SUMMARY
Elucidating the target or mechanism of action of potential drugs in the discovery pipeline, is an integral component of most programs. For antibacterial compounds, generation of resistant mutants followed by whole genome sequencing has often been successful in uncovering the proteins involved in regulating compound activation, uptake, efflux and importantly, target processes. When this process succeeds, we are quick to declare a target. In a study reported by Sing et al (in press), the combination of resistant mutant generation, whole genome sequencing and recombineering to identify the target of a Mycobacterium tuberculosis growth inhibitor, pointed to a mechanism involving a scaffolding protein, Wag31, involved in polar elongation of mycobacterial cells. Time-lapse microscopy and electron microscopy confirmed the view that this inhibitor resulted in interruption of nascent cell wall biosynthesis. However, co-expression as well as regulated titration of the putative Wag31 target demonstrated that the wild-type allele was dominant and showed no synergy with the inhibitor. The most plausible explanation from their results was that this inhibitor interfered with the interaction of Wag31 with one of its interacting partners in the elongation complex.
Current clinically used antibacterials have all been discovered in phenotypic screens against whole cells using in vitro screening assays with the exception of pyrazinamide that was discovered in mouse infection studies(Zhang & Mitchison, 2003). The successes of whole cell screens contrasted to the failure of target-based screens to deliver leads to feed the drug development pipeline, have encouraged large-scale screening programs(VanderVen et al., 2015) (Mak et al., 2012) (Christophe et al., 2009) (Grant et al., 2013) (Stanley et al., 2012) (Ananthan et al., 2009) (Maddry et al., 2009) (Ballell et al., 2013) (Warrier et al., 2015) (Tantry et al., 2015) over the last decade in efforts to develop new drugs to combat the ever-prevalent scourge, tuberculosis.
Tuberculosis remains the leading killer from an infectious disease worldwide despite the availability of a chemotherapeutic regimen that can achieve greater than 90% cure rates under clinically optimal settings(Zhang & Mitchison, 2003). The current treatment regimen for drug sensitive tuberculosis entails 2 months of treatment with isoniazid, rifampicin, pyrazinamide and ethambutol followed by a 4-month continuation phase with isoniazid and rifampicin. Lack of patient adherence due to side effects associated with these drugs, the pill burden and socioeconomic factors have contributed to disease relapse and emergence of drug resistance. Development of new drugs with better safety indexes and/or new drugs that will significantly shorten the duration of treatment would be predicted to lead to higher compliance and thus some hope of combatting this global threat. The defining features of a drug that would lead to treatment shortening are poorly understood. Conceptually it could be argued that a treatment shortening regimen would contain a drug that has a mechanism of action different from those engaged by current anti-tuberculars which comprise the current extended treatment regimen. In this respect, linezolid, a drug targeting the ribosome in a mechanism distinct from first and second line anti-tubercular agents, showed unexpected clinical efficacy(Lee et al., 2015) considering its static activity in both in vitro and in vivo models. Its utility in shortening treatment duration remains to be established since its application is hampered by mitochondrial toxicity-related adverse events. Newer drugs such as bedaquiline and delamanid have novel mechanisms of action and have passed regulatory approval based on clinical efficacy but are currently restricted for use in drug-resistant tuberculosis(Gler et al., 2012) (Diacon et al., 2014). Activity against slowly- or non-replicating persisting mycobacteria has also been identified as an important hallmark of a drug that would lead to treatment shortening(Nathan & Barry, 2015) based on the hypothesis that the bacteria that persist in the face of months of chemotherapy are those that slowly replicate and/or persist in large caseous or necrotic lesions(Chen et al., 2014). In the current treatment regimen, rifampicin and pyrazinamide were the two drugs that were key to the development of current short-course (6 month) chemotherapy(Fox et al., 1999), with pyrazinamide showing selective activity against M. tuberculosis in various in vitro models of non-replicating persistence(Alnimr, 2015) with unproven clinical relevance. In contrast, drugs that target biosynthesis of the mycolyl-arabinogalactan component of the cell wall including isoniazid, ethionamide, ethambutol and thiacetazone, have limited efficacy under in vitro models of non-replicating persistence and thus are likely to have limited efficacy against the recalcitrant bacteria in caseous and necrotic granulomas. Despite this, biosynthesis of peptidoglycan remains an attractive drug target as evidenced by the cidality of meropenem/clavulanate against non-replicating M. tuberculosis(Hugonnet et al., 2009).
Studies exploring the mechanism of action of hits from conventional whole cell screens do not allow researchers to pre-select their targets of choice. The process of taking a hit from a screen to a pre-clinical candidate is long and arduous with chemistry efforts often surpassing biological studies aimed at understanding the mechanism of action of the hits that have passed initial formal hit assessment assays. Thus the biology often lags behind as seen with pretomanid (Mukherjee & Boshoff, 2011)and SQ109(Sacksteder et al., 2012) where a lead was identified prior to the deciphering of the mechanism of action of these candidates. Although target identification is not absolutely essential in the drug development pipeline, it remains an important milestone. The identification of a target can assist rational drug design strategies, it enables identification of alternative scaffolds that may overcome medicinal chemistry liabilities encountered with the original scaffold, enables in-depth understanding of potential obstacles such as mechanisms of resistance and it allows target validation in the appropriate in vivo models while hits or leads are optimized for the desired ADME-T properties. Only favorable pharmacokinetic and pharmacodynamic properties allow an informed decision to be made about the lead in question based on results from in vivo efficacy studies. The typical process of drug target identification involves generation of resistant mutants(Ioerger et al., 2013) wherein mutations in the target have allowed the identification of the mechanism of action of drugs such as bedaquiline(Andries et al., 2005), Q203(Pethe et al., 2013) and the benzothiazinones(Makarov et al., 2009). For agents such as griselimycin, target amplification in the genome confirmed the target of the natural product(Kling et al., 2015). Similarly, resistance to an indazole sulfonamide scaffold was only observed in mutants that resulted in gene amplification, with subsequent structural studies providing an explanation: the inhibitor’s interactions were almost exclusively made with the cofactor in the active site of the target, inosine monophosphate dehydrogenase(Park et al., 2016). In some cases, the target is refractory to mutations that confer resistance. For anti-tubercular compounds such as cyclomarin, the ClpC1 protease component was identified by a chemical proteomics approach wherein the proteins that bound the inhibitor were identified using cyclomarin-linked beads(Schmitt et al., 2011). Intriguingly, resistance-conferring mutations in clpC1 were identified for another natural product, lassomycin, that targets the ClpC1P1P2 protease(Gavrish et al., 2014), emphasizing the importance of the nature of association of an inhibitor with its target in the ability of target mutation to confer resistance. Resistance mutations could not be identified for SQ109 and its mechanism of action was deciphered by macromolecular incorporation assays and analyses of trehalose mycolate lipid profiles that suggested the MmpL3 transporter as the target. This was further supported by mmpL3 mutants raised to other putative MmpL3 inhibitors, although direct inhibition or engagement of MmpL3 was never demonstrated(Tahlan et al., 2012).
The challenges in drug target identification are highlighted in the elegant study by Singh & Dhar and colleagues (in press). An amino-pyrimidine-sulfonamide (APYS) inhibitor of M. tuberculosis growth was identified in a whole cell screen. The pipeline of drug target identification through generation of resistant mutants according to typical drug target identification programs showed that all resistant mutants had amino acid substitutions in the C-terminal coiled coil domain of the Wag31 protein with their role in conferring resistance confirmed by recombineering. Wag31, the mycobacterial homolog of DivIVA, is an essential scaffolding protein that is localized at the bacterial poles and that orchestrates polar growth in actinobacteria through its interaction with proteins involved in DNA segregation and cell wall synthesis(Donovan & Bramkamp, 2014). This protein is also found in other gram-positive bacteria but, in bacteria such as Bacillus subtilis, interacts with other proteins involved in cytokinesis. Thus, polar growth does not occur in all bacteria that express this protein(Donovan & Bramkamp, 2014). The phosphorylation state of Wag31 in mycobacteria, and its subsequent interaction with cognate binding partners, is regulated by the essential serine/threonine kinases PknA and PknB involved in regulating cell division, thereby controlling the rate of polar peptidoglycan biosynthesis(Jani et al., 2010). Not only is the identification of a mechanism of action centered around Wag31 function a novel drug target, but its central role in coordinating cell division and new peptidoglycan biosynthesis is particularly attractive. Accordingly, treatment of cells with the APYS inhibitor resulted in cell lysis, reminiscent of that seen during inhibition of peptidoglycan biosynthesis (Fig. 1). If a picture paints a thousand words, the insightful use of time-lapse microscopy provided visual support of a mechanism of action centered around Wag31. This technique revealed that APYS inhibitor treatment resulted in swelling primarily at the old pole but that this was followed by division, after which both the daughter cell containing the old pole as well as the sibling containing the new pole, lyzed. Survivors always emerged from daughter cells containing the new pole. In contrast, down-regulation of Wag31 expression resulted in swelling of the new pole, likely because the old pole already contains pre-assembled Wag31.
Figure 1.
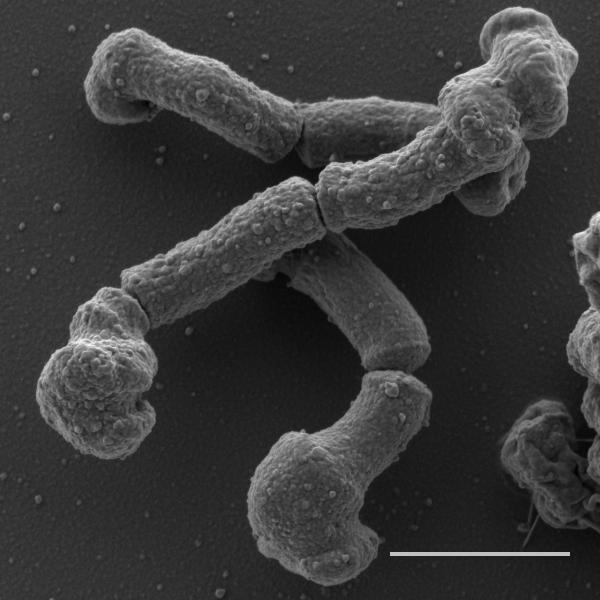
Scanning electron microscopy image showing the polar deformation of M. tuberculosis cells prior to cell lysis after 24 hours of exposure to the APYS inhibitor. The polar swelling is particularly prominent at the old pole. Scale bar is 1μM. Imagine courtesy of Gaëlle S. Kolly.
The researchers, however, hit a road-block when trying to establish that Wag31 was the target for this inhibitor. Expression of a wild-type allele in a resistant mutant conferred drug susceptibility and, conversely, expression of a wag31Q201R resistant allele in trans in wild-type cells did not confer resistance, showing that the wild-type allele was dominant. Regulated expression of Wag31 using a tetracycline-regulated promoter driving the native copy of wag31 also showed no synergy between protein down-regulation and APYS treatment and also did not shift the minimum inhibitory concentration of the inhibitor. The finding that none of the wag31 mutants showed a defect in growth argues against changes in Wag31 function affecting activity of downstream targets. The only conclusion from these results was that the APYS inhibitor affects interaction of Wag31 with one of its many binding partners. The finding that all amino acid substitutions mapped to the hydrophobic side of the C-terminal helix of Wag31 supports this notion. Unfortunately, the lack of titration of Wag31 expression with APYS susceptibility complicates interpretation of the nature of this protein-protein interaction. There are many binding partners for Wag31 of which only a few have been identified to date. For example, AccA3, an essential acetyl-CoA carboxylase involved in lipid biosynthesis for both cytoplasmic membrane and cell wall production, has been shown to interact with Wag31(Xu et al., 2014), yet APYS did not affect lipid biosynthesis, arguing against a role for AccA3 in the mechanism of action of this inhibitor. Other interacting partners for Wag31 include the penicillin-binding protein FtsI, CwsA and PknB(Mukherjee et al., 2009) (Plocinski et al., 2012) although, besides the phosphorylation of Thr73 by PknB, the exact nature of these interactions is not clearly defined.
What emerges from this work is the strong message that “discovering a drug target” carries a far deeper significance than what is reported from studies where the events unfold following their logical, anticipated course. Singh & Dhar et al (in press) used approaches that are consistently used in the traditional framework of drug discovery and combined these with time-lapse imaging studies which confirmed the interpretation that their hit inhibited the Wag31 component of the mycobacterial elongation complex. However, an analysis of protein level titration indicated that this inhibitor had a more complex effect of the protein machinery of the macromolecular protein machinery that directs elongation during cell growth. This study only emphasizes the complexities of the biological studies that support the drug discovery pipeline and challenges the framework of experiments designed to identify mechanisms of action in the “one inhibitor-one protein target” model that has been the mold for so long.
ACKNOWLEDGEMENTS
This work was funded by the Intramural Research Program of NIAID.
REFERENCES
- Alnimr AM. Dormancy models for Mycobacterium tuberculosis: A minireview. Brazilian journal of microbiology : [publication of the Brazilian Society for Microbiology] 2015;46:641–647. doi: 10.1590/S1517-838246320140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan S, Faaleolea ER, Goldman RC, Hobrath JV, Kwong CD, Laughon BE, Maddry JA, Mehta A, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Shindo N, Showe DN, Sosa MI, Suling WJ, White EL. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis. 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, Gonzalez R, Lozano S, Huss S, Santos-Villarejo A, Martin-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuinan-Blanco M, Lavandera JL, Perez-Herran E, Gamo-Benito FJ, Garcia-Bustos JF, Barros D, Castro JP, Cammack N. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem. 2013;8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RY, Dodd LE, Lee M, Paripati P, Hammoud DA, Mountz JM, Jeon D, Zia N, Zahiri H, Coleman MT, Carroll MW, Lee JD, Jeong YJ, Herscovitch P, Lahouar S, Tartakovsky M, Rosenthal A, Somaiyya S, Lee S, Goldfeder LC, Cai Y, Via LE, Park SK, Cho SN, Barry CE., 3rd PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Science translational medicine. 2014;6:265ra166. doi: 10.1126/scitranslmed.3009501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, Lee SY, Kang S, Seo MJ, Park EJ, Skovierova H, Pham H, Riccardi G, Nam JY, Marsollier L, Kempf M, Joly-Guillou ML, Oh T, Shin WK, No Z, Nehrbass U, Brosch R, Cole ST, Brodin P. High content screening identifies decaprenyl-phosphoribose 2' epimerase as a target for intracellular antimycobacterial inhibitors. PLoS pathogens. 2009;5:e1000645. doi: 10.1371/journal.ppat.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacon AH, Pym A, Grobusch MP, de los Rios JM, Gotuzzo E, Vasilyeva I, Leimane V, Andries K, Bakare N, De Marez T, Haxaire-Theeuwes M, Lounis N, Meyvisch P, De Paepe E, van Heeswijk RP, Dannemann B, T.C.S. Group Multidrug-resistant tuberculosis and culture conversion with bedaquiline. The New England journal of medicine. 2014;371:723–732. doi: 10.1056/NEJMoa1313865. [DOI] [PubMed] [Google Scholar]
- Donovan C, Bramkamp M. Cell division in Corynebacterineae. Frontiers in microbiology. 2014;5:132. doi: 10.3389/fmicb.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946-1986, with relevant subsequent publications. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 1999;3:S231–279. [PubMed] [Google Scholar]
- Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J, Lewis K. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chemistry & biology. 2014;21:509–518. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, Gao M, Awad M, Park SK, Shim TS, Suh GY, Danilovits M, Ogata H, Kurve A, Chang J, Suzuki K, Tupasi T, Koh WJ, Seaworth B, Geiter LJ, Wells CD. Delamanid for multidrug-resistant pulmonary tuberculosis. The New England journal of medicine. 2012;366:2151–2160. doi: 10.1056/NEJMoa1112433. [DOI] [PubMed] [Google Scholar]
- Grant SS, Kawate T, Nag PP, Silvis MR, Gordon K, Stanley SA, Kazyanskaya E, Nietupski R, Golas A, Fitzgerald M, Cho S, Franzblau SG, Hung DT. Identification of novel inhibitors of nonreplicating Mycobacterium tuberculosis using a carbon starvation model. ACS chemical biology. 2013;8:2224–2234. doi: 10.1021/cb4004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, O'Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, Murphy KC, Boshoff HI, Mizrahi V, Rubin EJ, Sassetti CM, Barry CE, 3rd, Sherman DR, Parish T, Sacchettini JC. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PloS one. 2013;8:e75245. doi: 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani C, Eoh H, Lee JJ, Hamasha K, Sahana MB, Han JS, Nyayapathy S, Lee JY, Suh JW, Lee SH, Rehse SJ, Crick DC, Kang CM. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC microbiology. 2010;10:327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, Konig C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Bronstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S, Muller R. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- Lee M, Cho SN, Barry CE, 3rd, Song T, Kim Y, Jeong I. Linezolid for XDR-TB--Final Study Outcomes. The New England journal of medicine. 2015;373:290–291. doi: 10.1056/NEJMc1500286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddry JA, Ananthan S, Goldman RC, Hobrath JV, Kwong CD, Maddox C, Rasmussen L, Reynolds RC, Secrist JA, 3rd, Sosa MI, White EL, Zhang W. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis. 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak PA, Rao SP, Ping Tan M, Lin X, Chyba J, Tay J, Ng SH, Tan BH, Cherian J, Duraiswamy J, Bifani P, Lim V, Lee BH, Ling Ma N, Beer D, Thayalan P, Kuhen K, Chatterjee A, Supek F, Glynne R, Zheng J, Boshoff HI, Barry CE, 3rd, Dick T, Pethe K, Camacho LR. A high-throughput screen to identify inhibitors of ATP homeostasis in non-replicating Mycobacterium tuberculosis. ACS chemical biology. 2012;7:1190–1197. doi: 10.1021/cb2004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Sureka K, Datta P, Hossain T, Barik S, Das KP, Kundu M, Basu J. Novel role of Wag31 in protection of mycobacteria under oxidative stress. Molecular microbiology. 2009;73:103–119. doi: 10.1111/j.1365-2958.2009.06750.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee T, Boshoff H. Nitroimidazoles for the treatment of TB: past, present and future. Future medicinal chemistry. 2011;3:1427–1454. doi: 10.4155/fmc.11.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Barry CE., 3rd TB drug development: immunology at the table. Immunological reviews. 2015;264:308–318. doi: 10.1111/imr.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Pacitto A, Bayliss T, Cleghorn LA, Wang Z, Hartman T, Arora K, Ioerger TR, Sacchettini J, Rizzi M, Donini S, Blundell TL, Ascher DB, Rhee KY, Breda A, Zhou N, Dartois V, Jonnala SR, Via LE, Mizrahi V, Epemolu O, Stojanovski L, Simeons FR, Osuna-Cabello M, Ellis L, MacKenzie CJ, Smith AR, Davis SH, Murugesan D, Buchanan KI, Turner PA, Huggett M, Zuccotto F, Rebollo-Lopez MJ, Lafuente-Monasterio MJ, Sanz O, Santos Diaz G, Lelievre J, Ballell L, Selenski C, Axtman M, Ghidelli-Disse S, Pflaumer H, Boesche M, Drewes G, Freiberg G, Kurnick MD, Srikumaran M, Kempf DJ, Green SR, Ray PC, Read KD, Wyatt PG, Barry Rd CE, Boshoff HI. Essential but not vulnerable: indazole sulfonamides targeting inosine monophosphate dehydrogenase as potential leads against Mycobacterium tuberculosis. ACS infectious diseases. 2016 doi: 10.1021/acsinfecdis.6b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK, Cechetto J, Christophe T, Lee H, Kempf M, Jackson M, Lenaerts AJ, Pham H, Jones V, Seo MJ, Kim YM, Seo M, Seo JJ, Park D, Ko Y, Choi I, Kim R, Kim SY, Lim S, Yim SA, Nam J, Kang H, Kwon H, Oh CT, Cho Y, Jang Y, Kim J, Chua A, Tan BH, Nanjundappa MB, Rao SP, Barnes WS, Wintjens R, Walker JR, Alonso S, Lee S, Kim J, Oh S, Oh T, Nehrbass U, Han SJ, No Z, Lee J, Brodin P, Cho SN, Nam K, Kim J. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nature medicine. 2013;19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- Plocinski P, Arora N, Sarva K, Blaszczyk E, Qin H, Das N, Plocinska R, Ziolkiewicz M, Dziadek J, Kiran M, Gorla P, Cross TA, Madiraju M, Rajagopalan M. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. Journal of bacteriology. 2012;194:6398–6409. doi: 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder KA, Protopopova M, Barry CE, 3rd, Andries K, Nacy CA. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future microbiology. 2012;7:823–837. doi: 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EK, Riwanto M, Sambandamurthy V, Roggo S, Miault C, Zwingelstein C, Krastel P, Noble C, Beer D, Rao SP, Au M, Niyomrattanakit P, Lim V, Zheng J, Jeffery D, Pethe K, Camacho LR. The natural product cyclomarin kills Mycobacterium tuberculosis by targeting the ClpC1 subunit of the caseinolytic protease. Angewandte Chemie. 2011;50:5889–5891. doi: 10.1002/anie.201101740. [DOI] [PubMed] [Google Scholar]
- Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS chemical biology. 2012;7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE, 3rd, Boshoff HI. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantry SJ, Degiacomi G, Sharma S, Jena LK, Narayan A, Guptha S, Shanbhag G, Menasinakai S, Mallya M, Awasthy D, Balakrishnan G, Kaur P, Bhattacharjee D, Narayan C, Reddy J, Naveen Kumar CN, Shandil R, Boldrin F, Ventura M, Manganelli R, Hartkoorn RC, Cole ST, Panda M, Markad SD, Ramachandran V, Ghorpade SR, Dinesh N. Whole cell screen based identification of spiropiperidines with potent antitubercular properties. Bioorganic & medicinal chemistry letters. 2015;25:3234–3245. doi: 10.1016/j.bmcl.2015.05.087. [DOI] [PubMed] [Google Scholar]
- VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, Wang T, Locher CP, Russell DG. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS pathogens. 2015;11:e1004679. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier T, Martinez-Hoyos M, Marin-Amieva M, Colmenarejo G, Porras-De Francisco E, Alvarez-Pedraglio AI, Fraile-Gabaldon MT, Torres-Gomez PA, Lopez-Quezada L, Gold B, Roberts J, Ling Y, Somersan-Karakaya S, Little D, Cammack N, Nathan C, Mendoza-Losana A. Identification of Novel Anti-mycobacterial Compounds by Screening a Pharmaceutical Small-Molecule Library against Nonreplicating Mycobacterium tuberculosis. ACS infectious diseases. 2015;1:580–585. doi: 10.1021/acsinfecdis.5b00025. [DOI] [PubMed] [Google Scholar]
- Xu WX, Zhang L, Mai JT, Peng RC, Yang EZ, Peng C, Wang HH. The Wag31 protein interacts with AccA3 and coordinates cell wall lipid permeability and lipophilic drug resistance in Mycobacterium smegmatis. Biochemical and biophysical research communications. 2014;448:255–260. doi: 10.1016/j.bbrc.2014.04.116. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2003;7:6–21. [PubMed] [Google Scholar]


