Abstract
The bacterium Myxococcus xanthus undergoes multicellular development when starved. Thousands of cells build mounds in which some differentiate into spores. This remarkable feat and the genetic tractability of Myxococcus provide a unique opportunity to understand evolution of gene regulatory networks (GRNs). Recent work has revealed a GRN involving interconnected cascades of signal-responsive transcriptional activators. Initially, starvation-induced intracellular signals direct changes in gene expression. Subsequently, self-generated extracellular signals provide morphological cues that regulate certain transcriptional activators. However, signals for many of the activators remain to be discovered. A key insight is that activators often work combinatorially, allowing signal integration. The Myxococcus GRN differs strikingly from those governing sporulation of Bacillus and Streptomyces, suggesting Myxococcus evolved a highly signal-responsive GRN to enable complex multicellular development.
Keywords: gene regulatory network, signal transduction, bacterial development, sporulation, Myxococcus xanthus
Myxococcus multicellular development is controlled by cascades of signal-responsive transcription factors
Myxobacteria are common soil inhabitants that often prey on other bacteria [1]. When prey become scarce, myxobacteria build multicellular structures in which some of the rod-shaped cells transform into ovoid spores. The spores are dormant and resist environmental insults, yet they remain receptive to nutrient signals and can germinate and resume growth. One species of myxobacteria, Myxococcus xanthus, has been studied as a model organism to understand cooperative motility, cell-cell signaling, and gene regulation during a multicellular developmental process. Myxococcus cells are long, flexible rods capable of gliding over solid surfaces. Swarms of these bacteria have been described as microbial wolfpacks that lyse a variety of other bacteria and digest their contents [2]. However, when nutrient sources are exhausted, Myxococcus cells coordinate their movements during a process called “aggregation” (see Glossary), forming mounds that contain tens of thousands of cells (Figure 1). Within a mound, some cells differentiate into spores (about 5%), completing the developmental process of fruiting body formation. The majority of cells lyse during the aggregation process [3], perhaps providing nutrients that delay and/or fuel sporulation of the minority. A sizable fraction of cells remain outside fruiting bodies as “peripheral rods” (about 15%) [4]. This may be a bet-hedging strategy evolved to rapidly exploit reappearance of a nutrient source. How individual cells in the starving population adopt different fates is largely unknown, but can be easily studied in this genetically tractable and exciting model for elucidating gene regulatory networks (GRNs).
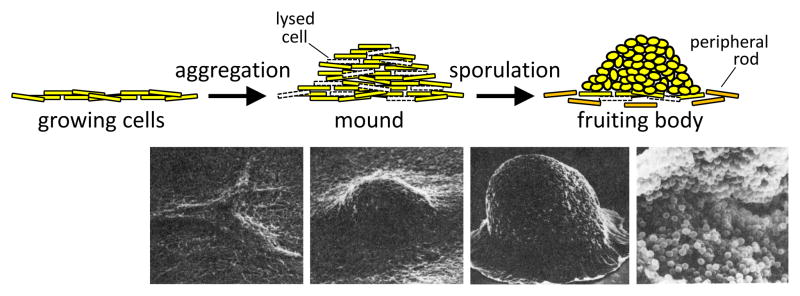
Figure 1. Myxococcus Fruiting Body Formation.
(A) Cartoon representation. When growing cells (yellow) become starved, they undergo aggregation and form a mound. Many cells lyse (dashed white cell outlines) during aggregation. Some of the rods differentiate into ovoid spores in the mound, resulting in a fruiting body. Other cells remain outside fruiting bodies as peripheral rods (orange). Depending on the strain and conditions, aggregation takes 1–2 days and spores form over the following 2–4 days. The process involves a much larger number of cells than depicted. (B) Scanning electron micrographs. The micrographs are aligned with the cartoon representation to show the sequence of morphological changes (left to right) from early aggregates [individual cells are barely visible as long (~5 μm), slender (~0.5 μm) rods] to mounds to fruiting bodies (~100 μm tall) that if cracked open reveal spores (~1 μm). Micrographs are from [87] with permission.
The complex multicellular developmental process of Myxococcus is governed by a GRN composed of signal-responsive transcription factors that act sequentially in cascades to control the timing of target gene expression [5]. An emerging theme is that some of the transcription factors also act combinatorially, providing signal integration, as is often seen in GRNs governing development of multicellular eukaryotes [6]. Recent work has also provided new insights into the signal inputs of the GRN as well as the functional ouputs of target genes. Based on analysis of DNA microarray data, the developmental GRN includes at least 410 up-regulated genes and 424 down-regulated genes, representing slightly more than 10% of the total genes [7]. This review will focus on the Myxococcus xanthus GRN, but will also include comparisons with other myxobacteria and with other model organisms, because their comparison provides unique insight into the evolution of GRNs governing bacterial development.
Overview of the GRN governing Myxococcus development
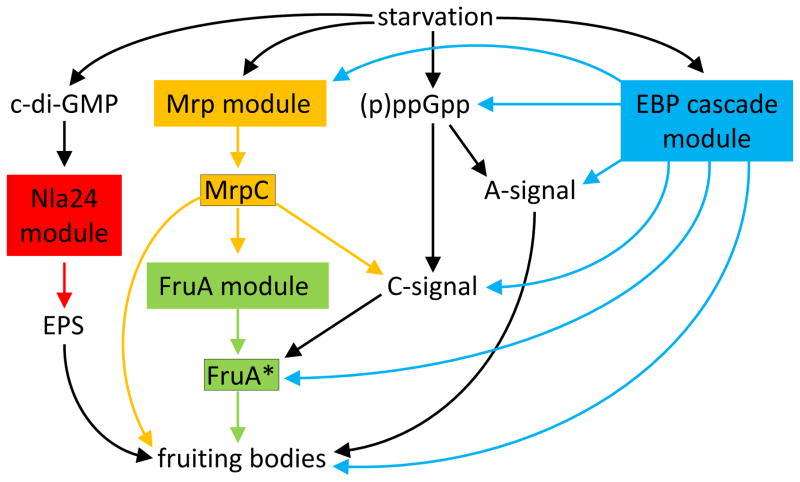
Previous reviews summarized the developmental GRN of Myxococcus in terms of three modules [5, 8]. This modular description will also be used here, with the addition of a fourth module that was discovered recently. The four modules – Nla24, Mrp, FruA, and an enhancer-binding protein (EBP) cascade – regulate key transcription factors (Figure 2). Starvation triggers the Nla24, Mrp, and EBP cascade modules, but the molecular signals are largely unknown. For the recently discovered Nla24 module, cyclic diguanylate (c-di-GMP) acts as a second messenger of the unknown starvation signal, and genes for exopolysaccharide (EPS) production are induced, which is necessary for fruiting body formation [9] (Figure 2). Starvation also causes another guanine nucleotide second messenger to be made in cells – RelA activity causes guanosine penta- and tetraphosphates [(p)ppGpp)] to accumulate when ribosomes stall due to amino acid limitation [10, 11] (Figure 2). This is called a “stringent response” and it is common in bacteria [12]. Stringent responses typically inhibit transcription of rRNA operons and up- or down-regulate transcription of many other genes. (p)ppGpp likely affects the transcriptome by binding to RNA polymerase and/or reducing the GTP level [13]. In Myxococcus, part of the (p)ppGpp-mediated stringent response is the production of extracellular A-signal and C-signal [10, 14–16] (Figure 2). The A-signal provides a measure of cell density [17]. If a quorum is reached, cells express early developmental genes and begin the aggregation process. Genetic evidence suggests that C-signal activates the transcription factor FruA (depicted as FruA* in Figure 2), facilitating aggregation [18–20], but many questions remain about C-signaling (Box 1). FruA* and the transcription factor MrpC (the output of the Mrp module) are believed to regulate late genes important for completion of the developmental process [21, 22] (Figure 2). As explained next, the EBP cascade module impacts many steps in the GRN (Figure 2).
Figure 2. Overview of the Gene Regulatory Network Governing Myxococcus Fruiting Body Formation.
Starvation triggers the Mrp (orange) and EBP cascade (blue) modules, and causes the second messengers c-di-GMP and (p)ppGpp to accumulate in cells. c-di-GMP binds to Nla24 (probably phosphorylated Nla24), the key transcription factor of the Nla24 module (red), activating genes for synthesis of exopolysaccharide (EPS) needed for fruiting body formation. (p)ppGpp causes extracellular A- and C-signals to be produced. A-signal provides quorum sensing for the decision to begin aggregation. C-signal is a short-range (possibly contact-dependent) signal that activates FruA (depicted as FruA*) (Box 1). FruA* and MrpC are transcription factor outputs of the FruA module (green) and the Mrp module, respectively, which separately and together regulate genes important for fruiting body formation. MrpC activates transcription of fruA. The EBP cascade stimulates the Mrp module, production of three signals, FruA* formation, and transcription of genes important for fruiting body formation. Feedback loops are omitted for simplicity. Adapted from [22].
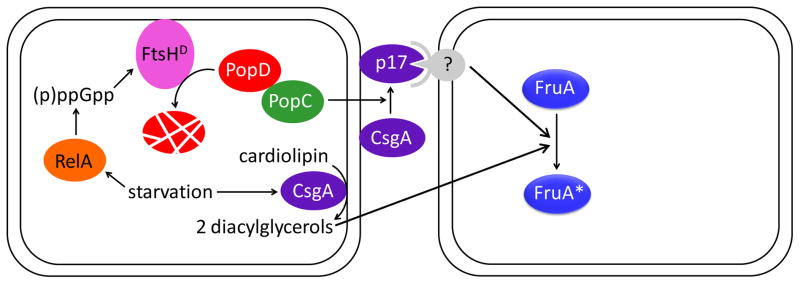
Box 1. C-signal and the Activation of FruA.
The identity of C-signal, its mode of transmission, and the mechanism by which it activates the FruA transcription factor are still under investigation.
C-signal appears to be a proteolytic fragment of CsgA and/or lipids produced by CsgA phospholipase activity. Involvement of CsgA in production of C-signal was inferred from isolation of csgA mutants whose development could be rescued by addition of wild-type cells [89, 90]. Purification of a factor from wild type capable of rescuing a csgA mutant identified a 17 kDa fragment of CsgA called C-factor or p17 [91, 92]. p17 was shown to be associated with the outer membrane [92], where it is produced from full-length CsgA (p25) by a protease called PopC [93] (Figure I, top pathway). Secretion of PopC is controlled by its inhibitor, PopD, which is degraded upon starvation in a RelA-dependent process that involves the protease FtsHD [15]. According to this model, p17 is C-signal. An alternate model is that cardiolipin phospholipase activity of intact CsgA releases diacylglycerols from the inner membrane that are C-signal [94] (Figure I, bottom pathway). This model is supported by analysis of a csgA suppressor mutation that causes overexpression of socA [95], which codes for a cardiolipin phospholipase [94], by mutational analysis ofcsgA [96], and by developmental rescue of a csgA socA mutant by a lipid fraction enriched in diacylglycerols from Myxococcus [94]. The two models for the identity of C-signal are not mutually exclusive.
Neither model for the identity of C-signal identifies a mode of transmission. p17 associated with the outer membrane of a donor cell would presumably require contact with a receptor on the surface of a recipient cell for transmission, but a receptor has not been identified. Diacylglycerols might passively diffuse through membranes, be actively transported across membranes, or be released by cell lysis. Presumably, these lipids would not travel far extracellularly, but they might not require cell-cell contact for transmission. Short-range or contact-dependent transmission of C-signal is supported by numerous studies [52–54].
The mechanism by which C-signal activates FruA is unknown. FruA is similar to response regulators of two-component signal transduction systems [19]. Typically, response regulators are phosphorylated by a protein kinase, but one has not been identified for FruA. Moreover, the receiver domain of FruA is atypical since it lacks key residues usually required for phosphorylation, and no change in DNA-binding by FruA after treatment with small-molecule phosphodonors was observed [21]. Hence, FruA may not be activated by phosphorylation as was originally proposed [18]. Many other mechanisms of posttranscriptional control are possible.
Figure I.
The GRN before and during Myxococcus aggregation
The EBP cascade module
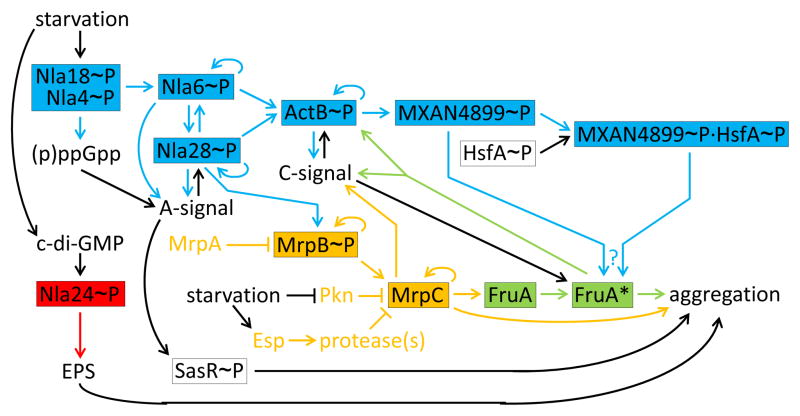
Figure 3 (Key Figure) shows early events during Myxococcus development in more detail. Starvation initiates a cascade of EBPs (shown in blue). These transcription factors typically bind to sites located about 100 base pairs upstream of promoters and activate transcription by σ54 RNA polymerase [23]. All EBPs in the cascade have a domain expected to be phosphorylated by a protein kinase in response to a signal, but the signals and some of the kinases remain to be identified [24]. For example, it is known that nla4 and nla18 mutants fail to induce (p)ppGpp and early developmental genes normally [25, 26], but little is known about how starvation presumably causes formation of Nla4~P and Nla18~P, or how these EBPs impact (p)ppGpp accumulation (Figure 3). Nla4~P and Nla18~P likely activate transcription of nla6 directly (since their DNA-binding domains bind to the promoter region), and likewise the phosphorylated forms of the other EBPs likely activate transcription of their targets directly [27, 28]. Combinatorial regulation by two or more EBPs (allowing signal integration) and positive autoregulation (providing signal amplification) are recurring themes in the EBP cascade (Figure 3). Perhaps the complexity of starvation sensing and signaling during aggregation demands combinatorial regulation, and EBPs evolved to fulfill this demand because their binding site position is flexible.
Figure 3 Key Figure. The Gene Regulatory Network before and during Myxococcus Aggregation.
Early events are shown in more detail than in Figure 2 and the same color scheme is used for the four modules (EBP cascade, blue; Nla24, red; Mrp, orange; FruA, green). Feedback loops, including autoregulatory ones, are also shown in this figure. Transcription factors are boxed. Arrows and lines with a barred end indicate positive and negative regulation, respectively. The EBP cascade is shown connected to the MrpC/FruA* cascade since Nla28~P appears to activate transcription of mrpAB (arrow from Nla28~P to MrpB~P) and MrpB~P appears to activate transcription of mrpC. (p)ppGpp is necessary for production of C-signal (Box 1), but this is omitted for clarity. FruA is activated posttranscriptionally by C-signal and by MXAN4899~P acting alone and/or in combination with HsfA~P (?), but the mechanisms of activation are unknown (Box 1). FruA* stimulates aggregation, enhancing short-range C-signaling (Box 1), which is depicted as positive feedback of FruA* directly on C-signal, for simplicity. See the text for details and references.
Consistent with the themes just noted, both Nla6~P and Nla28~P positively autoregulate, regulate each other, and regulate A-signal and ActB~P [27] (Figure 3). In addition, Nla28~P regulates MrpB~P [5], a component of the Mrp module that is also an EBP. All this regulation by Nla6~P and Nla28~P appears to involve transcriptional activation. In addition, Nla6~P appears to directly activate asgE [28], explaining why a nla6 mutant fails to make A-signal [29]. However, the mechanisms of reciprocal regulation between Nla28~P and A-signal are unknown (Figure 3). A-signal is a mixture of peptides and amino acids produced by secreted proteases, which (as noted above) may provide quorum sensing as a checkpoint prior to initiation of aggregation [17]. A-signal not only regulates the operon encoding Nla28 and its kinase [30], A-signal regulates most genes induced early in development [31]. Some of this regulation involves another EBP, SasR~P (Figure 3), whose cognate protein kinase, SasS, presumably senses A-signal [32]. However, direct targets of SasR~P have not been identified – this connection between A-signal and aggregation needs further investigation.
Farther downstream in the EBP cascade is ActB~P (Figure 3). It positively autoregulates, likely activates transcription of MXAN4899, and positively regulates C-signal [27, 33]. Interestingly, ActB~P and C-signal engage in reciprocal regulation [33, 34], as seen for Nla28~P and A-signal (Figure 3). Again, the mechanisms are unknown – the evidence for reciprocal regulation comes from phenotypic characterization of actB and csgA (the gene for C-signal production) mutants [33, 34]. The findings that both A-signal and C-signal feed back into the EBP cascade indicate that regulation of the cascade is linked to production and transmission of self-generated extracellular signals, coupling the GRN to morphological cues (i.e., cell density via A-signal and aggregation via C-signal).
The farthest downstream EBP known to be part of the cascade is MXAN4899~P (Figure 3). A null mutation in MXAN4899 appears to impair C-signal-dependent activation of FruA (i.e., formation of FruA*, as depicted in Figure 3) [35]. More recently, MXAN4899 was shown to form heterodimers with another EBP, HsfA, and combinatorially regulate genes involved in secondary metabolite production; whether HsfA~P is also involved in FruA* formation is unknown [36] (Figure 3). HsfA is present in growing cells [37], so it does not appear to be part of the EBP cascade in terms of its upstream regulation. A null mutation in hsfA impairs aggregation more strongly than a null mutation in MXAN4899 [36], but this phenotype may also reflect a separate role for HsfA~P in the activation of lonD (also called bsgA) early in development since LonD protease activity is necessary for aggregation [37]. A null mutation in lonD impairs expression of early developmental genes [38], but how LonD fits into the GRN is unknown [24].
To fully understand the role of the EBP cascade in the GRN, the molecular signals, cognate protein kinases, and target genes for each EBP must be elucidated.
The Nla24 module
This module was discovered recently [9], so much remains to be learned about its role in the GRN. So far, starvation has been shown to induce transcription of dmxB, which encodes a diguanylate cyclase that boosts the cellular level of c-di-GMP early in development. These events appear to be under control of the Dif chemosensory system [9], which responds not only to starvation, but to type IV pili of neighboring cells and lipids (perhaps released from lysing cells) [39]. c-di-GMP binds to Nla24 [9], an EBP encoded at a locus for EPS biosynthesis [40] but whose cognate protein kinase is unknown. Assuming that Nla24 is phosphorylated, it appears that a c-di-GMP·Nla24~P complex activates transcription of certain eps genes during development, increasing EPS production to enhance aggregation and sporulation [9] (Figure 3). c-di-GMP controls EPS production in many bacteria, and in some cases c-di-GMP regulation is coordinated with regulation by other second messengers and/or quorum sensors [41]. It will be interesting to see whether c-di-GMP regulation of the Nla24 module is integrated with control by other signals shown in Figure 3 or that remain to be discovered.
The Mrp module
The Mrp module includes three Mrp proteins and at least two signal transduction pathways (Pkn and Esp) that respond to starvation (Figure 3, orange). MrpA is a potential kinase of MrpB, but comparison of mrpA and mrpB mutant phenotypes suggests that MrpA acts primarily as a phosphatase of MrpB~P [42]. MrpB~P is an EBP that positively regulates transcription of the mrpAB operon (depicted as positive autoregulation of MrpB~P in Figure 3) and of mrpC, which encodes a transcription factor in the cyclic-AMP receptor protein (CRP) family [42]. MrpC is the key output of the Mrp module (Figure 2). MrpC appears to autoactivate [42, 43], activate transcription of fruA [44], and regulate a large number of genes, in many cases combinatorially with FruA* [21, 22] (Figure 3).
As with other modules in the GRN, starvation is an important input into the Mrp module (Figure 3). Starvation not only has an impact via the EBP cascade (i.e., Nla28~P appears to activate transcription of the mrpAB operon, which is depicted in Figure 3 as Nla28~P positively regulating MrpB~P) [5], but starvation also affects MrpC posttranslationally in two ways – starvation relieves phosphorylation of MrpC by the Pkn8 and Pkn14 cascade of protein kinases, allowing MrpC to bind DNA with higher affinity [43, 45] and starvation enhances MrpC proteolysis via the complex Esp signal transduction system [46, 47]. The molecules sensed by the Pkn and Esp pathways are unknown. These signals dictate the pace of development by controlling the activity and concentration of MrpC in cells. Mutations in components of the Pkn or Esp pathways can precociously elevate the MrpC level, resulting in earlier aggregation and premature sporulation outside of fruiting bodies [45, 46]. Starving cells engaging in aggregation accumulate more MrpC than non-aggregating cells in a population [3]. These observations suggest that MrpC is a master transcriptional regulator of aggregation and sporulation. In agreement, recent work suggests that the MrpC level also serves as a checkpoint for developmental progression – addition of nutrients during aggregation causes rapid proteolysis of MrpC and blocks commitment to sporulation [48]. Therefore, it is important to elucidate both the regulation and the targets of MrpC.
To begin finding targets of MrpC, a ChIP-seq study of developing Myxococcus was performed, and showed that MrpC binds to the promoter regions of nearly 300 genes known to be up- or down-regulated during development [22]. These include genes for many protein kinases and transcription factors, and genes involved in signal production, motility, and spore formation. However, these potential direct targets of MrpC require further investigation (e.g., RNA analysis of an mrpC mutant) before they can be incorporated into the GRN.
The FruA module
How C-signal activates FruA is a crucial question (Box 1), because activated FruA (depicted as FruA*) is the key output of the FruA module (Figures 2 and 3, green). Despite uncertainty about the nature of FruA*, and how C-signal and MXAN4899~P (alone or in combination with HsfA~P) stimulate FruA* formation (Figure 3), the role of FruA* in the GRN can be inferred from mutant phenotypes. A null mutation in MXAN4899 delays aggregation, but eventually mounds form, although they are larger and less regular in shape than normal [35]. A fruA mutant is even more defective for aggregation [19], suggesting that a low level of FruA* in the MXAN4899 mutant is sufficient to allow expression of some genes involved in aggregation. These aggregation genes presumably impact methylation of FrzCD [18, 20, 49], a component of the Frz chemosensory system that controls the gliding movements of cells [50, 51]. Aggregation brings cells into proximity, enhancing C-signaling [52–54] (Box 1). This is depicted as positive feedback of FruA* directly on C-signal in Figure 3 (for simplicity). A second positive feedback loop of FruA* on C-signal involves the act operon [34] (shown in Figure 3 as positive regulation of ActB~P). MrpC also regulates C-signal production positively [55] (Figure 3). These positive feedback loops presumably ensure that the FruA* level rises in aggregated cells, as has been shown for FruA by immunoblot [3]. Rising levels of C-signal and FruA* appear to differentially regulate genes and ensure that aggregation precedes sporulation [56–58].
Combinatorial control by MrpC and FruA*
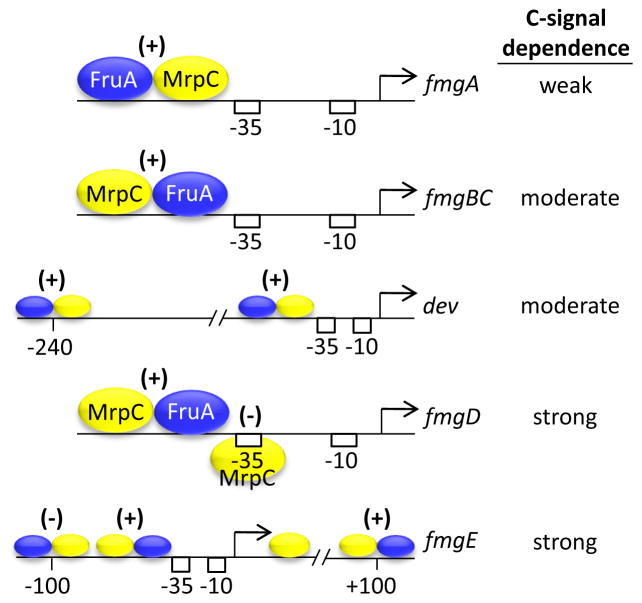
By studying the regulation of several C-signal-dependent genes, it was discovered that purified MrpC and FruA bind cooperatively to promoter regions of many genes important for normal aggregation and/or sporulation [21, 22, 59–62]. Figure 4 shows the arrangement of MrpC and FruA binding sites in five promoter regions that exhibit differential dependence on C-signal for expression. In all five cases, cooperative binding of MrpC and FruA* immediately upstream of the promoter −35 region appears to positively regulate transcription [21, 59–62]. Occupancy at this site is proposed to compete with occupancy by MrpC or the combination of MrpC and FruA* at nearby sites in the two promoter regions that depend most strongly on C-signal [59, 61]. Presumably, C-signal-dependent activation of FruA eventually causes the level of FruA* to rise sufficiently to activate transcription of strongly C-signal-dependent genes, which are expressed late during development [38]. In this way, the arrangement and affinity of binding sites for MrpC and FruA* may explain the observed differences in C-signal dependence and timing of expression of genes during development.
Figure 4. Cooperative Binding of MrpC and FruA at Promoter Regions May Explain Differential Dependence on C-signal.
The two transcription factors bind in different arrangements immediately upstream of the promoter −35 and −10 sequences, and activate transcription (+). In the fmgD promoter region, a second MrpC binding site overlaps the promoter −35 sequence and the FruA binding site, which is proposed to cause stronger dependence on C-signal activation of FruA (column at right) in order to overcome the negative effect of MrpC (−). Larger regions around the dev and fmgE promoters show the approximate position (relative to the transcriptional start site) and the effect on promoter activity (positive, +; negative, −) of distal cooperative binding sites. The negative regulatory site at −100 may compete with the fmgE promoter proximal site for cooperative binding of MrpC and FruA*, explaining the strong dependence on C-signal.
Combinatorial regulation by MrpC and FruA* appears to ensure that only starving cells (capable of accumulating MrpC) in close proximity in a mound (capable of C-signaling to activate FruA) commit to spore formation. As noted above, addition of nutrients during aggregation causes rapid proteolysis of MrpC and blocks commitment to sporulation [48]. Aggregating cells accumulate more MrpC, FruA, and C-signal than non-aggregating cells [3], but how these subpopulations relate to the three fates of cells (spore, peripheral rod, lyse) is unclear. Methods to measure gene expression in individual cells and track their fate during development are urgently needed. Developmental lysis (the fate of most cells) was proposed to be a programmed event mediated by MazF RNase activity [63], but recent work showed that MazF impacts lysis only of mutants with a defect in the outer membrane secretin PilQ [3, 64]. Whether lysis is a programmed event or a stochastic response to starvation will be exciting to investigate.
The GRN governing Myxococcus sporulation
Some of the C-signal-dependent genes under combinatorial control of MrpC and FruA* (Figure 4) are important for aggregation and/or sporulation [21, 60–62] (Figure 5). Among these genes, some in the dev operon powerfully impact sporulation. Null mutations in the devTRS genes reduce sporulation more than 100-fold [65, 66]. Surprisingly, it is not the loss of devTRS that inhibits sporulation of these mutants, but rather overexpression of devI, the first gene in the operon [67]. In devTRS mutants, transcription from the dev promoter is increased about 10-fold [66–68]. Further insight came from sequencing natural isolates – most were found to lack a functional dev promoter and devI [67]. Curiously, however, the strains lacking the dev promoter and devI still formed a normal number of spores, unlike the devTRS mutants. These observations suggested that devI overexpression in devTRS mutants might cause their sporulation defect. In agreement, null mutations in devI restore sporulation of devTRS mutants [67] (Rajagopalan and Kroos, unpublished). Also, devI mutants form spores about 6 h earlier than normal (Rajagopalan and Kroos, unpublished). It appears that DevI codes for a small protein that inhibits sporulation if overexpressed and delays sporulation when expressed normally (Figure 5). Elucidation of the mechanism of inhibition by DevI may shed light on the early steps of cellular shape change during spore formation. The dev operon is a CRISPR-Cas system that might protect cells from phage infection during development [68]. The lack of a functional dev promoter and devI in most natural isolates suggests this CRISPR-Cas system only recently became functional in a minority of strains that acquired a promoter [67], perhaps in niches where delayed sporulation and protection from phage infection proved advantageous.
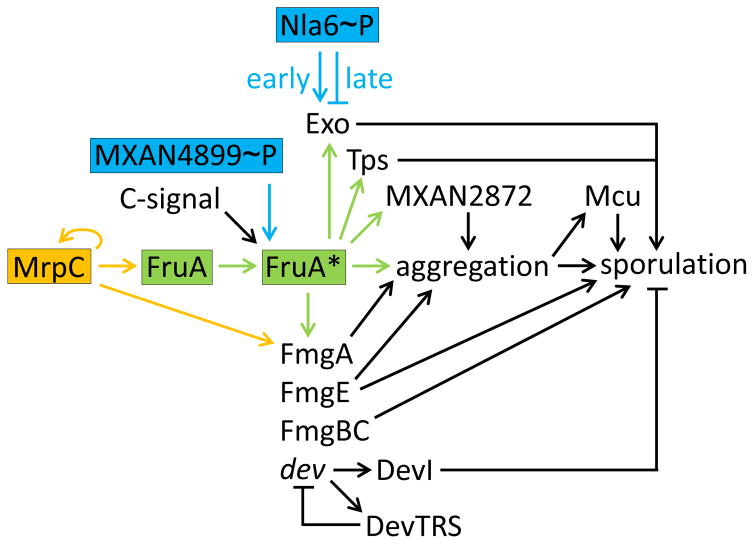
Figure 5. The Gene Regulatory Network Governing Myxococcus Sporulation.
The same color scheme is used as in Figures 2 and 3 for the Mrp (orange) and FruA (green) modules, but less detail about early events is shown. The MrpC/FruA* transcription factor cascade is emphasized. C-signal and MXAN4899~P (possibly with HsfA~P, Figure 3) activate FruA by unknown mechanisms (Box 1). Cells brought into proximity during aggregation are proposed to engage in efficient C-signaling that serves as a morphological cue and results in a rising level of FruA*, which separately and in combination with MrpC activates genes whose products promote further aggregation and eventually sporulation. Overexpression of DevI in devTRS mutants inhibits sporulation, perhaps by inhibiting Exo expression [69, 88] (not shown). See the text for details and references.
Sporulation involves cellular shape change and building a protective coat at the cell surface. The cytoskeletal protein MreB is important for the rod-to-spore transition and its reversal upon germination [69]. Presumably, these transitions involve extensive remodeling of the cell wall peptidoglycan. A few candidates for enzymes that might remodel peptidoglycan during sporulation emerged from genome-wide expression profiling during chemically-induced sporulation, which circumvents aggregation [70], but more work is needed to determine whether the corresponding genes are required for starvation-induced sporulation and, if so, how they fit into the GRN. The expression profiling experiment also led to genes whose products help build the spore coat. In a follow-up study, Exo and Nfs proteins were shown to be involved in forming the polysaccharide coat that encases the spore [69]. Nfs proteins form a complex that is moved around the spore surface by the Agl motor, distributing Exo-secreted glycan strands [71]. The Agl motor also powers gliding motility earlier in development during aggregation, and it powers motility for predation when prey are available [72]. Regulation of the nfs operon is not understood well enough to place Nfs in the GRN, but the exo operon is likely under direct combinatorial control of Nla6~P and FruA* (Figure 5). Nla6~P appears to activate exo transcription early in development, then repress it during aggregation [28], and FruA* appears to activate it just before sporulation [73].
FruA* also appears to regulate an abundant protein associated with the spore coat (Tps) [19] and a putative flavin adenine dinucleotide-binding monooxygenase (MXAN2872) that stimulates aggregation [74] (Figure 5). MXAN2872 was identified in a genetic screen for regulators of the muc operon [74], which encodes a chaperone/usher protein secretion system necessary for sporulation [75]. MXAN2872 appears to regulate muc indirectly by stimulating aggregation [74]. How MXAN2872 regulates aggregation is unknown. Further studies will likely reveal a novel mechanism controlling aggregation, a process we know little about currently.
The developmental GRN in other myxobacteria
Comparative genomic analysis so far has indicated that the developmental GRN is conserved in close relatives of M. xanthus, but not in more distant relatives. Three myxobacteria in the same suborder as M. xanthus have orthologs of proteins in the EBP cascade and the Mrp module (except Pkn8), and they have orthologs of FruA, CsgA (C-signal), and Nla24 [7, 76] (Kroos, unpublished). In contrast, two myxobacteria in different suborders than M. xanthus lack orthologs of most of these proteins (except MrpC, Pkn14, and Nla24). These differences could reflect divergent evolution from a common ancestor capable of development. Alternatively, the gene differences could reflect convergent evolution from a common ancestor incapable of development. Distinguishing between these possibilities will be facilitated by comparison of additional genome sequences and of GRNs characterized by genomic and proteomic approaches.
Comparison of the Myxococcus, Bacillus, and Streptomyces developmental GRNs
In two other experimental paradigms, GRNs governing bacterial development have been studied extensively. One GRN governs endospore formation by Bacillus subtilis [77]. The other is a composite of work on three species of Streptomyces (coelicolor, griseus, and venezuelae), which form chains of spores [78]. Although the developmental processes of Bacillus and Streptomyces differ morphologically from each other and from Myxococcus, and the processes evolved separately, the processes share some features – starvation initiates the process, hundreds of genes are regulated using multiple signals, and some cells lyse (or adopt other fates) while others differentiate into dormant spores that withstand insults and germinate when nutrients become available. Comparing the three GRNs provides unique insight into the constraints and flexibility of strategies to regulate a given developmental process.
Early events in the Bacillus GRN are controlled by a phosphorelay in which protein kinases autophosphorylate and initiate a chain of protein phosphotransfers that culminates in phosphorylation of Spo0A, a transcription factor that activates or represses many genes [77]. The level of Spo0A~P can differ between cells in a population and determine cell fate accordingly, both indirectly by down-regulating the AbrB and SinR global repressors, and directly by activating sporulation genes[77]. Neither Streptomyces nor Myxococcus have a phosphorelay, and little is known about cell fate determination. Both use guanine nucleotide second messengers to respond to starvation. In Streptomyces, decreasing c-di-GMP upon starvation appears to inactivate BldD, a global repressor of developmental genes [78].
A striking difference among the GRNs is the prevalence of EBPs in Myxococcus and their absence from Bacillus and Streptomyces. This difference was noted in an earlier comparison of the Myxococcus and Bacillus GRNs, as was the presence of autoregulatory, feed-forward, and feedback loops in both GRNs [79]. Such loops provide signal amplification, combinatorial control, and checkpoints that ensure proper temporal and spatial gene expression. In the Streptomyces GRN, several key transcription factors autoregulate [80], but combinatorial control and feedback loops are so far less apparent. An exception is combinatorial control of late sporulation genes by BldM·WhiI heterodimers, both of which are atypical response regulators [78], like Myxococcus FruA (Box 1).
The dominant role played by the interconnected EBP and MrpC/FruA* cascades in the Myxococcus GRN is replaced by cascades of σ factors in the Bacillus GRN [77]. Two cell types form during Bacillus endosporulation – the mother cell and the forespore. Three signaling pathways between the two cell types regulate their distinct σ factor cascades. Anti-σ factors modulate σ activity globally in each cell type [81, 82], while auxiliary transcription factors fine-tune the expression of target genes [79]. In the Streptomyces GRN, mixed cascades of σ factors and other transcription factors play the dominant role, with modulation of the activity of some σ factors by anti-σ factors [78, 80]. In contrast, very little is known about involvement of σ and anti-σ factors in the Myxococcus GRN, except that EBPs presumably activate transcription by σ54 RNA polymerase and that the genome encodes 52 σ factors in the σ70 family and at least 30 anti-σ factors [83, 84]. These potential components of the developmental GRN present intriguing opportunities for future exploration.
Concluding Remarks and Future Perspectives
The GRN governing Myxococcus development primarily involves two interconnected cascades (EBP and MrpC/FruA*) of signal-responsive transcription factors that often act combinatorially (Figure 3). Two additional pathways impact aggregation through Nla24~P and SasR~P, EBPs that respond to c-di-GMP and A-signal, respectively. Starvation induces a stringent response that generates the intracellular second messenger (p)ppGpp, which stimulates production of extracellular A- and C-signals. A-signal appears to be peptide signaling for the purpose of quorum sensing, providing both feedback to the EBP cascade via Nla28~P and input to SasR~P. C-signal appears to be a proteolytic fragment of CsgA and/or lipids produced by CsgA phospholipase activity, which provides a measure of cell proximity (i.e., aggregation), feeds back to the EBP cascade via ActB~P, and activates FruA. The mechanisms by which the A- and C-signals affect EBPs and FruA remain to be elucidated, as do the molecular signals and mechanisms that affect MrpC and many of the EBPs (see Outstanding Questions).
Outstanding Questions Box.
How is starvation detected? Specifically, how do Nla4~P and Nla18~P respond to starvation by regulating (p)ppGpp accumulation and early gene expression? Also, how is activity of MrpC regulated in response to starvation by the Pkn kinase cascade and the Esp signal transduction/proteolytic system, and when nutrients are added to developing cells prior to commitment to sporulation?
What molecular signals control activity of downstream EBPs in the cascade, how do they do so via cognate kinase/phosphatase proteins, and what are the outputs?
How are the A- and C-signals produced and what are their effects on the network? In particular, how does A-signal regulate most early genes and how does C-signal affect FruA activity?
How are cell fates specified? Is cellular lysis a programmed event during development and in any case what role does it play? What determines whether a cell remains outside fruiting bodies as a peripheral rod? What molecular events constitute commitment to sporulation? Are there genes under cooperative control of MrpC and FruA* that are up-regulated and together with MreB cause the rod-to-spore transition? How does DevI inhibit sporulation?
What ecological and other forces drove (and continue to drive) evolution of the highly signal-responsive Myxococcus developmental GRN and how does this compare with the GRNs of other myxobacteria and other organisms that undergo development?
The Myxococcus developmental GRN exhibits features commonly found in developmental GRNs, including those of Bacillus and Streptomyces – positive autoregulation provides signal amplification that drives development forward, combinatorial control allows signal integration that ensures proper timing of gene expression, and feedback loops reinforce developmental progression and in some cases couple the GRN to morphological cues. However, a distinctive feature of the Myxococcus GRN (as compared with those of Bacillus and Streptomyces) is the prominent role played by EBPs, all expected to be phosphorylated by protein kinases in response to signals and most if not all expected to activate transcription by σ54 RNA polymerase, whereas other σ factors, anti-σ factors, and global repressors, so far, appear to play less conspicuous or no roles. It appears that a GRN with many signals and much signal integration evolved to enable tens of thousands of Myxococcus cells to coordinate their movements and differentiation so that spore-filled fruiting bodies form. Comparatively speaking, the GRNs of Bacillus, Streptomyces, and perhaps even distantly-related myxobacteria, appear to be less signal-intensive, presumably because these GRNs evolved to govern less complex developmental processes. Each Myxococcus fruiting body is a population capable of surviving starvation and other insults, or being transported to a more favorable environment where spores will germinate and produce a colony for cooperative feeding. Understanding the ecology and evolution of Myxococcus and other myxobacteria in terms of their GRNs is an emerging area of research [24, 76, 85, 86] and a significant challenge (see Outstanding Questions). To meet this challenge, new systematic and quantitative experimental approaches will need to be coupled with computational methods to build molecular models of the GRNs that can rapidly predict their output under many different conditions, leading to novel testable hypotheses.
Supplementary Material
Trends Box.
In Myxococcus xanthus, a stringent response to starvation causes (p)ppGpp to accumulate, and recently a diguanylate cyclase was discovered that boosts c-di-GMP early in development. These second messengers initiate the signaling through the GRN.
Recent work found that a cascade of signal-responsive enhancer-binding proteins (EBPs) connects transcriptionally to a cascade involving MrpC and FruA, transcription factors that respond to starvation and short-range C-signaling, respectively. Signal integration is achieved by cooperative binding of MrpC and FruA to promoter regions.
Emerging genome sequences reveal surprising diversity among myxobacterial species, suggestive of divergent or convergent evolution of developmental GRNs.
The prominence of EBPs and other signal-responsive transcriptional activators in the Myxococcus GRN contrasts with the lack of these types of activators in the Bacillus and Streptomyces sporulation GRNs, suggesting Myxococcus development demanded evolution of a highly signal-responsive GRN.
Acknowledgments
I would like to thank members of my group, past and present, for stimulating discussions. I am grateful to David Arnosti and Amy Ralston for insightful comments on the manuscript. The work of my group on Myxococcus xanthus and Bacillus subtilis and is supported by NSF grant MCB-1411272 and NIH grant GM43585, respectively, and my salary is supported partly by Michigan State University AgBioResearch.
Glossary
- Aggregation
Myxococcus cells move to a particular location and pile on top of each other.
- Anti-sigma (σ) factor
a protein that binds to a sigma factor and inhibits its activity.
- Autoregulation
a gene product regulates the gene encoding it.
- Bet-hedging strategy
cells adopt different fates and the differentiated cells have optimal fitness under different conditions, improving the long-term fitness of the population.
- Cascade (of transcription factors)
one transcription factor activates transcription of a second transcription factor, which may in turn activate transcription of a third transcription factor, etc.
- Combinatorial (transcription factor activity)
two or more transcription factors regulate the same gene, possibly by forming heterodimers, binding cooperatively to DNA, or binding separately.
- Cyclic diguanylate (c-di-GMP)
a second messenger signal used in many bacteria.
- Endospore
a spore forms inside a mother cell.
- Enhancer-binding protein (EBP)
a transcription factor capable of binding DNA distal from a promoter, hydrolyzing ATP, and activating transcription by σ54 RNA polymerase; sometimes called NtrC-like activator (Nla) or σ54 activator.
- Forespore
the forming endospore.
- Gene regulatory network (GRN)
a description of the positive and negative effects of genes and/or proteins on synthesis and/or activity of each other, often including effects of signaling molecules as well.
- Global repressor
a direct negative regulator of transcription of many genes.
- Quorum sensing
cells produce extracellular, diffusible signal molecules and detect them to measure cell-population density, and regulate gene expression accordingly.
- Response regulator
a protein typically with an N-terminal receiver domain that is phosphorylated on an aspartate residue, causing a conformational change that activates a C-terminal output domain (a DNA-binding domain if the protein is a transcription factor).
- Second messenger
an intracellular signaling molecule produced in response to another signal, in this case starvation.
- Sigma (σ) factor
a subunit of bacterial RNA polymerase that directs the enzyme to particular promoters for transcription. Bacteria typically have multiple σ factors.
- Sporulation
the process by which a growing cell becomes a dormant, resistant spore.
- Stringent response
amino acid limitation or other stress conditions lower the cellular concentration of charged tRNA, causing ribosomes to stall and RelA to associate with those ribosomes and synthesize pppGpp, which can be converted to ppGpp by phosphohydrolase activity.
- Type IV pili
fibers that in Myxococcus can be extended from the cell surface, adhere to EPS on the surface of another cell or on the substratum (i.e., a “slime trail” left by passage of another cell), and then retracted to pull the cell (this form of movement is called “S motility”).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014. [Google Scholar]
- 2.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee B, et al. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol. 2012;194:3058–3068. doi: 10.1128/JB.06756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connor KA, Zusman DR. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajagopalan R, et al. Developmental gene regulation. In: Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014. pp. 105–126. [Google Scholar]
- 6.Davidson EH. Genomic Regulatory Systems. San Diego: Academic Press; 2001. [Google Scholar]
- 7.Huntley S, et al. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol Biol Evol. 2010;28:1083–1097. doi: 10.1093/molbev/msq292. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser D, et al. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb Perspect Biol. 2010;2:a000380. doi: 10.1101/cshperspect.a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skotnicka D, et al. A minimal threshold of c-di-GMP is essential for fruiting body formation and sporulation in Myxococcus xanthus. PLoS Genet. 2016;12:e1006080. doi: 10.1371/journal.pgen.1006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris BZ, et al. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haugen SP, et al. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford EW, Jr, Shimkets LJ. The Myxococcus xanthus socE and csgA genes are regulated by the stringent response. Mol Microbiol. 2000;37:788–799. doi: 10.1046/j.1365-2958.2000.02039.x. [DOI] [PubMed] [Google Scholar]
- 15.Konovalova A, et al. A RelA-dependent two-tiered regulated proteolysis cascade controls synthesis of a contact-dependent intercellular signal in Myxococcus xanthus. Mol Microbiol. 2012;84:260–275. doi: 10.1111/j.1365-2958.2012.08020.x. [DOI] [PubMed] [Google Scholar]
- 16.Konovalova A, et al. Two intercellular signals required for fruiting body formation in Myxococcus xanthus act sequentially but non-hierarchically. Mol Microbiol. 2012;86:65–81. doi: 10.1111/j.1365-2958.2012.08173.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuspa A, et al. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellehauge E, et al. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa M, et al. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 20.Sogaard-Andersen L, et al. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 21.Mittal S, Kroos L. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc Natl Acad Sci USA. 2009;106:1965–1970. doi: 10.1073/pnas.0808516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson M, et al. Transcription factor MrpC binds to promoter regions of many developmentally-regulated genes in Myxococcus xanthus. BMC Genomics. 2014;15:1123. doi: 10.1186/1471-2164-15-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev. 2012;76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bretl DJ, Kirby JR. Molecular mechanisms of signaling in Myxococcus xanthus development. J Mol Biol. 2016;428:3805–3830. doi: 10.1016/j.jmb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Diodati ME, et al. Nla18, a key regulatory protein required for normal growth and development of Myxococcus xanthus. J Bacteriol. 2006;188:1733–1743. doi: 10.1128/JB.188.5.1733-1743.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossa F, et al. The Myxococcus xanthus Nla4 protein is important for expression of stringent response-associated genes, ppGpp accumulation, and fruiting body development. J Bacteriol. 2007;189:8474–8483. doi: 10.1128/JB.00894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giglio KM, et al. A cascade of coregulating enhancer binding proteins initiates and propagates a multicellular developmental program. Proc Natl Acad Sci USA. 2011;108:E431–E439. doi: 10.1073/pnas.1105876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giglio KM, et al. The enhancer binding protein Nla6 regulates developmental genes that are important for Myxococcus xanthus sporulation. J Bacteriol. 2015;197:1276–1287. doi: 10.1128/JB.02408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caberoy NB, et al. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J Bacteriol. 2003;185:6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarwar Z, Garza AG. The Nla28S/Nla28 two component signal transduction system regulates sporulation in Mxyococcus xanthus. J Bacteriol. 2012;194:4698–4708. doi: 10.1128/JB.00225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuspa A, et al. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan H. Multicellular development and gliding motility in Myxococcus xanthus. Curr Opin Microbiol. 2003;6:572–577. doi: 10.1016/j.mib.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Gronewold TM, Kaiser D. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 34.Gronewold TM, Kaiser D. Mutations of the act promoter in Myxococcus xanthus. J Bacteriol. 2007;189:1836–1844. doi: 10.1128/JB.01618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelsbak L, et al. Enhancer-binding proteins with an FHA domain and the σ54 regulon in Myxococcus xanthus fruiting body development. Proc Natl Acad Sci USA. 2005;102:3010–3015. doi: 10.1073/pnas.0409371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volz C, et al. Enhancer binding proteins act as hetero-oligomers and link secondary metabolite production to myxococcal development, motility, and predation. Chem Biol. 2012;19:1447–1459. doi: 10.1016/j.chembiol.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Ueki T, Inouye S. Transcriptional activation of a heat-shock gene, lonD, of Myxococcus xanthus by a two component histidine-aspartate phosphorelay system. J Biol Chem. 2002;277:6170–6177. doi: 10.1074/jbc.M110155200. [DOI] [PubMed] [Google Scholar]
- 38.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 39.Zusman DR, et al. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 40.Lancero H, et al. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology. 2004;150:4085–4093. doi: 10.1099/mic.0.27381-0. [DOI] [PubMed] [Google Scholar]
- 41.Romling U, et al. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H, Shi W. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J Bacteriol. 2001;183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nariya H, Inouye S. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol. 2006;60:1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- 44.Ueki T, Inouye S. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc Natl Acad Sci USA. 2003;100:8782–8787. doi: 10.1073/pnas.1533026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nariya H, Inouye S. Identification of a protein Ser/Thr kinase cascade that regulates essential transcriptional activators in Myxococcus xanthus development. Mol Microbiol. 2005;58:367–379. doi: 10.1111/j.1365-2958.2005.04826.x. [DOI] [PubMed] [Google Scholar]
- 46.Higgs PI, et al. EspA, an orphan hybrid histidine protein kinase, regulates the timing of expression of key developmental proteins of Myxococcus xanthus. J Bacteriol. 2008;190:4416–4426. doi: 10.1128/JB.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schramm A, et al. Intra- and inter-protein phosphorylation between two hybrid histidine kinases controls Myxococcus xanthus developmental progression. J Biol Chem. 2012;287:25060–25072. doi: 10.1074/jbc.M112.387241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajagopalan R, Kroos L. Nutrient-regulated proteolysis of MrpC halts expression of genes important for commitment to sporulation during Myxococcus xanthus development. J Bacteriol. 2014;196:2736–2747. doi: 10.1128/JB.01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sogaard-Andersen L, Kaiser D. C factor, a cell-surface-associated intercellular signaling protein, stimulates the cytoplasmic Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaimer C, et al. Chemosensory signaling controls motility and subcellular polarity in Myxococcus xanthus. Curr Opin Microbiol. 2012;15:751–757. doi: 10.1016/j.mib.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keilberg D, Sogaard-Andersen L. Regulation of bacterial cell polarity by small GTPases. Biochemistry. 2014;53:1899–1907. doi: 10.1021/bi500141f. [DOI] [PubMed] [Google Scholar]
- 52.Kroos L, et al. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 53.Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 54.Kim SK, Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990;4:896–905. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 55.Sun H, Shi W. Analyses of mrp genes during Myxococcus xanthus development. J Bacteriol. 2001;183:6733–6739. doi: 10.1128/JB.183.23.6733-6739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SK, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruse T, et al. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol Microbiol. 2001;40:156–168. doi: 10.1046/j.1365-2958.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 58.Li SF, et al. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 59.Lee J, et al. Combinatorial regulation of fmgD by MrpC2 and FruA during Myxococcus xanthus development. J Bacteriol. 2011;193:1681–1689. doi: 10.1128/JB.01541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mittal S, Kroos L. Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development. J Bacteriol. 2009;191:2753–2763. doi: 10.1128/JB.01818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Son B, et al. Combinatorial regulation by MrpC2 and FruA involves three sites in the fmgE promoter region during Myxococcus xanthus development. J Bacteriol. 2011;193:2756–2766. doi: 10.1128/JB.00205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell A, et al. Combinatorial regulation of the dev operon by MrpC2 and FruA during Myxococcus xanthus development. J Bacteriol. 2015;197:240–251. doi: 10.1128/JB.02310-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 64.Boynton TO, et al. Characterization of Myxococcus xanthus MazF and implications for a new point of regulation. Mol Microbiol. 2013;87:1267–1276. doi: 10.1111/mmi.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boysen A, et al. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J Bacteriol. 2002;184:1540–1546. doi: 10.1128/JB.184.6.1540-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajagopalan R, et al. devI is an evolutionarily young negative regulator of Myxococcus xanthus development. J Bacteriol. 2015;197:1249–1262. doi: 10.1128/JB.02542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viswanathan P, et al. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol. 2007;189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muller FD, et al. Spore formation in Myxococcus xanthus is tied to cytoskeleton functions and polysaccharide spore coat deposition. Mol Microbiol. 2012;83:486–505. doi: 10.1111/j.1365-2958.2011.07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller FD, et al. Global transcriptome analysis of spore formation in Myxococcus xanthus reveals a locus necessary for cell differentiation. BMC Genomics. 2010;11:264. doi: 10.1186/1471-2164-11-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wartel M, et al. A versatile class of cell surface directional motors gives rise to gliding motility and sporulation in Myxococcus xanthus. PLoS Biol. 2013;11:e1001728. doi: 10.1371/journal.pbio.1001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrebi R, et al. An evolutionary link between capsular biogenesis and surface motility in bacteria. Nat Rev Microbiol. 2015;13:318–326. doi: 10.1038/nrmicro3431. [DOI] [PubMed] [Google Scholar]
- 73.Ueki T, Inouye S. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J Biol Chem. 2005;280:32279–32284. doi: 10.1074/jbc.M507191200. [DOI] [PubMed] [Google Scholar]
- 74.Cao S, et al. Identification of a putative flavin adenine dinucleotide-binding monooxygenase as a regulator for Myxococcus xanthus development. J Bacteriol. 2015;197:1185–1196. doi: 10.1128/JB.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leng X, et al. Evidence that a chaperone-usher-like pathway of Myxococcus xanthus functions in spore coat formation. Microbiology. 2011;157:1886–1896. doi: 10.1099/mic.0.047134-0. [DOI] [PubMed] [Google Scholar]
- 76.Huntley S, et al. Genome evolution and content in the myxobacteria. In: Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014. pp. 30–50. [Google Scholar]
- 77.Tan IS, Ramamurthi KS. Spore formation in Bacillus subtilis. Environ Microbiol Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bush MJ, et al. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol. 2015;13:749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 79.Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Ann Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- 80.McCormick JR, Flardh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36:206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bradshaw N, Losick R. Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. eLife. 2015;4:e08145. doi: 10.7554/eLife.08145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serrano M, et al. Dual-specificity anti-sigma factor reinforces control of cell-type specific gene expression in Bacillus subtilis. PLoS Genet. 2015;11:e1005104. doi: 10.1371/journal.pgen.1005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kroos L, Inouye S. Transcriptional regulatory mechanisms during Myxococcus xanthus development. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. ASM Press; Washington, D.C: 2008. pp. 149–168. [Google Scholar]
- 84.Abellon-Ruiz J, et al. The CarD/CarG regulatory complex is required for the action of several members of the large set of Myxococcus xanthus extracytoplasmic function sigma factors. Environ Microbiol. 2014;16:2475–2490. doi: 10.1111/1462-2920.12386. [DOI] [PubMed] [Google Scholar]
- 85.Velicer GJ, et al. Whence comes social diversity? Ecological and evolutionary analysis of the myxobacteria. In: Yang Z, Higgs P, editors. Myxobacteria: genomics, cellular and molecular biology. Caister Academic Press; Norfolk, UK: 2014. pp. 1–29. [Google Scholar]
- 86.Cao P, et al. How myxobacteria cooperate. J Mol Biol. 2015;427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuner JM, Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982;151:458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Licking E, et al. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J Bacteriol. 2000;182:3553–3558. doi: 10.1128/jb.182.12.3553-3558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hagen DC, et al. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 90.Shimkets LJ, et al. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci USA. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim SK, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 92.Lobedanz S, Sogaard-Andersen L. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 2003;17:2151–2161. doi: 10.1101/gad.274203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rolbetzki A, et al. Regulated secretion of a protease activates intercellular signaling during fruiting body formation in M. xanthus. Dev Cell. 2008;15:627–634. doi: 10.1016/j.devcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Boynton TO, Shimkets LJ. Myxococcus CsgA, Drosophila Sniffer and human HSD17B10 are cardiolipin phospholipases. Genes Dev. 2015;29:1903–1914. doi: 10.1101/gad.268482.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee K, Shimkets L. Cloning and characterization of the socA locus which restores development to Myxococcus xanthus C-signaling mutants. J Bacteriol. 1994;176:2200–2209. doi: 10.1128/jb.176.8.2200-2209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee BU, et al. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 1995;9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.