Summary
In an era where natural product biosynthetic gene clusters can be rapidly identified from sequenced genomes, it is unusual for the biosynthesis of an entire natural product class to remain unknown. Yet, the genetic determinates for pyrroloquinoline alkaloid biosynthesis have remained obscure despite their abundance and deceptive structural simplicity. In this work, we have identified the biosynthetic gene cluster for the ammosamides A-C, pyrroloquinoline alkaloids from Streptomyces sp. CNR-698. Through direct cloning, heterologous expression and gene deletions we have validated the ammosamide biosynthetic gene cluster and demonstrated that these seemingly simple molecules are derived from a surprisingly complex set of biosynthetic genes that are also found in the biosynthesis of lymphostin, a structurally related pyrroloquinoline alkaloid from Salinispora and Streptomyces. Our results implicate a conserved set of genes driving pyrroloquinoline biosynthesis, which consist of genes frequently associated with ribosomal peptide natural product biosynthesis, and whose exact biochemical role remains enigmatic.
Keywords: ammosamide, biosynthesis, coenzyme, F420, halogenation, lantibiotic dehydratase, lymphostin, natural products, peptide, pyrroloquinoline, ribosomal, RiPP, secondary metabolite
eTOC Blurb
Ribosomally synthesized posttranslationally modified peptides (RiPPs) comprise a vast array of potent, bioactive natural products. Now, Jordan and Moore have connected a collection of conserved cryptic RiPP genes to biosynthesis of lymphostin and the ammosamides, characteristic members of the entire class of pyrroloquinoline alkaloids whose biosynthesis was previously unknown.
Introduction
Microbial genome sequencing and genome mining have revealed a much greater capacity for secondary metabolite biosynthesis within the genomes of microbial organisms.(Bentley et al., 2002; Udwary et al., 2007; Weber, 2014; Zarins-Tutt et al., 2015; Ziemert et al., 2016) This latent capacity is often encoded within so-called ‘orphaned gene clusters’ for which the chemical product has not been identified. Yet, for many of these orphaned biosynthetic gene clusters (BGCs), gross structure predictions are possible in the absence of chemical information, especially for the most well characterized biosynthetic systems, such as polyketides, and non-ribosomal and ribosomal peptides.(Boddy, 2014) Although such classes of molecules continue to yield important drug leads, special attention has recently been given to non-canonical BGCs, where structure defining features cannot be predicted solely from sequence information.(Cimermancic et al., 2014) Much how the discovery and biochemical characterization of assembly-line biosynthesis has led to the genome mining revolution,(Challis, 2008; Corre and Challis, 2009; Walsh and Fischbach, 2010) deciphering the biosynthetic logic of non-canonical biosynthetic pathways will not only yield novel natural products and biochemistry, but also ‘inform’ the bioinformatic tools of future genome mining endeavors.
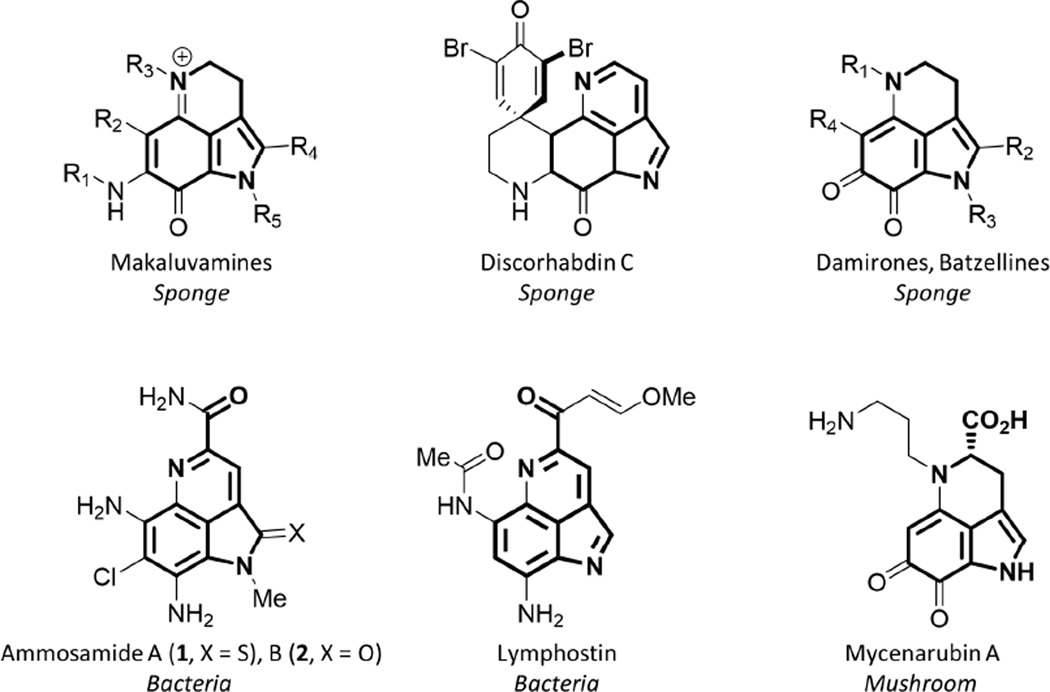
Alkaloids bearing a pyrrolo[4, 3,2-de]quinolone core have been the subject of numerous natural product discovery and total synthesis programs for decades because of their potent biological activity and unique structural features (Figure 1).(Antunes et al., 2005) The vast majority of these molecules have been isolated from marine sponges (over 80 examples), which have traditionally been intractable to genetic tools and laboratory cultivation. Consequently, aside from predictions that the pyrroloquinoline core is derived from tryptophan,(Hu et al., 2011) the genes for pyrroloquinoline biosynthesis have not been identified. In 2011, we verified the role of tryptophan in lymphostin biosynthesis and reported on the end-stage biosynthetic events leading to the production of lymphostin in three sequenced strains of the genus Salinispora.(Miyanaga et al., 2011) In that work, the O-methylation and two-carbon malonyl extension were attributed to the SAM-dependent methyltransferase LymB and the hybrid non-ribosomal peptide synthetase-polyketide synthase (NRPS-PKS) LymA, respectively. But there was no indication that neighboring genes at the lym genetic locus were responsible for the production of the pyrroloquinoline core.
Figure 1.
Structures of pyrroloquinoline alkaloids from varied sources. Pyrrolo[4, 3,2-de]quinolone core predicted to be derived from tryptophan indicated in bold.
Besides the Salinispora-derived lymphostins, only one other example of bacterial pyrroloquinoline alkaloids has been described. In 2009, Fenical and co-workers reported the isolation and characterization of ammosamides A (1) and B (2) from the marine bacterium Streptomyces sp. CNR-698.(Hughes et al., 2009) As the ammosamides share the same core structural features of lymphostin (Figure 1), they provide a unique opportunity to explore pyrroloquinoline alkaloid biosynthesis in a distinct genomic context. Herein we show that the marine bacteria-derived pyrroloquinoline alkaloids lymphostin and the ammosamides are derived from a highly non-canonical biosynthetic pathway that more closely resembles the genetic features of ribosomally synthesized, post-translationally modified peptide (RiPP) natural products.(Arnison et al., 2013)
Results and Discussion
Identification of the Ammosamide (amm) Biosynthetic Gene Cluster: Features of Ribosomal Peptide Biosynthesis
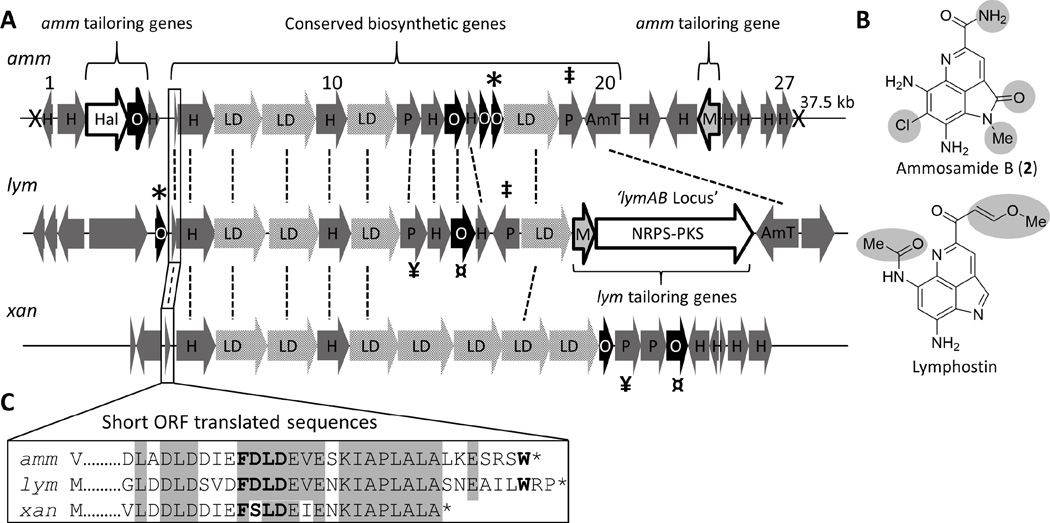
The sequenced and assembled genome of Streptomyces sp. CNR-698 (GenBank AZXC00000000) was acquired from the Joint Genome Institute. Due to a lack of genetic markers for pyrroloquinoline alkaloid biosynthesis, we were drawn to the chloro-substitution on the ammosamides as a means to identify the amm genetic locus in CNR-698. Enzymatic halogenation is often a hallmark secondary metabolism, and having previously established the role of tryptophan in lymphostin biosynthesis, we searched for homologues of the FAD-dependent tryptophan halogenase RebH (Yeh et al., 2005) in the CNR-698 draft genome. Indeed, a BLAST (Altschul et al., 1990; Boratyn et al., 2013) search revealed a single genomic scaffold carrying a tryptophan halogenase homologue (amm3, Figure 2). We further identified a SAM-dependent methyltransferase (amm23) 30 kb downstream of amm3, which we suspected could be responsible for the N-methylation on the ammosamides.
Figure 2.
A. Alignment of the ammosamide (amm) and lymphostin (lym) biosynthetic gene clusters. Related orphaned gene cluster (xan) from S. xanthophaeus also included. ORFs of high sequence homology are connected with a dotted line. TAR cloning sites for amm are indicated with “X”. Conserved genes are indicated with brackets and tailoring genes are outline in black. B. For ammosamide B and lymphostin, structural features derived from tailoring genes in their respective pathways are highlighted in grey. C. Aligned translated sequences of the short ORFs in amm, lym, and xan pathways. FNLD motif is in bold. Abbreviations: H = hypothetical protein; LD = lantibiotic dehydratase; P = peptidase; O = oxidoreductase; Amt = amido transferase; M = methyl transferase; Hal = halogenase. Pairs of ‡ * ¤ ¥ indicate homologues.
Sandwiched between the flavin-dependent halogenase (amm3) and SAM-dependent methyltransferase (amm23) genes are a collection 20 predicted open reading frames (ORFs), 14 of which are highly conserved based upon sequence identity and overall gene organization to ORFs of unknown function found upstream of the lymAB genetic locus in Salinispora (Figure 2A, see Table 1 for a complete list of predicted gene functions). Notably, the peripheral ORFs putatively responsible for ammosamide-specific modifications (Figure 2B), including halogenation (amm3), oxidation (amm4), and methylation (amm23), flank a core set of ORFs shared between the two BGCs. Strikingly, these ORFs include four lantibiotic dehydratases (LD) and two peptidases, genes traditionally associated with RiPP biosynthesis. RiPP BGCs, on the other hand, typically harbor just one or two LDs, as in the case of class II–IV systems and class I systems, respectively.(Chatterjee et al., 2005) When we queried the Conserved Domain Architecture Retrieval Tool (CDART),(Geer et al., 2002) it was revealed that each of the amm LDs are truncated, lacking both a C-terminal cyclase domain that is typical of Class II–IV LDs and a SpaB_C domain that is common amongst Class I LanB systems (LanBs are typically accompanied by standalone LanC cyclases). Furthermore, in the few cases where truncated LanBs lack a SpaB_C domain, as in the case of thiopeptide biosynthesis, a standalone SpaB_C-like protein is found within the gene cluster. However, we did not observe SpaB or LanC cyclase homologues in the amm gene cluster. (Zhang et al., 2012) (Li and Kelly, 2010) Despite vague features relating the amm and lym LDs to lanthipeptide and thiopeptide biosynthesis, the lym and amm LDs appear to be members of a larger group of small LanBs, many of which have no known function.(Ortega et al., 2015)
Table 1.
Predicted function of open reading frames (ORFs) in the amm locus of Streptomyces sp. CNR-698. ORFs capture by transformation associated recombination (TAR) are named amm1–27 and highlighted in grey. For homologues of amm ORFs that are associated with other reported biosynthetic gene clusters, the corresponding natural product is indicated in the “Notes” column.
| Name | Predicted Function | Size (a.a.) | Predicted Function of Closest Homologue | % ID | Notes |
|---|---|---|---|---|---|
| amm1 | (2Fe-2S)-Binding protein | 157 | HP, Streptomyces sp. CNT302 | 98% | |
| amm2 | HP | 486 | HP, Amycolatopsis japonica | 55% | |
| amm3 | Tryptophan halogenase (FAD) | 695 | Microbispora carollina, tryptophan-5-halogenase | 60% | Microbisporicin |
| amm4 | F420 Oxidase | 146 | Amycolatopsis thermoflava, F420 Oxidase | 42% | |
| amm5 | HP | 199 | HP, Streptomyces aurantiacus | 68% | |
| amm6 | HP | 56 | HP, Salinispora, multispecies | 68% | Lymphostin |
| amm7 | HP | 614 | HP, Salinispora, multispecies | 55% | Lymphostin |
| amm8 | LD | 806 | LD, Salinispora, multispecies | 54% | Lymphostin |
| amm9 | LD | 913 | LD, Salinispora, multispecies | 61% | Lymphostin |
| amm10 | HP | 524 | HP, Salinispora, multispecies | 42% | Lymphostin |
| amm11 | LD | 832 | LD, Salinispora, multispecies | 51% | Lymphostin |
| amm12 | P | 419 | P, Salinispora, multispecies | 46% | Lymphostin |
| amm13 | HP | 393 | HP, Salinispora, multispecies | 39% | Lymphostin |
| amm14 | FAD-dependent oxidase | 363 | Salinispora, multispecies, FAD-dep oxidase | 55% | Lymphostin |
| amm15 | HP | 225 | HP, Salinispora, multispecies | 73% | Lymphostin |
| amm16 | Flavoprotein | 199 | Salinispora, multispecies, flavoprotein | 54% | |
| amm17 | Flavoprotein | 212 | Salinispora, multispecies, flavin reductase | 54% | Lymphostin |
| amm18 | LD | 936 | LD, Salinispora multispecies, | 48% | Lymphostin |
| amm19 | P (M20 dipeptidase) | 395 | P (M20 dipeptidase), Salinispora, multispecies | 62% | Lymphostin |
| amm20 | Asparagine synthase | 612 | Salinispora, multispecies, asparagine synthase | 61% | Lymphostin |
| amm21 | T | 526 | T, Kibdelosporangium sp. MJ126-NF4 | 57% | |
| amm22 | HP | 525 | HP, Nocardia vinacea | 43% | |
| amm23 | MT | 356 | MT, Nocardiopsis sp. NRRL B-16309 | 45% | |
| amm24 | TetR regulator | 201 | TetR, Nocardia brevicatena | 56% | |
| amm25 | HP | 254 | Saccharothrix sp. NRRL B-16348, glyoxalase | 50% | |
| amm26 | HP | 284 | HP, Streptomyces sp. CNT302 | 99% | |
| amm27 | HP | 131 | HP, Streptomyces sp. CNT302 | 99% |
(Abbreviations: NC = not captured, HP = hypothetical protein, LD = lantibiotic dehydratase, P = peptidase, MT = Methyltransferase, T = Transporter)
Included at the beginning of the large conserved biosynthetic operon for amm is a short ORF (amm6) (Figure 2C) that encodes a peptide bearing a C-terminal tryptophan residue and bears a striking resemblance to RiPP precursor peptides. RiPP biosynthesis often begins with a short peptide sequence consisting of two domains: a N-terminal ‘leader’ sequence that bears recognition elements for posttranslational modification, and a ‘core’ peptide sequence that is posttranslationally modified prior to proteolytic removal of the leader peptide. Not only is amm6, including the C-terminal tryptophan residue, highly conserved among all producers of lymphostin and the ammosamides, but amm6 and its lymphostin counterpart also contain a conserved ‘FNLD’ amino acid motif (FDLD for lym and amm), which are common leader peptide recognition elements among Class I lantibiotics.(Chatterjee et al., 2005; van der Meer et al., 1994) Despite the similarities to lanthipeptide biosynthesis, no cysteine residues are present in either of the predicted precursor peptides from the amm or lym pathways. Cysteine residues are often highly suggestive of posttranslational modifications involving sequential serine and threonine dehydration events, followed by cyclization with cysteine to form thioether bridges, a common feature of lanthipeptide biosynthesis.(Arnison et al., 2013) The potential for such chemistry is evidently absent from ammosamide biosynthesis.
TAR Cloning and Heterologous Expression of amm: Ammosamides C is the Dominant Pathway Product
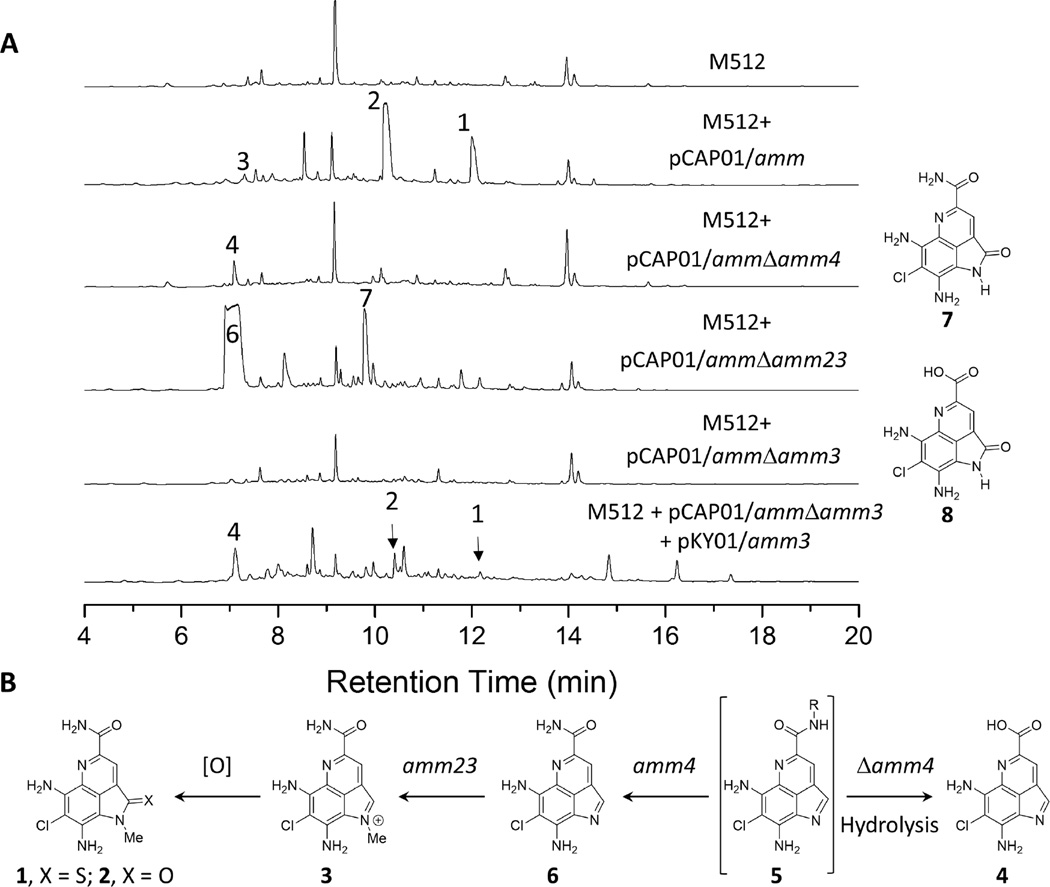
To validate whether the amm biosynthetic locus is indeed involved in the construction of the ammosamides and to probe the function of key genes, we directly captured the amm BGC (See Table 1) by transformation-associated recombination (TAR) in yeast.(Yamanaka et al., 2014) The resulting TAR-assembled ‘pCAP01/amm’ vector was transferred to E. coli followed by conjugation and chromosomal integration into Streptomyces coelicolor M512.(Gomez-Escribano and Bibb, 2012, 2014) Cultures of S. coelicolor M512-pCAP01/amm yielded ammosamides A and B (Figure 3) with 3–4 fold higher production (17 mg each) than the native producer, CNR-698.(Hughes et al., 2009) We further observed after only two days of growth the production of large quantities of ammosamide C (3), the reduced precursor to A and B. Although ammosamide C is only a minor component of ethyl acetate extracts from 12-day cultures, the liquid chromatography mass spectrometry (LCMS) analysis of aqueous aliquots from both pCAP01/amm and CNR-698 cultures revealed that ammosamide C is the predominant product of the amm pathway (Figure S1). Furthermore, up to 134 mg/L of ammosamide C can be isolated from four day cultures of pCAP01/amm. We also observed that large quantities of ammosamide C persists for one to two days prior to the formation of either ammosamide A and B. The sluggish final oxidative step suggested that it may proceed under non-enzymatic conditions. Previous work by MacMillan and co-workers showing that a wide range of amidine derivatives of the ammosamides can be generated from ammosamide C supports our observation.(Pan et al., 2013)
Figure 3.
A. LCMS analysis of S. coelicolor M512 mutants integrated with pCAP01/amm and gene deleted mutated vectors. Chromatograms at 254 nm are provided above. Peaks for ammosamides A (1), B (2) and C (3) and shunt products are indicated with compound numbers. B. Proposed biosynthetic scheme for the final stages of the ammosamide biosynthesis. Compounds 7 and 8, are oxidized analogues of 4 and 6, respectively, are thought to arise from ambient oxidation as their production is culture scale and extraction condition dependent. Compound 5 was not identified, but is implied based this biosynthetic scheme. Peaks were verified by retention time with respect to a positive control (pCAP01/amm), UV absorbance, and mass spectrometry.
Gene Deletions of amm4 and amm23: N-Methylation and Oxidative Primary Amide Synthesis are Late-Stage Modifications to the Pyrroloquinoline Core
Despite evidence suggesting a non-enzymatic final oxidation, it was tempting to speculate that the predicted coenzyme F420-dependent oxidase (amm4), which is unique to the amm BGC, could be responsible for accelerating the final oxidation step. However, upon deletion of amm4, extracts of M512-pCAP01/ammΔamm4 showed no signs of ammosamides A-C. We instead identified and characterized a new highly polar shunt product, ammosamaic acid 4, that bears a carboxylic acid moiety (Figure 3), implicating amm4 in primary amide biosynthesis. Typically, oxidative primary amide bond formation involves copper-dependent peptidylglycine-α-amidating monooxygenases.(Kulathila et al., 1999; Merkler et al., 1999; Prigge et al., 1997; Silakowski et al., 1999) However, such chemistry is not documented for F420-dependent oxidases, where the 5-deazaflavin cofactor is a functional equivalent of nicotinamide.(Walsh, 1986) Although many actinobacteria have the capacity for F420 biosynthesis,(Selengut and Haft, 2010) there are limited examples of coenzyme F420 in secondary metabolism,(Greening et al., 2016) with the most notable being its role in the final reductive step in oxytetracycline biosynthesis.(Wang et al., 2013)
The production of 4 by M512-pCAP01/ammΔamm4 implies a secondary amide precursor resembling 5 (Figure 3B), such that in the absence of amm4, a competing hydrolysis is required to yield 4. However, under no circumstances could we identify precursors such as 5 in extracts of the wild type or mutants. The absence of methylated products in extracts of pCAP01/ammΔamm4 suggested that N-methylation by amm23, a predicted SAM-dependent methyltransferase, takes place after the formation of the primary amide. To test this hypothesis, we inactivated amm23 and evaluated the methyltransferase knockout mutant M512-pCAP01/ammΔamm23, which yielded two new desmethyl ammosamide analogues 6 and 7.
Overall, heterologous expression of pCAP01/amm and the subsequent gene knockouts support the final biosynthetic steps described in Figure 3B, where amide bond formation followed by methylation are preludes to the final non-enzymatic oxidation step. Under all circumstances, only halogenated products could be isolated from M512-pCAP01/amm and various mutants, suggesting that chlorination occurs at an earlier stage in the biosynthesis.
Genetic Manipulations of amm3: A Flavin-Dependent Tryptophan Halogenase Related to MibH, a RiPP Pathway Halogenase
In consideration of the shared genetic features of the respective lym and amm gene clusters (Figure 2), we hypothesized that the halogenase Amm3 chlorinates tryptophan or an advanced pyrroloquinoline intermediate, such that deletion mutants might yield non-halogenated products.(Zehner et al., 2005) Deletion of amm3 abolished production of the ammosamides and any related pyrroloquinoline intermediates (Figure 3A). This observation is consistent with known FADH2-dependent tryptophan halogenases that have been biochemically characterized, where halogenation of free tryptophan is often the beginning of biosynthesis, and chemical complementation of tryptophan halogenase knockouts can rescue production.(Zeng and Zhan, 2011) However, cultures of M512-pCAP01/ammΔamm3 could not be rescued when supplemented with 6-chlorotryptophan (Figure S2). Only when genetically complemented with pKY01/amm3 under the control of the constitutively active promotor ermE could ammosamide production be restored (Figure 3A).(Yamanaka et al., 2012) Genetic complementation also yielded compound 4 as the dominant product, suggesting that gene deletion of amm3 may have disrupted the expression of the downstream oxidase amm4. Incidentally, larger-scale cultures of M512-pCAP01/ammΔamm3 + pKY01/amm3 also provided 8, a presumed byproduct of ambient oxidation of 4.
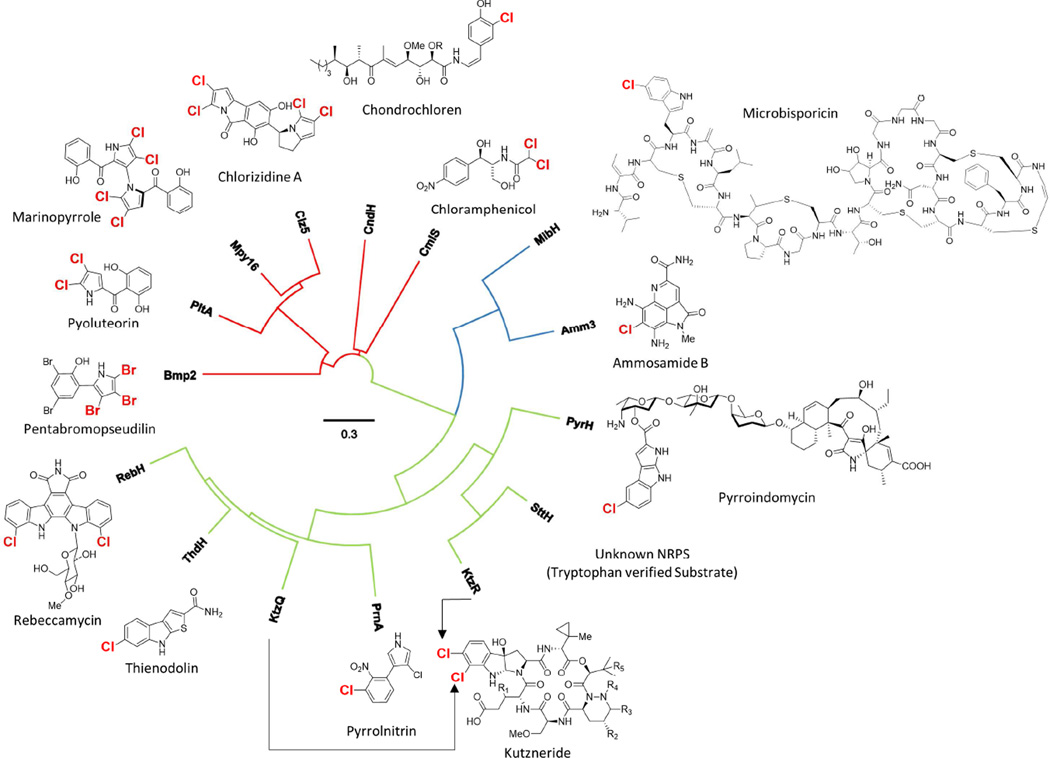
A closer bioinformatics analysis revealed that amm3 is unusual among characterized FADH2-dependent halogenases, which typically fall into two categories; those that utilize free substrates and those that utilize an acyl carrier protein (ACP) bound substrate. However, the amm biosynthetic gene cluster lacks any ORF resembling an ACP gene. The closest homologue of Amm3 based upon sequence identity (61%) is MibH, a tryptophan-5-halogenase involved in the biosynthesis of microbisporicin, a RiPP-class peptide antibiotic from Microbispora carollina.(Foulston and Bibb, 2010) Phylogenetic analysis shows that Amm3 and MibH are distinct from all other flavin-dependent halogenases.(Figure 4). Given that microbisporicin is a RiPP product, and the strict limitations of aminoacyl-tRNA substrates available for ribosomal peptide biosynthesis, tryptophan halogenation via MibH is posttranslational. The implication that Amm3 similarly chlorinates a peptide substrate is intriguing.
Figure 4.
Neighbor-Joining tree of flavin-dependent halogenases associated with characterized secondary metabolites. On each of the corresponding secondary metabolite, the site of halogenation is indicated. Red branches correspond to halogenases that modify acyl carrier protein bound substrates, and green branches correspond to enzymes that halogenate free substrates. The blue branch is occupied by Amm3, described in this work, and MibH, a halogenase that likely modifies a tryptophan embedded in a peptide substrate. MUSCLE alignment and phylogenetic tree were generated using Geneious 8.1.5.
Genetic Manipulations of Conserved Biosynthetic Genes: RiPP-Related ORFs are Essential for Ammosamide Production
The presence of four putative lantibiotic dehydratases (amm8, -9, -11, and -18) in the amm cluster is further suggestive that ammosamide may be derived from a peptide precursor. Individual in-frame gene deletions of all four LDs in the amm pathway resulted in the complete loss of production of the ammosamides and any detectable shunt products (Figure S3), supporting the LDs role in ammosamide biosynthesis. Similarly, deletion of each of the conserved flavin-dependent oxidoreductase genes (amm14, -16, and -17), the peptidase amm19, and one of the four hypothetical genes (amm7) resulted in a complete loss of ammosamide production (Figure S3). The absence of detectable shunt products from these mutants suggests their involvement in the requisite oxidative steps to convert peptidic tryptophan to the ammosamides.
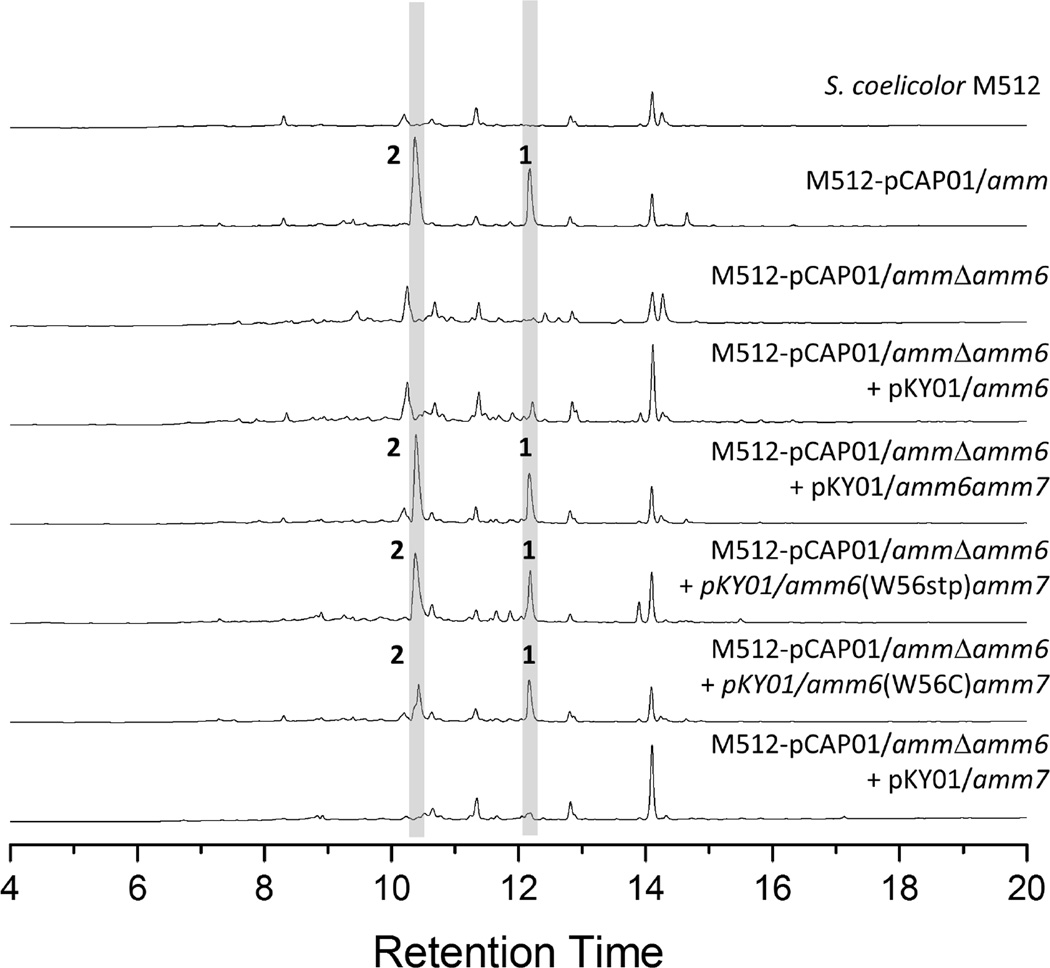
Genetic Manipulations of amm6: A short ORF encoding a 56 a.a. Peptide is Essential for Biosynthesis
Having established the necessity of RiPP-related genes in the amm pathway, we were particularly interested in the predicted 56-amino acid peptide encoded by amm6. Based upon the conserved C-terminal tryptophan and leader peptide recognition elements (Figure 2C), we imagined a scenario where the encoded peptide delivers a C-terminal tryptophan residue for posttranslational modification by pathway enzymes. Deletion of amm6 and consequent loss of ammosamide production support its role for biosynthesis (Figure 5). With an amm6 knockout mutant in hand, we next set out to probe the requirement of the conserved C-terminal tryptophan residue by attempting to genetically complement M512-pCAP01/ammΔamm6 with plasmids carrying amm6 mutants where the tryptophan residue had been replaced. However, ammosamide production could not be restored even with a positive control of native amm6 (Figure 5). Upon closer examination, we identified a region of DNA downstream of amm6 that encompasses the 5’ end of amm7 that includes a large inverted repeat with a predicted RNA secondary structure (ΔG = −57.7 kcal/mol.). Such RNA secondary structures are described in lanthipeptide pathways and putatively serve to control the stoichiometry of the precursor peptide relative to downstream modifying enzymes.(Foulston and Bibb, 2010; McAuliffe et al., 2001)
Figure 5.
Gene deletion of amm6 and genetic complementation of pCAP01/ammΔamm6 with various pKY01 constructs. pKY01 is an integrative plasmid for the expression of gene products under the control of a the ermE promotor. Peaks for ammosamides A (1) and B (2) produced by S. coelicolor-M512 pCAP01/amm are indicated. The absence of ammosamide A and B was verified by retention time with respect to the positive control (pCAP01/amm), UV absorbance, and mass spectrometry. Related metabolites could not be identified by UV absorbance or extracting masses.
In order to preserve the implicit relationship between amm6 and amm7 while still placing the expression of both genes under the control of the ermE promotor, we cloned amm6-amm7 from a single PCR product into the pKY01 vector. Integration of pKY01/amm6amm7 into the pCAP01/ammΔamm6 mutant restored ammosamide production (Figure 5). However, variants of pKY01/amm6amm7 where the terminal tryptophan was removed (pKY01/amm6(W56stp)amm7) or mutated to a cysteine residue (pKY01/amm6(W56C)amm7) also restored production of the ammosamides (Figure 5). This experiment demonstrated that the C-terminal tryptophan residue is not a strict requirement for ammosamide biosynthesis and thereby not the source of its pyrroloquinoline core structure. In contrast, integration of pKY01/amm7 alone into the pCAP01/ammΔamm6 mutant did not restore production, verifying that the genetic complementation with pKY01/amm6amm7, or related tryptophan substituted constructs, were not simply a result of over expressing functional amm7.
These results raise the question: How does a highly conserved, but not strictly necessary, tryptophan residue relate to a predicted precursor peptide sequence that appears to play an essential role in the biosynthesis? Several examples in the literature offer clues to how this may work. It is a common feature of RiPP biosynthesis that substrate recognition, via the leader peptide recognition elements, is spatially segregated from catalysis for peptide modifying enzymes.(Koehnke et al., 2015; Oman and van der Donk, 2010) Consequently, there are a growing number of examples of RiPP pathway enzymes where the leader sequence need not be connected to the core peptide sequence, and trans activation of the modifying enzyme by the discrete leader peptide drives product formation.(Goto et al., 2014; Khusainov and Kuipers, 2012; Oman et al., 2012) Whether the peptide product of amm6, lacking a tryptophan residue, is similarly able to trans activate amm pathway enzymes, which in turn utilize the cellular pool of free tryptophan, is the subject of ongoing in vitro biochemical investigations.
Orphaned Biosynthetic Gene Clusters Bearing Features of the amm and lym Pathways
The lym and amm pathways are highly unusual with several distinct genetic features that enable the identification of either pathway, as well as potentially related pathways, through similarity searches and genome mining tools such as antiSMASH 3.0.(Weber et al., 2015) We have identified homologues of Amm6 from gram-positive and gram-negative bacteria that all share a highly conserved amino acid sequence toward the C-terminus (Figure S4A). However, in many cases, the Amm6 homologues are missing the final 7–9 amino acids that includes the conserved tryptophan, which is a common feature of the lym and amm gene clusters. Rather, the truncated Amm6 homologues are often associated with other biosynthetic gene clusters apparently unrelated to lym or amm. Scant evidence exists for how these Amm6 homologues relate to biosynthetic gene clusters in their genetic neighborhood. However, recent studies with Myxococcus xanthus point to an amm6 homologue, MXAN_1609, which is found upstream of an NRPS gene cluster (MXAN_1608-1598), and is regulated by MXAN4899, a bacterial enhancer binding protein responsible for regulating the production of multiple secondary metabolites(Volz et al., 2012).
With regard to the remainder of the amm gene cluster, we have identified an orphaned biosynthetic gene cluster similar to amm and lym from Streptomyces xanthophaeus NRRL B-3004, a terrestrial bacterium not known to produce pyrroloquinoline type molecules. The ‘xan’ cluster maintains the same gene order and identity of the ORFs comprising the beginning of the shared lym and amm biosynthetic ORFs, including a short ORF encoding a peptide similar to amm6, which bears a ‘FNLD’ leader peptide motif, but lacks a C-terminal tryptophan residue (Figure 2C, for a complete list of xan ORFs see Table S1). In addition to the four LDs found in the amm and lym clusters, the xan cluster features three additional predicted LDs. Although one or two LDs are a requirement for lanthipeptide biosynthesis, the presence of multiple LDs is atypical, further underscoring the obscurity of these biosynthetic gene clusters. Other gene clusters encompassing multiple, clustered LDs include two predicted lanthipeptide pathways from Bacillus halodurans C-125 and Desmospora sp. 8437, each of which contain up to seven LDs (Figure S4). As in the amm, lym and xan pathways, the LDs associated with the Bacillus and Desmospora pathways are also truncated, lacking both a C-terminal cyclase domain and a SpaB_C domain. Incidentally, the orphaned lanthipeptide pathway from B. halodurans also includes a short ORF encoding a peptide with high sequence homology to the lym, amm, and xan pathways (Figure S4). Too little is known about these predicted lanthipeptide biosyntheses and if or how their respective biosynthetic products relate to pyrroloquinoline alkaloids, like lymphostin and the ammosamides.
In summary, we have identified the ammosamide BGC (amm) in Streptomyces sp. CNR-698, which bears a striking resemblance to the biosynthetic pathway for the closely related pyrroloquinoline alkaloid, lymphostin. Our analysis points to a conserved set of ORFs of unknown function that are responsible for pyrroloquinoline core biosynthesis in each pathway. Through direct cloning and heterologous expression of the amm pathway, and targeted gene deletions of amm tailoring genes, we reconstructed the final stages of ammosamide biosynthesis and provided four new ammosamide analogues. These results also revealed several rare biosynthetic features, including a predicted F420-dependent oxidase involved in primary amide biosynthesis and an unusual tryptophan halogenase phylogenetically related to a flavin-dependent halogenase involved in RiPP natural product biosynthesis. Perhaps most surprising, the genes shared between the amm and lym pathways consist of genes normally associated with RiPP natural product biosynthesis, including four predicted lantibiotic dehydratases and a short ORF encoding a putative RiPP pathway precursor peptide. Through targeted gene deletions of all four amm associated LDs and the gene (amm6) encoding the putative precursor peptide, we have established their requirement for biosynthesis. However, it remains unclear whether the peptide encoded by amm6 supplies a substrate for posttranslational modification, or modulates the activity of pathway enzymes.
Significance
The lym and amm pathways are unlike any biosynthetic gene clusters described to date, exemplifying the genetic diversity that drives small molecule biosynthesis. Although the lym and amm pathways currently represent only two examples of an unprecedented biosynthesis, the connection of lymphostin and the ammosamides to the broader class of pyrroloquinoline alkaloids is intriguing. Likely, many more opportunities to explore the biosynthesis of pyrroloquinoline alkaloids will come from marine sponges, which are the dominant source of these natural products.(Antunes et al., 2005) But, like many sponge-derived natural products, the pyrroloquinolines may be products of symbiotically associated bacteria, the genetic information of which is increasingly becoming accessible through metagenomic sequencing.(Hentschel et al., 2012) Insights from bacterial pyrroloquinoline biosynthesis may enlighten a general paradigm for the assembly of many pyrroloquinoline alkaloids, enabling future studies of sponge derived pyrroloquinolines and further expanding our basic understanding of the chemical diversity afforded through unconventional biosynthetic pathways.
Experimental Procedures
General
General methods including chemical and biological reagents and instrumentation employed in these studies can be found in the online supporting information.
Direct Capture of the amm gene cluster using transformation associated recombination
To directly capture the 37.5 kb region encompassing the ammosamide (amm) biosynthetic gene cluster, we followed the general procedure described by Yamanaka et al.(Yamanaka et al., 2014) Details specific to the TAR capture vector for the amm biosynthetic gene cluster can be found in the online supporting information.
Genetic Manipulations of pCAP01/amm
All genetic manipulations of pCAP01/amm were carried out using λ-Red recombination-mediated PCR targeted gene deletion, as described by Gust et al.(Gust et al., 2003) Individual gene disruption cassettes (for amm3, -4, -7, -8, -9, -11, -14, -16, -17 -18, -19, -23) were generated via PCR amplification of the apramycin resistance cassette (aac(3)IV) with flanking FRT sites from pMXT19(Tang et al., 2015). Primers were designed with 39 nucleotides matching the adjacent sequences of the targeted gene. Gene replacement by λ-Red recombination was achieved in E. coli BW25113/pKD20 carrying the pCAP01/amm construct. Mutated pCAP01/amm constructs were confirmed by restriction digest (NcoI) and PCR and then transferred to E. coli BT340 for FLP recombinase-mediated excision of the apramycin resistance gene. The corresponding knockouts, with FLP scar were verified by restriction digest and sequencing prior to transfer into Streptomyces coelicolor M512.
Gene deletion of amm6 was carried out in a similar manner as described in Yamanaka et al.(Yamanaka et al., 2012) An aac(3)IV cassette was amplified from pIJ773 by PCR using primers containing two NdeI (CATATG) restriction sites directly adjacent to the 39 nucleotides matching the region flanking amm6, such that the introduction of CATATG preserves the start codon of amm7 for in-frame gene replacement. Replacement of amm6 as described above was achieved in E. coli BW25113/pKD20 carrying pCAP01/amm. Excision of the apramycin resistance cassette from the mutant plasmid by NdeI digestion, followed by gel purification and ligation, yielded pCAP01/ammΔamm6.
Integration of Wild-Type and Mutant pCAP01/amm in S. coelicolor M512
Included in the pCAP01 vector backbone are the ϕC31 integrase gene (int) and the corresponding attachment site (attP) for integration into S. coelicolor. pCAP01/amm and mutant variants were introduced to E. coli ET12567 by electroporation and transferred to S. coelicolor M512 by triparental conjugation facilitated by E. coli ET-12567/pUB307. Kanamycin (Kan) resistant colonies were grown until sporulation on mannitol soy flour (MS) agar, with Kan and nalidixic acid (Nal), then plated for a second round of selection on Kan and Nal MS agar plates. Colonies of M512 with chromosomal insertion of pCAP01/amm were verified by PCR (gDNA isolated from 5-day tryptic soy broth (TSB) cultures). Exconjugants were screened using primer pairs corresponding to the sites of gene deletion and the resulting PCR product validated by Sanger sequencing. Spore stocks, which were used for all subsequent cultures, were created from a 3rd round of selection on Kan-Nal MS agar plates. For each mutant 3–4 individual colonies were selected for secondary metabolite profiling.
General Method for Genetic Complementation
The pKY01 plasmid, an integrative plasmid with ermE promotor and apramycin resistance cassette, was used for genetic complementation experiments, as previously described.(Yamanaka et al., 2012) pKY01 constructs were created using either Gibson Cloning or by traditional cloning using the NdeI and HindIII restriction sites of pKY01. The assembled plasmids were validated by restriction digestion and sequence prior to transfer to E. coli ET-12567 for triparental conjugation into designated S. coelicolor M512-pCAP01/amm mutants (see above). Exconjugants were selected on MS agar with apramycin (Apra) and Nal, and the colonies allowed to grow to sporulation twice. Primers F-pKY01Sc and R-pKY01Sc were used for screening exconjugants from isolated gDNA and Sanger sequencing of the corresponding PCR product. Spore stocks, which were used for all subsequent cultures, were created from a 3rd round of selection on Kan-Nal MS agar plates. For each mutant 3–4 individual colonies were selected for secondary metabolite profiling.
Design of pKY01/amm6amm7 Mutants Used for Genetic Complementation
Due to the overlapping amm6 tryptophan codon (TGG) and amm7 start codon (ATG), we were limited to mutations converting tryptophan to either a stop codon (W56stp) or cysteine (W56C) residue. These mutants are depicted as ‘pKY01/amm6(W56stp)amm7’ and ‘pKY01/amm6(W56C)amm7’ in the manuscript. As a consequence of amm6-amm7 overlap, mutating the tryptophan codon to a stop codon (TGG→TGA) also converts a valine residue on amm7 to leucine. Similarly, mutating the tryptophan codon to a cysteine codon (TGG→TGC) converts the valine residue on amm7 to methionine. These perturbations do not appear to affect the outcome of the experiments.
Secondary Metabolite Profiling of S. coelicolor M512-pCAP01/amm and mutants
Cultures of S. coelicolor M512-pCAP01/amm and mutant strains were started from 20 µL spores germinated in 200 µL 2× YT (yeast extract, tryptone) then added to 3 mL TSB with Nal and Kan. TSB precultures were grown for 5–7 days at 30 °C, prior to using 0.3 mL to inoculate 15 mL R5 production media without antibiotics.(Kieser et al., 2000) Production cultures were grown for 12 days at 30 °C and analyzed using the following method: Cultures were centrifuged to pellet M512 cells and the resulting supernatant loaded onto a 1 gram C18 solid phase extraction column that was washed, conditioned and pre-equilibrated at 5% acetonitrile (MeCN). The column was then washed with six column volumes of the 5% HPLC grade acetonitrile to remove salts and polar compounds, and then the entire column flushed with 9 column volumes of 85% MeCN. The eluted contents were frozen and the solvent was removed by lyophilization. The lyophilized residue was then dissolved in 0.4 mL HPLC grade MeOH and analyzed by LCMS using a Phenomenex Kinetex 2.6 µm XB-C18 100 Å, 150 × 4.6 mm column (0–3 min isocratic 10% MeCN, 3–23 minutes 10–100% MeCN; mobile phase buffered with 0.1% formic acid).
Secondary Metabolite Profiling of S. coelicolor M512-pCAP01/ammΔamm6
Cultures of S. coelicolor M512-pCAP01/ammΔamm6, including those genetically complemented with pKY01 constructs, were started from 20 µL spores germinated in 200 µL 2× YT then added to 3 mL TSB with Nal and Apra. TSB precultures were grown for 5–7 days at 30 °C prior to using 1 mL to inoculate 50 mL of R5 media without antibiotics. Cultures were grown for 12 days the extracted 3 times with 50 mL ethyl acetate and concentrated. The resulting residue was dissolved in 50 µL DMSO then diluted with 450 µL MeCN. This solution was filtered through a 50 µL plug of C18 silica to remove lipophilic components followed by 500 µL MeCN. The eluate was then analyzed by LCMS using a Phenomenex Kinetex 2.6 µm XB-C18 100 Å, 150 × 4.6 mm column (0–3 min isocratic 10% MeCN, 3–23 minutes 10–100% MeCN; mobile phase buffered with 0.1% formic acid).
Time-Course of Ammosamides A-C Production
Cultures S. coelicolor M512-pCAP01/amm in R5 media, or Streptomyces sp. CNR-698 in A1 were sampled daily starting one day following inoculation from TSB preculture or A1 preculture, respectively. Aliquots were diluted with an equal volume of HPLC grade MeOH and the 1:1 mixtures were vortexed for 1 minute then centrifuged at maximum speed for 5 minutes on a benchtop centrifuge. The supernatant was then filtered through a 0.2 micron filter and analyzed by LCMS using a Phenomenex Luna 5 µm C18(2) 100 Å, 100 × 4.6 mm column (0–3 min isocratic 10% MeCN, 2–10 min 10–100% MeCN, 10–12 min 100% MeCN; mobile phase buffered with 0.1% formic acid).
Chemical Complementation of M512-pCAP01/ammΔamm3
Secondary metabolite profiling for chemical complementation experiments was performed as above with R5 media supplemented with 6-Chloro-D,L-tryptophan. R5 media solutions were prepared by adding 6-Chloro-D,L-tryptophan (ChemImpex International, Cat. 20931), or L-tryptophan (control) to a final concentration of 4 mM, prior to autoclave sterilization. The R5 media was then used as previously as above for secondary metabolite profiling. Production cultures were extracted using C18 solid phase extraction and analyzed by LCMS using a Phenomenex Kinetex 2.6 µm XB-C18 100 Å, 150 × 4.6 mm column (0–3 min isocratic 10% MeCN, 3–23 minutes 10–100% MeCN; mobile phase buffered with 0.1% formic acid).
Production and Isolation of Ammosamide C from M512-pCAP01/amm
A 50 mL culture of R5 media was inoculated with 1 mL preculture of M512-pCAP01/amm (5 days, TSB media) and allowed to grow for 4 days prior to liquid extraction with n-butanol (3×50 mL). The extracts were concentrated, dissolved in water and loaded onto a 1 gram C18 solid phase extraction column. Red pigment began to elute with 10% MeCN. The mobile phase was switched to 100% MeOH and the combined eluate was concentrated in vacuo. The resulting residue was then purified by preparative HPLC using an Phenomenex Synergi 10 µm Hydro-RP 80, 250 × 21.2 mm preparative HPLC column (0–1 min isocratic 40% MeOH, 1–17 min 40–65% MeOH; mobile phase buffered with 0.1% trifluoroacetic acid) to yield 6.4 mg. LCMS and H-NMR analysis matched ammosamide C, which previously reported by Hughes et al.(Hughes and Fenical, 2010)
Production and Isolation of Compounds 4 and 8 from M512-pCAP01/ammΔamm3 + pKY01/amm3
Both mutants, M512-pCAP01/ammΔamm4 and M512-pCAP01/ammΔamm3 + pKY01/amm3, produce compound 4. However, for the isolation 4 we chose to use M512-pCAP01/ammΔamm3 + pKY01/amm3 due to consistently higher titers. Two 1L cultures of M512-pCAP01/ammΔamm3 + pKY01/amm3 were grown for 12 days prior to centrifugation to remove cell debris. The resulting pink colored supernatant could not be extracted with ethyl acetate or Amberlite XAD7HP resin. Thus, the entire 2 liters were passed through a bed of C18 silica gel. The resulting bed was washed thoroughly with water and the pink pigments eluted with 10–80% MeCN. The eluate was then lyophilized and the resulting crude material dissolved in 1:1 DMSO:MeOH for preparative HPLC purification with an Agilent Pursuit XRS 5 µm C18 100×21.2 mm preparative HPLC column (0–1 min 5% MeCN isocratic, 1–25 min 5–50% MeCN at 15 mL/min with a mobile phase buffered with 0.1% trifluoroacetic acid). Please see supporting items for UV-Vis, HPLC, MS and NMR spectra.
Compound 4 was further purified with on a Phenomenex Luna 5µm C8(2) 250×100 mm semi-preparative column (0–20 min, 10–13% MeCN at 4.5 mL/min with a mobile phase buffered with 0.1% trifluoroacetic acid) to yield 3.4 mg of 4. 1H-NMR (500 MHz, DMSO) δ 12.56 (s, 1H), 8.58 (s, 1H), 8.20 (s, 1H), 7.62 (s, 4H). 13C-NMR (126 MHz, DMSO) δ 166.2, 148.8, 147.9, 147.2, 140.9, 138.3, 135.6, 132.5, 122.3, 119.4, 100.0. Exact mass calculated for 4 [C11H7ClN4O2]H+ requires m/z = 263.0330, found 263.0356.
Compound 8 was further purified on a Phenomenex Luna 5µm C8(2) 250×100 mm semi-preparative column (0–20 min, 15–18% MeCN at 4.5 mL/min with a mobile phase buffered with 0.1% trifluoroacetic acid) to yield 0.8 mg of 8. 1H-NMR (599 MHz, DMSO) δ 10.20 (s, 1H), 8.32 (s, 1H), 6.57 (s, 2H), 6.37 (s, 2H). 13C-NMR (126 MHz, DMSO) δ 166.1, 165.2, 140.7, 132.9, 132.3, 131.7, 120.6, 118.0, 105.0, 104.4. Exact mass calculated for 8 [C11H7ClN4O3]H+ requires m/z = 279.0279, found 279.0314. The 1H- and 13C-NMR spectra matched synthetically prepared 8 (see Figure S5, and supporting experimental).
Production and Isolation of Compounds 6 and 7 from M512-pCAP01/ammΔamm23
A 1LM512-pCAP01/ammΔamm23 in R5 media was extracted three times ethyl acetate (3×1L) and the combined organics were dried with brine and anhydrous Mg2SO4 then concentrated in vacuo. The resulting residue was then dissolved in 1:1 DMSO:MeOH and purified by preparative HPLC with an Agilent Pursuit XRS 5 C18 100×21.2 mm preparative HPLC column (0–1 min 5% MeCN isocratic, 1–20 min 5–25% MeCN, at 15 mL/min with a mobile phase buffered with 0.1% trifluoroacetic acid). Both 6 and 7 were then further purified on a Phenomenex Luna 5µm C8(2) 250×100 mm semi-preparative column (5–25% MeCN at 4.5 mL/min with a mobile phase buffered with 0.1% trifluoroacetic acid) to yield 9.6 mg of 6 and 0.3 mg of 7. Please see supporting items for UV-Vis, HPLC, MS and NMR spectra.
Compound 6 1H-NMR (500 MHz, DMSO) δ 8.93 (s, 1H), 8.80 (s, 1H), 8.54 (s, 1H), 7.75 (s, 1H). 13C-NMR (126 MHz, DMSO) δ 165.9, 150.9, 148.3, 144.0, 136.9, 132.1, 128.3, 120.7, 119.6, 112.8, 98.4. Exact mass calculated for 6 [C11H8ClN5O]H+ requires m/z = 262.0490, found 262.0496.
Compound 7 1H-NMR (500 MHz, DMSO) δ 10.15 (s, 1H), 8.85 (s, 1H), 8.31 (s, 1H), 7.61 (s, 2H), 6.59 (s, 2H), 6.18 (s, 2H). 13C NMR (126 MHz, DMSO) δ 166.3, 165.1, 144.5, 140.1, 131.7, 131.4, 131.1, 120.0, 115.5, 104.4, 103.7. Exact mass calculated for 7 [C11H8N502]H+ requires m/z = 278.0439, found 278.0421. 1H- and 13C-NMR matched synthetic 7. The 1H- and 13C-NMR spectra matched synthetically prepared 7 (see Figure S5, and supporting experimental).
Highlights.
Pyrroloquinoline alkaloid biosynthesis is driven by cryptic conserved genes
Heterologous expression confirms the ammosamide gene cluster
Ribosomal peptide biosynthesis genes are required for an unprecedented biosynthesis
Acknowledgments
This work was funded by NIH grant R01-GM085770 to B.S.M. and a NIH postdoctoral fellowship to P.A.J. (5F32CA174333). Genome sequencing was conducted by the U.S. Department of Energy Joint Genome Institute and supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank our UCSD colleagues P. R. Jensen and W. Fenical for providing Streptomyces sp. CNR-698, C. C. Hughes for providing authentic standards, and B. M. Duggan and A. A. Mrse for providing NMR assistance. We are grateful for the helpful assistance from K. Yamanaka, E. N. Fielding and K. Chang.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
P.A.J. conducted the experiments. P.A.J. and B.S.M. designed the experiments and wrote the manuscript.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antunes EM, Copp BR, Davies-Coleman MT, Samaai T. Pyrroloiminoquinone and related metabolites from marine sponges. Nat. Prod. Rep. 2005;22:62–72. doi: 10.1039/b407299p. [DOI] [PubMed] [Google Scholar]
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Boddy CN. Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. Journal of Industrial Microbiology & Biotechnology. 2014;41:443–450. doi: 10.1007/s10295-013-1368-1. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. Genome mining for novel natural product discovery. J. Med. Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and Mode of Action of Lantibiotics. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Cimermancic P, Medema, Marnix H, Claesen J, Kurita K, Wieland Brown, Laura C, Mavrommatis K, Pati A, Godfrey, Paul A, Koehrsen M, Clardy J, et al. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre C, Challis GL. New natural product biosynthetic chemistry discovered by genome mining. Nat. Prod. Rep. 2009;26:977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- Foulston LC, Bibb MJ. Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:13461–13466. doi: 10.1073/pnas.1008285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: Protein Homology by Domain Architecture. Genome Research. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Chapter Fourteen - Streptomyces coelicolor as an Expression Host for Heterologous Gene Clusters. In: David AH, editor. Methods in Enzymology. Academic Press; 2012. pp. 279–300. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. Journal of Industrial Microbiology & Biotechnology. 2014;41:425–431. doi: 10.1007/s10295-013-1348-5. [DOI] [PubMed] [Google Scholar]
- Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. One-Pot Synthesis of Azoline-Containing Peptides in a Cell-free Translation System Integrated with a Posttranslational Cyclodehydratase. Chem. Biol. 2014;21:766–774. doi: 10.1016/j.chembiol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Greening C, Ahmed FH, Mohamed AE, Lee BM, Pandey G, Warden AC, Scott C, Oakeshott JG, Taylor MC, Jackson CJ. Physiology, Biochemistry, and Applications of F-420- and F-o-Dependent Redox Reactions. Microbiol. Mol. Biol. Rev. 2016;80:451–493. doi: 10.1128/MMBR.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Micro. 2012;10:641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- Hu J-F, Fan H, Xiong J, Wu S-B. Discorhabdins and Pyrroloiminoquinone-Related Alkaloids. Chem. Rev. 2011;111:5465–5491. doi: 10.1021/cr100435g. [DOI] [PubMed] [Google Scholar]
- Hughes CC, Fenical W. Total Synthesis of the Ammosamides. J. Am. Chem. Soc. 2010;132:2528–2529. doi: 10.1021/ja9106572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CC, MacMillan JB, Gaudencio SR, Jensen PR, Fenical W. The Ammosamides: Structures of Cell Cycle Modulators from a Marine-Derived Streptomyces Species. Angew. Chem., Int. Ed. 2009;48:725–727. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusainov R, Kuipers OP. When the Leader Gets Loose: In Vivo Biosynthesis of a Leaderless Prenisin Is Stimulated by a trans-Acting Leader Peptide. Chem Bio Chem. 2012;13:2433–2438. doi: 10.1002/cbic.201200437. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, David AH. Practical Streptomyces Genetics. John Innes Foundation; 2000. [Google Scholar]
- Koehnke J, Mann G, Bent AF, Ludewig H, Shirran S, Botting C, Lebl T, Houssen WE, Jaspars M, Naismith JH. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat Chem Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathila R, Merkler KA, Merkler DJ. Enzymatic formation of C-terminal amides. Nat. Prod. Rep. 1999;16:145–154. doi: 10.1039/a801346b. [DOI] [PubMed] [Google Scholar]
- Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat. Prod. Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- McAuliffe O, O'Keeffe T, Hill C, Ross RP. Regulation of immunity to the two-component lantibiotic, lacticin 3147, by the transcriptional repressor LtnR. Molecular Microbiology. 2001;39:982–993. doi: 10.1046/j.1365-2958.2001.02290.x. [DOI] [PubMed] [Google Scholar]
- Merkler DJ, Glufke U, Ritenour-Rodgers KJ, Baumgart LE, DeBlassio JL, Merkler KA, Vederas JC. Formation of nicotinamide from nicotinuric acid by peptidylglycine alpha-amidating monooxygenase (PAM): A possible alternative route from nicotinic acid (niacin) to NADP in mammals. J. Am. Chem. Soc. 1999;121:4904–4905. [Google Scholar]
- Miyanaga A, Janso JE, McDonald L, He M, Liu H, Barbieri L, Eustaquio AS, Fielding EN, Carter GT, Jensen PR, et al. Discovery and Assembly-Line Biosynthesis of the Lymphostin Pyrroloquinoline Alkaloid Family of mTOR Inhibitors in Salinispora Bacteria. J. Am. Chem. Soc. 2011;133:13311–13313. doi: 10.1021/ja205655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oman TJ, Knerr PJ, Bindman NA, Velásquez JE, van der Donk WA. An Engineered Lantibiotic Synthetase That Does Not Require a Leader Peptide on Its Substrate. J. Am. Chem. Soc. 2012;134:6952–6955. doi: 10.1021/ja3017297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan E, Oswald NW, Legako AG, Life JM, Posner BA, MacMillan JB. Precursor-directed generation of amidine containing ammosamide analogs: ammosamides E-P. Chemical Science. 2013;4:482–488. doi: 10.1039/C2SC21442C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: The structure of peptidylglycine alpha-hydroxylating monooxygenase. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- Selengut JD, Haft DH. Unexpected Abundance of Coenzyme F-420-Dependent Enzymes in Mycobacterium tuberculosis and Other Actinobacteria. J. Bacteriol. 2010;192:5788–5798. doi: 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silakowski B, Schairer HU, Ehret H, Kunze B, Weinig S, Nordsiek G, Brandt P, Blocker H, Hofle G, Beyer S, et al. New lessons of combinatorial biosynthesis from myxobacteria - The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 1999;274:37391–37399. doi: 10.1074/jbc.274.52.37391. [DOI] [PubMed] [Google Scholar]
- Tang X, Li J, Millán-Aguiñaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM, Moore BS. Identification of Thiotetronic Acid Antibiotic Biosynthetic Pathways by Target-directed Genome Mining. ACS Chemical Biology. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer JR, Rollema HS, Siezen RJ, Beerthuyzen MM, Kuipers OP, de Vos WM. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- Volz C, Kegler C, Müller R. Enhancer Binding Proteins Act as Hetero-oligomers and Link Secondary Metabolite Production to Myxococcal Development, Motility, and Predation. Chem. Biol. 2012;19:1447–1459. doi: 10.1016/j.chembiol.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Walsh C. Naturally Occuring 5-Deazaflavin Coenzymes - Biological Redox Roles. Accounts Chem. Res. 1986;19:216–221. [Google Scholar]
- Walsh CT, Fischbach MA. Natural Products Version 2.0: Connecting Genes to Molecules. J. Am. Chem. Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Bashiri G, Gao X, Sawaya MR, Tang Y. Uncovering the Enzymes that Catalyze the Final Steps in Oxytetracycline Biosynthesis. J. Am. Chem. Soc. 2013;135:7138–7141. doi: 10.1021/ja403516u. [DOI] [PubMed] [Google Scholar]
- Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. International Journal of Medical Microbiology. 2014;304:230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Ryan KS, Gulder TAM, Hughes CC, Moore BS. Flavoenzyme-Catalyzed Atropo-Selective N,C-Bipyrrole Homocoupling in Marinopyrrole Biosynthesis. J. Am. Chem. Soc. 2012;134:12434–12437. doi: 10.1021/ja305670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Garneau S, Walsh CT. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarins-Tutt JS, Barberi TT, Gao H, Mearns-Spragg A, Zhang L, Newman DJ, Goss RJM. Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2015;33:54–72. doi: 10.1039/c5np00111k. [DOI] [PubMed] [Google Scholar]
- Zehner S, Kotzsch A, Bister B, Sussmuth RD, Mendez C, Salas JA, van Pee K-H. A Regioselective Tryptophan 5-Halogenase Is Involved in Pyrroindomycin Biosynthesis in Streptomyces rugosporus LL-42D005. Chem. Biol. 2005;12:445–452. doi: 10.1016/j.chembiol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Zeng J, Zhan J. Characterization of a tryptophan 6-halogenase from Streptomyces toxytricini. Biotechnology Letters. 2011;33:1607–1613. doi: 10.1007/s10529-011-0595-7. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yu Y, Vélasquez JE, van der Donk WA. Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes - a review. Nat. Prod. Rep. 2016 doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]