Abstract
Background
Female urinary microbiota (FUM) are associated with urgency urinary incontinence (UUI) and response to UUI medication. FUM of women with stress urinary incontinence (SUI) has not been described.
Objective
Study the cross-sectional relationships between FUM features and demographic and clinical characteristics of women undergoing SUI surgery.
Design, Setting, and Participants
Pre-operative urine specimens were collected from women without urinary tract infection and were available from 197 women (174 voided, 23 catheterized) enrolled in a multi-center prospective randomized trial, the Value of Urodynamic Evaluation (ValUE) study. Demographic and clinical variables were obtained including SUI and UUI symptoms, menopausal status, and hormone use.
Outcome Measurements and Statistical Analysis
The bacterial composition of the urine was qualitatively assessed by sequencing the bacterial 16S rRNA gene. Phylogenetic relatedness and microbial alpha diversity were compared to demographics and symptoms using generalized estimating equation models.
Results
The majority of 197 urine samples (86%) had detectable bacterial DNA. Bacterial diversity was significantly associated with higher BMI (p=0.02), increased Medical, Epidemiologic, and Social Aspects of Aging (MESA) urge index score (p=0.04), and hormonal status (p<0.001). No associations were detected with SUI symptoms. Increased diversity was also associated with a concomitant lower frequency of Lactobacillus in hormone-negative women.
Conclusions
Women undergoing SUI surgery have detectable urinary microbiota. This cross-sectional analysis revealed that increased diversity of the microbiota was associated with UUI symptoms, hormonal status and BMI. In contrast, the FUM was not associated with stress urinary incontinence symptoms.
Keywords: Microbiome, Stress Urinary Incontinence, Urgency Urinary Incontinence, Estrogen, Bladder
INTRODUCTION
The influence of the human microbiota on health and disease is increasingly appreciated in a variety of medical fields [1] These microbial communities are often described by their predominant organism, the diversity of organisms within the community and the amount of those organisms [2–4].
The female urinary microbiota (FUM), composed of resident bladder bacteria, was recently recognized when bacterial DNA and low levels of live bacteria were detected in catheterized urine specimens considered “sterile” by standard urine culture [5–7]. Enhanced urine culture techniques have provided clear evidence that FUM microbes are alive; unlike standard urine culture protocols, these enhanced culture techniques provide the appropriate conditions for growth for a wide range of microbes [6, 8]. The living microbial community within the female bladder may provide insight into a variety of common urinary disorders, including urinary incontinence and urinary tract infections. The presence and response to urgency urinary incontinence (UUI) treatment appears related to FUM diversity and/or composition in adult women with UUI [2, 9]. There is also an association between the FUM and risk of urinary tract infection (UTI) following urinary tract surgery [10] or instrumentation [5, 11]. However, there is a lack of information regarding the FUM of adult women with stress urinary incontinence (SUI). The two most common forms of bothersome urinary incontinence (UUI and SUI) often coexist in adult women, especially those seeking surgical treatment for SUI. Information concerning the FUM has the potential to further develop the phenotype of adult women affected by urinary incontinence, with the hope of improving the targeting of treatment in order to improve overall outcomes.
The National Institutes of Health sponsored a large, multi-center, clinical trial of women with uncomplicated SUI planning surgery and previously established a biorepository of urine samples collected for various scientific purposes [12]. In this sub-study, we describe the FUM analysis using 16S rRNA gene sequencing to characterize the cross-sectional relationships between FUM parameters and demographic and clinical characteristics of adult women undergoing surgery for SUI.
MATERIALS AND METHODS
Subject Recruitment and Urine Collection
The Value of Urodynamic Evaluation (ValUE) study was an Institutional Review Board (IRB) approved, multi-center prospective randomized trial comparing surgical outcomes using 2 strategies for pre-surgical testing: multichannel urodynamic testing versus standardized basic office evaluation. [12, 13]. Briefly, adult women were eligible if they reported symptoms of SUI ≥3 months, had a post-void residual <150 mL, a negative urinalysis/standard urine culture, clinical assessment of urethral mobility, desire for SUI surgery, a positive provocative stress urinary test and a qualifying Medical, Epidemiologic, and Social Aspects of Aging (MESA) questionnaire [13, 14] subscale score (stress > urge). Demographic and clinical characteristics were obtained by self-report including hormonal status, which was categorized by the study team into the hormone group that most appropriately described the patient’s hormone use: pre-menopausal, post-menopausal (with or without self-reported, current exogenous hormone use) or uncertain about status.
Participants in the main study provided written consent to contribute a single baseline urine specimen to the biorepository. Urine specimens were collected prior to surgery by a standard protocol and tested by dipstick to exclude UTI at study entry. Specimens were centrifuged at 500–1500g for 10 minutes and the supernatant dispensed into ten 2cc Eppendorf tubes, which were frozen at −80°C until shipped on dry ice to the National Institute of Diabetes and Digestive and Kidney diseases (NIDDK) biorepository. Available baseline urine specimens with sufficient volume for the planned studies were shipped to Loyola University Chicago on dry ice, and stored at −80°C until processed for sequence analysis. Samples from 197 of the 630 (31%) ValUE participants were used in this analysis; most (174) samples had been obtained by clean catch, with the remaining 23 by catheterization. Analyses for this report were approved by The Loyola University Chicago IRB.
16S rRNA gene sequencing
Microbial composition was determined by sequencing the variable 4 (V4) region of the bacterial 16S rRNA gene, as described [2, 6, 9, 11]. The V4 region is ~250 bp, ideal for MiSeq sequence technology (Illumina), and sufficient to classify most bacteria to the family or genus level [15–18]. DNA isolation was performed in a laminar flow hood to avoid contamination. Genomic DNA was extracted from 1 ml of urine, using validated protocols [2, 6, 19]. The V4 region was amplified by a two-step polymerase chain reaction (PCR), using modified universal primers 515F and 806R, as described [2, 6]. Extraction negative controls (no urine) and PCR negative controls (no template) were included to assess contribution of extraneous DNA from reagents. Final PCR products were purified from unincorporated nucleotides and primers using Qiaquick PCR purification kit (Qiagen, Valencia, CA) and Agencourt AMPure XP-PCR magnetic beads (Beckman coulter). Purified samples were normalized to equal DNA concentration, as determined by Nanodrop spectroscopy (Thermo Scientific, Waltham, MA). The sample library and PhiX sequencing control library (Illumina) were denatured and added to the 2x250 bp sequencing reagent cartridge, according to manufacturer’s instructions (Illumina).
Sequence processing
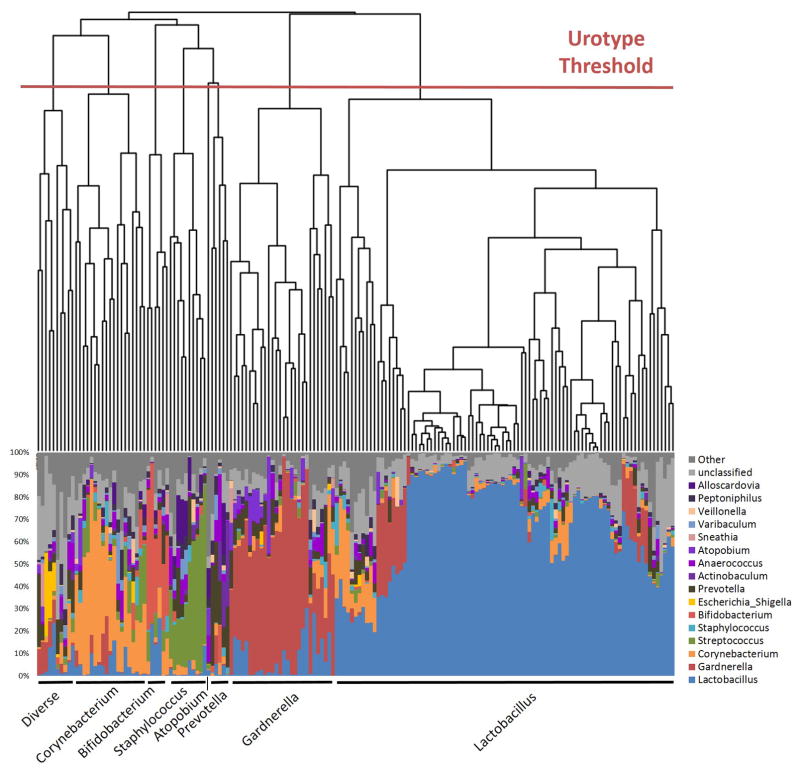
Each specimen was sequenced in duplicate and classified by phylogenetic diversity as measured by Bray-Curtis dissimilarity. A phylogenetic tree was generated and compared to percent total classified reads (relative abundance) at each taxonomic level (phyla, class, order, family, genus). For a genus level example, see Figure 1.
Figure 1. Using phylogenetic similarity to determine similar profiles (aka urotypes).
For each taxonomic level (Phylum, Class, Order, Family, Genus), all samples were compared to each other using Bray-Curtis similarity, which produces a phylogenetic tree, or dendrogram, in which shorter branches link similar samples, and longer branches link more dissimilar samples. Therefore, each tree can be divided into groups or clades. When aligned to relative sequencing abundance, the clades of each tree separate by the identity of the predominant organism. Below is one example, the genus classification from the first replica. The urotype indicates the clades that fall into the same urotype. Each urotype is named for the predominant genus. If no one genus is predominant, then the urotype is considered non-predominant. All corresponding graphs for each replica and each taxonomic level can be found in supplementary Figure S1.
Genus Identification from the first sequencing replica set
Each major branch or clade (termed urotype) in the phylogenetic tree was named for the predominant classified taxon (e.g., Lactobacillus). When there was no predominant taxon, we used the term “non-predominant” to describe the urotype [2, 9, 20]. MiSeq sequence reads were processed following mothur’s MiSeq SOP at http://www.mothur.org/wiki/MiSeq_SOP [18], with minor modifications. Mothur software (version 1.34.4) [21] was used to process raw reads and, using default mothur parameters, to remove low quality and chimeric sequences. Taxonomic classification from phylum to genus level of sequence reads was performed by the RDP Classifier (version 2.5) [22] using the default 0.8 confidence threshold. The sampling depth for this analytic set was set at 2000 reads; the Kolmogorov–Smirnov test confirmed that when subsampling depth exceeded 2000 reads, the distribution of subsampled and original reads distribution had >95.9% similarity amongst all samples.
Most (171/197) samples had detectable DNA with 338,000 and 340,000 reads subsampled for replicates 1 and 2, respectively. The 26 samples without detectable DNA following PCR amplification were classified as “below the detection threshold.” Due to read depths less than 2000, two samples from replica 1 and one sample from replica 2 were also classified as “below the detection threshold”, for a total of 28 in replica 1 and 27 in replica 2. Using mothur’s built-in average-linkage clustering algorithm, the cleaned high-quality sequences were clustered into species level operational taxonomic units (OTUs) based on the commonly used 97% similarity cutoff, resulted in 2579 and 3082 OTUs for replicas 1 and 2, respectively. We used the resultant OTU count table and the R package vegan [23, 24] to determine the Chao1 richness estimate, Pielou evenness index, and Shannon diversity index, which accounts for both richness and evenness, two measures of microbial diversity. Richness is a measure of the total number of unique taxa within a given individual, but does not take into account the distribution of those taxa. In contrast, evenness is a measure of distribution, or equality of representation, of taxa within an environment. Samples “below the detection threshold” lack diversity measurements; these were excluded from subsequent diversity comparisons.
Statistical analysis
Generalized Estimating Equations (GEE), extensions of Generalized Linear Models that account for correlation between replicas, were used to describe associations between demographic and clinical factors with diversity measurements after adjusting for genus urotype. A gamma distribution with a log link was assumed for Shannon, Chao, and Peilou diversity measurements, due to their skewed nature. To be inclusive, we did not make adjustments for Type I error in the GEE analyses when determining potential clinical and demographic associations with microbiota characteristics. Only results from the lowest detected resolution level (i.e., genus) are reported. There was insufficient sample size and power to compare urotypes between catheterized and voided samples. All statistical analyses were conducted using SAS v9.4 (SAS Software Cary, NC) and statistical significance was assessed at the α=0.05 level.
RESULTS
The demographic and clinical characteristics of the 197 participants we studied (Table 1) were similar to those of the overall trial population [12]. Most of these participants were non-Hispanic Caucasian (79%) and currently married (74%). The mean age of the subset was 51 (SD:9.7) years. Forty-two percent of women were pre-menopausal, 31% post-menopausal without current exogenous hormone use, and 18% were using exogenous hormones; the remaining 10% were “unsure” of their status. Consistent with the entrance requirements for the trial [12], women reported stress predominant UI with a median MESA stress index score of 78 (IQR:59–89) and 76% reporting urinary leakage every day and/or night. Concomitant urinary symptoms were common at time of trial enrollment; the median MESA urgency urinary incontinence index score was 33 (IQR:17–50). As only 21 participants had an urgency index of zero, dichotomous group comparisons by urgency index were not performed.
Table 1.
Baseline Clinical and Demographic Characteristics of VALUE Participants Assessed for Urinary Microbiota
| Demographics | N=197 |
|---|---|
| Age [Mean (SD)] | 51 (9.7) |
| Body Mass Index (kg/m2) [Mean (SD)] | 29 (5) |
| Race/Ethnicity* | |
| Hispanic | 23 (12%) |
| Non-Hispanic Caucasian | 156 (79%) |
| African-American | 9(5%) |
| Other | 9(5%) |
| Education | |
| Less High School | 6 (3%) |
| High School/GED | 39 (20%) |
| Some College | 58 (29%) |
| Completed 4 Years of College | 53 (27%) |
| Graduate/Professional Degrees | 41(21%) |
| Self Reported Hormonal Status* | |
| Pre-Menopausal | 82 (42%) |
| Post-Menopausal lacking exogenous hormones | 61 (31%) |
| Post-Menopausal or Uncertain about status on Exogenous hormones | 35 (18%) |
| Uncertain about status lackingexogenous hormones | 19 (10%) |
| Ever Pregnant | 190 (96%) |
| Number of Pregnancies, Mdn(Range) | 3 (0–10) |
| Vaginal Parity, Mdn (Range) | 2 (0–7) |
| History of Smoking | 66 (33%) |
| Currently Smoking | 20 (10%) |
| Currently Married | 145 (74%) |
| Prior Pelvic Surgeries | 151 (77%) |
| Prior Non-surgical Treatment | 122 (62%) |
| Symptom Severity | |
| MESA score a | |
| Stress Index, Mdn(IQR) | 78(59–89) |
| Urge Index, Mdn(IQR) | 33 (17–50) |
| Frequency of Urine Leakage | |
| Less than once a month | 0 |
| A few times a month | 11 (6%) |
| A few times a week | 36 (18%) |
| Every day and/or night | 150 (76%) |
| Voiding Phase Dysfunction | 7 (4%) |
| Suspected Intrinsic Sphincter Deficiency | 39 (20%) |
| Urine Measures | N=167 |
| Specific Gravity, Mdn(IQR) | 1 (1.01–1.02) |
| Urine pH, Mdn(IQR) | 6 (5–7) |
| Glucose positive n (%) | 6 (4%) |
| Blood | |
| Negative | 110 (70%) |
| Trace (Non–hemolyzed) | 11 (7%) |
| Moderate (Non-hemolyzed) | 5 (3%) |
| Trace | 11 (7%) |
| Small (+) | 6 (4%) |
| Moderate (++) | 6 (4%) |
| Large (+++) | 9 (6%) |
| Protein | 25 (16%) |
| DIVERSITY OUTCOME MEASURES[Mean (SD)]b | |
| Shannon | 1.86 (0.97) |
| Chao | 124.08 (59.35) |
| Pielou | 0.43 (0.19) |
Total number of SUI participants is 197.
Mean (SD) or N(%) reported unless otherwise specified.
Mdn=Median; SD=Standard deviation; IQR = Interquartile Range;
Stress Index and Urge Indices were calculated using the MESA questionnaire.
Race/Ethnicity and Hormonal status per subject report
Least Squares Means- adjusted for correlation between both replicas
Figure 1 displays a representative phylogenetic tree and histogram for replica 1 of the remaining samples classified at the genus level. Together, the phylogenetic tree and histogram show how samples cluster into urotypes. Supplementary Figure S1 displays similar representations for each replica at other taxonomic levels.
Replicas 1 and 2 yielded similar results (Table 2). The only major difference involved samples of the Lactobacillus urotype, the most common urotype in both replicas (replica 1: 46%, 90/197; replica 2: 37%, 72/197). The difference in Lactobacillus urotype frequency between replicas was mirrored by an inverse difference in the frequency of the “non-predominant” urotype (replica 1: 5%, 10/197; replica 2: 12%, 23/197). This inverse relationship was primarily caused by phylogenetic reorganization and therefore re-classification of the most diverse samples in the Lactobacillus urotype as members of the “non-predominant” urotype.
Table 2. Urotype frequency across all taxonomic levels and between replicas.
Using Bray-Curtis dissimilarity, each dataset was divided into clades (row). A urotype is defined as a clade that is predominated by a single organism, or labeled as “non-predominant” when no single organism predominates. Here, the frequency of urotype identification is shown for each taxonomic classification for both replicas. Urine is a low biomass niche and some samples had so little DNA that the signal could not be distinguished from background, these samples are labeled as “below detection level”.
| A. Replica 1: Frequency of Sequencing Urotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Phylum | Class | Order | Family | Genus | |||||
| Below Detection Level | 14% (28/197) | Below Detection Level | 14% (28/197) | Below Detection Level | 14% (28/197) | Below Detection Level | 14% (28/197) | Below Detection Level | 14% (28/197) |
|
| |||||||||
| Non-predominant | 4.6% (9/197) | Non-predominant | 11% (22/197) | Non- predominant | 5.1% (10/197) | ||||
|
| |||||||||
| Firmicutes | 41% (80/197) | Bacilli | 43% (84/197) | Lactobacillales | 48% (94/197) | Lactobacillaceae | 35% (68/197) | Lactobacillus | 46% (90/197) |
| Firmicutes/Actinobacteria | 15% (29/197) | Bacilli/Mixed | 4.1% (8/197) | Streptococcaceae | 2.0% (4/197) | Streptococcus | 4.6% (9/197) | ||
|
| |||||||||
| Actinobacteria | 6.6% (13/197) | Actinobacteria | 23% (46/197) | Bifidobacteriales | 15% (30/197) | Bifidobacteriaceae | 24% (48/197) | Bifidobacterium | 3.6% (7/197) |
| Actinobacteria/Mixed | 18% (35/197) | Actinobacteria/Mixed | 6.1% (12/197) | Gardnerella | 14% (28/197) | ||||
| Actinomycetales | 10% (20/197) | Corynebacteriaceae | 9.1% (18/197) | Corynebacterium | 10% (19/197) | ||||
| Atopobium | 0.5% (1/197) | ||||||||
|
| |||||||||
| Bacteroidetes | 3.0% (6/197) | Bacteroidetes | 2.0% (4/197) | Bacteroidales | Prevotellaceae | 2.0% (4/197) | Prevotella | 2.5% (5/197) | |
|
| |||||||||
| Proteobacteria | 1.0% (2/197) | Gammaproteobacteria | 1% (2/197) | Enterobacteriales | 0.5% (1/197) | Enterobacteriacea | 2.5% (5/197) | ||
|
|
|||||||||
| Firmicutes | Clostridia | 6.1% (12/197) | Clostridia | 7.6% (15/197) | |||||
|
|
|||||||||
| Fusobacterium | 2.0% (4/197) | ||||||||
| B. Replica 2: Frequency of Sequencing Urotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Phylum | Class | Order | Family | Genus | |||||
| Below Detection Level | 14% (27/197) | Below Detection Level | 14% (27/197) | Below Detection Level | 14% (27/197) | Below Detection Level | 14% (27/197) | Below Detection Level | 13% (27/197) |
|
| |||||||||
| Non-predominant | 12% (24/197) | Non-predominant | 12% (23/197) | Non- predominant | 12% (23/197) | ||||
|
| |||||||||
| Firmicutes | 53% (104/197) | Bacilli | 39% (77/197) | Lactobacillales | 44% (87/197) | Lactobacillaceae | 38% (74/197) | Lactobacillus | 37% (72/197) |
| Firmicutes/Actinobacteria | 17% (34/197) | Bacilli-Mixed | 8.1% (16/197) | Streptococcaceae | 12% (23/197) | Streptococcus | 2.5% (5/197) | ||
| Clostridiales | 7.1% (14/197) | ||||||||
|
| |||||||||
| Actinobacteria | 13% (25/197) | Actinobacteria | 24% (48/197) | Bifidobacteriales | 16% (31/197) | Bifidobacteriaceae | 23% (45/197) | Bifidobacterium | 2.0% (4/197) |
| Actinobacteria/Mixed | Actinobacteria/Mixed | 13% (26/197) | Gardnerella | 18% (35/197) | |||||
| Actinomycetales | 14% (28/197) | Corynebacteriaceae | 4.1% (8/197) | Corynebacterium | 7.1% (14/197) | ||||
|
| |||||||||
| Bacteroidetes | 2.0% (4/197) | Prevotellaceae | 2.5% (5/197) | Prevotella | 1.5% (3/197) | ||||
|
| |||||||||
| Proteobacteria | 1.5% (3/197) | Gammaproteobacteria | 0.5% (1/197) | Enterobacteriacea | 0.5% (1/197) | ||||
|
|
|||||||||
| Betaproteobacteria | 1.0% (2/197) | ||||||||
The results of the GEE analyses before and after adjustment for urotype were similar, thus only adjusted results are presented (Table 3). We used two types of microbial diversity measurements: richness (total number of unique taxa) and evenness (equality of representation of taxa within an environment). Richness, as estimated by Chao1, was significantly associated only with urine pH (p=0.03). In contrast, richness and evenness, as measured together by the Shannon index, were significantly associated with MESA urge index score (p=0.04), BMI (p=0.02) and hormonal status (p<0.001) (Table 3). For a 10% increase in MESA urge index score, the Shannon index increased by 0.03 units (p=0.04). On average, a 10-unit increase in BMI was associated with a 0.1-unit increase in Shannon diversity (p=0.02). Post-menopausal women not on exogenous hormones had a Shannon index 0.23 units higher than did pre-menopausal women (p=0.004).
Table 3. Association between Microbiota Characteristics and Demographic.
Factors or Symptom Severity adjusted for Genus urotype Generalized Estimating Equations (GEE), assuming a gamma distribution and log link, were used to assess the association between each demographic or symptom measurement for three separate microbial diversity measurements after adjusting for urotype at the genus level. P-values are not adjusted for type I error. Diversity measurements include Shannon diversity (richness and evenness), Chao diversity (richness only) and Peilou (evenness only).
| Community Diversity Gamma GEE Models Adjusted for Genus Urotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Shannon | Chao1 | Peilou | |||||||
| Demographics | Beta | SE | p-value | Beta | SE | p-value | Beta | SE | p-value |
| BMI | 0.01 | 0.006 | 0.02 | 0.006 | 0.005 | 0.18 | 0.01 | 0.005 | 0.02 |
| Race/Ethnicity* | 0.53 | 0.25 | 0.52 | ||||||
| Hispanic | 0.07 | 0.11 | 0.55 | 0.05 | 0.08 | 0.58 | 0.04 | 0.09 | 0.66 |
| Non-Hispanic Caucasian | Ref | ||||||||
| African-American | −0.15 | 0.22 | 0.51 | 0.08 | 0.09 | 0.36 | −0.17 | 0.20 | 0.40 |
| Other | −0.30 | 0.30 | 0.27 | −0.28 | 0.15 | 0.05 | −0.25 | 0.23 | 0.28 |
| Education | 0.51 | 0.31 | 0.59 | ||||||
| Less High School | 0.039 | 0.19 | 0.84 | 0.18 | 0.08 | 0.02 | −0.01 | 0.16 | 0.96 |
| High School/GED | −0.03 | 0.11 | 0.77 | 0.11 | 0.09 | 0.23 | −0.03 | 0.09 | 0.70 |
| Some College | Ref | ||||||||
| Completed 4 Years of College | −0.12 | 0.09 | 0.18 | 0.003 | 0.07 | 0.97 | −0.10 | 0.08 | 0.21 |
| Graduate/Professional Degrees | −0.16 | 0.11 | 0.15 | 0.07 | 0.08 | 0.34 | −0.14 | 0.10 | 0.17 |
| Hormonal Status* | <0.001 | 0.32 | <0.001 | ||||||
| Pre-Menopausal | Ref | ||||||||
| Post-Menopausal lacking exogenous hormones | 0.23 | 0.08 | 0.004 | 0.08 | 0.07 | 0.25 | 0.21 | 0.07 | 0.02 |
| Post-menopausal or Uncertain about exogenous hormonal status | −0.28 | 0.14 | 0.05 | −0.07 | 0.08 | 0.39 | −0.26 | 0.12 | 0.04 |
| Uncertain about exogenous hormonal status | 0.21 | 0.09 | 0.02 | −0.03 | 0.10 | 0.73 | 0.22 | 0.07 | 0.003 |
| Ever Pregnant | −0.21 | 0.15 | 0.16 | −0.04 | 0.10 | 0.73 | −0.18 | 0.12 | 0.22 |
| Smoking History | −0.16 | 0.07 | 0.04 | −0.07 | 0.06 | 0.21 | −0.13 | 0.06 | 0.06 |
| Currently Married | 0.10 | 0.08 | 0.21 | −0.04 | 0.06 | 0.54 | 0.11 | 0.07 | 0.11 |
| Prior Pelvic Surgeries | −0.14 | 0.07 | 0.08 | −0.02 | 0.06 | 0.72 | −0.13 | 0.07 | 0.07 |
| Prior Non-surgical Treatment | 0.03 | 0.08 | 0.66 | −0.02 | 0.06 | 0.73 | 0.04 | 0.07 | 0.54 |
|
| |||||||||
| Symptom Severity | |||||||||
|
| |||||||||
| MESA scorea | |||||||||
| Stress Index | 0.002 | 0.002 | 0.28 | 0.0003 | 0.002 | 0.87 | 0.002 | 0.002 | 0.31 |
| Urge Index | 0.003 | 0.002 | 0.04 | 0.002 | 0.001 | 0.18 | 0.003 | 0.001 | 0.04 |
| Frequency of Leakage | 0.08 | 0.61 | 0.04 | ||||||
| A few times a month | −0.38 | 0.24 | 0.11 | −0.05 | 0.09 | 0.53 | −0.37 | 0.21 | 0.07 |
| A few times a week | −0.15 | 0.10 | 0.12 | 0.05 | 0.07 | 0.30 | −0.15 | 0.08 | 0.06 |
| Every day and/or night | Ref | ||||||||
|
| |||||||||
| Urine Characteristics | |||||||||
| Protein | 0.18 | 0.10 | 0.09 | 0.08 | 0.08 | 0.38 | 0.14 | 0.08 | 0.09 |
| Specific Gravity | −0.18 | 0.01 | 0.32 | −0.10 | 0.01 | 0.31 | −0.14 | 0.008 | 0.32 |
| Urine pH | −0.06 | 0.04 | 0.11 | −0.07 | 0.03 | 0.03 | −0.05 | 0.04 | 0.13 |
| Glucose | −0.07 | 0.19 | 0.70 | −0.13 | 0.11 | 0.25 | −0.03 | 0.15 | 0.82 |
| Blood | 0.40 | 0.69 | 0.29 | ||||||
| Negative | Ref | ||||||||
| Trace (Non-hemolyzed) | −0.08 | 0.16 | 0.60 | −0.06 | 0.14 | 0.66 | −0.06 | 0.14 | 0.69 |
| Moderate (Non-hemolyzed) | −0.001 | 0.25 | 0.10 | 0.0853 | 0.13 | 0.50 | 0.008 | 0.24 | 0.97 |
| Trace | 0.18 | 0.11 | 0.09 | 0.16 | 0.10 | 0.10 | 0.18 | 0.10 | 0.08 |
| Small (+) | 0.21 | 0.13 | 0.11 | −0.10 | 0.12 | 0.43 | 0.23 | 0.09 | 0.01 |
| Moderate (++) | 0.39 | 0.18 | 0.03 | 0.13 | 0.15 | 0.39 | 0.32 | 0.14 | 0.03 |
| Large (+++) | 0.10 | 0.13 | 0.45 | 0.01 | 0.07 | 0.84 | 0.10 | 0.11 | 0.38 |
MESA Questionnaire - SUI and UUI index scores each ranging from 0–100 with higher scores indicating greater symptom severity)
Race/Ethnicity and Hormonal status per subject report
Because these parameters did not associate with richness (Chao1), we tested evenness alone, as measured by Peilou diversity (Table 3). Evenness was significantly associated with MESA urge index score (p=0.04), BMI (p=0.02) and estrogen status (p<0.001). For a 10% increase in the MESA urge index score, the Pielou diversity increased by 0.03 units (p=0.04). On average, a 10-unit increase in BMI was associated with a 0.1-unit increase in Peilou diversity. Finally, post-menopausal women had a 0.21 unit higher diversity measurement compared to pre-menopausal women (p=0.02). Since these results were similar to those observed with the Shannon index, we conclude that increased community evenness associates with UUI symptoms, BMI and especially hormonal status.
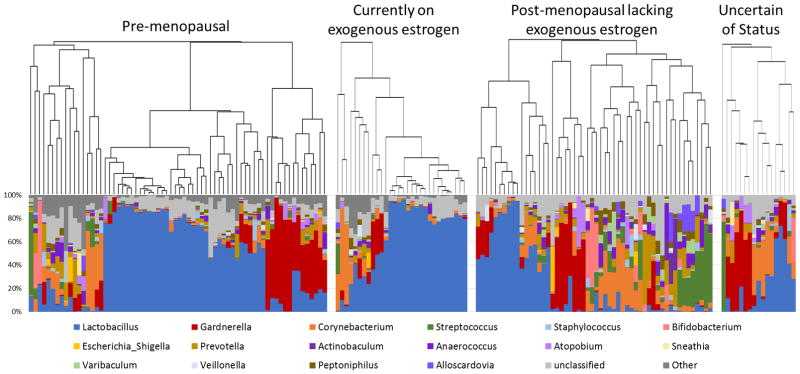
Because community evenness associates strongly with hormonal status, we constructed a visual comparison of microbial diversity subdivided by hormonal use (Figure 2). Compared to other groups, the hormone-positive women (pre-menopausal and post-menopausal on exogenous hormones) had a higher frequency of Lactobacillus or Gardnerella urotypes (66%) and a lower frequency of ‘non-predominant’ urotypes, while the hormone-negative women (post-menopausal not on exogenous hormones) had a lower frequency of Lactobacillus or Gardnerella urotypes (38%) and greater frequency of ‘non-predominant’ urotypes (p<0.001).
Figure 2. Phylogenetic diversity and urotype distribution between estrogen status.
Relative abundance of the microbial community at the genus level for each of the 4 estrogen groups. Each bar is a separate individual with the percent of total classified reads to the genus level represented on the y-axis. Phylogenetic relatedness as measured by Bray-Curtis dissimilarity is depicted in the dendrograms above each group.
The full cohort is separated by hormone status: pre-menopausal, post-menopausal (with or without self-reported, current exogenous hormones) or uncertain about hormonal status. Estrogen positive groups (pre-menopausal and those currently on exogenous estrogen) have a greater prevalence of Lactobacillus-predominant individuals (blue) than the estrogen negative group. The estrogen negative group has a greater number of non-predominant (multi-colored) profiles compared to the estrogen positive populations.
DISCUSSION
Main findings
In this study of women undergoing surgery for uncomplicated SUI, the presence of UUI symptoms appears related to increased microbial evenness, indicating that the FUM of women with UUI symptoms was less likely to be predominated by a single microbe. A similar relationship was observed between microbial evenness and both hormone status and BMI. The clinical impact of these findings is significant, given the common coexistence of UUI in women who undergo surgical SUI treatment. While considerable uncertainly persists about the effect of SUI surgery on pre-existing UUI symptoms and each individual patient’s risk of developing de novo UUI symptoms following SUI surgery, the microbiota may provide useful information for clinical phenotyping of patients prior to surgery. Without comparison to an age and hormone-status matched continent control group, we cannot conclude that women with “pure” SUI are like continent women. However, it does appear the presence and characteristics (i.e. evenness) of the FUM relate to UUI in this clinically relevant cohort of women undergoing SUI surgery. Our findings will require further research to validate and clarify the physiological mechanisms.
We observed this association between microbial evenness and UUI symptoms despite the possibility of vulvo-vaginal contamination in these voided urine samples. The FUM detected in the pre-surgical voided urine samples of this cohort was similar to those assessed in catheterized samples obtained from other cohorts of women [2, 9, 11]. To ensure appropriate data interpretation of voided sample data, therefore, we recommend prior information of FUM composition from women of a similar cohort using catheter collection methods that obtain urine directly from the bladder.
Research implications
Our finding of a relationship between microbial evenness to hormonal status may provide clues for further study. Menopausal women not on exogenous hormones had increased microbial evenness and their FUM was less likely to be predominated by a single microbe. These results suggest that predominance (most often by Lactobacillus species) is typical of hormone-positive women and that loss of that predominance might be associated with UUI symptoms. Increased microbial diversity, in particular community evenness, positively correlated with UUI and BMI symptoms, but not with SUI symptoms. BMI is considered a contributing factor to urgency symptoms [25], and the FUM of UUI women have been reported to be associated with BMI [2]. Obesity is also associated with an increase in circulating estrogens, emphasizing the complex nature of vaginal and urinary health [26, 27]. Further study of BMI and the FUM are needed.
It is well known that estrogen receptors are found throughout the lower urinary tract, supporting the likelihood that estrogen has a role in optimizing bladder function [28–30]. Intriguingly, the use of intravaginal estrogen has been reported to improve the lower urinary tract symptoms of urinary urgency, frequency, SUI, UUI and UTI [31]. Direct effects of estrogen on the bladder could include maintenance of bladder structural and functional integrity via bladder wall thickness, expression of vascular epithelial growth factor (VEGF), and effects on β3-adrenoreceptor activity, as described in rats [32, 33]. Given that use of intravaginal estrogen correlated with an increase in Lactobacillus species and a decrease in anaerobic bacteria [34], it might also influence the FUM.
Although we did not assess the vaginal microbiota of VaLUE subjects, we note the FUM’s similarity to published data regarding the vaginal microbiota. We acknowledge the possibility that the vulvo-vaginal flora contributes to bacterial diversity. However, it is biologically plausible that both these adjacent anatomical sites experience microbial alterations related to estrogen status. The bladder is a low biomass niche that contains many organisms similar to those of the vagina, including Lactobacillus, Gardnerella, and a diverse set of anaerobes [2, 6, 7]. The relationship between these two biological niches deserves further study.
Strengths and Limitations
Our study has a number of strengths, including rigorous participant characterization, multi-site recruitment, cutting-edge sequencing techniques and state-of-the art analytic approaches. The study has limitations associated with the cross-sectional study design. The study would have benefitted from controls matched for age, BMI and estrogen status, paired vaginal samples and/or longitudinal sampling, further details regarding hormone status, a higher proportion of participant samples, increased proportion of catheterized specimens and concurrent enhanced urine cultures [6]. Also, the known relationship between estrogen status and aging may mask a biological relationship that is rightfully attributed to only one of these variables. A larger study that includes women with various forms of urinary incontinence and matched controls will be needed.
CONCLUSIONS
Women undergoing SUI surgery have detectable urinary microbiota that may be of value for phenotyping patients. This cross-sectional analysis revealed that diversity of the microbiota was associated with UUI symptoms, hormonal status and BMI.
Supplementary Material
SUMMARY.
In pre-surgical SUI patients, urinary microbiota are associated with UUI symptoms, hormonal status and BMI, but not SUI-specific symptoms.
Acknowledgments
We acknowledge and thank Meghan Pearce PhD MPH helping to establish the DNA extraction and 16S rRNA sequencing protocol, Amy Rosenfeld PhD for performing the sequencing, and Eddi Lin BS and Qunfeng Dong PHD for the initial bioinformatics analysis.
FUNDING
This study and is registered at www.clinicaltrials.gov as NCT00803959 and was supported by NIH grants U01 DK58229 and R21 DK097435 (to AJW and LB). Loyola University Chicago Stritch School of Medicine’s research computing facility was developed through grant funds awarded by the Department of Health and Human Services as award number 1G20RR030939-01. Our funding sources had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript. The NIDDK Urinary Incontinence Treatment Network and investigators were supported by cooperative agreements from the National Institute of Diabetes and Digestive and Kidney Diseases, U01 DK58225, U01 DK58229, U01 DK58234, U01 DK58231, U01 DK60379, U01 DK60380, U01 DK60393, U01 DK60395, U01 DK60397, and DK60401. Supported was also provided by the National Institute of Child Health and Human Development and Office of Research in Women’s Health, NIH.
Glossary of Terms
- DNA Amplicon Sequencing
Sequencing is a process of determining the precise order of nucleotides within a DNA molecule. Amplicon sequencing refers to the sequencing of a short stretch of DNA amplified from the genome or sample
- 16S rRNA gene sequencing
Sequencing the 16S ribosomal RNA gene is a common amplicon sequencing method used to accurately classify bacteria present in a given sample
- Sequencing Reads
The individual sequences obtained from a given sample
- OTU
Operational taxonomic unit is used to classify groups of closely related sequencing reads
- RDP classifier
A naive Bayesian classifier that rapidly and accurately provides taxonomic assignments from domain to genus
- Phylogenetic tree
A phylogenetic tree is a branching diagram that shows the evolutionary or community relationships between samples
- Alpha Diversity
A measurement of diversity of a single site or sample. Compared to Beta diversity which is the measurement between sites or samples. Alpha diversity include measurement of species richness and evenness. Richness is a measure of the total number of unique taxa within a given individual, but does not take into account the distribution of those taxa. In contrast, evenness is a measure of distribution, or equality of representation, of taxa within an environment
- Pielou evenness index
An alpha diversity measurement for evenness only
- Chao 1 richness
An alpha diversity measurement for richness only
- Shannon diversity index
An alpha diversity measurement for both richness and evenness
- Urotype
A urotype is defined as a group on the phylogenetic tree (i.e., clade) that is predominated by a single organism, or labeled as “non-predominant” when no single organism predominates
Footnotes
Reprints will not be available.
CONFLICT OF INTEREST: The following authors report no disclosures: Thomas-White, Kliethermes, Moalli, Norton, Richter, Zimmern, Kusek. The following authors report disclosures: E. Lukacz: Uroplasty (grant), Pfizer (research support), Up to Date (author royalties) L. Rickey: Analytica (consultant), Health Monitor Netowrk (consultant), Up to Date (author royalties), H. Richter: Kimberly Clark (consultant), UpToDate (royalties)
Wolfe: Astellas Scientific and Medical Affairs (research support) Brubaker: Up to Date (editorial royalties)
AUTHOR ROLES Krystal J. THOMAS-WHITE: Conception and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis
Stephanie KLIETHERMES: Analysis and interpretation of data, critical revision of the manuscript for important intellectual content, statistical analysis
Leslie RICKEY: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Emily LUKACZ: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Holly E. RICHTER: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Pamela MOALLI: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Philippe ZIMMERN: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Peggy NORTON: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
John W. KUSEK: Acquisition of data, critical revision of the manuscript for important intellectual content, obtaining funding
Alan J. WOLFE: Conception and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtaining funding, supervision
Linda BRUBAKER: Conception and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtaining funding, supervision
References
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearce MM, et al. The Female Urinary Microbiome: a Comparison of Women with and without Urgency Urinary Incontinence. MBio. 2014;5(4) doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce MM, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. 2015;213(3):347e1–347e11. doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brubaker L, et al. Urinary bacteria in adult women with urgency urinary incontinence. Int Urogynecol J. 2014 doi: 10.1007/s00192-013-2325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilt EE, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(3):871–6. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe AJ, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50(4):1376–83. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price TK, et al. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol. 2016 doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas-White KJ, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J. 2015 doi: 10.1007/s00192-015-2847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fok CS, et al. Day of surgery urine cultures identify urogynecologic patients at increased risk for postoperative urinary tract infection. J Urol. 2013;189(5):1721–4. doi: 10.1016/j.juro.2012.11.167. [DOI] [PubMed] [Google Scholar]
- 11.Pearce MM, et al. The female urinary microbiota in urgency urinary incontinence. Am J Obstet Gynecol. 2015 [Google Scholar]
- 12.Nager CW, et al. Design of the Value of Urodynamic Evaluation (ValUE) trial: A non-inferiority randomized trial of preoperative urodynamic investigations. Contemp Clin Trials. 2009;30(6):531–9. doi: 10.1016/j.cct.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nager CW, et al. A randomized trial of urodynamic testing before stress-incontinence surgery. N Engl J Med. 2012;366(21):1987–97. doi: 10.1056/NEJMoa1113595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzog AR, Fultz NH. Prevalence and incidence of urinary incontinence in community-dwelling populations. J Am Geriatr Soc. 1990;38(3):273–81. doi: 10.1111/j.1532-5415.1990.tb03504.x. [DOI] [PubMed] [Google Scholar]
- 15.Ghyselinck J, et al. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One. 2013;8(8):e71360. doi: 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen TW, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347(6218):170–5. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubinak JL, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun. 2015;6:8642. doi: 10.1038/ncomms9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozich JJ, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S, et al. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7(3):e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brubaker L, Wolfe AJ. The new world of the urinary microbiota in women. Am J Obstet Gynecol. 2015 doi: 10.1016/j.ajog.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Team, R.D.C. R: a language and environment for statistical computing. R Foundation for Statistical Computing; 2014. [Google Scholar]
- 24.Jari Oksanen FGB, Kindt Roeland, Legendre Pierre, O’Hara RB, Simpson Gavin L, Solymos Peter, Henry M, Stevens H, Wagner Helene. Package ‘vegan’. Community Ecology Package, Version, 2.2–2.1. 2008. [Google Scholar]
- 25.Subak LL, et al. Weight loss to treat urinary incontinence in overweight and obese women. N Engl J Med. 2009;360(5):481–90. doi: 10.1056/NEJMoa0806375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tworoger SS, et al. Birthweight and body size throughout life in relation to sex hormones and prolactin concentrations in premenopausal women. Cancer Epidemiol Biomarkers Prev. 2006;15(12):2494–501. doi: 10.1158/1055-9965.EPI-06-0671. [DOI] [PubMed] [Google Scholar]
- 27.De Pergola G, et al. Inhibitory effect of obesity on gonadotropin, estradiol, and inhibin B levels in fertile women. Obesity (Silver Spring) 2006;14(11):1954–60. doi: 10.1038/oby.2006.228. [DOI] [PubMed] [Google Scholar]
- 28.Cipullo LM, et al. Pharmacological approach to overactive bladder and urge urinary incontinence in women: an overview. Eur J Obstet Gynecol Reprod Biol. 2014;174:27–34. doi: 10.1016/j.ejogrb.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Robinson D, et al. Oestrogens and overactive bladder. Neurourol Urodyn. 2014;33(7):1086–91. doi: 10.1002/nau.22464. [DOI] [PubMed] [Google Scholar]
- 30.Robinson D, Toozs-Hobson P, Cardozo L. The effect of hormones on the lower urinary tract. Menopause Int. 2013;19(4):155–62. doi: 10.1177/1754045313511398. [DOI] [PubMed] [Google Scholar]
- 31.Rahn DD, et al. Vaginal estrogen for genitourinary syndrome of menopause: a systematic review. Obstet Gynecol. 2014;124(6):1147–56. doi: 10.1097/AOG.0000000000000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y, et al. Effects of steroid hormones on morphology and vascular endothelial growth factor expression in female bladder. Urology. 2009;73(6):1210–7. doi: 10.1016/j.urology.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara S, et al. Estrogen levels influence beta-3-adrenoceptor-mediated relaxation of the female rat detrusor muscle. Urology. 2002;59(4):621–5. doi: 10.1016/s0090-4295(01)01583-7. [DOI] [PubMed] [Google Scholar]
- 34.Brotman RM, et al. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause. 2014;21(5):450–8. doi: 10.1097/GME.0b013e3182a4690b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.