Abstract
Development of the adaptive immune system is dependent on V(D)J recombination, which forms functional antigen receptor genes through rearrangement of component gene segments. The V(D)J recombinase, consisting of recombination-activating proteins RAG1 and RAG2, guides the initial DNA cleavage events to the recombination signal sequence (RSS), which flanks each gene segment. Although the enzymatic steps for RAG-mediated endonucleolytic activity was established over two decades ago, only recently have high resolution structural studies of the catalytically active core regions of the RAG proteins shed light on conformational requirements for the reaction. While outstanding questions remain, we have a clearer picture for how RAG proteins function in generating the diverse repertoires of antigen receptors, the underlying foundation of the adaptive immune system.
Keywords: V(D)J recombination, RAG1, RAG2, lymphocyte development
The basis of the V(D)J Recombination reaction
Our adaptive immune system is a critical line of defense against the onslaught of pathogenic organisms and viruses that our bodies are exposed to on a daily basis. B and T cell populations expressing diverse antigen receptors, which have the collective ability to recognize a vast array of foreign antigens, mediate the immune response. The diverse receptors are formed during lymphocyte development through shuffling of individual gene segments in the antigen receptor loci in a process known as V(D)J recombination (reviewed in (1)). Antigen receptor gene segments are termed V (variable), D (diversity), and J (joining) and are marked for potential recombination events by a flanking recombination signal sequence (RSS). The RSS consists of a conserved heptamer and nonamer sequence separated by 12 or 23 base pairs (Figure 1A). Recombination preferentially occurs between two segments flanked by RSSs with differing spacer lengths, a restriction referred to as the 12/23 rule. The 12/23 rule maintains the correct ordered assembly of the gene segments to yield joined V-D-J gene segments, or V-J in antigen receptor loci lacking D segments. Thus, based on the arrangement of the gene segments and the type of flanking RSS (12-RSS or 23-RSS), the 12/23 rule serves to prevent joining of the same class of gene segment (i.e. V-V) or incorrect combinations (i.e. V-J in gene loci containing D segments). In certain cases an additional restriction termed the ‘Beyond 12/23 rule’ exists, which applies additional constraints on the recombination reaction, as reviewed by Schatz and Swanson (1).
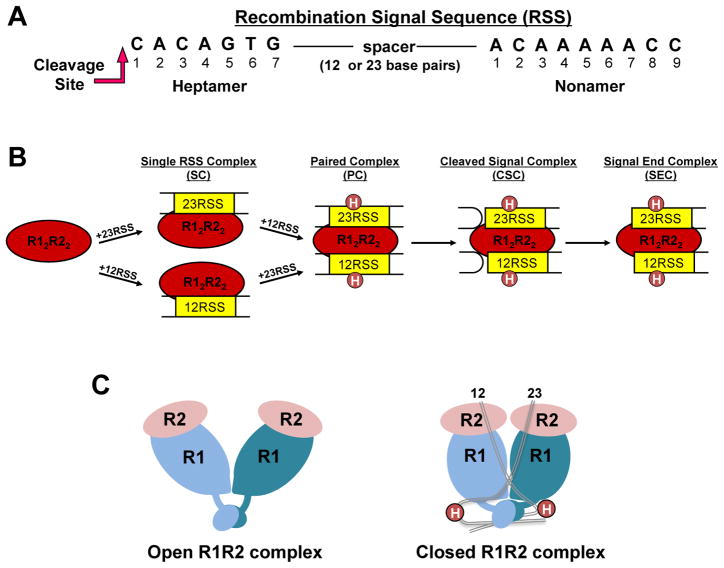
Figure 1. RAG-RSS complexes formed during V(D)J recombination.
(A) Consensus heptamer and nonamer sequences in the RSS. Only the DNA strand initially nicked by the RAG proteins is shown. Nicking occurs 5′ to the heptamer at the position indicated. (B) RAG complexes formed in the V(D)J recombination reaction. R1 and R2 refer to RAG1 and RAG2, respectively. Circles labeled H (in the PC, CSC, and SEC) refer to HMGB1 or HMGB2. (C) Cartoons representing structural models of R12R22 complexes. The DNA-free ‘apo’ and 12/23-RSS-bound R12R22 complexes are proposed to be in “open” and “closed” conformations, respectively (18).
The recombination activating gene proteins, RAG1 and RAG2, initiate V(D)J recombination by generating DNA double strand breaks (DSBs) at the border of the gene segment and flanking RSS through a two step nicking and hairpin formation mechanism (1). RAG-mediated DNA DSBs occur in the context of a paired complex (PC), with the RAG proteins simultaneously bound to a 12-RSS and a 23-RSS with the intervening DNA looped out (Figure 1B). Both RAG proteins are required for DNA cleavage activity, with RAG1 containing the active site residues, as well as the RSS specific binding sites (1,2). The role for RAG2 is less clear, but may function to activate RAG1 for sequence-specific binding and cleavage (3,4), and also provide additional DNA binding capability (5). Following RAG-mediated DNA cleavage the appropriate DNA ends are joined by the action of ubiquitous DNA repair factors that function in nonhomologous DNA end-joining (NHEJ) (see Glossary) (reviewed in (6–8)). Erroneous RAG-mediated DNA cleavage, either at RSS-like sites or non-B form structures, as well as mistakes in DNA repair, are known to result in chromosomal translocations with increased risk for development of certain leukemias and lymphomas (reviewed in (9,10)).
The pathway towards assembly of the PC has been the subject of intensive research (reviewed in (1)). These collective studies show that the RAG1 and RAG2 proteins form a complex in the absence of DNA, and bind the RSSs as a single heterotetrameric complex that consists of two subunits each of RAG1 and RAG2, referred to as the RAG12RAG22 complex (Figure 1B). The RAG proteins bound to a single RSS (either 12-RSS or 23-RSS) is referred to as an SC (Single RSS Complex). The second RSS is subsequently recruited to the SC to form the PC. Formation of the full complement of RAG proteins and RSSs also requires the presence of the high mobility group proteins, HMGB1 or HMGB2 (1). Nicking may occur in either the SC or PC, although the extent of nicking in the SC is impeded for less canonical RSSs (11). In contrast, hairpin formation occurs only in the context of the PC containing both a 12-and 23-RSS (1). Thus, completion of DNA DSBs in V(D)J recombination is constrained to the PC, ensuring coupled cleavage at both RSSs. Subsequent to DNA cleavage, the interaction of the RAG proteins with the coding ends in the cleaved signal complex (CSC; Figure 1B) properly channels the DNA ends to classical NHEJ DNA repair factors (reviewed in (12)).
Origin of RAG proteins
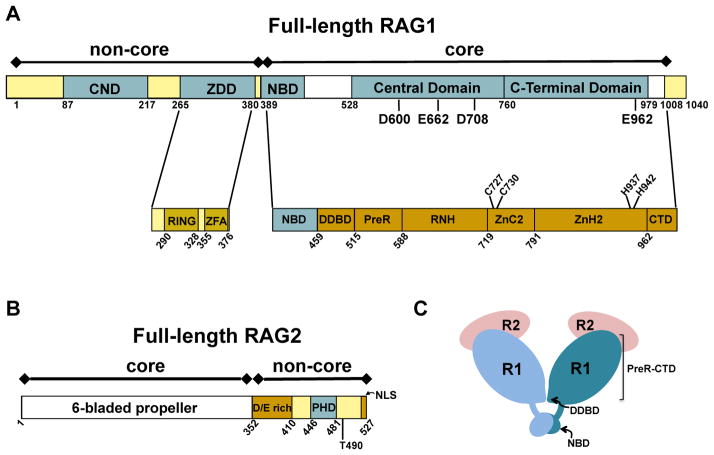
Both RAG proteins are large multi-domain proteins that consist of core regions (Box 1) and non-core regions (Box 2, Figure 2A&B), where the enzymatically ‘dispensable’ non-core regulates the activity of the catalytically-essential core regions during V(D)J recombination (13,14). It has long been proposed that the introduction of RAG genes into jawed vertebrates (gnathostomes), and the resulting split antigen receptor genes, occurred through the insertion of a RAG-like transposon into an ancient antigen receptor gene (15). Specifically, it is posited that the RAG-like transposon was flanked on each side by a 12-RSS and 23-RSS, encoded RAG-like proteins, and its insertion into the receptor-like gene eventually led to the current germline configuration of the antigen receptor loci with multiple gene segments bordered by 12- or 23-RSSs. This proposal is bolstered by the close juxtaposition of intron-less RAG1 and RAG2 genes, as well as previous findings that purified core RAG proteins can indeed complete the transposition reaction by catalyzing strand transfer of RSS signal ends into target DNA (15). Moreover, the recent discovery of a transposon, termed protoRAG, in lancelets fits the necessary criteria of an expected RAG-like transposon (16). Together, the overwhelming evidence indicates that the emergence of our current adaptive immune system resulted from the domestication and repurposing of a RAG-like transposon up to 600 million years ago.
Box 1. Features of the core RAG proteins.
Previous biochemical studies indicated that core RAG1 contained three topologically independent domains, including the nonamer binding domain (NBD), the central domain, and the C-terminal domain (Figure 2A). Each of these domains recognizes the RSS and/or recapitulated endonucleolytic activity of intact core RAG1 on certain DNA substrates (2,61). In the core RAG12RAG22 crystal structure, core RAG1 was denoted as containing seven separate structural modules (17), with modules 3–5 (PreR, RNH, and ZnC2) and modules 6–7 (ZnH2 and CTD) roughly corresponding to the central and C-terminal domains, respectively (Figure 2A).
It was previously shown that core RAG1 coordinated Zn2+ and Mg2+ ions (1,2). In accordance, the cryo-EM structures revealed that the DNA cleavage active site in RAG1 (referred to as a DDE motif) coordinates two Mg2+ ions. The two metal ions are ligated by residues D620, E684, D730, and E984 in zebrafish RAG1, corresponding to D600, E662, D708, and E962, respectively, in murine RAG1 (Figure 2A). The DDE motif active site is a shared feature amongst a large family of transposases and integrase enzymes (62). Additionally, the core RAG12RAG22 crystal structure reveals a single zinc binding site in core RAG1 coordinated by two pairs of ligands, a Cys pair (C727 and C730) and a His pair (H937 and H942) (17). It was previously shown that an active one zinc (1-Zn) form of the protein could be formed by dialysis in EDTA (63), indicating that the catalytically essential zinc ion is tightly coordinated.
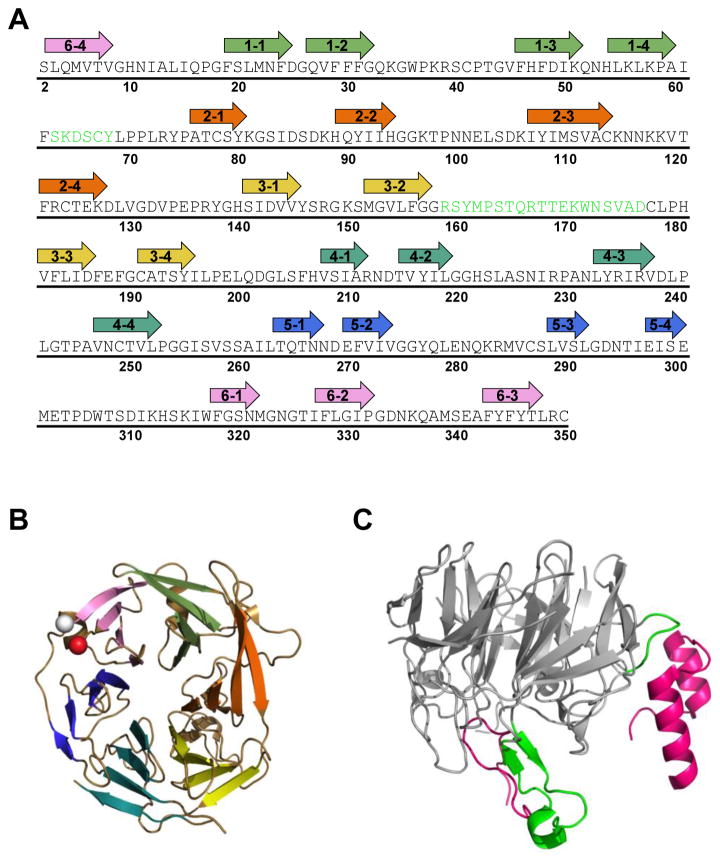
As expected from previous computer modeling studies (1), high resolution structures show that this region of the protein is a six-bladed propeller structure (Figure 2B). Interfacial contacts in the core RAG12RAG22 complex are contributed by extended loops from each protein (17). The interfacial loop regions of RAG1, residues 537-553 and 750-782, were shown in biochemical studies to be important in complex formation with core RAG2 (64–66). In the crystal structure, the three N-terminal beta-propeller blades of core RAG2 mediate the majority of interactions with RAG1 (Figure 3). A major portion of the interface is constructed by a loop between the two middle beta strands of blade 3 that form extensive interactions with the first interfacial loop in RAG1 (residues 537-553). The first blade and the intervening loop between blades 1 and 2 in the RAG2 propeller structure contact the second interfacial loop region in RAG1 (residues 750-782).
Box 2. Zinc rich non-core domains in RAG1 and RAG2.
A prominent feature in the N-terminal non-core region of RAG1 is the zinc binding dimerization domain (ZDD) (Figure 2A). Point mutations within this domain, particularly at zinc coordinating residues, lead to decreased V(D)J recombination activity (13). Subdomains within the ZDD includes a RING finger and a C2H2 zinc finger motif, referred to as ZFA (zinc finger A) (59). The RING finger in the ZDD functions as an E3 ubiquitin ligase mediating autoubiquitination and/or ubiquitination of Histone H3 (30,31,37,67–70). Additionally, RAG1 may act with other E3 ubiquitin ligases as part of a larger multisubunit complex (71).
An additional zinc-binding domain in non-core RAG1 is the central non-core domain (CND) (35), which is N-terminal to the ZDD (Figure 2A). The positioning of the CND to the ZDD in non-core RAG1 is conserved in all vertebrate RAG1 sequences and in some, but not all, invertebrate RAG1-like sequences (72–74). An homologous sequence to the CND, termed the Chapa domain, is present in Chapaev transposases, which have a similar domain architecture as RAG1 (72). It was determined previously that the CND was a structurally independent domain capable of tightly coordinating up to two zinc ions (35), and mutation of putative zinc-coordinating Cys residues resulted in severe defects in V(D)J recombination activity (75). The CND is highly positively charged and binds double-stranded DNA nonspecifically, placing it in position to serve as an accessory DNA interacting module to RSS-flanking DNA (35).
Non-core RAG2 also contains a zinc-binding domain, termed the plant homology domain (PHD), which coordinates two zinc ions and is located between residues 414-487 (Figure 2B). This domain specifically binds histone H3 trimethylated on Lysine 4 (47,48), with the binding affinity modestly increased if Arginine residue 2 is also symmetrically dimethylated (47). High resolution structures of the RAG2 PHD have been determined in the unbound state, as well as bound to N-terminal histone H3 peptides methylated on either K4 alone, or on both R2 and K4 (47,48,76). In the bound complexes, methylated K4 binds within a hydrophobic ‘channel’ on the surface of the RAG2 PHD structure, which is lined with residues Y415, M443, and W453 (47). Mutation of these residues leads to severe defects in in vivo V(D)J recombination activity (48,77).
Figure 2. Schematics of domains and subdomains in the RAG proteins.
Core and non-core regions are denoted above the bar diagram of each RAG protein. (A) Full length murine RAG1. Non-core regions in RAG1 include an N-terminal region (residues 1-388) and a short C-terminal region (residues 1009-1040). Topologically-independent domains are shown as blue boxes. In non-core RAG1 these include the central non-core domain (CND) and zinc dimerization domain (ZDD). A crystal structure of the ZDD showed two sub-domains, the RING finger and zinc finger A (ZFA). Domains in core RAG1 include the nonamer binding domain (NBD), the Central domain, and the C-terminal domain. The core region contains multiple modules, as labeled in the lower bar. The three DDE active site residues (D600, D708, and E962), a putative fourth active site residue (E662), and zinc-coordinating ligands (C727, C730, H937, and H942) are labeled in the RAG1 bar diagrams. (B) Full length murine RAG2. The core region is a 6-bladed propeller domain. Non-core RAG2 contains an acidic region, labeled as D/E rich, as well as a plant homology domain (PHD) that specifically binds the trimethylated lysine residue of histone H3K4me3. Positions of residue T490 and the C-terminal nuclear localization signal (NLS) are indicated. (C) Cartoon depicting the core RAG12RAG22 complex. RAG1 (R1) subunits are in light and dark blue, and RAG2 (R2) subunits are in pink. The domains (NBD and DDBD) that form the RAG1 dimer interface are labeled. The remaining subdomains (PreR to CTD) compose the catalytically active region of core RAG1.
Core regions of RAG1 and RAG2: Domain architecture, complex interface, metal binding, and RSS interactions
Core RAG1-RAG2 Crystal Structure
Recent high-resolution structures of RAG1 and RAG2 core regions obtained from crystallography and cryo-electron microscopy (cryo-EM) studies have provided much anticipated views into the catalytically active V(D)J recombinase complex (17,18). The crystal structure showed that the overall shape of the core RAG12RAG22 complex resembles a ‘Y-shaped’ structure (17) (Figures 1C & 2C). Although the crystals used to determine the structure of the murine core RAG proteins contained 12/23 RSS substrates, electron densities for the DNA fragments were not visible (17).
The core RAG1 structure revealed seven separate structural modules ((17), shown schematically in Figure 2A). The stalk of the ‘Y’ consists of two core RAG1 DNA binding and dimerization domains (NBD and DDBD) that provide the RAG1 homodimerization interface (Figure 2C). Each branch of the ‘Y’ consists of the remaining modules of core RAG1 (PreR, RNH, ZnC2, ZnH2, and CTD), and contains the active site for DNA cleavage (see Box 1). The PreR provides a spacer region between the DDBD and the RNH, where the RNH contains the majority of the active site. The ZnC2 and ZnH2 modules contribute ligands to a catalytically-essential zinc-binding site, and the CTD provides additional residues to the active site, as well as residues that contribute to RAG1 dimerization.
Core RAG2 is positioned at the tip of each branch of the ‘Y’ and assumes a six-bladed beta-propeller structure (17) (Figure 3). The majority of RAG2 point mutations defective in DNA cleavage activity, previously identified from immunodeficient patients or from biochemical studies (5,19,20), are located at the RAG1-RAG2 interface or within the interior of core RAG2. However, several lysine and arginine residues that were deficient in either DNA cleavage or joining (5,20) are solvent exposed in the core RAG12RAG22 crystal structure, and as discussed below, directly contribute to DNA binding interactions.
Figure 3. Core RAG2 topology and interface with RAG1.
(A) Murine core RAG2 sequence. Arrows are shown above residues that constitute the beta strands in the beta-propeller structure. The first number in each arrow corresponds to the blade number (1 to 6). The second number refers to the strand (1 to 4) in each blade, where strand 1 and 4 are at the interior and exterior sides of the blade, respectively, according to the orientation of the structure shown in panel B. Residues in green are positioned at the interface with RAG1 in the crystal structure of RAG12RAG22. (B) Ribbon diagram of murine core RAG2 (from PDB ID: 4WWX). The N- and C-termini are shown as white and red spheres, respectively. Each propeller blade is color coded according to the arrows in panel A, where blades 1 to 6 are in light green, orange, yellow, light blue, dark blue, and pink, respectively. (C) The core RAG2 structure rotated by 90° relative to panel B. Residues in the interface with RAG1 are shown in green, and correspond to the residues in green in panel A. The murine core RAG1 loops (residues 537-553 and 750-782) that form the majority of the interface with core RAG2 are shown as magenta ribbons.
Structures of Core RAG-RSS complexes
While electron densities for the RSSs were not apparent in the core RAG12RAG22 crystallography studies, cryo-EM structural models containing both the 12-RSS and 23-RSS were recently determined using zebrafish RAG1 and RAG2 (18). These include symmetrized and non-symmetrized PC models consisting of core RAG regions bound to nicked 12 and 23 RSSs. In addition, a signal end complex (SEC) containing the core RAG proteins bound to 12 and 23-RSS substrates blunt ended at the 5′ end of the heptamer (signal ends) was determined. Zebrafish and mouse core RAG1 and core RAG2 regions are highly conserved, with 78% and 53% identity, respectively. Notably, the residue numbering in RAG1 is offset between the zebrafish and mouse sequences due to deletions/insertions in the N-terminal non-core regions. Thus, for clarity in the discussion below, residue numbers refer to the murine sequence of the RAG proteins unless otherwise specified.
The ‘non-symmetrized’ PC is of particular interest, as this model includes the entire core RAG regions and all nucleotides in the nicked 12/23 RSS substrates. The NBD intertwined dimer bound to the 12/23 RSS nonamers aligns well with a previous crystal structure of the isolated NBD complexed with RSS nonamers (17,18,21). Interestingly, the NBD dimer does not share the same two-fold axis of symmetry as the remainder of the complex, and is preferentially oriented toward the 12-RSS (18). This asymmetry provides a plausible explanation for the 12/23 rule, since the tilting of the NBD toward the 12-RSS nonamer in the PC would preclude symmetrical binding of two identical DNA substrates.
The overall shape of the PC and SEC cryo-EM structures (18) differs from the core RAG12RAG22 crystal structure (17) in that the branches of the ‘Y’ are in closer proximity. Notably, the crystal structure resembles in overall shape a DNA-free form of a low-resolution cryo-EM core RAG12RAG22 structure (18). Thus, binding the 12- and 23-RSSs is proposed to lead to a large conformational change from an ‘open’ to a ‘closed’ form (shown schematically in Figure 1C), which decreases the distance between the tips of the ‘Y’ by >25 Å (18). In the 12/23-RSS-bound closed conformation of the cryo-EM structures, additional contacts are formed between the RAG proteins (18). Specifically, an alpha helix in the RAG1 C-terminal domain that includes residues 827-835 in mouse RAG1 interacts with the RAG2 subunit from the opposite ‘Y’ branch by forming contacts with residues 308-314 (located on the intervening loop between propeller blades 5 and 6). Additional interactions in the closed conformation are contributed from loops in RAG1 and RAG2 that were unresolved in the crystal structure, and include RAG1 residues 608-615 and RAG2 residues 333-338 in propeller blade 6.
In the non-symmetrized PC, both RSSs assume bent conformations (18), with the 12-RSS and 23-RSS bent by ~60° and ~120°, respectively. These results are consistent with previous studies that RSS substrates complexed to the RAG proteins are strongly bent, as determined using fluorescence resonance energy transfer (FRET) experiments that obtained distances between fluorophores incorporated into RSS substrates, and an atomic force microscopy (AFM) study that directly visualized the contours of RSS-containing substrates bound to RAG proteins (22–24). Furthermore, the resolutions of the cryo-EM structures are sufficient to show distortions in the RSSs, including a widening of the minor grooves in the heptamers of each RSS and base unpairing at the RSS/coding junction. Significantly, two separate DNA bases are completely flipped out of the DNA helix, including the 5′ C of the heptamer (heptamer base 1 in Figure 1A) and the base opposite the 3′OH of the coding end. As previously predicted, this latter base flipping would facilitate nucleophilic attack of the 3′OH to the opposing phosphate group to yield hairpin formation (25,26). The flipped bases are mainly stabilized by van der Waals interactions with the sidechains of M847 and R848 from RAG1. As a consequence of this interaction, the 3′-OH group is positioned to attack the opposing phosphate group on the complementary strand to yield hairpin formation and complete the reaction for DNA DSB formation.
RAG-DNA contacts
With the cryo-EM SEC and PC structures (18), as well as the crystal structure of the isolated NBD bound to RSS nonamer (21), a more complete picture of the RAG-DNA contacts formed in the 12/23 complexes can now be appreciated. The majority of sequence-specificity to the RSS nonamer is contributed by the NBD, a helical rich domain containing an AT-hook motif at its N-terminal end (reviewed in (15)). Mutations in the NBD lead to severe combined immunodeficiency (T−B−SCID) as a result of the complete absence of V(D)J recombination activity (27). In the NBD-RSS nonamer structure (21), residues 389-393 constitute the AT-hook that binds the minor groove of the nonamer and forms sequence specific contacts with nonamer bases 5–7 (numbering as in Figure 1A). Notably, each NBD subunit contacts both RSS nonamer molecules in the complex, strongly indicating that the bridging of two RSS nonamers by the NBD dimer contributes to PC formation (21). Interactions with the RSS spacer are mediated by residues in the NBD and DDBD that primarily form non-sequence-specific contacts with the phosphate backbone, consistent with the poor conservation of the RSS spacer (18).
The cryoEM SEC and PC structures reveal sequence-specific interactions of RAG1 with the RSS heptamer. Consistent with the finding that the isolated central domain binds specifically to the heptamer (2,28), the corresponding region in core RAG1 (RNH) forms several contacts with the heptamer, with base-specific interactions between E649 and heptamer base C3. The C-terminal domain also forms extensive contacts with the heptamer, which are primarily mediated by residues C-terminal to the E962 active site residue. Each core RAG1 subunit bridges across the RSS heptamers in the PC and SEC complexes, with residues from the central and C-terminal domains forming contacts with both RSS heptamers.
RAG1 residues 977-979 appear to widen the RSS heptamer’s minor groove resulting in a distortion of the B-form DNA helix. Additionally, residues 891, 895, and 901 in the C-terminal domain contact the 5′ C that is flipped outward from the DNA helix. RAG2 does not directly contact the heptamer, although its presence has previously been shown to increase the specific interaction of RAG1 for the heptamer (3). This may occur via RAG2 affecting the conformation of RAG1 (3) and/or suppressing its nonspecific DNA binding ability (4).
The coding DNA forms contacts with both the central and C-terminal domains of RAG1. Residues flanking RAG1 active site residues in the central domain are positioned to interact with the phosphate backbone near the coding end. Residues in the C-terminal domain, in particular M847, stabilize the opposing base from the nicked site that is flipped out of the DNA helix. Lastly, basic and polar residues within the first two beta-propeller blades in core RAG2 directly contact the phosphate backbone of the coding DNA, including a single stranded region at the coding-heptamer junction. Previous studies supported the role for basic residues in RAG2 forming DNA contacts in the PC (5,20).
Overall, the asymmetric PC cryo-EM structure provides a plausible basis for how hairpin formation, and thus coupled cleavage, requires PC formation (18). This necessitates extensive bridging interactions between each RAG1 subunit and both the nonamer and heptamer of the two RSSs. In addition, the nicked 12-RSS and 23-RSS substrates in the ‘closed’ conformation of the asymmetric PC contain significant structural distortions and single stranded regions at the heptamer-coding border. With these conformational properties, the asymmetric PC appears primed to carry out the final hairpin formation step at each RSS in a coupled manner.
Non-core domains in the RAG proteins
Zinc-binding domains constitute the N-terminal RAG1 non-core region
Non-core RAG1 consists of the N-terminal 385 residues, which is composed of multiple zinc-binding motifs (Figure 2A). In addition, residues 1009-1040 at the C-terminal end of the protein comprise a smaller non-core region. Non-core RAG1 is dispensable for DNA cleavage activity, although its presence increases the efficiency and fidelity of V(D)J recombination, as well as regulating RAG DNA cleavage activity (reviewed in (13,29)). Regions within non-core RAG1 have been implicated in interacting with proteins linked to the V(D)J recombination process and DNA repair, including Histone H3 (30,31), Ku (32), and MDC1 (33), as well as proteins that are not clearly correlated with RAG1 function, including the RNA splicing protein SF3A2 (34).
Two major domains in the N-terminal region of non-core RAG1 include the zinc dimerization domain (ZDD) and the central non-core domain (CND) (Figure 2A and Box 2). The CND and ZDD are separated by a spacer of 47 residues that is predicted to be in a disordered conformation (35), and which includes two regions of consecutively positively charged residues termed BII and BIII from residues 221-224 and 243-249, respectively (36). Although the specific role of this spacer toward RAG1 function is not known, the high density of positively charged residues in this spacer region would lend itself to DNA binding. Further this region includes K233, which is targeted for ubiquitylation by the RAG1 RING finger, a modification that has been shown to increase RAG-mediated DNA cleavage activity (37).
The localization and stability of RAG2 is regulated by its non-core region
Non-core RAG2 consists of approximately one third of the full length protein, and includes residues 352-527 (Figure 2B). The presence of the non-core region of RAG2 facilitates long range recombination reactions (38–42), as well as mediates its nuclear localization and association with specific histone-modified chromatin (43–46).
Non-core RAG2 contains a plant homology domain (PHD) (Box 2) that binds histone H3 trimethylated on Lysine 4 (H3K4me3) (47,48). The interaction of RAG2 and H3K4me3 increases V(D)J recombination activity (49) via multiple proposed mechanisms including providing a chromatin-docking site for RAG2 (50,51), activating RAG-mediated DNA cleavage (52,53), stabilizing the post-cleavage complex (54), and suppressing the RAG2 non-core region’s ability to autoinhibit DNA cleavage activity (55).
Besides the PHD, non-core RAG2 contains an acidic hinge region (residues 352-410) that has been shown to facilitate interaction with Histone H3 and to stabilize the PC complex (40,56). Additional features in non-core RAG2 include the conserved, phosphorylatable T490 residue (57), as well as a nuclear localization signal (NLS) at the C-terminal end of the full length protein (43,45). Phosphorylation of T490 by CDK2 leads to degradation of RAG2 at the G1/S border of the cell cycle, thereby helping to restrict V(D)J recombination events to G1 (57). In addition, cells expressing the RAG2 T490A mutant demonstrate a block in the nuclear export of full length RAG2 in cells untreated (44,46) and treated with genotoxic stressors (46).
Molecular modeling RAG1 domains
High resolution structures of the core RAG proteins have shown for the first time the overall fold of the proteins, as well as interfacial regions between macromolecular components of the V(D)J recombinase complexes. A major challenge going forward will be to determine the structural and functional relationships of the non-core to the core regions in the RAG12RAG22 complex. Obtaining the spatial orientation of the non-core domains would provide important clues as to their relative contributions to the activity and regulation of the RAG proteins. For example, such information would help in determining how non-sequence-specific DNA-binding abilities of the CND, autoinhibitory mechanisms of non-core RAG2, and E3 ubiquitin ligase activities of the RING finger contribute to V(D)J recombination.
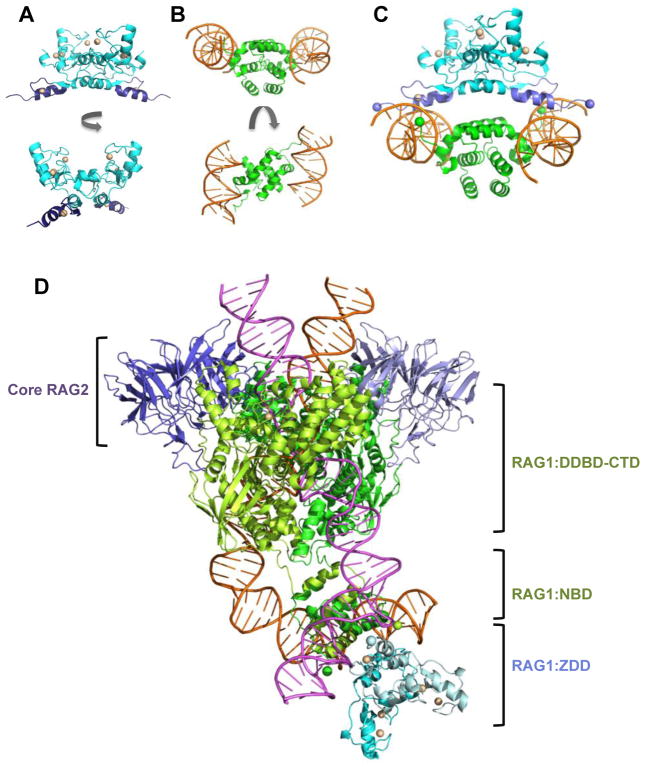
We can also build on the current structures to predict the overall topology of these large multi-domain proteins. For example, as the ZDD borders the N-terminal side of the NBD in the RAG1 sequence, a fairly simple structural model for the ZDD plus NBD can be built using the available high resolution structural data of the ZDD (58) and NBD:RSS nonamer (21) (Figure 4). Docking of the two structures is aided by the fact that the ZDD and the isolated NBD form homodimeric domains, as expected from solution studies (21,59). In the ZDD dimer, the C-termini of the subunits are separated by ~80 Å, and the N-termini of the NBD dimeric subunits are separated by ~60 Å. A simple model in which the respective ends are joined is not possible, since eight intervening residues (residues 381-388) between the ZDD and NBD are not included in either crystal structure. As a result, numerous orientations between the ZDD and NBD are possible, which differ in the angle of rotation of each domain relative to the other about a shared two-fold axis. However, the presence of the bound DNA in the NBD:RSS nonamer structure leads to a preferred biologically-relevant model for the orientation between the ZDD and NBD, based largely on the position of the zinc finger A (ZFA) modules in the combined structure (Figure 4C&D). In this orientation, the alpha-helices of each ZFA insert into the major grooves of each RSS nonamer with the sidechains of conserved charged and polar residues in the ZFA alpha-helix positioned closely to nucleotides at the 5′ end of the RSS nonamer (bases 2–4 in the nonamer as numbered in Figure 1A). The juxtaposition of ZFA with the 5′ end of the nonamer in the docked model is consistent with previous DNA footprinting studies on RAG-RSS binding interactions, which showed that full length versus core RAG1 induced increased structural distortion at the 5′ end of the nonamer in the PC (60). In this model, the eight intervening residues between the ZDD and NBD connect the two domains by looping over the DNA strands (connecting the green and purple spheres in Figure 4C), with the combined domains forming a pincer clamp around each RSS nonamer. This orientation could function to increase the stability of the RAG-RSS interaction, particularly in the 12/23 PC, and may account in part for previous findings that core RAG1 forms less stable 12/23 PCs as compared to full length RAG1 (60).
Figure 4. Models of the combined RAG1 domains ZDD and NBD bound to RSS nonamers.
(A) Ribbon diagram of the ZDD structure in two orientations, generated using PDB ID: 1RMD. The ZDD subunits are in blue with the ZFA regions in purple. The zinc ions are light yellow spheres. (B) Ribbon diagram of the structure consisting of an NBD dimer bound to 2 RSS nonamers is shown in two orientations, generated using PDB ID: 3GNA. The NBD subunits and RSS nonamers are in green and orange, respectively. (C) Docked model of the structures in panels A and B are shown. The N-termini of NBD and C-termini of ZDD are shown as green and purple spheres, respectively. (D) Model of the ZDD (PDB ID: 1RMD) docked to the non-symmetrized PC complex (PDB ID: 3JBW), in which the relative orientation of the ZDD to the RSS nonamers is the same as in panel C. Core RAG2 subunits are in purple, core RAG1 subunits are in light and dark green, and non-core RAG1 ZDD subunits are in cyan and light blue. The positions of core RAG2, non-core RAG1 ZDD, core RAG1 NBD, and the region containing the remaining core RAG1 modules (DDBD-CTD) are as indicated. The 12-RSS and 23-RSS are pink and orange, respectively. The two-fold axis of symmetry in the ZDD was modeled to coincide with the two-fold axis for the NBD dimer. Ribbon diagrams were produced using The PyMOL Molecular Graphics System, Version 1.6.9, Schrödinger, LLC.
Simple docking models of the ZDD-NBD regions provides a testable hypothesis for how a non-core region of RAG1 could function in concert with the DNA cleavage active core regions to maintain fidelity of the V(D)J recombination reaction. Specifically, the ZFA subdomains may form additional contacts with the 5′ end of the RSS nonamer, and increase the stability of the RAG-RSS paired complex. Furthermore, the interaction of the ZFA modules with the RSS nonamer (in the context of the PC) may induce conformational changes in the RING finger subdomains, given that the RING and ZFA subdomains form a solvent excluded interface in the ZDD structure (58). Formation of the PC would then be translated to the RING finger subdomains in the non-core region of RAG1, thereby affecting E3 ubiquitin ligase activity.
Concluding Remarks
Recent high resolution structure have brought the core regions of the RAG proteins into focus including their overall folds and the macromolecular interfaces in the V(D)J recombinase complexes. Further, these structures have provided insight for the first time into the basis for coupled hairpin formation and conformational restrictions behind the 12/23 rule. Building on the core structures to determine the structures of the individual RAG proteins, as well as the single RSS complex, will provide a deeper understanding of the progression of conformations necessary for RAG-mediated endonucleolytic activity (see Outstanding Questions).
Outstanding Questions.
What are the structures of each RAG protein in the absence of the other? Along those lines, what is the effect of core RAG2 on the conformation of core RAG1, and do these structural changes activate RAG1 for DNA cleavage activity?
Do the RAG proteins induce structural changes in the RSS in the single RSS complex, and if so, do these changes promote nicking activity?
How do non-core RAG regions promote stability of the PC and the CSC complexes?
What proteins are targeted for ubiquitination by non-core RAG1, and how does this activity affect V(D)J recombination?
How does the interaction of RAG2 PHD with H3K4me3 promote long-range recombination, and suppress autoinhibition?
What is the molecular basis for the coordination of RAG-mediated DNA cleavage with DNA repair through the NHEJ pathway?
How do RAG proteins fit in with the interplay of processes that regulate the cellular and chromatin environment necessary for V(D)J recombination? Specifically, how do RAG proteins coordinate with factors that prepare for and respond to RAG activity during V(D)J recombination?
How are RAG protein activity, stability, and localization properties regulated during the DNA damage response?
What are the mechanistic differences between RAG-mediated canonical versus aberrant V(D)J recombination reactions?
As the functions of the non-core regions of the RAG proteins are crucial to regulated and canonical activity of the RAG proteins in developing lymphocytes, it will be important to further elucidate the structure and function of these regions in the context of the full length proteins. Along those lines, many questions remain regarding the interplay of the RAG proteins with the assembly of cellular factors that prepare for and respond to RAG activity during V(D)J recombination, and how defects in these processes lead to aberrant recombination events (see Outstanding Questions). Continued structural studies, along with a wide range of investigational avenues currently being pursued, will be crucial to understanding these intriguing proteins that play such an essential role in our immune system.
Trends.
The mechanistic basis for RAG-mediated endonucleolytic activity in V(D)J recombination has been illuminated by high resolution structures of the catalytically essential core regions of each RAG protein. Together, the structures indicate that a series of conformational changes in the RAG proteins and the bound RSSs facilitate coupled cleavage of a paired 12- and 23-RSS.
The non-core regions of RAG1 and RAG2 regulate V(D)J recombination by affecting cell cycle-dependent degradation, cellular localization, and the stability of RSS-RSS complexes during the course of the reaction.
It is increasingly apparent that legitimate V(D)J recombination events in developing lymphocytes are driven by RAG protein interactions with modified histones, as well as proper coordination with certain DNA repair factors.
Acknowledgments
I would like to thank David Schatz and William Rodgers for a critical reading of the manuscript. This work was supported by funds from the National Institutes of Health (AI-094141 and AI-126001), the Oklahoma Center for Advancement in Science and Technology (HR14-109), and the Presbyterian Health Foundation.
Glossary
- C2H2 zinc finger
The C2H2 zinc finger is a common zinc-binding motif of approximately 30 residues in length. Folding of this motif is dependent on coordination of a single zinc ion by two cysteine and two histidine residues. The structure of classical C2H2 zinc fingers consists of a two-stranded beta sheet followed by an alpha helix.
- CDK2
Cyclin dependent kinase 2 is activated during the G1/S phase of the cell cycle upon binding specific cyclins (Cyclin A or Cyclin E), and its activity is important for cell cycle progression.
- Cryo-EM
Cryo-electron microscopy is a method in which three dimensional structures of macromolecules, which are embedded in vitreous ice, are determined. It is an alternate approach to x-ray crystallography in which moderate to high resolution structures of large macromolecules can be solved.
- H3K4me3
Histone H3 trimethylated on Lysine 4 is a posttranslational modification that is formed at the transcription start sites of active genes in eukaryotic cells. This modification serves as a docking site for proteins containing domains that specifically binds with the trimethylated K4 residue of histone H3.
- NHEJ
Nonhomologous DNA end-joining is a ubiquitous DNA repair pathway that joins DNA ends without the requirement for an homologous copy of undamaged DNA. NHEJ is particularly important for repair of DNA double strand breaks that form during the G1 phase of the cell cycle.
- PHD
Plant homology domain is a two-zinc coordinating domain that typically functions to recognize methylated lysine residues. Many chromatin-interacting proteins contain a PHD motif that specifically recognizes certain methylated histone proteins, such as H3K4me3 or H3K9me3.
- RING finger
The RING finger is a two-zinc binding domain, in which the polypeptide interleaves between the coordinated zinc ions to result in a cross-braced structure. RING fingers generally function as E3 ubiquitin ligases by recruiting substrate proteins to an E2 ubiquitin-conjugating enzyme to be mono- or poly-ubiquitylated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 2.De P, Rodgers KK. Putting the pieces together: Identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 3.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Gwyn LM, De P, Rodgers KK. A Non-Sequence-Specific DNA Binding Mode of RAG1 Is Inhibited by RAG2. J Mol Biol. 2009;387:744–758. doi: 10.1016/j.jmb.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 6.Malu S, Malshetty V, Francis D, Cortes P. Role of non-homologous end joining in V(D)J recombination. Immunol Res. 2012;54:233–246. doi: 10.1007/s12026-012-8329-z. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishana M, Raghavan SC. Role of recombination activating genes in the generation of antigen receptor diversity and beyond. Immunology. 2012;137:271–281. doi: 10.1111/imm.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee JK, Schatz DG. Synapsis alters RAG-mediated nicking at Tcrb recombination signal sequences: implications for the “beyond 12/23” rule. Mol Cell Biol. 2014;34:2566–2580. doi: 10.1128/MCB.00411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth DB. V(D)J Recombination: Mechanism, Errors, and Fidelity. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0041-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones JM, Simkus C. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz) 2009;57:105–116. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- 14.Horowitz JE, Bassing CH. Noncore RAG1 regions promote Vbeta rearrangements and alphabeta T cell development by overcoming inherent inefficiency of Vbeta recombination signal sequences. J Immunol. 2014;192:1609–1619. doi: 10.4049/jimmunol.1301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng G, Schatz DG. Regulation and Evolution of the RAG Recombinase. Adv Immunol. 2015;128:1–39. doi: 10.1016/bs.ai.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, Le Petillon Y, Liu X, Chen S, Schatz DG, Xu A. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez CA, Ptaszek LM, Villa A, Bozzi F, Sobacchi C, Brooks EG, Notarangelo LD, Spanopoulou E, Pan ZQ, Vezzoni P, Cortes P, Santagata S. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–5664. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu JX, Kale SB, Yarnal Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 21.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlicek JW, Lyubchenko YL, Chang Y. Quantitative analyses of RAG-RSS interactions and conformations revealed by atomic force microscopy. Biochemistry. 2008;47:11204–11211. doi: 10.1021/bi801426x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciubotaru M, Surleac MD, Metskas LA, Koo P, Rhoades E, Petrescu AJ, Schatz DG. The architecture of the 12RSS in V(D)J recombination signal and synaptic complexes. Nucleic Acids Res. 2015;43:917–931. doi: 10.1093/nar/gku1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciubotaru M, Trexler AJ, Spiridon LN, Surleac MD, Rhoades E, Petrescu AJ, Schatz DG. RAG and HMGB1 create a large bend in the 23RSS in the V(D)J recombination synaptic complexes. Nucleic Acids Res. 2013;41:2437–2454. doi: 10.1093/nar/gks1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundy GJ, Hesse JE, Gellert M. Requirements for DNA hairpin formation by RAG1/2. Proc Natl Acad Sci U S A. 2007;104:3078–3083. doi: 10.1073/pnas.0611293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 27.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Xu K, Deng A, Fu X, Xu A, Liu X. An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1. Proc Natl Acad Sci U S A. 2014;111:397–402. doi: 10.1073/pnas.1318843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao J, Rothschild G, Basu U. Ubiquitination events that regulate recombination of immunoglobulin Loci gene segments. Front Immunol. 2014;5:100. doi: 10.3389/fimmu.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones JM, Bhattacharyya A, Simkus C, Vallieres B, Veenstra TD, Zhou M. The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol Lett. 2011;136:156–162. doi: 10.1016/j.imlet.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Deng Z, Liu H, Liu X. RAG1-mediated ubiquitylation of histone H3 is required for chromosomal V(D)J recombination. Cell Res. 2015;25:181–192. doi: 10.1038/cr.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic Acids Res. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coster G, Gold A, Chen D, Schatz DG, Goldberg M. A dual interaction between the DNA damage response protein MDC1 and the RAG1 subunit of the V(D)J recombinase. J Biol Chem. 2012;287:36488–36498. doi: 10.1074/jbc.M112.402487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maitra R, Sadofsky MJ. A WW-like module in the RAG1 N-terminal domain contributes to previously unidentified protein-protein interactions. Nucleic Acids Res. 2009;37:3301–3309. doi: 10.1093/nar/gkp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arbuckle JL, Rahman NS, Zhao S, Rodgers W, Rodgers KK. Elucidating the domain architecture and functions of non-core RAG1: The capacity of a non-core zinc-binding domain to function in nuclear import and nucleic acid binding. BMC Biochem. 2011;12:23. doi: 10.1186/1471-2091-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spanopoulou E, Cortes P, Shih C, Huang CM, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 37.Singh SK, Gellert M. Role of RAG1 autoubiquitination in V(D)J recombination. Proc Natl Acad Sci U S A. 2015;112:8579–8583. doi: 10.1073/pnas.1510464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-toDJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 39.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Curry JD, Schlissel MS. RAG2’s non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic Acids Res. 2008;36:5750–5762. doi: 10.1093/nar/gkn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaumeil J, Micsinai M, Ntziachristos P, Roth DB, Aifantis I, Kluger Y, Deriano L, Skok JA. The RAG2 C-terminus and ATM protect genome integrity by controlling antigen receptor gene cleavage. Nat Commun. 2013;4:2231. doi: 10.1038/ncomms3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corneo B, Benmerah A, Villartay JP. A short peptide at the C terminus is responsible for the nuclear localization of RAG2. Eur J Immunol. 2002;32:2068–2073. doi: 10.1002/1521-4141(200207)32:7<2068::AID-IMMU2068>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 44.Mizuta R, Mizuta M, Araki S, Kitamura D. RAG2 is down-regulated by cytoplasmic sequestration and ubiquitin-dependent degradation. J Biol Chem. 2002;277:41423–41427. doi: 10.1074/jbc.M206605200. [DOI] [PubMed] [Google Scholar]
- 45.Ross AE, Vuica M, Desiderio S. Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells. Mol Cell Biol. 2003;23:5308–5319. doi: 10.1128/MCB.23.15.5308-5319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodgers W, Byrum JN, Sapkota H, Rahman NS, Cail RC, Zhao S, Schatz DG, Rodgers KK. Spatio-temporal regulation of RAG2 following genotoxic stress. DNA Repair (Amst) 2015;27:19–27. doi: 10.1016/j.dnarep.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng G, Maman Y, Resch W, Kim M, Yamane A, Qian J, Kieffer-Kwon KR, Mandal M, Ji Y, Meffre E, Clark MR, Cowell LG, Casellas R, Schatz DG. RAG Represents a Widespread Threat to the Lymphocyte Genome. Cell. 2015;162:751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu C, Ward A, Bettridge J, Liu Y, Desiderio S. An autoregulatory mechanism imposes allosteric control on the v(d)j recombinase by histone h3 methylation. Cell Rep. 2015;10:29–38. doi: 10.1016/j.celrep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G, Dhar K, Swanson PC, Levitus M, Chang Y. Real-time monitoring of RAG-catalyzed DNA cleavage unveils dynamic changes in coding end association with the coding end complex. Nucleic Acids Res. 2012;40:6082–6096. doi: 10.1093/nar/gks255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coussens MA, Wendland RL, Deriano L, Lindsay CR, Arnal SM, Roth DB. RAG2’s acidic hinge restricts repair-pathway choice and promotes genomic stability. Cell Rep. 2013;4:870–878. doi: 10.1016/j.celrep.2013.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desiderio S. Temporal and spatial regulatory functions of the V(D)J recombinase. Semin Immunol. 2010;22:362–369. doi: 10.1016/j.smim.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 59.Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 60.Kumar S, Swanson PC. Full-length RAG1 promotes contact with coding and intersignal sequences in RAG protein complexes bound to recombination signals paired in cis. Nucleic Acids Res. 2009;37:2211–2226. doi: 10.1093/nar/gkp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De P, Peak MM, Rodgers KK. DNA cleavage activity of the V(D)J recombination protein RAG1 is autoregulated. Mol Cell Biol. 2004;24:6850–6860. doi: 10.1128/MCB.24.15.6850-6860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 63.Gwyn LM, Peak MM, De P, Rahman NS, Rodgers KK. A zinc site in the C-terminal domain of RAG1 is essential for DNA cleavage activity. J Mol Biol. 2009;390:863–878. doi: 10.1016/j.jmb.2009.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byrum JN, Zhao S, Rahman NS, Gwyn LM, Rodgers W, Rodgers KK. An interdomain boundary in RAG1 facilitates cooperative binding to RAG2 in formation of the V(D)J recombinase complex. Protein Sci. 2015;24:861–873. doi: 10.1002/pro.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ko JE, Kim CW, Kim DR. Amino acid residues in RAG1 responsible for the interaction with RAG2 during the V(D)J recombination process. J Biol Chem. 2004;279:7715–7720. doi: 10.1074/jbc.M311471200. [DOI] [PubMed] [Google Scholar]
- 66.Zhang YH, Shetty K, Surleac MD, Petrescu AJ, Schatz DG. Mapping and Quantitation of the Interaction between the Recombination Activating Gene Proteins RAG1 and RAG2. J Biol Chem. 2015;290:11802–11817. doi: 10.1074/jbc.M115.638627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grazini U, Zanardi F, Citterio E, Casola S, Goding CR, McBlane F. The RING domain of RAG1 ubiquitylates histone H3: a novel activity in chromatin-mediated regulation of V(D)J joining. Mol Cell. 2010;37:282–293. doi: 10.1016/j.molcel.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 70.Simkus C, Makiya M, Jones JM. Karyopherin alpha 1 is a putative substrate of the RAG1 ubiquitin ligase. Mol Immunol. 2009;46:1319–1325. doi: 10.1016/j.molimm.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kassmeier MD, Mondal K, Palmer VL, Raval P, Kumar S, Perry GA, Anderson DK, Ciborowski P, Jackson S, Xiong Y, Swanson PC. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 2012;31:945–958. doi: 10.1038/emboj.2011.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 2015;10:20. doi: 10.1186/s13062-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roman CA, Cherry SR, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1−/− B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- 76.Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 77.Couedel C, Roman C, Jones A, Vezzoni P, Villa A, Cortes P. Analysis of mutations from SCID and Omenn syndrome patients reveals the central role of the Rag2 PHD domain in regulating V(D)J recombination. J Clin Invest. 2010;120:1337–1344. doi: 10.1172/JCI41305. [DOI] [PMC free article] [PubMed] [Google Scholar]