Abstract
While protein glycosylation has been reported in several spirochetes including the syphilis bacterium Treponema pallidum and Lyme disease pathogen Borrelia burgdorferi, the pertinent glycan structures and their roles remain uncharacterized. Herein, we report a novel glycan with an unusual chemical composition and structure in the oral spirochete Treponema denticola, a keystone pathogen of periodontitis. The identified glycan of mass 450.2 Da is composed of a monoacetylated nonulosonic acid (Non) with a novel extended N7 acyl modification, a 2-methoxy-4,5,6-trihydroxy-hexanoyl residue in which the Non has a pseudaminic acid configuration (L-glycero-L-manno) and is β-linked to serine or threonine residues. This novel glycan modifies the flagellin proteins (FlaBs) of T. denticola by O-linkage at multiple sites near the D1 domain, a highly conserved region of bacterial flagellins that interact with Toll-like receptor 5. Furthermore, mutagenesis studies demonstrate that the glycosylation plays an essential role in the flagellar assembly and motility of T. denticola. To our knowledge, this novel glycan and its unique modification sites have not been reported previously in any bacteria.
Keywords: Spirochete, Treponema denticola, flagella, glycosylation
Graphical abstract
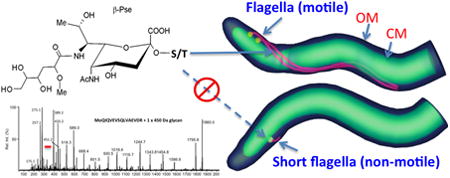
Introduction
Spirochetes are a group of medically important but poorly understood bacteria, some of which cause human illnesses such as Lyme disease (Borrelia burgdorferi), leptospirosis (Leptospira interrogans), and syphilis (Treponema pallidum) [for recent reviews, see references (Radolf et al., 2012, Giacani & Lukehart, 2014, Evangelista & Coburn, 2010)]. Spirochetes are recognized for their distinct cell morphology and unusual means of motility - they are either flat-waved or corkscrew-shaped and swim by means of rotating flagella that are located between the outer membrane and the peptidoglycan layer (Li et al., 2000b, Charon et al., 2012). Thus, spirochetal flagella are referred to as endoflagella or periplasmic flagella (PFs). Several studies have shown that the PFs are not only essential for cell motility and morphology but are also directly linked to the pathogenicity of spirochetes [for recent review, see reference (Charon et al., 2012)]. For example, aflagellated mutants of B. burgdorferi are non-motile, rod-shaped instead of flat-waved, and unable to establish infection in mice (Li et al., 2010, Sultan et al., 2013, Sultan et al., 2015, Li et al., 2000b, Lin et al., 2015). Flagella-deficient mutants of L. interrogans are non-motile, lack the distinctive hook-shaped ends, and are unable to cause infection in hamsters, a standard experimental model of leptospirosis (Lambert et al., 2012, Wunder et al., 2016, Fontana et al., 2016).
PFs are structurally similar to the flagella of other bacteria, as each consists of a basal body-motor complex, a hook, and a filament (Charon et al., 2012, Zhao et al., 2014). The filament is a long, thin, helical structure that functions as a propeller and is essential for bacterial locomotion (Berg & Anderson, 1973). In most bacteria, flagellar filaments consist of a single polymeric protein known as flagellin (FliC in Escherichia coli and Salmonella enterica) (Samatey et al., 2001). However, the PF filament has an unusual structure and protein composition, and it is one of the most complex bacterial flagellar filaments thus far analyzed [for review, see references (Li et al., 2000b, Charon et al., 1992)]. Specifically, in most spirochete species, the PFs are comprised of at least one sheath protein (FlaA) and one to three core proteins (FlaB1-3). The FlaA proteins (ranging in size from 37 to 44 kDa) are similar between species, based on their amino acid sequences and antigenic cross-reactivity (Norris et al., 1988, Norris, 1993). The FlaA proteins are likely exported to the periplasmic space via the type II secretion pathway, as their N-termini contain a typical peptidase I cleavage site (Pugsley, 1993). The FlaA proteins form a sheath around the core proteins, determine the diameter and helicity of the PFs, and are important for cell motility (Li et al., 2000a, Li et al., 2008). In contrast, the FlaB proteins are exported to the periplasmic space through the flagellar type III secretion apparatus (Erhardt et al., 2010). The FlaB proteins are generally 30 to 41 kDa in size and immunologically cross-reactive (both within a given species and also between species) (Norris et al., 1988, Norris, 1993). For spirochetes that have multiple core proteins, such as Brachyspira hyodysenteriae (the causative agent of swine dysentery) (Koopman et al., 1992), the function of FlaB proteins overlaps such that neither one of these proteins is essential for the intact PF structure and cell motility (Li et al., 2000a, Li et al., 2008). The FlaB proteins form a core structure that is sheathed by FlaA. There is no sequence similarity or antigenic cross-reactivity between the FlaA and FlaB proteins (Norris et al., 1988, Norris, 1993).
The genus Treponema includes both commensal and pathogenic spirochetes, some of which cause human illnesses such as yaws, pinta, and syphilis (Giacani & Lukehart, 2014, Radolf, 1996). Certain Treponema species are also implicated in polymicrobial infections in humans and animals, such as periodontitis and bovine digital dermatitis [for reviews, see (Evans et al., 2016, Giacani & Lukehart, 2014)]. Since a majority of treponemal species cannot be reliably cultivated in vitro, the oral spirochete Treponema denticola, which can be cultivated, has been used as a model organism to understand the biology and pathogenicity of the treponemes (Anand et al., 2013, Chi et al., 1999, Bian et al., 2015). T. denticola is an obligatory anaerobic, facultative, and highly motile bacterium, which is implicated in both periodontal and endodontic infections [for review, see references (Ellen & Galimanas, 2005, Dashper et al., 2011)]. As revealed by electron microscopy and cryo-electron tomography (cryo-ET), T. denticola has two PFs that arise from each end of the cell and extend toward the center of the cell where they overlap (Izard et al., 2008, Izard et al., 2009). Similar to those in other spirochetes, the T. denticola PFs are essential for cell shape and motility; flagella-deficient mutants are non-motile and less helical at the regions where the PFs interact with the cell cylinder (Li et al., 1996, Ruby et al., 1997, Limberger et al., 1999). Motility also contributes to the pathogenicity of T. denticola; non-flagellated mutants are unable to penetrate oral epithelial cells (Lux et al., 2001).
The PF filament of T. denticola consists of at least one sheath protein, FlaA, and three core proteins designated as FlaB1, FlaB2, and FlaB3 (Ruby et al., 1997). These flagellin proteins share a high sequence similarity (> 67% identity) to their counterparts in T. pallidum (Norris et al., 1988, Norris, 1993). Wyss et al. reported that the flagellin proteins of T. pallidum and pathogenic oral Treponema spp. are glycosylated (Wyss, 1998). Flagellar glycosylation was also reported in other spirochetes, including B. hyodysenteriae (Li et al., 1993) and Spirochaeta aurantia (Brahamsha & Greenberg, 1988). However, their chemical features, structures, and roles in motility and flagellation remain uncharacterized.
Flagella glycosylation has been demonstrated in numerous motile bacteria and archaea and it has been found that such a post-translational modification affects flagellar assembly, motility, as well as virulence in some pathogenic bacteria [for reviews, see references (Logan, 2006, Merino & Tomas, 2014)]. For instance, the flagellins of Campylobacter jejuni and Campylobacter coli are glycosylated with O-linked pseudaminic acid (Pse) or its acetamidino derivative PseAm (Logan et al., 2002). Mutant strains which could no longer synthesize Pse were unable to assemble intact flagella, were non-motile and less virulent (Howard et al., 2009, Guerry et al., 2006). Similar to Campylobacter, the flagellins of Helicobacter pylori are also modified with Pse and the glycan modification is also essential for flagellar assembly, motility, and virulence (Josenhans et al., 2002, Schirm et al., 2003). In this study, we used genetic, biochemical, and structural approaches to elucidate the structure of a novel glycan that modifies T. denticola flagellins. We demonstrate that glycosylation is required for flagellar assembly and consequent motility.
Results
Analysis of T. denticola PFs
The genome of T. denticola encodes six putative flagellin proteins – three FlaA (TDE1408, 236 aa; TDE1409, 246 aa; and TDE1712, 349 aa) and three FlaB proteins (TDE1004, 286 aa; TDE1475, 285 aa; and TDE1477, 286 aa) (Seshadri et al., 2004). However, Ruby et al., found only four of these proteins (one FlaA and three FlaBs) in the isolated flagellar filaments of T. denticola (Ruby et al., 1997). To address this discrepancy, the PFs were isolated from the wild-type strain and analyzed in detail. Four protein bands were visualized by SDS-PAGE (Fig. 1A). The top band (approximate 37.0 kDa) reacted to T. denticola FlaA antiserum and the lower bands (around 31.0 kDa) were recognized by the T. pallidum FlaB antibody. Of note, the size of FlaA (TDE1712) was smaller than its predicted molecular weight (MW, 39.3 kDa), suggesting that it is posttranslationally processed prior to assembly. Along with this prediction, a 22 aa signal peptide was identified at its N-terminus (Fig. S1). The MWs of the three FlaB proteins are very close to each other and could not be fully separated by 1D SDS-PAGE (Fig. 1A). To decipher the filament protein composition, the isolated PFs were separated by 2D-gel electrophoresis. In addition to FlaA, three FlaB proteins were clearly visualized on the 2D gels (Fig. 1B) and could be detected by T. pallidum FlaB antiserum (Fig. 1C). These results indicate that the flagellar filament of T. denticola comprises one FlaA and three FlaB proteins, which corroborates the previous report (Ruby et al., 1997). Based on their sequences, MWs, and isoelectric points (pIs), these filament proteins were assigned: FlaA (TDE1712, pI 5.39, 39.3 kDa), FlaB1 (TDE1477, pI 5.4, 31.3 kDa), FlaB2 (TDE1004, pI 6.54, 31.6 kDa), and FlaB3 (TDE1475, pI 5.3, 30.9 kDa).
Fig. 1. The flagellin proteins of T. denticola are glycosylated.

(A) SDS-PAGE (left panel) and immunoblotting (right panel) analyses of isolated PFs from the wild-type strain. 2-D electrophoresis (B) and immunoblotting (C) analyses of the isolated PFs from the wild-type strain. Purified PFs from the wild-type strain were analyzed using 2D-gel electrophoresis, followed by glycosylation staining (GS) (D) and lectin blots with ConA (E) and LFA (F). For the immunoblotting, the blots were probed with the T. pallidum FlaB antiserum (αFlaB) and T. denticola FlaA antiserum (αFlaA). For the lectin blots, the blots were probed with either ConA (Concanavalin A) or LFA (Limax Favus Agglutinin). A (FlaA); B1 (FlaB1); B2 (FlaB2); and B3 (FlaB3).
The FlaB proteins of T. denticola are glycosylated
Wyss previously reported that the flagellar proteins of pathogenic Treponema species are glycosylated (Wyss, 1998). To determine if this is the case for T. denticola, the PFs were isolated from the wild-type strain and analyzed by SDS-PAGE and 2D-gel electrophoresis followed by glycosylation staining as well as lectin blotting analyses with ConA and LFA. The three FlaBs, but not FlaA, stained positive for glycosylation (Fig. 1D). In addition, FlaB1 and FlaB2 reacted with both ConA and LFA (Fig. 1E, F). LFA is a lectin that specifically recognizes sialic acid-like sugar moieties (Miller et al., 1982), suggesting that the FlaB proteins may be modified with sialic acid-like sugars. Of note, FlaB3 was not visibly reactive with ConA and LFA (Fig. 1E, F), but this may be due to its relatively low level in the PFs and the sensitivity of the lectin blotting analysis. We used densitometry analysis to measure the level of FlaB3 relative to the other three flagellin proteins and found that the average stoichiometry of these four proteins is: FlaA (5.0): FlaB1 (3.5): FlaB2 (3.5): FlaB3 (1.0).
To confirm that the observed glycosylated products are indeed the filament core proteins, the genes encoding the three FlaBs were deleted by allelic exchange mutagenesis as illustrated in Fig. S2. The PFs were isolated from these individual mutants (ΔflaB1, ΔflaB2, and ΔflaB3) and subjected to 2D-gel electrophoresis, followed by Coomassie blue and glycosylation staining. As shown in Fig. 2, the cognate gene products were abolished in each of these mutants (Fig. 2A-C), and the pertinent glycosylated products were absent, as shown by glycosylation staining (Fig. 2D-F). For example, the glycosylated product tentatively assigned as FlaB1 was not detected in the PFs isolated from the ΔflaB1 mutant. The same results were observed in the ΔflaB2 and ΔflaB3 mutants. Based on these results, we concluded that the three filament core proteins (FlaB1, FlaB2, and FlaB3) are glycosylated. This deletion mutagenesis analysis also confirms the gene assignment of the observed PF proteins (e.g., FlaB1 is encoded by TDE1477).
Fig. 2. The three filament core proteins are glycosylated.

The PFs isolated from three flaB deletion mutants (ΔflaB1, ΔflaB2, and ΔflaB3) were separated by 2D-gel electrophoresis, followed by Coomassie staining (A, B, C) and glycosylation staining (D, E, F). A (FlaA); B1 (FlaB1); B2 (FlaB2); and B3 (FlaB3).
The FlaB proteins are O-glycosylated
Protein glycosylation occurs as either O-linkage or N-linkage. Some N-linked glycoproteins can be deglycosylated with PNGase F (Kim & Leahy, 2013) and O-linked glycans can be chemically removed through β-elimination with alkaline (Peter-Katalinic, 2005). To determine the glycan linkage in the FlaB proteins, deglycosylation assays with either PNGase F or by β-elimination were carried out using the PFs isolated from the ΔflaA mutant to avoid any potential influence of the outer sheath during the treatments. PNGase F treatments under different conditions had no evident impact on the flagellin glycosylation (Fig. S3A). In contrast, β-elimination appeared to remove the glycoreactive moieties so that no glycosylated FlaB proteins were detected by glycosylation staining (lower panel, Fig. S3B). Of note, after the treatment, the FlaB proteins were moderately degraded; whereas a non-specific protein associated with the PFs remained unaffected (top band, Fig. S3B), suggesting that glycosylation may affect the stability of FlaB within the assembled filaments. Nevertheless, these degraded products were still reactive with the FlaB antiserum (middle panel, Fig. S3B). Collectively, these results suggest that the FlaB proteins are modified with O-linked glycans.
Mass spectrometry (MS) analysis of the flagellin proteins
As shown in Fig. 1, the MWs of three FlaB proteins are very similar. In order to separate them and prepare pure FlaB proteins for LC-MS/MS analysis, two double mutants were constructed ΔflaAflaB1 and ΔflaAflaB2, which allowed us to prepare the pure FlaB1 from ΔflaAflaB2 and FlaB2 and FlaB3 proteins from ΔflaAflaB1 (Fig. S3). The FlaB proteins were separated by SDS-PAGE, excised, digested with trypsin and then analyzed by using nLC-MS/MS, as previously described (Logan et al., 2002, Schoenhofen et al., 2006a). An initial MASCOT database search was conducted and many of the MS/MS spectra were assigned to unmodified tryptic peptides from all three of the FlaB proteins. Manual inspection of the fragment ion patterns in the remaining, unassigned MS/MS spectra identified 3-4 peptides from each of the FlaB proteins, all of which are modified with a novel 450.2 Da moiety (m/z 451.2 in Fig. 3 and Table 1). Moreover, many of the fragment ions in the lower region of the MS/MS spectra arise from the 450.2 Da modification (highlighted with a “◆” in Fig. 3c) and some have been observed previously in the MS/MS fragment ion spectra of the nonulosonic acids (i.e., pseudaminic acid and legionaminic acid) that decorate the flagellin of many bacteria. Therefore, it was hypothesized that the 450.2 Da moiety is a modified nonulosonic acid glycan. All of the identified glycopeptides contain a serine (S) and/or a threonine (T) residue (insets in Fig. 3 and Table 1) and have no N-linkage sequon (NXS or NXT, where X is any amino acid other than proline) (Rao & Bernd, 2010), indicative of O-linked glycosylation. Furthermore, no evidence of the presence of any other glycan modification was found. Also, of note, glycosylation staining suggested that the sheath protein FlaA is not glycosylated (Fig. 1D). To confirm this observation, the FlaA band on the SDS-PAGE gels was excised, digested with trypsin, and then analyzed by nLC-MS/MS as described above. All MS/MS spectra were assigned to tryptic peptides from the FlaA protein sequence, and no evidence of glycosylation was observed (Fig. S5).
Fig. 3. Representative glycopeptide nLC-MS/MS spectra from three FlaB proteins, each modified with the 450.2 Da glycan.
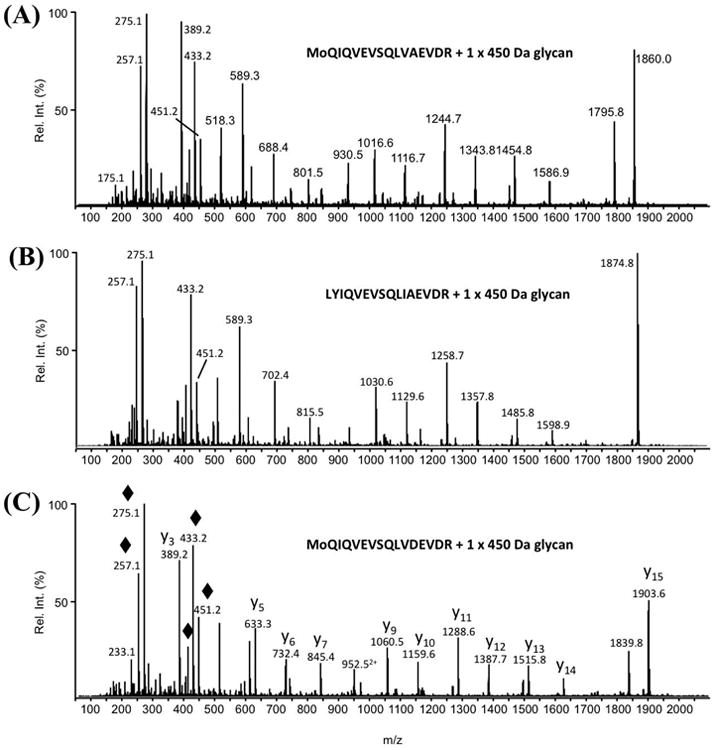
(A) MS/MS spectrum of MH22+ ion at m/z 1155.6 from T108-123 of FlaB1, (B) MS/MS spectrum of MH22+ ion at m/z 1163.1 from T108-123 of FlaB2, and (C) MS/MS spectrum of MH22+ ion at m/z 1177.6 from T108-123 from FlaB3. The amino acid sequences of the three glycopeptides are presented in the insets. “Mo” indicates the presence of oxidized methionine. By way of example, the major peptide fragment ions are identified in the MS/MS spectrum presented in panel c (“y” fragment ions start at the C-terminal amino acids). The mass differences between the peptide fragment ions were used to determine the amino acid sequence of this glycopeptide. The lower region of each MS/MS spectrum is dominated by the m/z 451.2 glycan oxonium ion and its related fragment ions (indicated with ◆ in panel c) that arose from multiple losses of water as well as the loss of the hexanoyl moiety.
Table 1.
Glycopeptides identified by nLC-MS/MS analysis of T. denticola flagellin tryptic digests.
| Protein | Amino acid seq. | Amino acid # | # of 450.2 Da modifications |
|---|---|---|---|
| FlaB1 | MQIQVEVSQLVAEVDR | 108-123 | 1 |
| IASSAQFNGMNMLTGR | 124-139 | 2 | |
| ETGENVVTGSMWFHIGANMDQR | 143-164 | 1 | |
| VYIGTMSAAALGIR | 167-180 | 1 | |
| FlaB2 | LYIQVEVSQLIAEVDR | 108-123 | 1 |
| IASHAQFNGMNMLTGR | 124-139 | 1,2 | |
| FAQETGENTVTASMWFHIGANMDQR | 140-164 | 1 | |
| AYIGTMTAK | 167-175 | 1 | |
| FlaB3* | MQIQVEVSQLVDEVDR | 108-123 | 1 |
| IASHAQFNGMNILTGR | 124-139 | 1,2 | |
| IYIGTMTATALGIIGAQQGGEDK | 163-185 | 1 |
Underlined sequences from FlaB1 and FlaB3 were isolated for NMR analysis.
The FlaB3 tryptic peptide, T140-160 (FAQDSVTGPMQLHVGANMDQR), was detected only as a non-glycosylated peptide; whereas the equivalent tryptic peptides from FlaB1 and FlaB2 were detected in glycosylated form only.
The distinct glycan fragment ion pattern in the glycopeptide LC-MS/MS spectra indicated that the 450.2 Da glycan consists of two components – a 274 Da sugar thought to be a monoacetylated nonulosonic acid (the m/z 275.1 oxonium ion in Fig. 3A-C) and an unknown 176 Da component. High-resolution mass spectrometry (HRMS) analysis performed with the LTQ-Orbitrap XL (Fig. 4) determined that the 274 Da sugar has the same mass as a monoacetylated nonulosonic acid (calculated m/z value for oxonium ion: m/z 275.1243; observed m/z value: m/z 275.1233 ± 0.0004). HRMS also indicated that the other component has an elemental composition of C7O5H12 (observed mass 176.0688 ± 0.0004 Da), which ruled out the possibility that it could be an uronic acid (calc. mass 176.0321 Da).
Fig. 4. High-resolution mass spectrometry analysis of the 450.2 Da glycan.
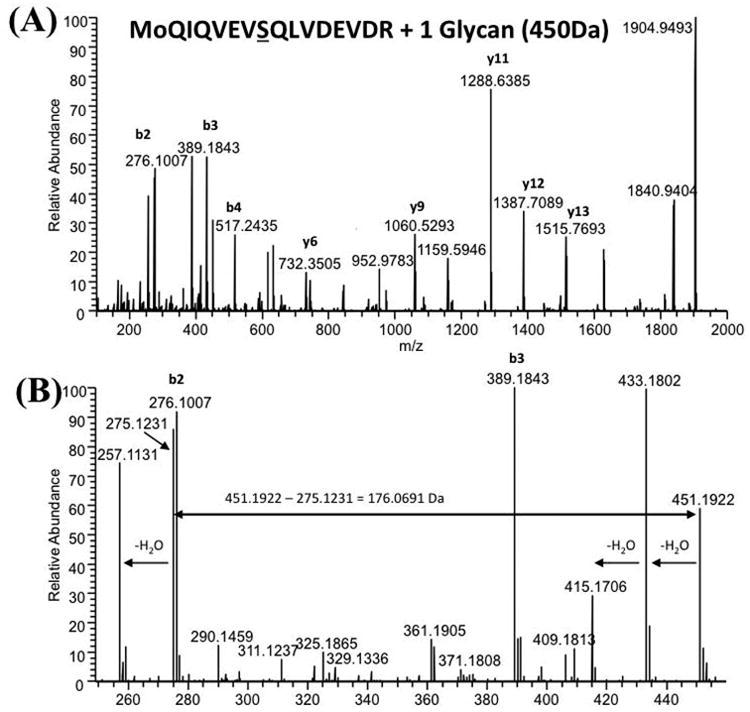
nLC-HCD MS/MS analysis of the T108-123 (see sequence in Table 1) FlaB3 glycopeptide ion at m/z 1177.6: (A) full m/z range and (B) zoomed-in area showing the glycan oxonium ions plus neighboring peptide fragment ions. The best fitting elemental formulae for the m/z 451.1922 and m/z 275.1231 were C18O11H31N2, C11O6H19N2, respectively. The mass of the second glycan component was determined by subtracting the nonulosonate fragment ion from the intact glycan ion (m/z 451.1922 – m/z 275.1231 = 176.0691 Da), and the best fitting elemental composition was C7O5H12.
NMR structural analysis of flagellin glycopeptides
To determine the structure of 450.2 Da glycan, 25 mg of PFs was isolated from the ΔflaA mutant, digested with proteinase K, and fractionated on a Biogel P10 column. 1H NMR spectra were recorded for all fractions. The fractions containing glycopeptides were further fractionated on a Zorbax C18 column and then analyzed by 1H NMR to identify glycopeptide-containing sub-fractions. Four glycopeptide containing sub-fractions were selected for 2D NMR analysis. Each sub-fraction contained the same sugar component, but they differed in terms of peptide compositions. NMR data for one of the sub-fractions (fraction #32) are presented in Fig. 5A and Table 2. Amino acids observed in this sub-fraction correspond to the peptide sequence of VSQLV found in FlaB3 (TDE1475) and FlaB1 (TDE1477) (Table 1). In addition to the signals of amino acids, the spectra contained spin systems of a 5,7-diamino-3,5,7,9-tetradeoxy-non-2-ulopyranosonic acid (Non), an acetyl group, and a component that we designated as ‘A’. The configuration of the pyranose ring of Non is identical to that of β-pseudaminic acid, as confirmed by signal positions and coupling constants of the structure's atoms matching what was previously reported (Knirel et al., 2003). An observed coupling constant J6,7 of 11 Hz was also characteristic of the Pse, and thus C-7 had the same configuration in Non as in Pse. The configuration of C-8 was not clear from NMR data. The position of the Non C-9 signal, which allows discrimination between Pse and 8-epi-pseudaminic acid (8-epi-Pse), was right in the middle between the signal positions of both isomers. The H-8 signal was quite far from that observed for Pse and closer to that reported for 8-epi-Pse (observed 4.38 ppm, 4.18 ppm in Pse, 4.25 ppm in 8-epi-Pse), although it should be acknowledged that the presence of a bulky acyl group on N-7 (see below) could significantly influence the NMR spectra. As a consequence, we have chosen to present the structure of the flagellin glycan in a Pse configuration (Fig. 5B) but do acknowledge the possibility that it may be 8-epi-Pse.
Fig. 5. 1H-13C HSQC spectrum of the glycopeptide from T. denticola and its 1H NMR spectrum.
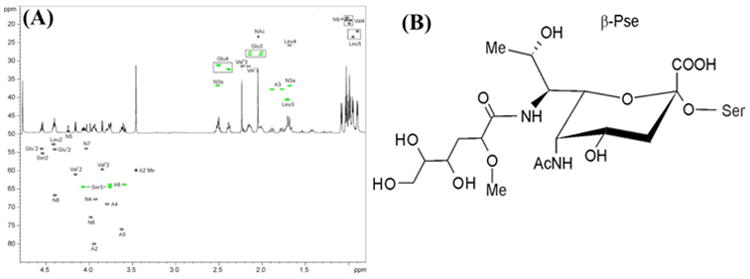
(A) CH signals are black, and CH2 are green. Nonulosonic acid signals are labeled as N. (B) Structure of the flagellar glycan with Pse configuration (L-glycero-L-manno) of the nonulosonic acid residue β-linked to serine (NB C-8 may have the 8-epi-Pse configuration).
Table 2.
NMR data for a flagellin glycopeptide VSQLV (fraction 32) from T. denticola. NAc: 175.9; 2.04; 23.4; Me: 3.46/59.8 ppm.
| Fr. 32 | H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 | H/C-7 | H/C-8 | H/C-9 |
|---|---|---|---|---|---|---|---|---|---|
| A | 3.94 | 1.77; 1.88 | 3.79 | 3.62 | 3.60; 3.76 | ||||
| 80.0 | 37.7 | 69.1 | 76.0 | 63.7 | |||||
| Pse | 1.68; 2.52 | 3.93 | 4.24 | 3.98 | 4.03 | 4.40 | 1.07 | ||
| J3a,4 11.8 | J4,5 ∼2 | J5,6 ∼2 | J67 11 | J78 2 | J89 6.5 | ||||
| 173.6 | 101.9 | 36.6 | 67.8 | 49.2 | 72.7 | 53.9 | 66.7 | 18.5 | |
| Ser | 4.54 | 3.76; 4.07 | |||||||
| 172.0 | 55.3 | 64.4 | |||||||
| Leu | 4.41 | 1.71 | 1.69 | 0.90 | 0.95 | ||||
| 177.6 | 52.8 | 40.5 | 25.7 | 21.9 | 23.4 | ||||
| Glu | 4.40 | 2.01; 2.13 | 2.38 | ||||||
| 173.9 | 54.1 | 28.2 | 32.2 | 179.0 | |||||
| Glu* | 4.55 | 2.00; 2.14 | 2.50 | ||||||
| 174.1 | 54.0 | 27.5 | 31.1 | 178.1 | |||||
| Val | 4.16 | 2.16 | 0.97 | 0.99 | |||||
| 174.6 | 61.0 | 31.3 | 18.8 | 19.8 | |||||
| Val* | 3.85 | 2.22 | 1.01 | 1.02 | |||||
| N-end | 170.3 | 59.6 | 31.2 | 18.2 | 18.9 |
N-5 of Non was acylated by an acetyl group as determined from the heteronuclear bond correlation (HMBC) between Ac C-1 and Non H-5. COSY, TOCSY and HSQC data for the component A indicated that this structure is a 2-methoxy-4,5,6-trihydroxy-hexanoyl residue (Fig. 5). No signals of its carboxyl group were observed in the HMBC spectrum, but accurate mass analysis by MS confirmed the elemental composition. The configurations of the three chiral atoms (C-2, C-4, and C-5) in A were not determined. This group has to acylate N-7 of Non, since there is no other location where it can be linked that agrees with the NMR and MS data. The linkage between Non and Ser was found from the HMBC between Non C-2 and Ser H-3. In summary, the NMR and MS data indicate that the structure is a novel C7-acylated pseudaminic acid derivative, as presented in Fig. 5B. There is a possibility that the Non sugar could be the structural isomer at C-8 of Pse, known as 8-epi-Pse.
A PseI homolog (TDE0960) is essential for flagellin synthesis
Pse and Pse-like glycans have been identified in several flagellated bacteria such as Campylobacter and Helicobacter species (Logan, 2006). In Campylobacter jejuni, the Pse biosynthetic genes are located within a large flagellar glycosylation locus that contains ∼50 genes. In contrast, the Pse biosynthetic genes of Helicobacter pylori are sporadically distributed around the genome (Schirm et al., 2003, Logan et al., 2002). BLAST searches of the T. denticola genome identified several putative Pse biosynthesis genes that are sporadically distributed on the genome. One of those homologs is PseI (TDE0960), a putative Pse synthase. TDE0960 has a 28% identity to the PseI protein of H. pylori and contains a conserved NeuB domain that is associated with bacterial sialic acid synthases (Fig. S6). To determine if TDE0960 is involved in FlaB glycosylation, the cognate gene was deleted, and the resultant mutant (Tde960mut) was cis-complemented using the method illustrated in Fig. S2. Immunoblotting analysis using a specific antibody against TDE0960 showed that the cognate gene product was abolished in the mutant and restored in the complemented strain (named Tde960com) (Fig. 6A). Glycosylation staining analysis revealed that the glycosylated FlaBs were detected in the wild-type and Tde960com strains but not in Tde960mut (Fig. 6B). Immunoblotting analysis with the FlaA and FlaB antibodies showed that the flagellin proteins were almost undetectable in the mutant (Fig. 6C). Interestingly, a trace amount of FlaA and FlaB proteins could still be detected in the mutant whole cell lysate when its loading amount was increased 8-fold (Fig. 6D). In contrast to the FlaA and FlaB proteins, the flagellar hook protein FlgE remains unaffected in the TDE960 mutant strain (Fig. 6C). It is worth noting that the MWs of the detected FlaBs in Tde960mut were lower than in the wild type (Fig. 6D), indicative of unmodified forms of FlaBs. Collectively, these data suggest that TDE0960 is essential for flagellin production and glycosylation in T. denticola.
Fig. 6. Characterizations of the TDE0960 mutant (Tde960mut) and its complemented strain (Tde960com).
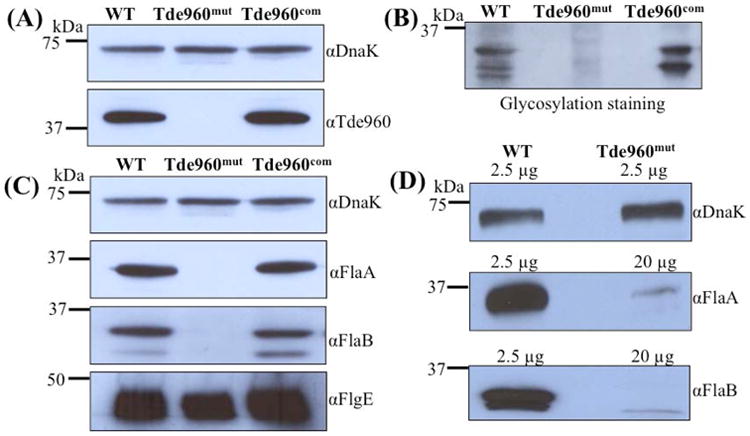
(A) Immunoblotting analysis of the wild-type, Tde960mut, and Tde960com strains. Equivalent amounts of whole cell lysates were analyzed by SDS-PAGE and then probed with a specific antibody against TDE0960. DnaK was used as a loading control. (B) Glycosylation staining analysis of the whole cell lysates. The analysis was carried out as described above. (C) The levels of FlaA and FlaBs were significantly decreased in the Tde960mut strain. Equivalent amounts of the whole cell lysates were analyzed by SDS-PAGE and then probed with the antibodies of DnaK, FlaA, FlaBs, or FlgE. (D) Different amounts of whole cell lysates (as indicated on the figure) were analyzed by SDS-PAGE and then probed with the FlaA and FlaB antibodies.
TDE0960 affects flagellin gene expression and protein stability
To elucidate the potential mechanisms that underlie the reduction of FlaA and FlaBs that was observed in Tde960mut, the levels of flagellin gene transcripts (flaA, flaB1, flaB2, and flaB3) were measured by qRT-PCR. Using the dnaK transcript (TDE0628) as an internal control, the relative levels of the four flagellin mRNAs in Tde960mut were measured. The results showed that the levels of flaA and flaB transcripts in the mutant were decreased by 50∼90% relative to those in the wild type, with the flaB1 mRNA being decreased by nearly 90% (Fig. 7A).
Fig. 7. Detection of flagellin gene mRNA by qRT-PCR and protein stability by turnover assays.

(A) The levels of flaA, flaB1, flaB2, and flaB3 transcripts were measured by qRT-PCR as previously described (Bian et al., 2013). The dnaK gene transcript was used as an internal control to normalize the qRT-PCR data. The results are expressed as Tde960mut flagellin gene transcript expression relative to the wild-type levels. Asterisks indicate that the difference between the wild-type and Tde960mut transcript levels was statistically significant at a P value of < 0.01 (two-way ANOVA). (B) Protein translation was arrested by adding spectinomycin to the cultures of the wild-type and Tde960mut strains. Samples were withdrawn at the indicated time points and analyzed by immunoblotting. The whole cell lysates of the wild-type (2.5 μg) and Tde960mut strains (20 μg) were separated by SDS-PAGE, then transferred to PVDF membranes, and finally probed with antibodies against DnaK, FlaA, or FlaBs. DnaK was used as a sample loading control. SuperSignal West Femto Maximum Sensitivity substrate was used to develop the immunoblots of Tde960mut.
Protein turnover assays were also carried out to monitor the stability of flagellin proteins in the mutant (Fig. 7B). After arresting protein synthesis with addition of spectinomycin, the levels of FlaA and FlaBs remained unchanged in the wild type during a course of 24 hrs (left panels). In contrast, in the Tde960mut mutant, the FlaA protein was decreased nearly 60% at 4 h and completely abolished by 24 h; and the levels of FlaB proteins were reduced by nearly 54% by 24 h compared to their levels in the wild type (right panels). Taken together, these results indicate that both flagellin gene transcription and protein stability are influenced by perturbation of the glycan biosynthetic pathway.
The Tde960mut mutant has short and truncated PFs
To determine if the reduction of FlaA and FlaBs in Tde960mut impairs the PF structure, the mutant cells were dissected in situ using cryo-ET. In the wild-type cells, two long PFs (average length >1,500 nm, n= 18 cells) arose from each cell pole, wrap around the cell cylinder, and extended towards the central region of the cell (Fig. 8A, B and video 1). In the mutant cells, although two PFs were visible at each cell pole, the flagellar filaments were much shorter (average length = 96 ± 36.5 nm, n=14 cells) than the PFs of the wild-type cells (Fig. 8C, D and video 2). In contrast to the filaments, the flagellar basal body and hook structures were unaffected, which is consistent with the immunoblotting results of FlgE that showed no change in its expression level (lower panel, Fig. 6C). These results indicate that the deletion of TDE0960 only impairs the level of FlaA and FlaB proteins and assembly of the filaments in T. denticola but does not affect the assembly of the flagellar hook.
Fig. 8. Cryo-ET analysis of the wild-type and Tde960mut strains.
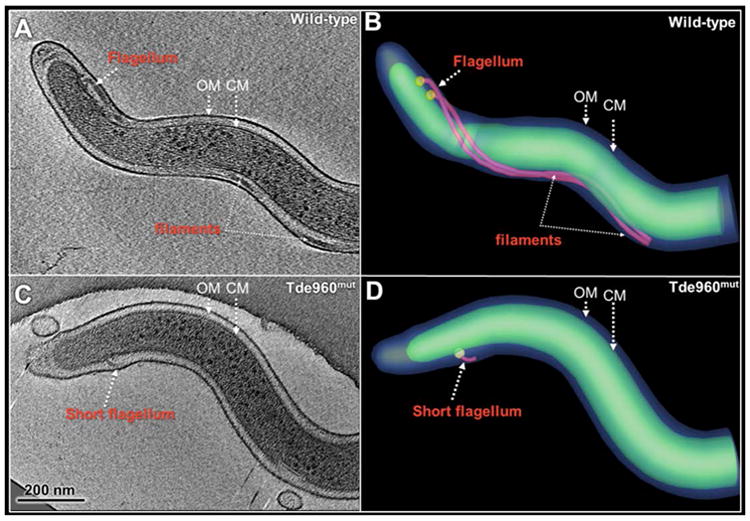
(A) A central slice of a typical tomographic reconstruction from a wild-type cell tip. PFs are observed in the periplasmic space between the outer membrane (OM) and the cytoplasmic membrane (CM). (B) The 3-D surface rendering of the reconstruction (A) shows two long PFs (colored in red) surrounding the cell body (green). (C) A central slice of a tomographic reconstruction from a Tde960mut cell. A short flagellum is attached to the CM. (D) The short flagellum is clearly shown in the surface rendering.
The Tde960mut mutant is non-motile and has altered cell shape
Microscopic observation and bacterial motion tracking analysis showed that Tde960mut failed to migrate in the growth medium containing 1% methylcellulose (videos 3 & 4), indicating that the mutant is non-motile. This proposition was further substantiated by bacterial swimming plate assays (Fig. 9A). The average diameter of swimming rings formed by Tde960mut (5.8 mm, n=5) was significantly smaller (P < 0.01) than that of the wild-type (12.75 mm, n=5) and Tde960com (12.80 mm, n=5) and was almost the same diameter as seen with Δtap1 (5.5 mm, n=5), a non-motile mutant of T. denticola (Limberger et al., 1999). Interestingly, SEM analysis revealed that the mutant cells are rod-shaped instead of the corkscrew-shaped wild-type and Tde960com strains (Fig. 9B). Taken together, these results indicate that the Tde960mut mutant is non-motile and has an altered cell shape, highlighting the important role of flagellar glycosylation in cell motility and morphogenesis.
Fig. 9. The Tde960mut mutant is non-motile and has an altered cell morphology.
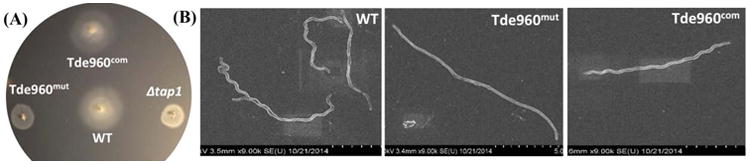
(A) Swimming plate assay of the wild-type, Tde960mut, and Tde960com strains. The assay was carried out on 0.35% agarose containing the TYGVS medium diluted 1:10 with PBS. The plates were incubated anaerobically at 37°C for 5 to 7 days to allow the cells to swim. Δtap1, a non-motile mutant, was used as a control to determine initial inoculum sizes (Limberger et al., 1999). (B) Scanning electron microscopy analysis of the wild-type, Tde960mut, and Tde960com strains. The cells were processed and observed using a Hitachi SU-70 scanning electron microscope at an acceleration voltage of 2.0 kV.
Discussion
In this study, we isolated PF filaments from T. denticola and analyzed the protein composition using SDS-PAGE, 2-D electrophoresis, and immunoblotting analyses. The results clearly demonstrate that the T. denticola flagellar filament consists of one sheath protein (FlaA) and three core FlaB proteins and that these proteins are immunologically cross-reactive to antiserum raised against the corresponding T. pallidum flagellin proteins (Fig. 1). The protein composition was further confirmed by targeted mutagenesis. Deletions of individual flagellin genes abolished the cognate gene products in the PFs isolated from the mutants (Fig. 2). Two putative FlaA homologs (TDE1408 and TDE1409) were not found in the isolated PFs. A BLAST search revealed that although these two proteins belong to the superfamily of FlaA (pfam04620), they have very low (E value > 1.5) sequence similarity to the FlaA protein (TDE1712) that we identified as a component of PFs. It is possible that these two proteins are components of PFs but their levels are too low to be detected. Alternatively, they may have a distinct role to play in motility but are not components of the isolated PFs. Future studies will investigate the role of these two genes in the motility of T. denticola.
After delineating the filament protein composition, we undertook a comprehensive study to characterize the flagellin structural proteins FlaA, FlaB1, FlaB2 and FlaB3. The results from these experiments provide conclusive proof that the FlaA sheath protein is not glycosylated while the three FlaB proteins are all modified with an identical, novel, O-linked glycan (Figs. 2-5). The glycopeptides identified for each FlaB protein were localized to two regions of the primary amino acid sequences that lie within the predicted D1 and D2/D3 domains based on the atomic model of the Salmonella flagellin protein FliC (Fig. 10) (Samatey et al., 2001). While the D2/D3 region is most likely exposed on the external surface of the assembled flagellar filament, the D1 domain is buried in polymerized flagellar filaments. Glycosylation within the D2/D3 domains has been observed in many other bacterial species that produce glycosylated flagellins (Goon et al., 2003, Schirm et al., 2003, Schirm et al., 2004, Verma et al., 2006), but this is the first identification of a glycosylation site within the conserved D1 domain (Samatey et al., 2001). A recent cryo-ET study of T. pallidum flagellum architecture suggested that the FlaA sheath forms a lattice structure covering the inner FlaB core (Liu et al., 2010). This lattice structure could provide surface access to the D2/D3 glycan modifications found on the FlaB proteins. While flagellar glycans in other bacterial pathogens have been shown to play a role in host pathogen interactions due to their location on the external surface of the assembled flagellar filaments [for reviews, see references (Logan, 2006, Nothaft & Szymanski, 2010)], it remains to be established what the biological roles of the glycans are on the T. denticola flagella and motility. It is possible that the glycosylation facilitates interactions with the FlaA sheath structure and/or increases the stability of flagellin proteins after they are secreted into the periplasmic space.
Fig. 10. The first two glycopeptides of FlaBs are localized within the D1 domain of flagellin protein.
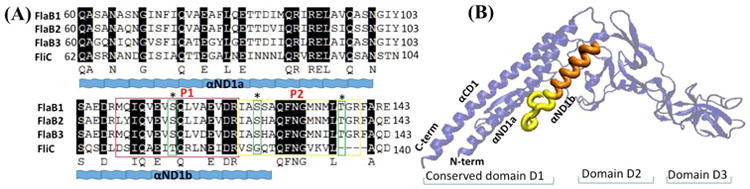
(A) Sequence alignment of flagellin D1 domains. Flagellin D1 domain sequences of three FlaBs and Salmonella FliC (1IO1) (Yoon et al., 2012, Samatey et al., 2001) were aligned by Clustal X. The secondary structures of α-helices (αND1a and αND1b) are shown as waves; color-boxed P1 (orange) and P2 (yellow) sequences are two glycopeptides identified in all FlaB proteins; and green color-coded residues represent three glycosylation residues. (B) Homology mapping of the glycopeptides. Based on the results from pairwise sequence alignment shown on (A), P1 and P2 were directly mapped to the corresponding sequences on the crystal structure of 1IO1 using homology mapping (Beaver et al., 2007). Both P1 and P2 are located within αND1b located within interface-B between FliC and TLR5 (Yoon et al., 2012).
As a keystone pathogen of periodontitis, T. denticola inhabits at the forefront of subgingival biofilms and directly encounters host innate immune responses (Ellen & Galimanas, 2005, Kurniyati et al., 2013). The innate immune response to flagellins is mediated by Toll-like receptor 5 (TLR5) (Hayashi et al., 2001, Smith et al., 2003), and structural studies have defined the precise domains and amino acid residues of both TLR5 and the D0/D1 domains of FliC that are involved in this interaction (Yoon et al., 2012). One of the identified sites of glycosylation present in all three T. denticola FlaBs (S116) lies between conserved residues E114 and Q117 of the αND1b region of this D1 domain (Fig. 10). Both residues were identified in the co-crystallization study of TLR5 with Salmonella FliC fragment CBLB502 as residues that interact via H bonding with the LRR9 loop groove of primary interface-B of TLR5. The LRR9 loop has been described as a “hot spot” in the TLR-FliC interaction (Yoon et al., 2012). It remains to be seen if the novel glycosylation identified on the T. denticola FlaB proteins at S116 perturbs this interaction and so affects the pattern recognition mediated by TLR5. If so, this could subsequently protect the pathogen from the host immune responses. The role of glycosylation in the interplay between the T. denticola flagellins and TLR5 receptor and its subsequent impact on the host immune responses can now be explored at the molecular level.
Flagellin glycosylation and Pse biosynthesis have been extensively studied in C. jejuni and H. pylori. A set of core enzymes (i.e., PseB, C, G, H, F, and I) has been shown to be required for the biosynthesis of Pse (Schoenhofen et al., 2006a, Schoenhofen et al., 2006b, Schoenhofen et al., 2006c). Genome mining efforts to identify the biosynthetic genes in T. denticola revealed that there is no comparable flagellar genetic locus. In addition, BLAST searches using the H. pylori and C. jejuni biosynthetic enzymes as queries identified only four Pse homologs in the genome of T. denticola – TDE0714 (PseB), TDE0725 (PseC), TDE0960 (PseI), and TDE1917 (PseF). Interestingly, these 4 homologs were also found in T. pallidum (Fraser et al., 1998), (TP0077, TP0078, TP0562, and TP0289), and they share high sequence identities (50∼70%) with their counterparts in T. denticola. These findings suggest that the flagellins of T. pallidum are probably modified with the same glycan as those of T. denticola. From a biosynthetic perspective, no homologs of PseH (N-acetyltransferase) and PseG (nucleotidase) were identified in either of the Treponema genomes. PseH is responsible for the N-4 acetylation of UDP-4-amino-4,6-dideoxy-β-l-AltNAc to form UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-AltNAc. PseH is responsible for removal of UDP from C1 of the PseH product to form 2,4-diacetamido-2,4,6-trideoxy- β-l-altropyranose (Liu et al., 2014, McNally et al., 2006, Ud-Din et al., 2015, Schoenhofen et al., 2006b). Although such an apparent absence of two key biosynthetic enzymes may simply be due to very low levels of sequence similarity with the Campylobacter and Helicobacter enzymes, it is tempting to speculate that a divergence in the biosynthetic pathway may have occurred between these organisms and Treponema spp., which is reflected in the novel acyl moiety identified at N7 in the final Pse sugar (N4 in the biosynthetic precursor sugar). This is currently under investigation.
Among the Pse homologs identified in Treponema genomes, TDE0960/TP0562 is the most conserved with 27∼28 % sequence identity to PseI (HP0178) of H. pylori and C. jejuni (Cj1317) (Fig. S6). Previous studies in H. pylori and C. jejuni have shown that PseI functions as a Pse synthase, which condenses 2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose with pyruvate to form Pse (McNally et al., 2006). To determine if this homolog (TDE0960) is involved in biosynthesis of the T. denticola glycan, we constructed a Tde960mut mutant and found that the deletion abolished flagellar glycosylation and filament assembly (Figs. 6-8). Of potential significance regarding the possible divergence in the Pse biosynthetic pathway in T. denticola is the inability to demonstrate restoration of motility via heterologous complementation of either the T. denticola TDE0960 mutant with the PseI from H. pylori or the H. pylori PseI mutant with TDE0960 (data not shown). The substrate specificity of the T. denticola and H. pylori PseI enzymes may have prevented effective cross complementation.
While previous publications had provided indirect evidence for PF glycosylation (Wyss, 1998, Brahamsha & Greenberg, 1988, Li et al., 1993), this study provides the first direct proof that the process of prokaryotic flagellar glycosylation extends to the periplasmic flagella of spirochetes. Furthermore, it expands the catalog of diverse prokaryotic nonulosonate sugar structures by identifying a novel monoacetylated Pse that carries a unique acyl moiety at N7. Both the biosynthetic pathway and biological function of this novel glycan can now be examined and will undoubtedly reveal new and exciting roles for the glycosylation process in both the assembly of periplasmic flagella and the biology of pathogenic spirochetes, in particular in oral Treponema spp and the syphilis spirochete T. pallidum.
Materials and Methods
Bacterial strains and culture conditions
T. denticola ATCC 35405 (wild-type) was used in this study (Seshadri et al., 2004). Cells were grown in tryptone-yeast extract-gelatin-volatile fatty acids-serum (TYGVS) medium at 37°C in an anaerobic chamber in the presence of 85% nitrogen, 10% carbon dioxide, and 5% hydrogen (Ohta et al., 1986). T. denticola isogenic mutants were grown with appropriate antibiotic(s) for selective pressure as needed: 50 μg/ml erythromycin and/or 20 μg/ml gentamicin. E. coli 5α strain (New England Biolabs, Ipswich, MA) was used for DNA cloning, and BL21-CodonPlus (DE3)-RIL (Agilent, Santa Clara, CA) was used for preparing recombinant proteins. E. coli strains were cultivated in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Periplasmic flagella (PFs) isolation
The PFs of T. denticola were isolated as previously described with some modifications (Miller et al., 2014). Briefly, 500 ml of the late-logarithmic-phase cultures (∼108 cells/ml) were harvested at 8,000×g for 20 min at 4°C. The cell pellets were washed four times with phosphate-buffered saline (PBS, pH 7.4) and once with T1 buffer (0.15 M Tris-HCl, pH 6.8). The final cell pellets were resuspended in 30 ml of T1 buffer. Then, 3 ml 10% Triton X-100 was slowly added, followed by incubation for 1 h at room temperature (RT). Three milliliters of 200 μg/ml mutanolysin (Sigma-Aldrich, St. Louis, MO) were slowly added, followed by addition of 300 μl T2 buffer (0.1 M Tris-HCl, pH 6.8). The mixture was incubated for 2 h at RT and then at 4°C overnight. After the incubation, 600 μl 0.1 M MgSO4 was added, followed by addition of 600 μl T2 buffer. The mixture was incubated for 5 min at RT. The cell suspension was centrifuged at 17,000×g for 15 min at 4°C. The supernatant containing PFs was collected, and 2 ml 20% PEG 8000 (Alfa Aesar, London, UK) in 1 M NaCl was added. The resultant sample was incubated for 30 min on ice. The precipitated PFs were centrifuged at 27,000×g for 30 min at 4°C. The resultant pellets were resuspended in an alkaline buffer (0.1 M KCl, 0.5 M sucrose, 0.1% Triton X-100, 50 mM sodium bicarbonate, pH 11) and incubated for 1 h on ice. The PFs were collected by ultracentrifugation at 80,000×g for 45 min at 4°C and washed once in 20 mM Tris-HCl (pH 8). The final PF pellets were resuspended in water and stored at 4°C for further analysis.
Electrophoresis
Two-dimensional (2D) gel electrophoresis was carried out as previously described (Veith et al., 2009). Equal amounts of purified PFs were resuspended in a rehydration buffer (5 M urea, 2 M thiourea, 2% CHAPS, 2% SB 3-10, 0.2% Bio-Lyte 3/10 Ampholyte, 40 mM Tris, and 0.0002% Bromophenol Blue) and then subjected to 17 cm long pH 5→8 linear IPG strips. The first dimension of isoelectric focusing (IEF) was performed using PROTEAN IEF (Bio-Rad Laboratories, Hercules, CA), followed by equilibration according to manufacturer's protocols. The second dimension separation was carried out using sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) as described previously (Li et al., 2012). The resultant gels were subjected to Coomassie blue staining, glycosylation staining, or immunoblotting analyses. The antibodies against T. pallidum FlaBs or T. denticola FlaA are described in previous publications (Ruby et al., 1997, Bian et al., 2015). The antibodies against T. denticola DnaK or TDE0960 were recently raised in rats as described below.
Lectin blot and glycosylation staining
Lectin blot analysis was performed as previously described (Kurniyati et al., 2013). Equal amounts of whole cell lysates or purified PFs were separated on SDS-PAGE or 2-D gels and then transferred to PVDF membranes. The blots were first blocked in 1× Carbo-Free blocking solution (Vector Laboratories, Burlingame, CA) containing 0.05% Tween-20 and then incubated with biotinylated Concanavalin A (ConA, Sigma-Aldrich) or Limax Favus Agglutinin (LFA) in an incubation buffer (0.2× Carbo-Free solution and 0.05% Tween-20) for 1 h at room temperature. The resulting blots were washed four times with PBS-T buffer (PBS, 0.05% Tween-20), followed by incubation with streptavidin-horseradish peroxidase conjugate. After the incubation, the blots were washed four times with PBS-T buffer and developed with the enhanced chemiluminescent luminol (ECL) detection system (Thermo Scientific). Glycosylation staining was performed using an ECL glycoprotein detection kit (GE Healthcare, Buckinghamshire, UK), according to the manufacturer's protocol.
Enzymatic and β-elimination deglycosylation assays
For enzymatic removal of glycan, the purified PFs were treated with a cocktail of PNGase F, O-glycosidase (New England Biolabs), and Clostridium perfringens neuraminidase NanH (Sigma-Aldrich), as previously described (Kurniyati et al., 2013). Briefly, 5 μg of purified PFs was boiled for 10 min to denature the flagella and then incubated with 0.005 U NanH, 25 U PNGase F, or 2000 U O-glycosidase in different combinations for 16 h at 37°C. The treated samples were subjected to SDS-PAGE and lectin blot analyses. β-elimination treatment of the purified PFs was carried out using GlycoProfile β-Elimination Kit (Sigma-Aldrich), according to the manufacturer's protocol. The treated samples were subjected to SDS-PAGE, immunoblotting, and glycosylation staining analyses.
Construction of vectors for deletion of flagellin genes (flaA, flaB1, flaB2, and flaB3)
The diagrams in Fig. S2 show how the entire open reading frames (ORFs) of flaA (TDE1712), flaB1 (TDE1477), flaB2 (TDE1004), and flaB3 (TDE1475) were deleted. Here, we used flaB1 as an example to describe how this vector (FlaB1∷ermB) was constructed and used to delete the targeted gene. The same method was used to construct FlaB2∷ermB, FlaB3∷ermB, and FlaA∷aacC1. To construct FlaB1∷ermB, the flaB1 upstream region and a previously described erythromycin B resistant cassette (ermB) (Goetting-Minesky & Fenno, 2010) were PCR amplified with primers P1/P2 and P3/P4, respectively, and then fused together with primers P1/P4, generating Fragment 1. The downstream region of flaB1 was PCR amplified with primers P5/P6, and then fused to Fragment 1 by PCR using primers P1/P6. The fused fragment was finally cloned into the pGEM-T easy vector (Promega, Madison, WI), generating FlaB1∷ermB (Fig. S1A). For the FlaA∷aacC1 vector, a previously modified gentamicin cassette (aacC1) (Bian et al., 2012) was used. The primers used here are listed in Table S1. These primers were synthesized by Integrated DNA Technologies (Coralville, IA). To delete flaB1, the FlaB1∷ermB vector was linearized and transformed into T. denticola wild-type competent cells via electroporation, as previously described (Kurniyati et al., 2013). The deletions were confirmed by PCR and immunoblotting. The resultant mutants were designated as ΔflaA, ΔflaB1, ΔflaB2, and ΔflaB3.
Construction of a TDE0960 deletion mutant and its complemented strain
The same method described above was used to construct TDE0960∷ermB (Fig. S2E), which was used to replace the entire ORF of TDE0960 with ermB. The vector TDE0960-aacC1 (Fig. S2F) was constructed to cis-complement the TDE0960 deletion mutant. To construct TDE0960-aacC1, the full length TDE0960 gene along with its upstream promoter region and the aacC1 cassette were PCR amplified with primers P21/P25 and P17/P18, respectively, and then fused together with primers P21/P18, generating Fragment 1. The downstream region of TDE0960 was PCR amplified with primers P26/P24 and then fused to Fragment 1 by PCR using primers P21/P24, generating TDE0960-aacC1 (Fig. S2F). The obtained DNA fragment was cloned into the pGEM-T Easy vector (Promega). For the complementation, TDE0960-aacC1 was transformed into TDE0960 mutant competent cells via electroporation. The complemented clones were screened and characterized, as previously described (Kurniyati et al., 2013). The primers used here are listed in Table S1.
Preparation of DnaK and TDE0960 antibodies
Truncated DnaK (TDE0628) (from aa 61 to 646) and the full-length TDE0960 recombinant protein were prepared to generate polyclonal antibodies in rats, as previously described (Kurniyati et al., 2013). Briefly, the DNA fragments encoding the DnaK and TDE0960 proteins were PCR amplified with primers P27/P28 and P29/P30, respectively, using Pfx DNA polymerase (Life Technologies, Grand Island, NY). The resultant dnaK PCR product was cloned into the pET200/D-TOPO expression vector (Life Technologies), which encodes a six-histidine (6xHis) tag at the N-terminus. The resulting plasmids were transformed into BL21-CodonPlus (DE3)-RIL (Agilent). For TDE0960, the resultant PCR product was cloned into the pGEMT-easy vector (Promega) and then cloned into pQE30 expression vector (Qiagen, Valencia, CA), which encodes 6xHis-tag at the N-terminus. The resulting plasmids were transformed into M15 (Qiagen). The expression of DnaK and TDE0960 were induced using 1 mM isopropyl β-d-1-thiogalactosidase (IPTG). The recombinant proteins were purified using Ni-NTA agarose (Qiagen) under native conditions according to the manufacturer's protocol. The purified proteins were then dialyzed in a buffer containing 20 mM Tris-HCl buffer (pH 8) at 4°C overnight using 3.0 kDa molecular weight cut-off Spectra/Por®dialysis bags (Spectrum Laboratories, Rancho Dominguez, CA). The concentrations of purified proteins were determined using a Bio-Rad Protein Assay Kit (Bio-Rad). Approximately 5 mg of purified recombinant protein was used to immunize the rats (2.5 mg for each animal). The primers used here are listed in Table S1.
Preparation of flagellar filament proteins for LC-MS/MS analysis
Purified PFs were separated using SDS-PAGE. Flagellar filament proteins were visualized by staining in 0.25 M potassium chloride, excised from the gels, and placed into 3.0 kDa molecular weight cut-off Spectra/Por® dialysis bags. PBS was then added, and the proteins were eluted from the gel matrix using SDS-PAGE running buffer. The eluted proteins were transferred into new 3.0 kDa molecular weight cut-off dialysis bags and dialyzed in PBS at 4°C overnight. The concentration of purified protein was determined using a Bio-Rad Protein Assay Kit (Bio-Rad). Approximately 100 μg purified flagellar filament proteins were digested and subjected to LC-MS/MS analysis.
LC-MS/MS analysis
All analyses were performed on tryptic digests of the purified flagellin proteins (Schirm et al., 2003). Briefly, the flagellin isolates were incubated overnight at 37°C with trypsin (Promega) at an approximate vol/vol ratio of 20:1 (protein:enzyme). The digests were analyzed by nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) using a NanoAquity UPLC system (Waters, Milford, MA) coupled to an Ultima hybrid quadruple time-of-flight (Q-Tof) mass spectrometer (Waters). The digests were injected onto an Acclaim PepMax100 C18 μ-precolumn (5 mm by 300 μm i.d.; Dionex/Thermo Scientific, Sunnyvale, CA) and resolved on a 1.7 μm BEH130 C18 column (100 μm by 100 mm i.d.; Waters, Milford, CA) using the following gradient conditions: hold at 1% mobile phase B (ACN, 0.1% formic acid) for 1 minute, 1% to 45% B in 36 min and 45% to 95% B in 2 min. The flow rate was 400 nL/min. MS/MS spectra were acquired on double, triple and quadruple charged ions and searched against the NCBInr database using the MASCOT search engine (Matrix Science, Ltd., London, United Kingdom). Glycopeptide LC-MS/MS spectra were interpreted by hand.
Exact mass analysis of the glycan oxonium ions was performed using nanoAquity UPLC coupled to a LTQ-Orbitrap XL hybrid mass spectrometer. The nanoLC conditions were similar to those described above. The Orbitrap resolution was set to 60,000 and HCD MS/MS spectra were acquired in a data-dependent manner (lock mass: polysiloxane at m/z 593.158120, isolation width 3.0 Da, normalized collision energy: 28.5 V, default charge state: 3, activation time: 30 msec). The exact m/z values of the glycan oxonium ions were measured to 4 decimal places. The mass accuracy of the measurements was assessed using the m/z values of adjacent peptide b and y fragment ions.
Large-scale glycopeptide purification for NMR analysis
To obtain glycan material devoid of protein backbone for structural analysis, 25 mg purified flagellin was digested with proteinase K at a ratio of 1:1 (Sigma) in 10 mM Na2PO4 pH 7.6 at 37°C for 48 hours. The proteinase K-digested material was lyophilized and resuspended in dH2O. The sample was then fractionated on a Biogel P10 column (2.5×80 cm, 1% acetic acid, RI detector). Each fraction was analyzed by 1H NMR. The glycopeptide-containing fraction was then applied to a Zorbax C18 column in a 0.1% TFA-80% acetonitrile gradient with UV detector at 220 nm. The fractions were collected and reexamined by 1H NMR for the presence of glycan.
NMR spectroscopy
NMR experiments were carried out on a Bruker AVANCE III 600 MHz (1H) spectrometer with 5 mm Z-gradient probe with acetone internal reference (2.225 ppm for 1H and 31.45 ppm for 13C) using standard pulse sequences cosygpprqf (gCOSY), mlevphpr (TOCSY, mixing time 120 ms), roesyphpr (ROESY, mixing time 500 ms), hsqcedetgp (HSQC), hsqcetgpml (HSQC-TOCSY, 80 ms TOCSY delay) and hmbcgplpndqf (HMBC, 70 or 100 ms long range transfer delay). Resolution was kept <3 Hz/pt in F2 in proton-proton correlations and <5 Hz/pt in F2 of H-C correlations. The spectra were processed and analyzed using the Bruker Topspin 2.1 program.
Bacterial swimming plate assay
The swimming plate assay was conducted as previously described (Limberger et al., 1999). Briefly, 3 μL of T. denticola cultures (109 cells/ml) were inoculated onto 0.35% agarose in 1:10 PBS-diluted TYGVS medium. The plates were incubated at 37°C for 5 to 7 days. The diameters of swim rings were measured in millimeters. Δtap1, a non-motile mutant of T. denticola (Limberger et al., 1999), was used as a negative control to determine the initial inoculum size. The average diameters of each strain were calculated from three independent plates. The results are represented as the mean of diameters ± standard error of the mean (SEM).
Scanning electron microscopy (SEM)
T. denticola cells were centrifuged, washed once with PBS, and then resuspended in fresh PBS. The resultant samples were subjected to SEM. Briefly, 10 μL of cell suspensions was settled on poly-L-lysine-coated round cover slips (BD BioSciences, San Jose, CA). Samples were fixed statically in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde, 0.075% ruthenium red, and 0.075 M lysine acetate for 1h at RT followed by three 10-min washes using 0.2 M sodium cacodylate buffer (pH 7.2) containing 0.075% ruthenium red at room temperature without agitation. The resultant samples were then dehydrated with a graded ethanol series (30%, 50%, 75%, 95%, and 100%) at RT for 10 min for each incubation, exchanged into 100% hexamethyl-disilazane, and allowed to air dry. The resultant samples were subjected to a Hitachi SU-70 scanning electron microscope at an acceleration voltage of 2.0 kV.
Cryo-electron tomography (cryo-ET)
T. denticola cultures were mixed with 10 nm gold markers, deposited onto freshly glow-discharged holey carbon grids, and then blotted with filter paper and plunge frozen in liquid ethane. The grids were imaged at -170°C on a 300 kV Technai F30 Polara (FEI) equipped with a K2 Summit direct electron detector (Gatan). Tilt series were collected in dose fractionation mode at a magnification of 9,400×, resulting in a final pixel size of 4.5 Å at the specimen level. Using Serial EM (Mastronarde, 2005), low-dose tilt series were collected at -8 μm defocus with a cumulative dose of ∼60 e-/Å2 distributed over 41 images and covering an angular range of -60° to +60°, with angular increments of 3°. We used MotionCorr and IMOD (Kremer et al., 1996) for drift correction and alignment. We then used Tomo3D (Agulleiro & Fernandez, 2015) to generate 3-D reconstructions. In total, 48 reconstructions from cell tips of three T. denticola strains (wild type, a TDE0960 deletion mutant and its complemented strain) were generated.
We used IMOD for 3-D visualization of the cell tips and built 3-D models for four objects: the outer membrane (blue), the inner membrane (green), the PFs (red) and motors (yellow). Each object is one or more groups of contours, and all contours of the same object share common attributes (e.g., the same color and line width). Except for the motors, we used open contours to represent three objects since the starting point and the ending point of the contour are not connected. The open contour was drawn in sections along the boundary of the object. The radii for contour points of the outer and inner membranes are varied because the cross sectional radii of these two objects are variable while the radius of the PFs is constant. After drawing contours, we meshed them by computing the surfaces of objects. For the motors, the scatter points were used to display a sphere in 3-D. After drawing the contours, we built the surface rendering of the whole cell tips.
Protein turnover assay
This assay was carried out as previously described (Zhang et al., 2012). T. denticola strains were first grown to late-logarithmic-phase (∼108 cells/ml). Following addition of 100 μg/ml spectinomycin to arrest protein synthesis, 5 ml T. denticola cultures were sampled at the indicated time points and then subjected to immunoblotting with the DnaK, FlaA, or FlaB antibodies. Immunoblots were developed using horseradish peroxidase-labeled secondary antibodies with Pierce® ECL Western Blotting Substrate Kit (Thermo Scientific). Densitometry of immunoreactive proteins in the blots was used to determine the relative amounts of proteins. Densitometry was measured using the Molecular Imager® ChemiDoc™ XRS Imaging system (Bio-Rad).
Quantitative RT-PCR (qRT-PCR)
Bacterial RNA was isolated as previously described (Kurniyati et al., 2013). Briefly, T. denticola cells were harvested in late-logarithmic phase (∼108 cells/ml). Total RNA was extracted using Trizol reagent (Sigma-Aldrich), following the manufacturer's instructions. The resultant samples were treated with Turbo DNase I (Ambion, Austin, TX) at 37°C for 2 h to eliminate genomic DNA contamination. The resultant RNA samples were re-extracted using acid phenol-chloroform, precipitated in isopropanol, and washed once with 70% ethanol. The RNA pellets were resuspended in RNase-free water. cDNA was generated from 1 μg of the purified RNA using AMV Reverse Transcriptase (Promega). qPCR was performed using iQ SYBR Green Supermix and a MyiQ thermal cycler (Bio-Rad). The dnaK (TDE0628) transcript, a house-keeping gene of T. denticola, was used as an internal control to normalize the qRT-PCR data, as previously described (Bian et al., 2013). The results were expressed as the normalized difference of the threshold cycle (ΔΔCT) between the wild type and mutants. The primers used here are listed in Table S1.
Homology modeling and mapping
Pairwise sequence alignments of FlaB proteins were conducted using Clustal X. Automodel module in Modeller 9v7 (Sali & Blundell, 1993) was applied to build homology models using Salmonella FliC (PDB: 1IO1) (Samatey et al., 2001) as a template. Based on the results from pairwise sequence alignments, conserved sequences were postioned to the corresponding sites of 1IO1 using an approach of homology mapping (Beaver et al., 2007).
Supplementary Material
Acknowledgments
We thank R. Limberger for providing the non-motile Tap1 mutant, C. Fenno for providing the FlaA antiserum, and L. Tessier for assistance with the mass spectrometry analysis. This research was supported by Public Health Service Grants DE023080 and AI078958 to C. Li, DE023431 to N. Charon, NIH grant AI087946 and Welch Foundation grant AU-1714 to J. Liu.
Footnotes
Authors' Contributions: KK and YT conducted genetics, biochemistry and bioinformatics studies. JFK, EV, and AR performed LC-MS/MS and NMR experiments. JW and JJ carried out the cryo-ET experiment. SML and CL designed the study and wrote the manuscript.
Conflict of Interests: The authors declare that they have no conflict of interests.
References
- Agulleiro JI, Fernandez JJ. Tomo3D 2.0--exploitation of advanced vector extensions (AVX) for 3D reconstruction. J Struct Biol. 2015;189:147–152. doi: 10.1016/j.jsb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Anand A, Luthra A, Edmond ME, Ledoyt M, Caimano MJ, Radolf JD. The major outer sheath protein (Msp) of Treponema denticola has a bipartite domain architecture and exists as periplasmic and outer membrane-spanning conformers. J Bacteriol. 2013;195:2060–2071. doi: 10.1128/JB.00078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver JE, Bourne PE, Ponomarenko JV. EpitopeViewer: a Java application for the visualization and analysis of immune epitopes in the Immune Epitope Database and Analysis Resource (IEDB) Immunome Res. 2007;3:3. doi: 10.1186/1745-7580-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- Bian J, Fenno JC, Li C. Development of a modified gentamicin resistance cassette for genetic manipulation of the oral spirochete Treponema denticola. Appl Environ Microbiol. 2012;78:2059–2062. doi: 10.1128/AEM.07461-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Liu X, Cheng YQ, Li C. Inactivation of cyclic Di-GMP binding protein TDE0214 affects the motility, biofilm formation, and virulence of Treponema denticola. J Bacteriol. 2013;195:3897–3905. doi: 10.1128/JB.00610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Tu Y, Wang SM, Wang XY, Li C. Evidence that TP_0144 of Treponema pallidum is a thiamine-binding protein. J Bacteriol. 2015;197:1164–1172. doi: 10.1128/JB.02472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahamsha B, Greenberg EP. Biochemical and cytological analysis of the complex periplasmic flagella from Spirochaeta aurantia. J Bacteriol. 1988;170:4023–4032. doi: 10.1128/jb.170.9.4023-4032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Greenberg EP, Koopman MB, Limberger RJ. Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res Microbiol. 1992;143:597–603. doi: 10.1016/0923-2508(92)90117-7. [DOI] [PubMed] [Google Scholar]
- Chi B, Chauhan S, Kuramitsu H. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infection and immunity. 1999;67:3653–3656. doi: 10.1128/iai.67.7.3653-3656.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashper SG, Seers CA, Tan KH, Reynolds EC. Virulence factors of the oral spirochete Treponema denticola. J Dent Res. 2011;90:691–703. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol. 2005;200038:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista KV, Coburn J. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol. 2010;5:1413–1425. doi: 10.2217/fmb.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NJ, Murray RD, Carter SD. Bovine digital dermatitis: Current concepts from laboratory to farm. Vet J. 2016;211:3–13. doi: 10.1016/j.tvjl.2015.10.028. [DOI] [PubMed] [Google Scholar]
- Fontana C, Lambert A, Benaroudj N, Gasparini D, Gorgette O, Cachet N, Bomchil N, Picardeau M. Analysis of a Spontaneous Non-Motile and Avirulent Mutant Shows That FliM Is Required for Full Endoflagella Assembly in Leptospira interrogans. PLoS One. 2016;11:e0152916. doi: 10.1371/journal.pone.0152916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Giacani L, Lukehart SA. The endemic treponematoses. Clin Microbiol Rev. 2014;27:89–115. doi: 10.1128/CMR.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetting-Minesky MP, Fenno JC. A simplified erythromycin resistance cassette for Treponema denticola mutagenesis. J Microbiol Methods. 2010;83:66–68. doi: 10.1016/j.mimet.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, Pattarini D, Majam G, Thibault P, Logan S. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, Karlyshev A, Kelly JF, Jones MA, Stevens MP, Logan SM, Wren BW. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infection and immunity. 2009;77:2544–2556. doi: 10.1128/IAI.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josenhans C, Vossebein L, Friedrich S, Suerbaum S. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol Lett. 2002;210:165–172. doi: 10.1111/j.1574-6968.2002.tb11176.x. [DOI] [PubMed] [Google Scholar]
- Kim MS, Leahy D. Enzymatic deglycosylation of glycoproteins. Methods Enzymol. 2013;533:259–263. doi: 10.1016/B978-0-12-420067-8.00019-2. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Shashkov AS, Tsvetkov YE, Jansson PE, Zahringer U. 5,7-diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv Carbohydr Chem Biochem. 2003;58:371–417. doi: 10.1016/s0065-2318(03)58007-6. [DOI] [PubMed] [Google Scholar]
- Koopman MB, Baats E, van Vorstenbosch CJ, van der Zeijst BA, Kusters JG. The periplasmic flagella of Serpulina (Treponema) hyodysenteriae are composed of two sheath proteins and three core proteins. J Gen Microbiol. 1992;138:2697–2706. doi: 10.1099/00221287-138-12-2697. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kurniyati K, Zhang W, Zhang K, Li C. A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola. Mol Microbiol. 2013;89:842–856. doi: 10.1111/mmi.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A, Picardeau M, Haake DA, Sermswan RW, Srikram A, Adler B, Murray GA. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infection and immunity. 2012;80:2019–2025. doi: 10.1128/IAI.00131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Corum L, Morgan D, Rosey EL, Stanton TB, Charon NW. The spirochete FlaA periplasmic flagellar sheath protein impacts flagellar helicity. J Bacteriol. 2000a;182:6698–6706. doi: 10.1128/jb.182.23.6698-6706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Kurniyati, Hu B, Bian J, Sun J, Zhang W, Liu J, Pan Y, Li C. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infection and immunity. 2012;80:3–13. doi: 10.1128/IAI.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Motaleb A, Sal M, Goldstein SF, Charon NW. Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol. 2000b;2:345–354. [PubMed] [Google Scholar]
- Li C, Wolgemuth CW, Marko M, Morgan DG, Charon NW. Genetic analysis of spirochete flagellin proteins and their involvement in motility, filament assembly, and flagellar morphology. J Bacteriol. 2008;190:5607–5615. doi: 10.1128/JB.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu H, Zhang K, Liang FT. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol Microbiol. 2010;75:1563–1576. doi: 10.1111/j.1365-2958.2010.07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dumas F, Dubreuil D, Jacques M. A species-specific periplasmic flagellar protein of Serpulina (Treponema) hyodysenteriae. J Bacteriol. 1993;175:8000–8007. doi: 10.1128/jb.175.24.8000-8007.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Gao L, Zhao X, Liu J, Norris SJ. Mutations in the Borrelia burgdorferi Flagellar Type III Secretion System Genes fliH and fliI Profoundly Affect Spirochete Flagellar Assembly, Morphology, Motility, Structure, and Cell Division. MBio. 2015;6:e00579–00515. doi: 10.1128/mBio.00579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Ud-Din AI, Roujeinikova A. Cloning, purification and preliminary crystallographic analysis of the Helicobacter pylori pseudaminic acid biosynthesis N-acetyltransferase PseH. Acta Crystallogr F Struct Biol Commun. 2014;70:1276–1279. doi: 10.1107/S2053230X14015398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SM. Flagellar glycosylation - a new component of the motility repertoire? Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol Microbiol. 2002;46:587–597. doi: 10.1046/j.1365-2958.2002.03185.x. [DOI] [PubMed] [Google Scholar]
- Lux R, Miller JN, Park NH, Shi W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infection and immunity. 2001;69:6276–6283. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Hui JP, Aubry AJ, Mui KK, Guerry P, Brisson JR, Logan SM, Soo EC. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J Biol Chem. 2006;281:18489–18498. doi: 10.1074/jbc.M603777200. [DOI] [PubMed] [Google Scholar]
- Merino S, Tomas JM. Gram-negative flagella glycosylation. Int J Mol Sci. 2014;15:2840–2857. doi: 10.3390/ijms15022840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Motaleb MA, Liu J, Hu B, Caimano MJ, Miller MR, Charon NW. Initial characterization of the FlgE hook high molecular weight complex of Borrelia burgdorferi. PLoS One. 2014;9:e98338. doi: 10.1371/journal.pone.0098338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Collawn JF, Jr, Fish WW. Purification and macromolecular properties of a sialic acid-specific lectin from the slug Limax flavus. J Biol Chem. 1982;257:7574–7580. [PubMed] [Google Scholar]
- Norris SJ. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Treponema pallidum polypeptide research Group. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ, Charon NW, Cook RG, Fuentes MD, Limberger RJ. Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J Bacteriol. 1988;170:4072–4082. doi: 10.1128/jb.170.9.4072-4082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Ohta K, Makinen KK, Loesche WJ. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infection and immunity. 1986;53:213–220. doi: 10.1128/iai.53.1.213-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter-Katalinic J. Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 2005;405:139–171. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD. Treponema. In: Baron S, editor. Medical Microbiology. Galveston (TX): 1996. [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RS, Bernd W. Do N-glycoproteins have preference for specific sequons? Bioinformation. 2010;5:208–212. doi: 10.6026/97320630005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JD, Li H, Kuramitsu H, Norris SJ, Goldstein SF, Buttle KF, Charon NW. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J Bacteriol. 1997;179:1628–1635. doi: 10.1128/jb.179.5.1628-1635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- Schirm M, Kalmokoff M, Aubry A, Thibault P, Sandoz M, Logan SM. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J Bacteriol. 2004;186:6721–6727. doi: 10.1128/JB.186.20.6721-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 2003;48:1579–1592. doi: 10.1046/j.1365-2958.2003.03527.x. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 2006a;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 2006b;16:8C–14C. doi: 10.1093/glycob/cwl010. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006c;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW, Motaleb MA. Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infection and immunity. 2013;81:2012–2021. doi: 10.1128/IAI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Sekar P, Zhao X, Manne A, Liu J, Wooten RM, Motaleb MA. Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infection and immunity. 2015;83:1765–1777. doi: 10.1128/IAI.03097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Din AI, Liu YC, Roujeinikova A. Crystal structure of Helicobacter pylori pseudaminic acid biosynthesis N-acetyltransferase PseH: implications for substrate specificity and catalysis. PLoS One. 2015;10:e0115634. doi: 10.1371/journal.pone.0115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Dashper SG, O'Brien-Simpson NM, Paolini RA, Orth R, Walsh KA, Reynolds EC. Major proteins and antigens of Treponema denticola. Biochim Biophys Acta. 2009;1794:1421–1432. doi: 10.1016/j.bbapap.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Verma A, Schirm M, Arora SK, Thibault P, Logan SM, Ramphal R. Glycosylation of b-Type flagellin of Pseudomonas aeruginosa: structural and genetic basis. J Bacteriol. 2006;188:4395–4403. doi: 10.1128/JB.01642-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder EA, Figueira CP, Benaroudj N, Hu B, Tong BA, Trajtenberg F, Liu J, Reis MG, Charon NW, Buschiazzo A, Picardeau M, Ko AI. A novel flagellar sheath protein, FcpA, determines filament coiling, translational motility and virulence for the Leptospira spirochete. Mol Microbiol. 2016 doi: 10.1111/mmi.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Flagellins, but not endoflagellar sheath proteins, of Treponema pallidum and of pathogen-related oral spirochetes are glycosylated. Infection and immunity. 1998;66:5751–5754. doi: 10.1128/iai.66.12.5751-5754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Tong BA, Liu J, Li C. A single-domain FlgJ contributes to flagellar hook and filament formation in the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol. 2012;194:866–874. doi: 10.1128/JB.06341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


