Abstract
Nucleotide excision repair (NER) is the key DNA repair system that eliminates the majority of DNA helix distorting lesions. RNA polymerase (RNAP) expedites the recognition of DNA damage by NER components via transcription-coupled DNA repair (TCR). In bacteria, a modified nucleotide ppGpp (“Magic Spot”) is a pleiotropic second messenger that mediates the response to nutrient deficiencies by altering the initiation properties of RNAP. In this review we will discuss newly elucidated roles of ppGpp in transcription elongation that couple this alarmone to DNA damage repair and maintenance.
Keywords: DNA repair, transcription elongation, backtracking, ppGpp, mutation rate
Introduction
In bacteria, NER begins when a complex of UvrA and UvrB proteins (UvrAB) binds to the site of DNA damage [1]. However, the massive welter of undamaged DNA compared to the low frequency of lesions presents a fundamental challenge to this process. The recruitment of NER factors to DNA in vivo is also complicated by molecular crowding and by the abundance of proteins normally associated with DNA. The SOS response system stimulates NER by increasing intracellular concentrations of NER enzymes and also by altering the global gene expression under genotoxic conditions [2].
Transcription-coupled repair (TCR) is a facilitator of the NER. TCR is initiated when adducts in the template DNA strand block elongating RNAP. Even though the halted RNAP can obscure the DNA lesions and inhibit repair, the two known TCR factors, Mfd and UvrD (representing two separate TCR pathways), use it as a signal for recognition of DNA damage sites. Mfd protein can dissociate RNAP from the site of damage by pushing it forward via its ATP-dependent translocase activity and recruit UvrA to initiate the repair process [3]. In the other TCR pathway, which has recently been discovered, the UvrD helicase pulls the stalled RNAP backwards, away from the site of damage, followed by UvrAB recruitment [4][5]. As a result, the transcribed strand of the gene is repaired more rapidly than the non-transcribed strand [6].
Nutrient deprivation triggers accumulation of guanosine 5’-diphosphate 3’-diphosphate (ppGpp) [7]. Build up of uncharged tRNAs at the A-site of the ribosome leads to ribosome pausing and activation of the ribosome-associated ppGpp synthase RelA [8][9]. ppGpp binds RNAP and, with the help of transcription factor DksA, destabilizes the open promoter complexes of stable RNA genes and ribosomal protein encoding genes [10][11]. This leads to a dramatic decrease in the rate of ribosome production, helping bacteria to conserve energy. This so called “stringent response” also enables fast recovery upon repletion of nutrients, contingent on degradation of ppGpp by the dedicated hydrolase SpoT. “Relaxed” (relA) mutants fail to make this adjustment and consequently resume growth slower during the recovery phase [12].
Following decades of profound insights into the molecular mechanics of the stringent response, we now begin to understand how ppGpp also helps to establish another major physiological condition - the response to genotoxic stress. Here we discuss how a single small molecule messenger mediates two different modes of stress responses with only partial overlapping outcomes.
ppGpp and transcription-replication conflicts
A link between ppGpp, RNAP and DNA damage repair was first established through genetic analysis [13][14][15]. A relA spoT (ppGpp0) mutation exacerbated the sensitivity of a ruvAC mutant (defective in Holliday structure branch migration and resolution) to UV radiation, but not to Mitomycin C. Additionally, a spoT1 (ppGpp++) mutation suppressed this sensitivity of ruvAC [13]. The difference between the two forms of insults is that UV mostly causes lesions on one strand of DNA at a given genomic location [3], whereas double strand breaks are a common damage signature of Mitomycin C. This implied that in the ruvAC mutant Holliday junctions are formed as part of a repair process distinct from recombination-dependent double strand break repair. In addition, the replication restart gene priA and the helicase encoding gene recG were found to be required for the suppressive effect of spoT1, suggesting stalling, regression and restart of a replication fork in ruvAC-mediated UV damage repair process [13]. Genetic screening for suppressor mutants to rescue this sensitivity identified point mutations in rpoB encoding the beta subunit of RNAP (collectively, this set of mutants was referred to as rpo*). ruvAC sensitivity could be suppressed by either rpo* or by spoT1 (ppGpp++), but no additive effect was seen when the two suppressing alleles were combined [13]. These results were interpreted to mean that rpo* mutants are functional mimics of ppGpp-bound RNAP. The recent identification of two ppGpp binding sites on RNAP [16] will help to determine whether rpo* mutations are true structural mimics of ppGpp-bound RNAP.
UV-induced cyclobutane pyrimidine dimers (CPD) present a formidable roadblock to elongating RNAP [17], which can also lead to replication fork stalling. Collectively, these results [13][14][15] supported a model where replication fork stalling, regression and subsequent RuvAC-dependent repair is more likely to occur when RNAP is not ppGpp-bound. Moreover, ppGpp binding to RNAP appears to prevent conflicts with the replisome, or at least mitigates the negative outcome of such collisions.
Further investigation showed that the rpo* mutants exhibited increased resistance to the antibiotic bicyclomycin (BCM) [18]. BCM targets the essential transcription termination factor Rho [19]. Much like blocking of RNAP by CPDs, inhibition of transcription termination stalls replication forks due to collisions with arrays of paused RNAPs. ppGpp binding to the elongating RNAP appears to relieve the genotoxic stress associated with such collisions.
As the replisome moves on DNA much faster than RNAP, these machineries are bound to collide frequently even under normal growth conditions. The orientation of transcription relative to replication influences how elongating RNAPs would interfere with replication [20][21][22]. The replication fork can encounter RNAP in either a head-on or co-directional orientation. In head-on oriented genes, RNAP elongates in the 3’>5’ direction along the lagging strand template, while the replicative helicase DnaB moves towards it on the same strand in the 5’>3’ direction. In co-directional oriented genes, RNAP and DNA polymerase (Pol III) are both moving on the leading strand template in the 3’>5’ direction. A noteworthy feature of bacterial genome organization is a strong bias for co-directional positioning of highly transcribed genes with respect to replication. For instance, in E. coli all seven rRNA operons are found in the co-directional orientation. As rRNA operons are the most highly transcribed sequences in the bacterial genome, they present the most substantial challenge for the replisome. The consequences of transcription-replication collisions in the head-on orientation were tested using a genetically engineered E. coli strains with inverted rRNA operons (INV strains) [20]. Under conditions allowing maximal transcription rate of rRNA, the INV strains now require 2 out of 3 accessory helicases for viability; the replisome associated helicase Rep, the RNAP associated helicase UvrD, and DinG [20]. The synthetic lethality of all 3 combinations of helicase pairs does not occur in wild type E. coli [23]. Nonetheless, even in the natural co-directional orientation, rrn genes are ‘hot-spots’ for replisome stalling, at least in B. subtilis [21]. Therefore, while head-on collisions between transcription and replication are more dangerous, co-directional collisions in highly transcribed genes have the potential to interfere with a replication fork as well.
In addition to the orientation of RNAP relative to the replisome, the pausing state of RNAP, and more specifically its backtracking state, affects the fate of transcription-replication conflicts. Unlike the replisome, elongating RNAP complexes have the ability to shuffle back and forth along the template strand - a universal phenomenon known as backtracking [24]. As it moves backward, the 3'-OH end of the growing transcript is displaced from the template into the secondary channel that can be occupied by transcription factors DksA, GreA or GreB. The ppGpp potentiator, DksA, plugs the secondary channel, thereby partially preventing backtracking [25]. The transcript cleavage factors GreA and GreB stimulate the RNAP-mediated cleavage of backtracked RNA, which generates a new 3'-OH in the RNAP active site, enabling transcription to resume [26]. Backtracking is also prevented by the trailing ribosome as it translates the emerging transcript [27].
A plasmid based system was designed to monitor transcription-replication collisions in vivo, where, an inducible promoter controls expression of a gene oriented either in the Head-on’ or ‘Co-directional’ manner with respect to replication [28]. Primer extension on plasmid DNA purified from bacterial cultures of various genetic backgrounds and in different conditions enabled measurement of single strand nicks and double strand breaks. In the co-directional orientation, when RNAP was backtracked, due to the lack of anti-backtracking transcription cleavage factors GreA and GreB, dsDNA breaks are readily detectable. The same was true for dsDNA breaks in chromosomal DNA [28]. These in vivo results were surprising considering that co-directional conflicts were thought to be benign [20]. However, they can be rationalized in light of a different study, in which co-directional conflicts were reconstituted with purified RNAP and the replisome [29]. In this experimental setup the replisome pauses and then readily displaces co-directional RNAP upon collision. However, in the process of terminating RNAP the replisome frequently uses the nascent RNA as a primer to resume leading strand synthesis. Therefore, a model was proposed in which dsDNA breaks caused by co-directional conflicts with backtracked RNAP is a two-step process. First, the use of the nascent RNA as a primer leads to a nick in the leading strand. Then, the next round of replication converts it into a double strand break [28][30]. Thus, with respect to genome integrity, the outcome of using the nascent RNA as a primer by the replisome depends on whether RNAP was in the backtracked state at the moment of collision. As most essential and active genes in bacteria are oriented co-directionally, such events are either rare under normal growth conditions or the initial damage (nicks) must be repaired very efficiently. Instances in which the nascent RNA is not used as a primer, the replication fork may collapse, which is considered to be the worst case scenario [31]. Interestingly, in a rpo* genetic background, the greA mutant no longer accumulates dsDNA breaks [28]. The straightforward explanation was that the rpo* elongation complex is unstable and easily removed by the replication fork [13]. This idea was supported by biochemical characterization of purified rpo* RNAP and ppGpp-bound wt RNAP, where a destabilizing effect of ppGpp on elongating RNAP was noticed [14][15]. In these studies the stability of elongation complexes was defined as the ability to recover multiple elongating wt RNAP molecules paused on a DNA template in vitro. With respect to rpo* RNAP, or ppGpp-bound wt RNAP, fewer elongating complexes were recovered as compared to wt RNAP. A complementary model that also explains the phenotypes of rpo* mutants is discussed in the following section.
ppGpp, backtracking, and NER
Because the transcribed strand is repaired faster than the non-transcribed strand of the same gene [6][32][33], these results imply that RNAP plays an active role in the repair process following genotoxic stress. Implicitly, it also suggests that various phenotypes of rpo* mutants are not simply due to instability of the elongating RNAP. To facilitate repair, RNAP must both unmask the site of damage and help recruit the NER complex. Recently the UvrD helicase was identified as a major factor in the new, Mfd-independent, TCR pathway (reviewed in [4][5]). Monomeric UvrD binds RNAP under normal growth conditions, essentially making RNAP an efficient scanner of DNA damage for NER [34][35]. In a fully reconstituted system, dimeric UvrD, with the help of the transcription elongation factor NusA, pulls the paused RNAP backwards, away from CPD. As UvrD and NusA, respectively, bind UvrB and UvrA directly [36][37], they can subsequently recruit the NER complex to the lesion site to initiate the repair process (Figure 1, Key Figure). The sensitivity of uvrD cells to genotoxic stress was greatly suppressed by the lack of anti-backtracking transcript cleavage factors GreAB or Mfd. The latter functions as the anti-backtracking translocase that can “push” backtracked RNAP forward [38] Likewise, slowing down “anti-backtracking” ribosomes by a sublethal dose of chloramphenicol also partially suppressed uvrD sensitivity to genotoxic stress [34] (see below). Therefore, in the absence of UvrD, the alternative means of promoting backtracking also benefit DNA repair.
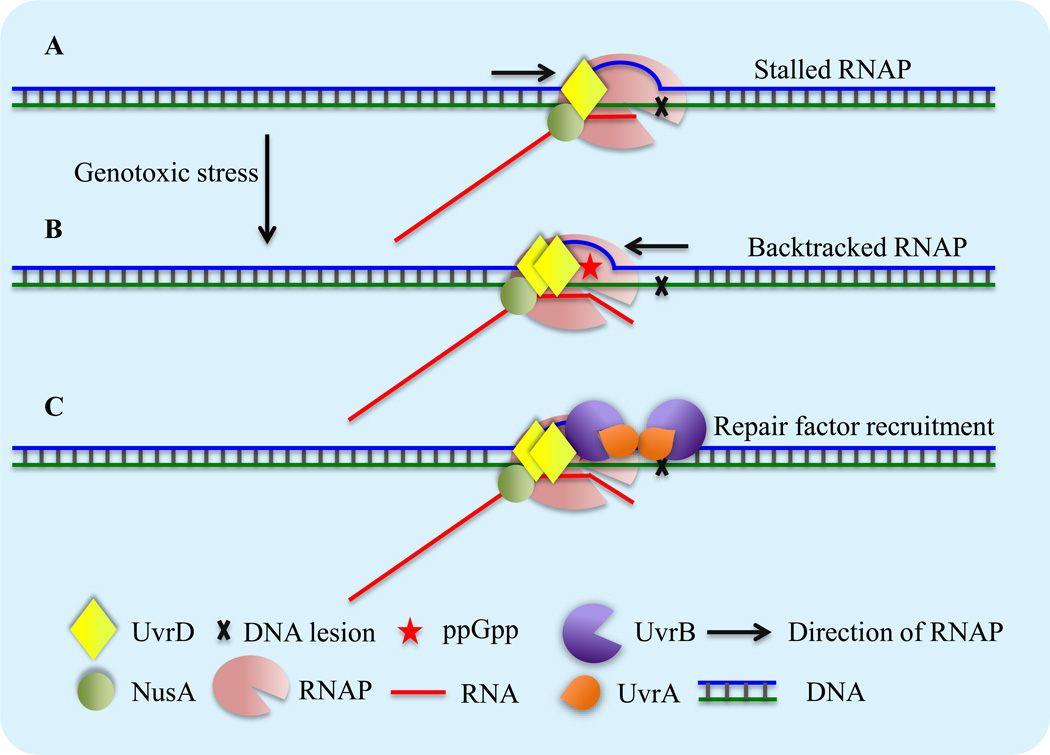
Figure 1, Key Figure. Schematics of UvrD/ppGpp-dependent TCR.
(A) When RNAP encounters a bulky DNA lesion in the template strand it stalls and obscures the access of repair enzymes to the site of damage.
(B) SOS response leads to transient accumulation of UvrD. ppGpp binds RNAP and renders it backtracking-prone. Active UvrD dimers then pull RNAP backwards to expose the lesions to the repair machinery.
(C) UvrD and NusA then recruit UvrA and UvrB to the lesion site to initiate the process of NER.
The protective phenotypes of rpo* mutants, the finding that rpo* are not simply unstable elongating RNAP mutants [28], and the RNAP modulating activity of ppGpp suggest a possible connection between ppGpp and TCR. Indeed, a recent study demonstrates that ppGpp is important for efficient TCR [33](Table 1). Moreover, it established ppGpp as an integral part of the UvrD-dependent TCR pathway. While ppGpp does not cause any substantial RNAP backtracking by itself in vitro, it renders RNAP backtracking-prone, thereby potentiating the pro-backtracking activity of UvrD (Figure 1, Key Figure) [33] In the same assay, rpo* mutant RNAP also potentiated UvrD-mediated backtracking in the absence of ppGpp. Addition of ppGpp to the rpo* mutant does not stimulate UvrD-mediated backtracking any further [33]. Thus, the rpo* mutation functionally mimics ppGpp-bound RNAP by rendering it more prone to UvrD-mediated backtracking. In agreement with this mechanism of action, the rpo* mutant was unable to suppress the sensitivity of uvrD to genotoxic stress. It is important to note that ppGpp is necessary, but not sufficient to potentiate the pro-backtracking activity of UvrD [33]. The SOS induced expression of UvrD is critical for increasing its cellular concentration. Only at a sufficiently high concentration of UvrD, do competent active helicase dimers form [39] Essentially, the pro-backtracking effect of ppGpp can explain most phenotypes of rpo* mutants.
Table 1.
Comparing ppGpp mediated response to starvation and genotoxic stress
| Amino acid starvation | Genotoxic Shock | |
|---|---|---|
| Common Inducer | Amino acid analogs (e.g: SHX) | DNA modifiers |
| Response signal | Uncharged tRNA at A-site | Unknown |
| Response sensor | RelA | RelA or SpoT |
| Peak Time (min) | ≤10 min | ≤15 min |
| Duration | Hours | Minutes |
| Fold induction ppGpp | ~70 | ~20 |
| Primary target | Initiating RNAP (stable RNA genes) | Elongating RNAP |
| Phenotype of relAspoT | Poor recovery from starvation | Poor survival of genotoxic stress |
greAB mutants are temperature sensitive, presumably due to excessive backtracking [40][41]. While the suppression of the greAB phenotype by the deletion of pro-backtracking uvrD [34] is logical, its suppression by rpo*[14] is less obvious. However, it is consistent with ambiguous nature of rpo* RNAP in vitro. On the one hand, the rpo* enzyme is slightly faster than the wild type RNAP and less responsive to certain strong pauses [28] suggesting that it backtracks less under non-stressed conditions (hence its suppression of the greAB phenotype). On the other hand, rpo* RNAP backtracks more readily in response to UvrD [33] These results suggest that rpo* has a weaker grip on DNA, i.e. it is looser than the wild type in terms of lateral stability. Indeed, according to the structural model of the ppGpp-RNAP complex [42], ppGpp causes RNAP to partially open its DNA-binding clamp. Considering that rpo* mimics ppGpp binding, such RNAP may also have a wider clamp, which would explain its wobbly character during elongation.
ppGpp-mediated backtracking: beyond UvrD
Genetic interaction between relA spoT and uvrD is epistatic when challenged with a high concentration of a DNA damaging agent [33]. This result corroborates the biochemical data arguing that UvrD and ppGpp act in the same DNA repair pathway. However, at a lower concentration of the drug, this genetic interaction becomes synergistic (V. Kamarthapu and E. Nudler, unpublished). It is likely that the DNA protective effect of ppGpp is not mediated exclusively through its potentiation of pro-backtracking activity of UvrD. Therefore, what other pathways targeted by ppGpp might contribute to genotoxic stress survival?
During amino acid starvation, ppGpp destabilizes transcription initiation complexes at stable RNA genes and ribosomal protein genes [10][11](Table 1), thereby adjusting ribosome numbers to a dwindling pool of charged tRNAs. We propose that the decreased transcription rate of the highly transcribed rRNA operons as well as the diminishing number of ribosomes per se contribute to the overall pro-backtracking effect of ppGpp.
The cooperative activity of multiple RNAP molecules moving along the same transcription unit reduces the chances of backtracking by each individual molecule within the array, thereby increasing the overall transcription rate and processivity [43][44]. Independent support for this model comes from a recent study that employed a novel next generation sequencing method: ribonuclease Native Elongating Transcript sequencing (rNET-seq) [45]. This adaptation of the original NET-seq [46] enables genome-wide mapping of backtracking type pauses [45]. In this approach, elongating RNAP and its associated native transcripts are isolated, treated with a mixture of ribonucleases, and the portion of nascent RNA protected by RNAP is then sequenced. The footprint length varies according to the pause type (pre-, post-translocated or backtracked). Approximately 50% of pauses were mapped to rRNA operons. This percentage is lower than the 98% of rRNA abundance typically reported in RNA-seq datasets, suggesting that rRNA transcription is generally pause-free, presumably because the tight arrays of elongating RNAPs suppress backtracking. Although the high rate of transcription at ribosomal genes is beneficial under normal growth conditions, it also presents a challenge for DNA repair (see the discussion on transcription-replication conflicts above). Hence, the increased level of backtracking due to ppGpp-mediated suppression of initiation at these operons should contribute to genomic stability under genotoxic stress (Figure 2A).
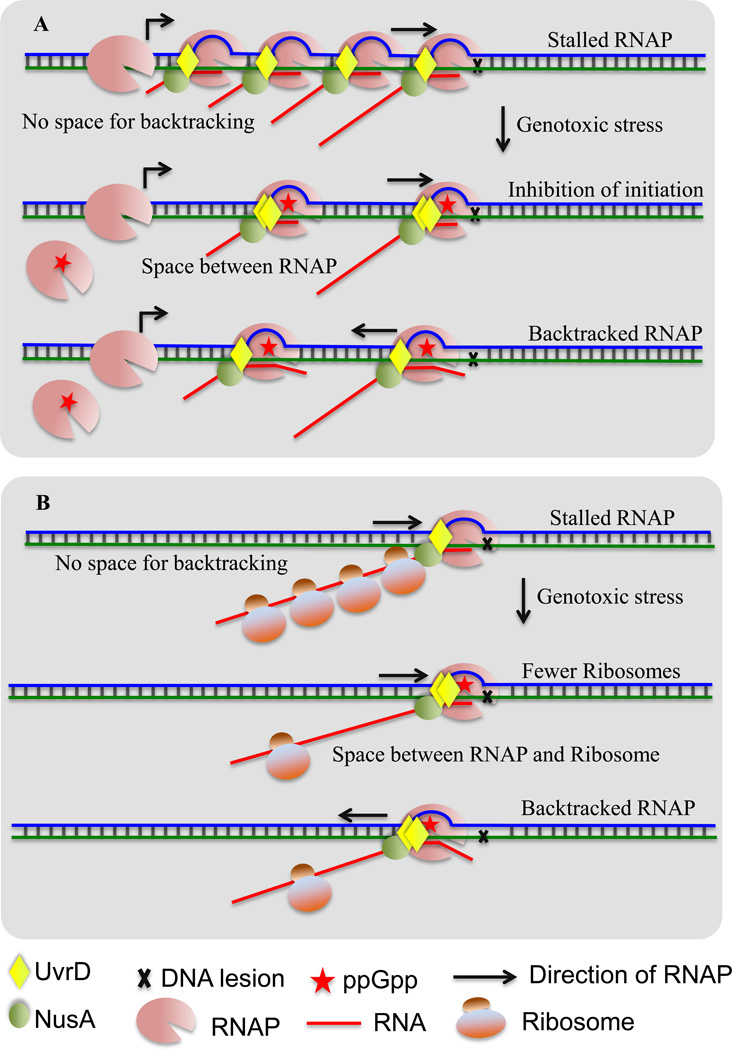
Figure 2. Possible indirect activities of ppGpp that promote backtracking.
(A) ppGpp may facilitate TCR at ribosomal operons during genotoxic stress by partially suppressing initiation at their promoters. Top: When RNAP encounters a lesion in the highly transcribed rRNA genes it cannot backtrack due to tightly packed arrays of RNAP molecules. Such arrays would also be resistant to Mfd- and Rho-dependent termination. Middle: Transient accumulation of ppGpp during genotoxic stress may partially inhibit initiation at rrn operons thereby reducing the number of elongating RNAPs. This should increase the space between adjacent RNAPs and create room for backtracking. Bottom: RNAP can now backtrack at the site of damage to enable TCR.
(B) ppGpp can promote TCR by controlling the number of ribosomes during genotoxic stress. Top: Ribosomes act as anti-backtracking factors [44]. Middle: ppGpp can diminish the number of ribosomes, thereby increasing the probability of uncoupling transcription from translation. Bottom: Fewer ribosomes increase the probability of backtracking.
Lowering the number of ribosomes appears to be yet another aspect of the ppGpp program meant to improve genotoxic stress survival. Support for this view comes from a suppressive interaction between genotoxic drugs and protein synthesis inhibitors [47]. Suppressive drug-drug interaction is seen in special instances where the combined effect of the two drugs on bacterial growth is weaker than that of the individual compounds. As multiple different pairs of drugs show this type of interaction, the specific chemical nature of one drug or another cannot account for this effect. It is likely that diminishing the capacity for protein synthesis contributes to survival of DNA damage. Indeed, decreasing the capacity for protein synthesis in E. coli by deleting 6 of the 7 naturally found copies of the rrn operons (encoding rRNA genes) eliminates the suppressive effect of sub-inhibitory concentrations of the macrolide spiramycin [48]. Also, this mutant strain was more resistant to genotoxic stress than the wild type. Therefore, the response of wild type E. coli to genotoxic stress is sub-optimal with respect to its capacity for protein synthesis. As such, elimination of ppGpp, the central negative regulator of protein synthesis, is expected to exacerbate this sub-optimal response. In agreement with this idea, the sensitivity of uvrD and relA spoT mutants to genotoxic stress can be partially suppressed by sub-inhibitory concentrations of chloramphenicol [33][34]. Note that chloramphenicol inhibits translation elongation, not initiation. Modest suppression of translation elongation per se is not expected to impact energy conservation, and as a result, to increase resistance to genotoxic stress. Because ribosomes are the major ant-backtracking factors [27] the partial uncoupling of translation from transcription by chloramphenicol should promote backtracking. Therefore, increasing the distance between RNAP and the trailing ribosomes constitutes yet another means of promoting backtracking by ppGpp leading to increased protection against genotoxic stress (Figure 2B).
To summarize, ppGpp promotes RNAP backtracking during stress by at least 3 different mechanisms. First, ppGpp facilitates UvrD-mediated backtracking by rendering RNAP backtracking prone. Second, ppGpp promotes backtracking at rRNA operons by lowering the density of RNAP molecules within those genes. Third, ppGpp promotes backtracking at coding sequences by diminishing the anti-backtracking effect of ribosomes moving behind RNAP.
ppGpp and the local mutation rates
ppGpp acts not only via RNAP to protect the bacterial genome. It also negatively regulates replication initiation in a SeqA (origin of replication sequestration factor) and Dam (DNA methylase) dependent manner [49][50]. Inhibition of replication initiation during genotoxic shock should lower the chances of mutation fixation. Therefore, ppGpp is expected to reduce the mutation rate during genotoxic stress by both enabling TCR-NER-based error-proof repair (over mutagenic translesion TLS polymerases [51]) and by reducing mutation fixation by inhibiting new rounds of replication. According to a comparative genomics study, TCR is an important force in shaping the mutational landscape of bacterial genomes [52][53]. Analysis of neutral SNPs from dozens of closely related E. coli strains revealed that the mutation rates vary locally and that highly expressed genes show lower mutation rates [52]. TCR alone could not be responsible for this protective effect, as both the transcribed and non-transcribed strands mutate at a lower rate within the highly expressed genes. However, the disproportion between C->T and G->A mutation rates on a given strand indicate that within those genes the transcribed strand, is indeed, more protected than the non-transcribed strand. Although the initial finding that highly expressed genes mutate at a lower rate has been challenged [54], a recent study has provided independent evidence for a lower mutation rate of such genes and suggested that TCR could be responsible [55]. This study introduced a novel next generation sequencing method – Maximal Depth Sequencing (MDS) to measure mutation rates in an error correcting high yield manner. Unlike previous methodologies used to measure bacterial mutation rates, MDS does not rely on selection [22] and does not require bacteria to grow for many generations to allow accumulation of mutations [56]. The error correction element is crucial because mutation rates in bacteria are lower than the rate of errors introduced by the high-fidelity PCR polymerases used in sequencing protocols. In MDS, the chromosomal DNA molecules are individually barcoded by adding a barcode to each original template molecule. Subsequent linear PCR dilutes any PCR introduced errors. Focusing the entire capacity of an Illumina high-sequencing machine on a few selected regions of interest (ROIs) makes the yield of MDS high enough to accurately measure bacterial mutation rates. The built-in linear PCR makes this method strand specific, and as such, ideal to study TCR. For the small group of ROIs chosen, the results from this study showed that highly expressed genes mutate slower. In addition, the mutation rate of the transcribed strand in one highly transcribed gene tested was significantly lower than that of the non-transcribed strand. MDS appears to be an ideal tool to study TCR and the effect of ppGpp on mutational rate within transcribed and non-transcribed ROIs under various growth and stress conditions. Future experiments may soon explain the relationship between the mutational rate and TCR.
Concluding remarks
In response to starvation, ppGpp reprograms gene expression by targeting RNAP initiation complexes. In contrast, in response to DNA damage ppGpp targets the elongating RNAP, rendering it backtracking prone. Together, ppGpp and RNAP-associated helicase UvrD, may constitute a major TCR pathway in bacteria. Activation of this pathway requires both induction of the SOS response to allow transient accumulation (and dimerization) of UvrD and also of ppGpp. Efficient, yet short-lived, backtracking of RNAP is likely to be crucial not only for TCR but also for resolution of inevitable transcription-replication conflicts. Despite the recent advancements in understanding of bacterial TCR and the role of ppGpp in this process many important questions remain (See Outstanding Questions Box). For example, what is the mechanism that triggers ppGpp accumulation in response to DNA damage? Is the UvrD/ppGpp-mediated TCR limited to NER or may be involved in other DNA repair pathways? Can TCR be mutagenic under certain stress conditions? We expect that new powerful technologies such as MDS and rNET-seq will complement the classical methodologies used to study TCR and accelerate the progress in this exciting field.
Outstanding questions.
What is the signal for ppGpp accumulation in response to DNA damage and what is the sensor?
Does ppGpp modulate any other processes or target the factors other than RNAP in response to DNA damage?
What happens to the transcription elongation complex following UvrD/ppGpp-mediated backtracking: transcription recovery or termination?
Is there DNA repair pathway(s) other than NER to be coupled to transcription?
Trends Box.
The alarmone ppGpp is a guardian of the bacterial genome.
ppGpp facilitates both resolution of transcription-replication conflicts and transcription-coupled repair (TCR).
Binding of ppGpp to elongating RNAP facilitates UvrD-mediated TCR by rendering RNAP backtracking prone.
Superposition of two major bacterial stress responses, the SOS response and the stringent response, elicits backtracking-dependent TCR.
Acknowledgments
The authors would like to thank the members of the Nudler lab for helpful comments and discussions. This work was supported by the NIH grant R01 GM107329 and by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganesan A, et al. Transcription-coupled DNA repair in prokaryotes. Prog. Mol. Biol. Transl. Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 2.Kreuzer KN. DNA damage responses in prokaryotes: regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb. Perspect. Biol. 2013;5:a012674. doi: 10.1101/cshperspect.a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancar A. Mechanisms of DNA Repair by Photolyase and Excision Nuclease (Nobel Lecture) Angew. Chem. Int. Ed Engl. 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 4.Epshtein V. UvrD helicase: an old dog with a new trick: how one step backward leads to many steps forward. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015;37:12–19. doi: 10.1002/bies.201400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamarthapu V, Nudler E. Rethinking transcription coupled DNA repair. Curr. Opin. Microbiol. 2015;24:15–20. doi: 10.1016/j.mib.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 7.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 8.Brown A, et al. Ribosome-dependent activation of stringent control. Nature. 2016;534:277–280. doi: 10.1038/nature17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loveland AB, et al. Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife. 2016;5 doi: 10.7554/eLife.17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemke JJ, et al. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barker MM, et al. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 12.Borek E, et al. Studies on a mutant of Escherichia coli with unbalanced ribonucleic acid synthesis. J. Bacteriol. 1956;71:318–323. doi: 10.1128/jb.71.3.318-323.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 14.Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trautinger BW, et al. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Ross W, et al. ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for Its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol. Cell. 2016;62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 18.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc. Natl. Acad. Sci. U. S. A. 2011;108:792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwiefka A, et al. Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry (Mosc.) 1993;32:3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]
- 20.Boubakri H, et al. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merrikh H, et al. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankar TS, et al. The nature of mutations induced by replication–transcription collisions. Nature. 2016;535:178–181. doi: 10.1038/nature18316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit M-A, . and Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002;21:3137–3147. doi: 10.1093/emboj/cdf317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perederina A, et al. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 26.Borukhov S, et al. Escherichia coli transcript cleavage factors GreA and GreB: functions and mechanisms of action. Methods Enzymol. 2001;342:64–76. doi: 10.1016/s0076-6879(01)42536-5. [DOI] [PubMed] [Google Scholar]
- 27.Proshkin S, et al. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta D, et al. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomerantz RT, O’Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wimberly H, et al. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat. Commun. 2013;4:2115. doi: 10.1038/ncomms3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons LA, et al. Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. J. Bacteriol. 2009;191:1152–1161. doi: 10.1128/JB.01292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide-excision repair of the lactose operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamarthapu V, et al. ppGpp couples transcription to DNA repair in E. coli. Science. 2016;352:993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gwynn EJ, et al. The conserved C-terminus of the PcrA/UvrD helicase interacts directly with RNA polymerase. PloS One. 2013;8:e78141. doi: 10.1371/journal.pone.0078141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen SE, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manelyte L, et al. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair. 2009;8:1300–1310. doi: 10.1016/j.dnarep.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J-S, et al. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 39.Maluf NK, et al. A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 40.Orlova M, et al. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toulmé F, et al. GreA and GreB proteins revive backtracked RNA polymerase in vivo by promoting transcript trimming. EMBO J. 2000;19:6853–6859. doi: 10.1093/emboj/19.24.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo Y, et al. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 44.Epshtein V, et al. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imashimizu M, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh P, et al. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 48.Bollenbach T, et al. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–718. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferullo DJ, et al. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods San Diego Calif. 2009;48:8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naiman K, et al. A defect in homologous recombination leads to increased translesion synthesis in E. coli. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martincorena I, et al. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- 53.Martincorena I, Luscombe NM. Non-random mutation: the evolution of targeted hypermutation and hypomutation. BioEssays News Rev. Mol. Cell. Dev. Biol. 2013;35:123–130. doi: 10.1002/bies.201200150. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Zhang J. No gene-specific optimization of mutation rate in Escherichia coli. Mol. Biol. Evol. 2013;30:1559–1562. doi: 10.1093/molbev/mst060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jee J, et al. Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature. 2016;534:693–696. doi: 10.1038/nature18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]