anti-proliferative and anti-invasive action of the thyroid hormone receptor beta. Oncotarget. 2014;5:2918–2933. doi: 10.18632/oncotarget.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Zhang H, Zhang P, Li J, Shan Z, Teng W. Upregulation of miR-2861 and miR-451 expression in papillary thyroid carcinoma with lymph node metastasis. Med Oncol. 2013;30:577. doi: 10.1007/s12032-013-0577-9. [DOI] [PubMed] [Google Scholar]
- 60.Long C, Jiang L, Wei F, Ma C, Zhou H, Yang S, Liu X, Liu Z. Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS One. 2013;8:e66676. doi: 10.1371/journal.pone.0066676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakazawa K, Dashzeveg N, Yoshida K. Tumor suppressor p53 induces miR-1915 processing to inhibit Bcl-2 in the apoptotic response to DNA damage. FEBS J. 2014;281:2937–2944. doi: 10.1111/febs.12831. [DOI] [PubMed] [Google Scholar]
- 62.Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan KG, Yin WF, Satharasinghe DA, Liew WC, Tan SW, et al. MiRNA transcriptome profiling of spheroid-enriched cells with cancer stem cell properties in human breast MCF-7 cell line. Int J Biol Sci. 2016;12:427–445. doi: 10.7150/ijbs.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16:374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 64.Xu C, Zhang L, Li H, Liu Z, Duan L, Lu C. MiRNA-1469 promotes lung cancer cells apoptosis through targeting STAT5a. Am J Cancer Res. 2015;5:1180–1189. [PMC free article] [PubMed] [Google Scholar]
- 65.Yu CC, Chen YW, Chiou GY, Tsai LL, Huang PI, Chang CY, Tseng LM, Chiou SH, Yen SH, Chou MY, et al. MicroRNA let-7a represses chemoresistance and tumourigenicity in head and neck cancer via stem-like properties ablation. Oral Oncol. 2011;47:202–210. doi: 10.1016/j.oraloncology.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Molina-Pinelo S, Gutiérrez G, Pastor MD, Hergueta M, Moreno-Bueno G, García-Carbonero R, Nogal A, Suárez R, Salinas A, Pozo-Rodríguez F, et al. MicroRNA-dependent regulation of transcription in non-small cell lung cancer. PLoS One. 2014;9:e90524. doi: 10.1371/journal.pone.0090524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ogony JW, Malahias E, Vadigepalli R, Anni H. Ethanol alters the balance of Sox2, Oct4, and Nanog expression in distinct subpopulations during differentiation of embryonic stem cells. Stem Cells Dev. 2013;22:2196–2210. doi: 10.1089/scd.2012.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeineddine D, Hammoud AA, Mortada M, Boeuf H. The Oct4 protein: More than a magic stemness marker. Am J Stem Cells. 2014;3:74–82. [PMC free article] [PubMed] [Google Scholar]
- 69.Liu CG, Lu Y, Wang BB, Zhang YJ, Zhang RS, Lu Y, Chen B, Xu H, Jin F, Lu P. Clinical implications of stem cell gene Oct-4 expression in breast cancer. Ann Surg. 2011;253:1165–1171. doi: 10.1097/SLA.0b013e318214c54e. [DOI] [PubMed] [Google Scholar]
- 70.Liu C, Cao X, Zhang Y, Xu H, Zhang R, Wu Y, Lu P, Jin F. Co-expression of Oct-4 and Nestin in human breast cancers. Mol Biol Rep. 2012;39:5875–5881. doi: 10.1007/s11033-011-1398-6. [DOI] [PubMed] [Google Scholar]
- 71.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 72.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung JW, Park SB, Lee SJ, Seo MS, Trosko JE, Kang KS. Metformin represses self-renewal of the human breast carcinoma stem cells via inhibition of estrogen receptor-mediated OCT4 expression. PLoS One. 2011;6:e28068. doi: 10.1371/journal.pone.0028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu J, Qin K, Zhang Y, Gong J, Li N, Lv D, Xiang R, Tan X. Downregulation of transcription factor Oct4 induces an epithelial-to-mesenchymal transition via enhancement of Ca2+ influx in breast cancer cells. Biochem Biophys Res Commun. 2011;411:786–791. doi: 10.1016/j.bbrc.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 75.Trosko JE. From adult stem cells to cancer stem cells: Oct-4 Gene, cell-cell communication, and hormones during tumor promotion. Ann NY Acad Sci. 2006;1089:36–58. doi: 10.1196/annals.1386.018. [DOI] [PubMed] [Google Scholar]
- 76.Wu F, Zhang J, Wang P, Ye X, Jung K, Bone KM, Pearson JD, Ingham RJ, McMullen TP, Ma Y, et al. Identification of two novel phenotypically distinct breast cancer cell subsets based on Sox2 transcription activity. Cell Signal. 2012;24:1989–1998. doi: 10.1016/j.cellsig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 77.Nagata T, Shimada Y, Sekine S, Hori R, Matsui K, Okumura T, Sawada S, Fukuoka J, Tsukada K. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer. 2014;21:96–101. doi: 10.1007/s12282-012-0357-y. [DOI] [PubMed] [Google Scholar]
- 78.Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH, et al. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci USA. 2009;106:1548–1553. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis-Wambi JS, Cunliffe HE, Kim HR, Willis AL, Jordan VC. Overexpression of CEACAM6 promotes migration and invasion of oestrogen-deprived breast cancer cells. Eur J Cancer. 2008;44:1770–1779. doi: 10.1016/j.ejca.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ihnen M, Kilic E, Köhler N, Löning T, Witzel I, Hagel C, Höller S, Kersten JF, Müller V, Jänicke F, et al. Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. J Clin Pathol. 2011;64:146–152. doi: 10.1136/jcp.2010.082602. [DOI] [PubMed] [Google Scholar]
- 81.Maraqa L, Cummings M, Peter MB, Shaaban AM, Horgan K, Hanby AM, Speirs V. Carcinoembryonic antigen cell adhesion molecule 6 predicts breast cancer recurrence following adjuvant tamoxifen. Clin Cancer Res. 2008;14:405–411. doi: 10.1158/1078-0432.CCR-07-1363. [DOI] [PubMed] [Google Scholar]
- 82.Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomes IM, Maia CJ, Santos CR. STEAP proteins: From structure to applications in cancer therapy. Mol Cancer Res. 2012;10:573–587. doi: 10.1158/1541-7786.MCR-11-0281. [DOI] [PubMed] [Google Scholar]
- 84.Sano H, Wada S, Eguchi H, Osaki A, Saeki T, Nishiyama M. Quantitative prediction of tumor response to neoadjuvant chemotherapy in breast cancer: Novel marker genes and prediction model using the expression levels. Breast Cancer. 2012;19:37–45. doi: 10.1007/s12282-011-0263-8. [DOI] [PubMed] [Google Scholar]
- 85.Li CF, MacDonald JR, Wei RY, Ray J, Lau K, Kandel C, Koffman R, Bell S, Scherer SW, Alman BA. Human sterile alpha motif domain 9, a novel gene identified as down-regulated in aggressive fibromatosis, is absent in the mouse. BMC Genomics. 2007;8:92. doi: 10.1186/1471-2164-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stone A, Valdés-Mora F, Gee JM, Farrow L, McClelland RA, Fiegl H, Dutkowski C, McCloy RA, Sutherland RL, Musgrove EA, et al. Tamoxifen-induced epigenetic silencing of oestrogen-regulated genes in anti-hormone resistant breast cancer. PLoS One. 2012;7:e40466. doi: 10.1371/journal.pone.0040466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cimino D, Fuso L, Sfiligoi C, Biglia N, Ponzone R, Maggiorotto F, Russo G, Cicatiello L, Weisz A, Taverna D, et al. Identification of new genes associated with breast cancer progression by gene expression analysis of predefined sets of neoplastic tissues. Int J Cancer. 2008;123:1327–1338. doi: 10.1002/ijc.23660. [DOI] [PubMed] [Google Scholar]
- 88.Ito Y, Motoo Y, Yoshida H, Iovanna JL, Takamura Y, Miya A, Kuma K, Miyauchi A. Decreased expression of tumor protein p53-induced nuclear protein 1 (TP53INP1) in breast carcinoma. Anticancer Res. 2006;26B:4391–4395. [PubMed] [Google Scholar]
- 89.Sorbello V, Fuso L, Sfiligoi C, Scafoglio C, Ponzone R, Biglia N, Weisz A, Sismondi P, De Bortoli M. Quantitative real-time RT-PCR analysis of eight novel estrogen-regulated genes in breast cancer. Int J Biol Markers. 2003;18:123–129. doi: 10.1177/172460080301800205. [DOI] [PubMed] [Google Scholar]
- 90.He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, Li K, Liu J, Chen J. HLA-G expression in human breast cancer: Implications for diagnosis and prognosis, and effect on allocytotoxic lymphocyte response after hormone treatment in vitro. Ann Surg Oncol. 2010;17:1459–1469. doi: 10.1245/s10434-009-0891-9. [DOI] [PubMed] [Google Scholar]
- 91.Jia Y, Liu H, Zhuang Q, Xu S, Yang Z, Li J, Lou J, Zhang W. Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol Rep. 2012;27:1865–1872. doi: 10.3892/or.2012.1701. [DOI] [PubMed] [Google Scholar]
- 92.Yin R, Zhang S, Wu Y, Fan X, Jiang F, Zhang Z, Feng D, Guo X, Xu L. MicroRNA-145 suppresses lung adenocarcinoma-initiating cell proliferation by targeting OCT4. Oncol Rep. 2011;25:1747–1754. doi: 10.3892/or.2011.1252. [DOI] [PubMed] [Google Scholar]
- 93.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 94.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cicatiello L, Mutarelli M, Grober OM, Paris O, Ferraro L, Ravo M, Tarallo R, Luo S, Schroth GP, Seifert M, et al. Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am J Pathol. 2010;176:2113–2130. doi: 10.2353/ajpath.2010.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ali HO, Arroyo AB, González-Conejero R, Stavik B, Iversen N, Sandset PM, Martínez C, Skretting G. The role of microRNA-27a/b and microRNA-494 in estrogen-mediated downregulation of tissue factor pathway inhibitor α. J Thromb Haemost. 2016;14:1226–1237. doi: 10.1111/jth.13321. [DOI] [PubMed] [Google Scholar]
- 97.Lee YM, Lee JY, Ho CC, Hong QS, Yu SL, Tzeng CR, Yang PC, Chen HW. miRNA-34b as a tumor suppressor in estrogen-dependent growth of breast cancer cells. Breast Cancer Res. 2011;13:R116. doi: 10.1186/bcr3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tan S, Ding K, Li R, Zhang W, Li G, Kong X, Qian P, Lobie PE, Zhu T. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res. 2014;16:R40. doi: 10.1186/bcr3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li YQ, Yan TB, Sun XG, Hu P, Zhang TC. Estrogen receptor α mediates proliferation of breast cancer MCF-7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J. 2014;281:927–942. doi: 10.1111/febs.12658. [DOI] [PubMed] [Google Scholar]
- 101.Zhang C, Zhao J, Deng H. 17β-estradiol up-regulates miR-155 expression and reduces TP53INP1 expression in MCF-7 breast cancer cells. Mol Cell Biochem. 2013;379:201–211. doi: 10.1007/s11010-013-1642-6. [DOI] [PubMed] [Google Scholar]
- 102.Masuda M, Miki Y, Hata S, Takagi K, Sakurai M, Ono K, Suzuki K, Yang Y, Abe E, Hirakawa H, et al. An induction of microRNA, miR-7 through estrogen treatment in breast carcinoma. J Transl Med. 2012;10(Suppl 1):S2. doi: 10.1186/1479-5876-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manavalan TT, Teng Y, Litchfield LM, Muluhngwi P, Al-Rayyan N, Klinge CM. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLoS One. 2013;8:e62334. doi: 10.1371/journal.pone.0062334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu Y, Xiao CH, Tan LD, Wang QS, Li XQ, Feng YM. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br J Cancer. 2014;110:724–732. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS, Yiu CC, Loo TY, Nakamura Y, Akahira J, Ishida T, et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J Pathol. 2012;227:357–366. doi: 10.1002/path.4019. [DOI] [PubMed] [Google Scholar]
- 106.Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012;12:29. doi: 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, Jones FE. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quann K, Jing Y, Rigoutsos I. Post-transcriptional regulation of BRCA1 through its coding sequence by the miR-15/107 group of miRNAs. Front Genet. 2015;6:242. doi: 10.3389/fgene.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fu XM, Zhou YZ, Cheng Z, Liao XB, Zhou XM. MicroRNAs: Novel players in aortic aneurysm. Biomed Res Int. 2015;2015:831641. doi: 10.1155/2015/831641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie LX. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin Chem Lab Med. 2012;50:1423–1428. doi: 10.1515/cclm-2011-0826. [DOI] [PubMed] [Google Scholar]
- 113.de Gonzalo-Calvo D, Davalos A, Montero A, Garcia-Gonzalez A, Tyshkovska I, Gonzalez-Medina A, Soares SM, Martínez-Camblor P, Casas-Agustench P, Rabadán M, et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol. 1985;119:124–134. doi: 10.1152/japplphysiol.00077.2015. [DOI] [PubMed] [Google Scholar]
- 114.Rodosthenous RS, Coull BA, Lu Q, Vokonas PS, Schwartz JD, Baccarelli AA. Ambient particulate matter and microRNAs in extracellular vesicles: A pilot study of older individuals. Part Fibre Toxicol. 2016;13:13. doi: 10.1186/s12989-016-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao D, Chen G, LiD, Wang X, Cao H, et al. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res Treat. 2015;154:423–434. doi: 10.1007/s10549-015-3591-0. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Y, Zhang D, Wang F, Xu D, Guo Y, Cui W. Serum miRNAs panel (miR-16-2*, miR-195, miR-2861, miR-497) as novel non-invasive biomarkers for detection of cervical cancer. Sci Rep. 2015;5:17942. doi: 10.1038/srep17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao Q, Deng S, Wang G, Liu C, Meng L, Qiao S, Shen L, Zhang Y, Lü J, Li W, et al. A direct quantification method for measuring plasma MicroRNAs identified potential biomarkers for detecting metastatic breast cancer. Oncotarget. 2016;7:21865–21874. doi: 10.18632/oncotarget.7990. [DOI] [PMC free article] [PubMed] [Google Scholar]