Abstract
Plant pathogens cause serious crop losses worldwide. Recent new studies demonstrate that spraying double-stranded RNAs (dsRNAs) and small RNAs (sRNAs) that target essential pathogen genes on plant surfaces confer efficient crop protection. This so-called spray-induced gene silencing (SIGS) strategy of disease control is potentially sustainable and environmentally friendly.
Keywords: Spray-induced gene silencing (HIGS), RNA interference (RNAi), small RNAs, RNA-based fungicide
Plant diseases caused by eukaryotic pathogens, such as fungi and oomycetes, have a devastating worldwide economic and agronomic effect on crop production. For example, Fusarium graminearum causes Fusarium head blight and Fusarium seedling blight in important cereal crops such as rice, maize, and wheat, as well as other crops such as soybean. Mycotoxins produced during the progression of these diseases are harmful to animals and humans and compromise food safety, putting a strain on the world grain industry. Current disease control methods are still mainly dependent on chemical sprays, which potentially have harmful environmental and health side-effects, and induce fungicide-resistant pathogen strains [1]. With the increasing world population, reduced farmland, and need for heightened global food security comes the need for new sustainable, effective, and environmentally-friendly solutions to control plant diseases. A new transgene-based host-induced gene silencing (HIGS) strategy, which involves host expression of hairpin RNAs or small RNAs targeting genes in interacting pathogens and pests, has been developed in multiple crop systems to effectively control diseases caused by insects, nematodes, fungi, and oomycetes [2]. However, HIGS is limited by several factors: the lack of available transformation protocols in many crop species, public concern for the production of genetically modified crops (GMOs), and the instability of engineered RNA silencing traits.
A recent study by Koch et al. published in the current issue of PLoS Pathogens demonstrated an effective RNA spraying method – the so called spray-induced gene silencing (SIGS) – for controlling F. graminearum infections on barley [3]. Koch et al. previously showed that Arabidopsis and barley ectopically expressing a double-stranded RNA (dsRNA) targeting three important fungal CYP51 genes which encode cytochrome P450 lanosterol C14α-demethylase significantly enhanced plant resistance to F. graminearum species by disrupting fungal membrane integrity [4]. In the current study, Koch et al. elegantly demonstrated that even spraying detached barley leaves with a 791-nt long CYP3-dsRNAs that contains complementary sequences to CYP51B, CYP51A, and CYP51C prior to fungal infection could effectively inhibit disease and yield much smaller lesions, indicating reduced disease development [3]. Moreover, the levels of the three CYP51 genes were also reduced as measured by reverse transcription quantitative PCR, indicating that the CYP51-targeting RNAs get into fungal cells to suppress the expression of fungal CYP51 genes [3]. They also demonstrated that spraying the RNA fragments of jellyfish green fluorescent protein (GFP) on barley leaves effectively silenced the expression of GFP in a GFP-expressing F. graminearum strain, suggesting that such SIGS is not sequence selective, potentially allowing for targeting of any essential genes in various interacting pathogens [3]. Thus, SIGS is a new innovative strategy for protecting crops from pathogen infection. Indeed, similar disease control was also observed in another study by Wang et al. who showed that when externally applying dsRNAs and small RNAs (sRNAs) targeting Dicer-like protein genes DCL1 and DCL2 of Botrytis cinerea on vegetables, fruits, and flower petals, grey mold disease was effectively suppressed [5]. Both Botrytis cinerea DCL proteins are required for generating sRNA effectors [6]. These studies suggest that such RNA-based disease control strategy is effective on both monocots and dicots. SIGS is powerful, fast, and environmentally friendly, which also circumvents the problems in creating GMOs.
Strikingly, SIGS also conferred resistance against F. graminearum in unsprayed distal leaf parts. The relative amounts of fungal CYP51 transcripts were also reduced in the unsprayed distal area, strongly suggesting that these dsRNAs were translocated into plant cells and tissues and the silencing signals were effectively spread to distal parts [3]. RNA uptake has been only observed in a few organisms, and most mechanistic studies were performed in nematodes [7]. Recently, Wang et al. also showed that the plant fungal pathogen Botrytis cinerea is capable of taking up external dsRNAs and sRNAs [5]. However, plant uptake of external RNA molecules was not reported. Here, Koch et al. have provided convincing evidence to show that green fluorescent dye ATTO 488-labeled dsRNAs were indeed taken up by barley cells when sprayed on the barley leaf surface and subsequently transported into other parts of the plants through the vasculature [3]. The labeled RNAs were observed in xylem, phloem parenchyma cells, companion cells, mesophyll cells, trichomes, and stomata. CYP3-dsRNAs were also detected by Northern blot analysis in both sprayed and non-sprayed leaf parts [3]. This is the first example of active RNA uptake by plant cells. Thus, the RNAs sprayed on the plant surfaces have at least two possible pathways to get into fungal cells (Figure 1): the RNAs are taken up by the plant cells first and then transferred into the fungal cells [3], and/or the RNAs are taken up by the fungal cells directly [5]. Fungal cells are likely to take up RNAs via both pathways spontaneously. Furthermore, Koch et al. showed that the F. graminearum DCL1 protein is required for CYP3-dsRNA processing and efficient SIGS in systemic leaf areas [3], indicating that translocated long dsRNAs are processed into sRNAs by fungal DCL1 proteins to induce silencing of fungal genes.
Figure 1. Two Possible Pathways of Silencing Fungal Genes Induced by dsRNA- and sRNA- Sprayed Plant Surfaces.
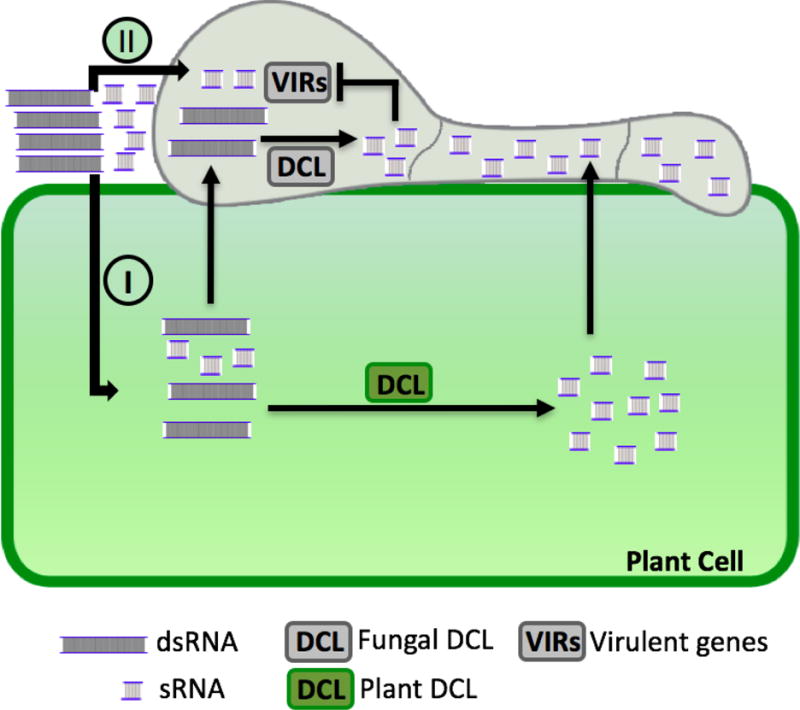
Pathway one: the external dsRNAs and sRNAs are taken up by the plant cells and then transferred into fungal cells (I). These dsRNAs are cleaved into sRNAs by either the plant DCL proteins or fungal DCL proteins. At the same time, the transferred dsRNAs and sRNAs in the plant cells also systemically spread and are transferred into fungal cells. The systemically spread dsRNAs are processed into sRNAs mainly by the fungal DCL protein. Pathway two: the external dsRNAs and sRNAs are directly taken up by the fungal cells (II), and the transferred dsRNAs are processed into sRNAs by the fungal DCL proteins.
To serve as an efficient disease control agent, a reasonable duration of efficacy is desired. The Northern blot analysis showed that the expression of CYP3-dsRNAs was not reduced even at 168 hours post spray of the local sprayed site, suggesting either these external RNAs were stable for seven days on the surface of the leaves and/or they were efficiently taken up and remained stable in the plant cells [3]. Consistent with this, Wang et al. also demonstrated that dsRNAs and sRNAs could protect vegetables and fruits against grey mold disease for up to 8 days [5].
Taken together, the study by Koch et al. highlight the effectiveness of this novel SIGS disease control method by targeting genes vital to fungal integrity or pathogenicity. Indeed, spraying RNAs were also used as ‘oral insecticides’ to control plant pests [8–10]. SIGS can be tailored to be highly specific for fungal genes without disrupting host gene expression and can potentially be developed against an unlimited range of pathogens or pests that have RNAi machinery. SIGS has opened an avenue of development of ‘RNA fungicides’ that are environmentally-friendly [3, 5], because they are made of nucleotides that are present in all life and will not leave toxic residues in the soil and environment. Furthermore, because such RNA fungicides are sequence based rather than structure based, they are likely to be sustainable and should not induce resistant or tolerant mutated pathogen strains.
Further research is needed to determine how these external RNAs are taken up by plant and fungal cells, and how these RNAs are transported from plant cells into fungal cells. Application strategies can be improved by mixing the RNAs with chemical reagents to stabilize the RNAs and thus increase the strength and duration of plant protection. Overall, this new generation of RNAi-based fungicides looks promising to meet our world’s increasing demands for increasing quality crop yields to feed the growing population.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bai G, Shaner G. Management and resistance in wheat and barley to fusarium head blight. Annu Rev Phytopathol. 2004;42:135–61. doi: 10.1146/annurev.phyto.42.040803.140340. [DOI] [PubMed] [Google Scholar]
- 2.Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol. 2012;13:519–29. doi: 10.1111/j.1364-3703.2011.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch A, et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS pathogens. 2016;12:e1005901. doi: 10.1371/journal.ppat.1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch A, et al. Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci USA. 2013;110:19324–9. doi: 10.1073/pnas.1306373110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature plants. 2016;2:16151. doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiberg A, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–23. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whangbo JS, Hunter CP. Environmental RNA interference. Trends in Genetics. 2008;24:297–305. doi: 10.1016/j.tig.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Gong L, et al. Testing insecticidal activity of novel chemically synthesized siRNA against Plutella xylostella under laboratory and field conditions. PloS one. 2013;8:e62990. doi: 10.1371/journal.pone.0062990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li HC, et al. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015;38:2277–2285. doi: 10.1111/pce.12546. [DOI] [PubMed] [Google Scholar]
- 10.San Miguel K, Scott JG. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag Sci. 2016;72:801–809. doi: 10.1002/ps.4056. [DOI] [PubMed] [Google Scholar]


