Abstract
A fundamental question in biology is how the extraordinary range of living organisms arose. In this theme issue, we celebrate how evolutionary studies on the origins of morphological diversity have changed over the past 350 years since the first publication of the Philosophical Transactions of The Royal Society. Current understanding of this topic is enriched by many disciplines, including anatomy, palaeontology, developmental biology, genetics and genomics. Development is central because it is the means by which genetic information of an organism is translated into morphology. The discovery of the genetic basis of development has revealed how changes in form can be inherited, leading to the emergence of the field known as evolutionary developmental biology (evo-devo). Recent approaches include imaging, quantitative morphometrics and, in particular, genomics, which brings a new dimension. Articles in this issue illustrate the contemporary evo-devo field by considering general principles emerging from genomics and how this and other approaches are applied to specific questions about the evolution of major transitions and innovations in morphology, diversification and modification of structures, intraspecific morphological variation and developmental plasticity. Current approaches enable a much broader range of organisms to be studied, thus building a better appreciation of the origins of morphological diversity.
This article is part of the themed issue ‘Evo-devo in the genomics era, and the origins of morphological diversity’.
Keywords: evolution, development, morphological diversity, homology, genomics, history of science
1. Introduction to the issue and a (very) short history of evo-devo
How the extraordinary range of different organisms of all shapes and sizes evolved is such a big question because it brings together many different fields of biology, including evolutionary biology, palaeontology, comparative anatomy and embryology, genetics and genomics. In this issue, we concentrate on morphology although other evolutionary changes in, for example, physiology and behaviour are also critical in ensuring that organisms are adapted to their environment [1].
Our understanding of the origin of morphological diversity has changed immeasurably over the past 350 years since the first publication of the Philosophical Transactions of The Royal Society in 1665 (see reference [2] for details of the history of the publication). Key concepts such as the theory of evolution and the laws of inheritance on which twenty-first century biology is founded had yet to come, let alone the discovery of the structure of deoxyribonucleic acid (DNA) and the sequencing of the human genome! (table 1). Development is central to understanding the origins of morphological diversity because it is the means by which the genetic information of an organism is translated into morphology. The first studies of embryos can be traced back to ancient Greek philosophers, and in the 1600s there were two competing schools of thought about how embryos developed. The preformation theory proposed that the egg contained a preformed organism, while the competing theory favoured the view, originating from Aristotle, that the parts of the body arose gradually during development, in a process known as epigenesis [3,4]. Supporters of this later theory included William Harvey, better known as the discoverer of circulation, who published a description of the development of chick embryos in 1651. Around the same time, a description of the structure of the bean seed was published by Nemehiah Grew. During this century, microscopes were invented allowing the observation of cells, eventually leading to the formulation of the cell theory that proposed that all organisms were composed of one or more cells and thus contributing to arguments against the preformation theory.
Table 1.
Timeline of some of the key discoveries over the past 350 years that have led to contemporary evo-devo.
| year | discovery |
|---|---|
| 1651 | Harvey publishes account of chick embryo development |
| late 1600s | Hooke and van Leeuwenhoek make and use microscopes and observe cells |
| 1665 | first publication of the Philosophical Transactions of the Royal Society |
| 1672 | Grew publishes account of the structure of bean seeds (Malphigi also publishes in same year) |
| 1828 and 1837 | Von Baer describes laws of development |
| 1838 | cell theory put forward by Schleiden and Schwann |
| 1843 | Owen defines the concepts of homology and analogy |
| 1859 | Darwin publishes ‘On the origin of species’ |
| 1865 and 1866 | Mendel proposes the laws of inheritance |
| 1866 | Haeckel puts forward the biogenetic law that ‘ontogeny recapitulates phylogeny’ |
| 1900 | Mendel's laws re-discovered |
| 1901 | Garrod proposes concept that genes produce enzymes |
| 1909 | Johanssen puts forward the concepts of genotype and phenotype |
| 1924 | Spemann and Mangold discover embryonic induction |
| 1941 | Beadle and Tatum show that genes direct the manufacture of proteins |
| 1942 | Waddington publishes a paper on canalization and the inheritance of acquired characteristics |
| 1944 | Avery, MacLeod and McCarty show that DNA is the information-carrying molecule |
| 1952 | Turing puts forward a reaction–diffusion model for morphogenesis |
| 1953 | Watson and Crick discover the structure of DNA |
| 1961 | Crick and co-workers publish the genetic code for proteins and Nirenberg and Matthieu identify the first codon |
| 1969 | Wolpert puts forward the theory of positional information |
| 1977 | Roberts and Sharpe discover alternative splicing |
| 1978 | Lewis publishes paper on ‘A gene complex controlling segmentation in Drosophila’ revealing how segment identity is specified |
| 1980 | Nüsslein-Volhard and Wieschaus publish paper on ‘Mutations affecting segment number and polarity in Drosophila’ |
| 1984 | McGinnis, Gehring and colleagues report conservation of homeobox genes across Metazoa |
| 1993 | Ambros and others discover microRNAs |
| 2001 | draft sequence of human genome published |
| 2002 | Davidson and collaborators describe the gene regulatory networks in early sea urchin development |
| 2012 | map of human genetic variation from 1092 human genomes published by the 1000 human genome consortium |
| 2012 | encyclopaedia of regulatory elements in the human genome ENCODE is published |
| 2014 | 50 000 year old Neanderthal genome sequenced |
| 2016 | map of genetic variation in the model plant Arabidopsis thaliana from 1135 genomes is published by the 1001 genomes consortium |
By the middle of the nineteenth century, the epigenetic theory of development had been accepted and in 1859 Darwin put forward his theory of evolution by natural selection and the concept of ‘descent with modification’ (table 1). The comparative zoologist Richard Owen defined homology and analogy, distinguishing between ‘Homologue. The same organ in different animals under every variety of form and function’ and ‘Analogue. A part or organ in one animal which has the same function as another part or organ in a different animal’ [5]. Darwin regarded embryology as being a key piece of evidence for evolution revealing homology owing to common ancestry. Karl Ernst Von Baer's studies on comparative embryology showed that embryos look much more similar than adult animals and become progressively more complex as development proceeds. These findings were used by Ernst Haeckel in the late 1800s to support the concept of recapitulation, which postulated that the stages of embryonic development represent the adult forms of ancestral or earlier-branching extant species. Haeckel encapsulated his ideas in the law that ‘ontogeny recapitulates phylogeny’ (the development of an individual organism recapitulates the evolutionary history of a species or group [4]), also putting forward the gastrea theory, which suggested that a two-layered gastrula was the ancestor of all multicellular animals. It soon became clear, however, that many observations did not obey his law and that embryos evolve.
In the early part of the twentieth century, the new field of genetics was emerging and this revolutionized the understanding of evolution, with genes being recognized as the basis for heritability. In 1909, Wilhelm Johannsen introduced the concepts of genotype—the inherited genetic constitution of an organism—and phenotype—the outward visible form of the organism and its anatomy and physiology. Although DNA had been first discovered in 1869 by Friedrich Miescher, no direct association was then made between heredity and this molecule. Following on the experiments of Griffith [6] where mice died after being injected with live but non-virulent bacteria along with heat-killed virulent bacteria, suggesting the presence of a transforming principle being picked up by the non-virulent bacteria, Avery et al. [7] showed that the transforming principle was absent when DNA was destroyed in the heat-killed bacteria but remained intact when proteins or RNA were degraded. Hershey & Chase [8] later confirmed DNA as the heritable molecule and the ensuing race to solving the structure of DNA culminated in Crick & Watson's [9] proposal of a complementary double helix structure, with nucleic acids adenine always paired with thymine and guanine paired with cytosine. Watson and Crick's paper, together with subsequent work by Crick, Brenner and others and also by Nirenberg and Matthaei (table 1), revealed how DNA is replicated and how genes encode proteins, which are the molecules that govern the behaviour of cells.
During the same period in the first half of the twentieth century, development played little part in evolutionary biology thinking [10]. Descriptive embryology had given way to experimental embryology. In the 1930s, Hans Spemann and Hilde Mangold carried out transplantation experiments on early amphibian embryos and discovered induction, thus showing very dramatically the importance of cell–cell interactions in embryonic development. However, efforts at that time to isolate the inducing factors were unsuccessful. Nevertheless, despite this focus on experimental embryology carried out on a few model organisms, several influential concepts were introduced that aimed to provide general developmental principles.
In the 1940s, Conrad Waddington proposed the concepts of canalization, a process that buffers developmental pathways against minor perturbations, and genetic assimilation, a process in which a phenotype elicited in response to the environment is selected for so that it is eventually taken over by the genotype (reviewed in [11,12]). Soon after, the mathematician Turing [13] suggested that a reaction–diffusion-type mechanism involving the interaction of chemical substances that he called morphogens might generate spatial patterns such as stripes and spots during morphogenesis. Some years later, Wolpert [14] used the idea of morphogens in his concept of positional information. According to this concept, pattern formation is a two-step process: first, cells are informed of their position within the developing organism thus acquiring a positional identity and then second, cells interpret this information in an appropriate fashion according to their position. Wolpert suggested that positional information could be specified by gradients of morphogens and famously used the pattern of different coloured stripes that make up the French Flag to illustrate the universality of his model, which as a consequence is often referred to as The French flag model. One of the predictions of positional information is that the mechanisms that inform cells of their position in embryos are widely conserved because it is the interpretation—which depends not only on position, but also on the genetic constitution of the organism—that gives rise to the differences in morphology. Positional information and reaction–diffusion are still the main models used to account for how morphology is generated during development (for discussion about these might interact, see reference [15] and for a recent hypothesis, see reference [16]).
The discovery of the genetic basis of development in the late 1970s through the work on Drosophila mutants by Edward Lewis and by Christiane Nüsslein-Volhard and Eric Weischaus [17,18] brought evolution and developmental biology back together again and this marked the emergence of the contemporary field of evo-devo [11,19]. The two main classes of developmentally important genes were found to be genes that encode secreted proteins that act as morphogens and/or are signalling molecules involved in cell–cell interactions and genes that encode proteins that act as transcription factors to control the expression of other genes. Surprisingly, it emerged that these developmental genes are evolutionarily ancient and highly conserved. Thus, for example, many of the genes encoding the main vertebrate signalling molecules were first discovered in Drosophila [18] but were subsequently found widely throughout the metazoan animal kingdom (as discussed by Babonis & Martindale [20]). Gene regulatory networks—complex networks of interactions between transcription factors and gene regulatory regions—for example, the gene regulatory network involved in early specification in sea urchin embryos [21]—and other regulatory networks involving signalling molecules and their signal transduction pathways were also found to be highly conserved. These comparative studies led to new concepts. The unexpected finding that Hox genes (genes that encode transcription factors with a particular DNA binding domain), for example, are conserved in different animals and that their expression in embryos appears to define relative position within the body plan led to the concept of the zootype [22], whereas the discovery that regulatory networks are highly conserved in the development of various structures in distantly related organisms led to the concept of deep homology ([23,24], discussed in this issue by Tschopp & Tabin [25]. This conservation also suggested that evolutionary novelties arise by modification of pre-existing regulatory networks. It was also possible to begin to look at the phylogeny of developmental genes, bringing an entirely new perspective and providing a starting point for understanding the evolution of morphological differences (discussed in this issue by Holland et al. [26]) and major transitions such as multicellularity (discussed in this issue by Babonis & Martindale [20]).
The field of evo-devo has been dramatically transformed by whole genome sequencing. In 1996, the bakers' yeast, Saccharomyces cerevisiae was the first eukaryotic species to have its genome sequenced [27]. The sequencing of the genomes of the fruit fly Drosophila melanogaster [28], the nematode Caenorharbditis elegans [29], the model plant Arabidopsis thaliana [30] and the mouse, Mus musculus [31], together with the release of the human genome sequence [32,33], expanded the understanding of many genes that constitute genetic pathways [34,35] and the intricate arrangement of cis-regulatory regions [36] that regulate development in multicellular organisms and phenotypic variations [37,38].
Lower costs and improvements in whole genome sequencing and the development of genome-wide transcriptome profiling techniques have allowed the sequencing of hundreds of genomes and gene expression profiles of an ever growing number of species and even multiple individuals within a species [39,40]. This has provided clues about the genomic changes that have had an impact on the evolution developmental programmes resulting in the vast phenotypic diversity observed between species. Many types of genomic changes have been found to be associated with phenotypic innovations; mutations involving the substitution of single nucleotides, chromosomal rearrangements as well as duplications and deletions of chromosomal segments and, although less frequent, whole genome duplications. All of these changes have contributed to the evolution of novel phenotypes by modifying proteins encoded by a gene or even resulting in novel genes altogether. Mutations that occur in cis-regulatory elements, which are segments along the DNA sequence that help regulate transcription of genes leading to changes in gene expression patterns, are increasingly recognized as a major source of phenotypic innovation [41]. The two most common cis elements are promoters, short sequences upstream of genes that include the transcription start site and the TATA-box, and enhancers, which can be located further away from the gene or downstream of it, including within introns. In addition, over the past 15 years, a myriad of non-coding, but functional RNAs have been discovered, including microRNAs and long non-coding RNAs that can regulate transcription [42] but more commonly translation [43] of protein coding genes. Mutations affecting non-coding genes can thus lead to changes in the repertoire of proteins that determines cell behaviour in developmental programmes, resulting in phenotypic evolution. Long non-coding RNAs have been associated with development and evolution but they are less well studied than shorter microRNAs and small RNAs [44].
2. The contemporary evo-devo landscape
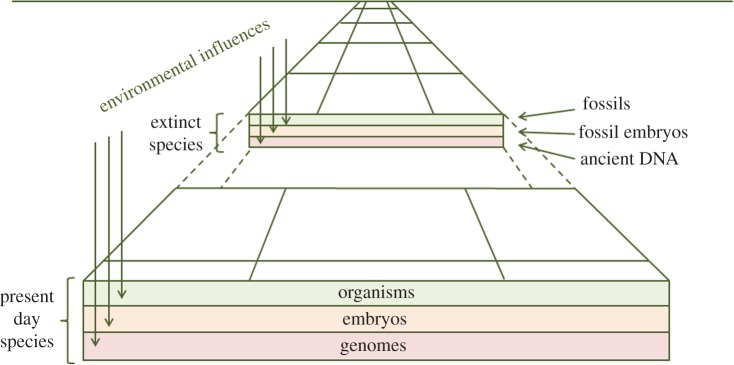
Figure 1 illustrates the contemporary landscape of evo-devo research that the eclectic collection of articles in this issue populates. This landscape consists of three layers, with adult morphology (depicted by the upper layer) being dictated by the genome (lower layer) and translated by development (middle layer). As in the past, phylogeny is inferred from comparative studies of living organisms but these comparisons can now be made at all three levels: comparative anatomy, comparative development and comparative genomics. The article by Arkhat Abzhanov and co-workers in this issue [45] on shape of the cranium in various species of birds illustrates a modern approach to comparative anatomy, while the article by Tucker [46] on the evolution of the mammalian middle ear illustrates how comparative development is currently being studied.
Figure 1.
The contemporary landscape of evo-devo research into the origins of morphological diversity. The diagram shows the perspective of evolutionary time and sections through the landscape depict the layers of research on present-day organisms and also those on extinct species. Environmental influences are also indicated. (Online version in colour.)
Phylogeny can also be inferred by examining the fossil record, and figure 1 incorporates the perspective of evolutionary time over which present-day organisms, embryos and genomes have evolved. Palaeontology classically provided information about the morphology of the ancestors of present-day organisms, and new fossils are still being discovered. Recent technological advances have made it possible also to obtain ancient genome and protein sequences from fossils as described by Orlando and co-workers in his article in this issue on human evolution ([47], see also table 1).
Figure 1 also indicates interactions with environmental factors whereby organisms with genotypes that produce better-adapted phenotypes are selected and maintained during evolution. Developmental stages will also be under selective pressure, but it is difficult to envisage how complex morphogenetic events such as gastrulation evolved and how intermediate forms could have been of selective advantage. Wolpert has argued that ‘reliability is the key demand on development’ [48].
It has been recognized for over 100 years that environmental factors can influence embryonic development (reviewed in [11]), so that markedly different phenotypes are produced by a single genotype. In this issue, Xu & Zhang [49] describe the striking developmental plasticity of brown planthoppers in which short-winged and long-winged hoppers develop depending on environmental factors, including temperature and population density. The basis of the alternative wing morphologies in these hoppers has been traced to the activity of two insulin receptors with opposite effects on wing bud growth. The same signalling pathway is also involved in the remarkable developmental plasticity seen in social insects such as bees [50]. In the case of the planthoppers, the evolutionary advantage of being able to produce long-winged insects that can disperse long distances when conditions become crowded seems clear but how this plasticity evolved is an intriguing question.
3. The organization of this theme issue
The issue begins with articles about various aspects of genomics, with Holland et al. [26] discussing gene duplication and Urrutia and co-workers [51] alternative splicing and their contributions to phenotypic innovation while Orlando and co-workers [47] outline the latest methodologies that allow analysis of fossil genomes. Tschopp & Tabin [25] consider the impact of genomics on the concept of deep homology. The other major sections illustrate how genomics and other approaches are being applied to gain insights into the evolutionary origins of morphological diversity. Major morphological transitions and innovations are considered first with articles on the origins of animal multicellularity from a classical perspective by Cavalier-Smith [52] and illustrating the impact of genomics by Babonis & Martindale [20] and an article on the evolution of land plants by Harrison [53]; the major transition that created the unique mammalian middle ear is discussed by Tucker [46]. The next section focuses on diversification and modifications of morphology as exemplified by tetrapod limbs by Saxena et al. [54], flowers by Pam Soltis and co-workers [55], cranial shape in birds by Abzhanov and co-workers [45] and wing coloration patterns in butterflies by Jiggins et al. [56]. The last section considers the relatively recent evolution of genetically determined morphological variation within a single species owing to either natural selection, using as examples, cavefish (article by Krishnan & Rohner [57]) and stickleback (article by Piechel & Marques [58]) or artificial selection, using dogs as an example (article by Elaine Ostrander and co-workers [59]) and ends with the article on developmental plasticity by Xu & Zhang [49].
The range of different organisms covered by these articles illustrates how genomics has extended the reach of evo-devo. In some cases, embryos of well-established model organisms such as mice and chickens can be used as proxies for testing and/or confirming hypotheses about the genetic basis for certain morphologies (see articles [46,59]) but also, as described in other articles in this issue, new model organisms such as liverworts [53] and butterflies [56] are being developed. An important advance has been the establishment of model systems that can be thought of as exemplifying ‘evolution in action’. The two examples discussed in this issue are the natural populations of cavefish [57] and stickleback [58] with diverse morphologies that live in different environments.
4. Recurring themes
(a). Genomic changes underlying origins of morphological diversity
The key question that crops up in many of the articles concerns the genomic changes that underlie the origins of morphological diversity. One change could involve mutations in the coding regions of genes leading to proteins with modified functions and hence to phenotypic changes. In dogs, as Elaine Ostrander and co-workers discuss [59], the huge range in the texture of the coats of different breeds is associated with different alleles of just a few genes and they report the discovery of a gene mutation that would be predicted to lead to loss of function of the encoded protein, in a new breed of hairless dog. Interestingly, the genes in dogs associated with differences in their coats and hairlessness had already been implicated in the development of hair/fur by studies in mice. Different alleles associated with other breed characteristics of dogs such as size and leg length have also been identified and a huge amount of information relating phenotype to genotype in dogs has been uncovered. It needs to be borne in mind, however that, in dogs, these phenotypes have been selected artificially rather than by natural selection and this may be why phenotypes arising from changes in coding regions are able to be selected.
Another class of genomic change that underlies morphological diversity is the creation of new genes by gene duplication. In this issue, Holland and co-workers argue that the importance of gene duplication as a mechanism for evolutionary change has been rather underplayed and give several examples from animal development where gene duplication followed by asymmetric divergence of the gene duplicates has created genes with new functions [26]. Duplication of individual genes or of the whole genome has occurred in both animal and plant evolution. Examples of gene duplication in the origins of land plants are described in this issue by Harrison [53]. In plants, such duplicated genes often take on antagonistic roles. It is interesting that the developmental plasticity in the brown planthoppers described in the article [49] is based on the opposite actions of the products of two closely related genes. The whole genome duplications that have occurred in the evolution of flowers are discussed by Soltis and co-workers [55]. As they point out, duplications of whole genomes allow complex intergenic interactions and therefore have the potential to produce greater morphological diversity than single-gene duplications. Another mechanism that creates new genes is by gene fusion and this is discussed in the article in this issue by Babonis & Martindale [20] in relation to the evolution of the genes that encode components of metazoan signalling pathways.
Recent comparative studies of transcriptome profiles identifying alternative splicing events suggest a potentially important role for this process in the evolution of phenotypic plasticity and phenotypic innovation, as reviewed by Bush et al. [51]. However, the adaptive relevance of transcript diversification mediated by alternative splicing remains unclear [60,61].
Another class of genomic change that underlies the origin of morphological diversity involves changes in cis-regulatory regions, so that a gene is expressed in a new context. Such changes are of particular relevance to evolution because they will not affect the existing gene function [10]. Several articles in the issue provide examples where the evolution of morphological diversity has been found to involve cis-regulatory regions. The stunning coloured patterns of the wings of the different species of Heliconius butterflies provide outstanding examples in which a gene is expressed in a new context and takes on a completely new function [56]. Striking examples of changes in cis-regulatory regions underlying morphological diversity are also found in natural populations of stickleback [58]. What is particularly exciting about the stickleback system is that it has been possible to prove experimentally that the genetic changes affect the function of cis-regulatory regions of key developmental genes and that these changes are responsible for the phenotypic differences (for a detailed example, see [62]). However, even in stickleback, as Piechel and Marques point out [58], it remains to be seen whether all the different morphological features seen in different populations are based on such changes. Sequence differences in a cis-regulatory region have also been suggested to underlie morphological diversification in vertebrate limbs (discussed in the article by Cooper and co-workers [54]). The cis-regulatory region implicated in this case controls expression of a gene encoding a key component of a signalling pathway specifically in the limb bud, and the differences in gene expression found in mice and cows have been suggested to explain the different numbers of digits in the two animals (for details, also see [63]).
(b). Homology
Another recurring theme throughout this issue is that of homology. Traditionally, the identification of homologous structures depended on comparative anatomy and/or embryology. As already mentioned, these approaches are still relevant today and can be extended by modern techniques. The article by Tucker [46] describes, for example, how genetic fate mapping in mouse embryos can reveal the origins of different tissues in a complex organ that may be relevant to assessing homology. The article also illustrates how detailed and thorough examination of embryonic development in different organisms can provide crucial insights. The main emphasis in this issue, however, is on homology revealed by comparisons of the genes involved in development of organs in different organisms.
The article by Tschopp & Tabin [25] explains the concept of deep homology and provides a recently discovered example of deep homology in which similarities in gene expression involving both different organs and the same organs in different organisms have been assessed using high-throughput sequencing (for details, also see [64]). They also refer to another example of structures with deep homology—the insect wing and the vertebrate limb—which share the same network of developmental genes. What is so surprising about this example is that these morphologically disparate organs would have traditionally been considered to be analogous rather than to be homologous. Examples of deep homology are also discussed in the article on land plant evolution, with the same genes being involved in the formation of ‘leaves’ and ‘root-like’ organs in different plants which fulfil the same function [53].
The eye is a particularly striking example of deep homology, with the gene encoding the same transcription factor being the ‘master’ gene for eye development in mice and flies, even though the anatomy of these organs and their embryonic development is completely different. Remarkably, when the mouse gene is expressed in the antennal imaginal disc of a fly, fly eye structures develop [65]. It seems as though this ‘master gene’ provides the information to make an organ that carries out a particular function and that the interpretation of this information—à la positional information—produces the appropriate anatomy.
One of the big challenges for the concept of positional information in general is to identify the genetic basis for interpretation that gives rise to different morphologies. This will involve looking for differences rather than similarities. There are an increasing number of genome-wide analyses aiming to identify differences in gene expression in developing organs of different organisms, including, for example, comparisons between mouse, chimpanzee and human (reviewed in [66]). Interpretation of positional information may often involve fine-tuning morphogenesis and the spatial coordination of various cell activities such as movement, adhesiveness and proliferation to produce for example the specific shapes of the bird cranium [45] or the bones in vertebrate limb digits [54] or the scale structure in Heliconius butterflies [56]. At first sight, the dissection of how such complex processes are fine-tuned seems very daunting but recent work has shown that morphogenesis can proceed—apparently spontaneously—given the right conditions. This has been shown dramatically, for example, by the generation of optic cup-like structures in aggregates of mouse embryonic stem cells that have been differentiated into retinal epithelial cells [67].
5. Final remarks
The articles in this issue illustrate beautifully (in some cases, quite literally!) the magnificent range of morphologically diverse living organisms being studied in the current evo-devo research landscape (figure 1). It is no accident that many are those that fascinated and intrigued Darwin—such the finch species of the Galapagos islands and the different breeds of dog. There is also growing interest in human evolution as novel tools become available to study it. In this issue, a detailed analysis of the human genome is described that has led to the discovery of a set of homeobox genes restricted to the genomes of placental mammals [26], whereas the article on ancient human genome sequences describes how these have already given fascinating insights into our ancestors and the origins of modern human populations [47]. Such studies, together with comparative genomics, hold out the promise of identifying the genetic basis for human-specific morphology (reviewed in [66]).
For each organism, the aim is to uncover the genetic basis of morphology and how this is translated during development, while the aim of the comparative studies of the different organisms is to illuminate steps in phylogeny. The phylogenetic steps discussed in this issue range from those involved in major transitions such as the acquisition of animal multicellularity and the evolution of land plants to those involved in the origins of morphological diversity in present-day populations of stickleback and cavefish. A long-standing question is whether evolution is based on changes in a few genes that have a large effect or on a combination of changes in many genes that each has a small effect. Interestingly, Harrison points out that single-gene mutations can produce a major changes in plant morphology relevant to evolutionary innovations [53], whereas the analysis of morphological characteristics of stickleback by Piechel & Marques [58] demonstrates that, similarly, changes in the activity of a few large effect genes are responsible for diversification. In addition, Jiggins et al. [56] point out that only four major genetic loci control most of the variation in wing coloration patterns in Heliconius butterflies.
Comparative studies on living organisms can be complemented by the fossil record and the way in which knowledge both from studies on fossils and existing organisms is being synthesized to understand how particular morphologies might have evolved is illustrated in many of the articles in this issue [25,53–55] (see also [68], for detailed discussion of the goal of integrating palaeontology and developmental biology). There are also the exciting prospects of being able to study not only fossil genomes [47], but also fossil embryos [69]. Finally, understanding the interactions with the environment which lead to adaptation and selection of organisms that are better suited to their environment brings in ecology. There is increasing interest in integrating ecology and developmental biology and in exploring the mechanisms involved in developmental plasticity with the emergence of a new field known as ecological evolutionary developmental biology [70]. The systems that represent ‘evolution in action’—cavefish and stickleback—are at the forefront here. For example, these systems have provided key information about the origins of the mutations that produce the novel phenotypes on which selection acts in nature, whether these are new mutations or whether they already existed at low frequency in the population. In stickleback, examples of both have been found [58], whereas in cavefish, there is evidence for a mechanism, as predicted by Waddington, that normally buffers genetic variation [57]. It is particularly satisfying that current evo-devo research is illuminating this and other classical concepts.
Biographies
Author profiles
 Cheryll Tickle is a Fellow of The Royal Society and currently Emeritus Professor at the University of Bath. She has a wide interest in the mechanisms involved in embryonic development, with her research focusing mainly on the development of the vertebrate limb. The chick wing has been her major experimental model, and she was involved in developing genomic resources for the chicken. Work from her laboratory has also included studies on the embryos of pythons, dogfish and stickleback. She has been a co-author on the last two editions of Lewis Wolpert's textbook, Principles of development.
Cheryll Tickle is a Fellow of The Royal Society and currently Emeritus Professor at the University of Bath. She has a wide interest in the mechanisms involved in embryonic development, with her research focusing mainly on the development of the vertebrate limb. The chick wing has been her major experimental model, and she was involved in developing genomic resources for the chicken. Work from her laboratory has also included studies on the embryos of pythons, dogfish and stickleback. She has been a co-author on the last two editions of Lewis Wolpert's textbook, Principles of development.
 Dr Araxi Urrutia is a senior lecturer at the University of Bath where she leads a research group working in the area of functional genomics. Araxi first arrived in the UK, after concluding her undergraduate degree at the National University of Mexico, to pursue her PhD studies. Following postdoctoral training in the USA, she took an academic post at the University of Bath in 2007 with funding from an L'Oreal UK Women in Science Fellowship and a Royal Society Dorothy Hodgkin Research Fellowship. Araxi also has a long-standing commitment towards gender equality and regularly gives talks and workshops on career advice for women. Through the analyses of transcriptome and genomic data, her projects aim to uncover how genes act in concert and change through time driving the evolution of complex phenotypes in eukaryotic systems.
Dr Araxi Urrutia is a senior lecturer at the University of Bath where she leads a research group working in the area of functional genomics. Araxi first arrived in the UK, after concluding her undergraduate degree at the National University of Mexico, to pursue her PhD studies. Following postdoctoral training in the USA, she took an academic post at the University of Bath in 2007 with funding from an L'Oreal UK Women in Science Fellowship and a Royal Society Dorothy Hodgkin Research Fellowship. Araxi also has a long-standing commitment towards gender equality and regularly gives talks and workshops on career advice for women. Through the analyses of transcriptome and genomic data, her projects aim to uncover how genes act in concert and change through time driving the evolution of complex phenotypes in eukaryotic systems.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Bertossa RC. 2011. Morphology and behaviour: functional links in development and evolution. Phil. Trans. R. Soc. B 366, 2056–2068. ( 10.1098/rstb.2011.0035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partridge L. 2015. Celebrating 350 years of Philosophical Transactions: life sciences papers. Phil. Trans. R. Soc. B 370, 20140380 ( 10.1098/rstb.2014.0380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horder T. 2010. History of developmental biology. In eLS. Chichester, UK: John Wiley & Sons, Ltd; ( 10.1002/9780470015902.a0003080.pub2) [DOI] [Google Scholar]
- 4.Wolpert L, Tickle C, Arias AM. 2015. Principles of development. Oxford, UK: Oxford University Press. [Google Scholar]
- 5.Boyden A. 1943. Homology and analogy: a century after the definitions of ‘homologue’ and ‘analogue’ of Richard Owen. Q. Rev. Biol. 18, 228–241. ( 10.1086/394676) [DOI] [Google Scholar]
- 6.Griffith F. 1928. The significance of pneumococcal types. J. Hygiene 27, 113–159. ( 10.1017/S0022172400031879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avery OT, MacLeod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79, 137–158. ( 10.1097/00003086-200010001-00002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershey AD, Chase M. 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39–56. ( 10.1085/jgp.36.1.39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson JD, Crick FH. 1953. Molecular structure of nucleic acids. Nature 171, 737–738. ( 10.1038/171737a0) [DOI] [PubMed] [Google Scholar]
- 10.Carroll SB. 2012. Endless forms most beautiful: the new science of evo devo and the making of the animal kingdom. London, UK: Hachette. [Google Scholar]
- 11.Gilbert SF. 2003. The morphogenesis of evolutionary developmental biology. Int. J. Dev. Biol. 47, 467. [PubMed] [Google Scholar]
- 12.Slack JM. 2002. Conrad Hal Waddington: the last renaissance biologist? Nat. Rev. Genet. 3, 889–895. ( 10.1038/nrg933) [DOI] [PubMed] [Google Scholar]
- 13.Turing AM. 1952. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72. ( 10.1098/rstb.1952.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47. ( 10.1016/S0022-5193(69)80016-0) [DOI] [PubMed] [Google Scholar]
- 15.Wolpert L. 1989. Positional information revisited. Development 107, 3–12. [DOI] [PubMed] [Google Scholar]
- 16.Green JB, Sharpe J. 2015. Positional information and reaction–diffusion: two big ideas in developmental biology combine. Development 142, 1203–1211. ( 10.1242/dev.114991) [DOI] [PubMed] [Google Scholar]
- 17.Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. ( 10.1038/276565a0) [DOI] [PubMed] [Google Scholar]
- 18.Nüsslein-Volhard C, Wieschaus E. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. ( 10.1038/287795a0) [DOI] [PubMed] [Google Scholar]
- 19.Bickel RD, Brisson JA. 2011. Evolution of development. eLS. Chichester, UK: Wiley & Sons Ltd ( 10.1002/9780470015902.a0001661.pub2) [DOI] [Google Scholar]
- 20.Babonis LS, Martindale MQ. 2017. Phylogenetic evidence for the modular evolution of metazoan signalling pathways. Phil. Trans. R. Soc. B 372, 20150477 ( 10.1098/rstb.2015.0477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson EH, et al. 2002. A genomic regulatory network for development. Science 295, 1669–1678. ( 10.1126/science.1069883) [DOI] [PubMed] [Google Scholar]
- 22.Slack JM, Holland PW, Graham CF. 1993. The zootype and the phylotypic stage. Nature 361, 490–492. ( 10.1038/361490a0) [DOI] [PubMed] [Google Scholar]
- 23.Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823. ( 10.1038/nature07891) [DOI] [PubMed] [Google Scholar]
- 24.Scotland RW. 2010. Deep homology: a view from systematics. Bioessays 32, 438–449. ( 10.1002/bies.200900175) [DOI] [PubMed] [Google Scholar]
- 25.Tschopp P, Tabin CJ. 2017. Deep homology in the age of next-generation sequencing. Phil. Trans. R. Soc. B 372, 20150475 ( 10.1098/rstb.2015.0475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland PWH, Marlétaz F, Maeso I, Dunwell TL, Paps J. 2017. New genes from old: asymmetric divergence of gene duplicates and the evolution of development. Phil. Trans. R. Soc. B 372, 20150480 ( 10.1098/rstb.2015.0480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goffeau A, Barrell BG, Bussey H, Davis R. 1996. Life with 6000 genes. Science 274, 546 ( 10.1126/science.274.5287.546) [DOI] [PubMed] [Google Scholar]
- 28.Adams MD, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. ( 10.1126/science.287.5461.2185) [DOI] [PubMed] [Google Scholar]
- 29.The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. ( 10.1126/science.282.5396.2012) [DOI] [PubMed] [Google Scholar]
- 30.Kaul S, et al. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. ( 10.1038/35048692) [DOI] [PubMed] [Google Scholar]
- 31.Chinwalla AT, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. ( 10.1038/nature01262) [DOI] [PubMed] [Google Scholar]
- 32.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 33.Venter JC, et al. 2001. The sequence of the human genome. Science 291, 1304–1351. ( 10.1126/science.1058040) [DOI] [PubMed] [Google Scholar]
- 34.de Bruijn S, Angenent GC, Kaufmann K. 2012. Plant ‘evo-devo’ goes genomic: from candidate genes to regulatory networks. Trends Plant Sci. 17, 441–447. ( 10.1016/j.tplants.2012.05.002) [DOI] [PubMed] [Google Scholar]
- 35.Graveley BR, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. ( 10.1038/nature09715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meireles-Filho AC, Stark A. 2009. Comparative genomics of gene regulation—conservation and divergence of cis-regulatory information. Curr. Opin. Genet. Dev. 19, 565–570. ( 10.1016/j.gde.2009.10.006) [DOI] [PubMed] [Google Scholar]
- 37.Copley RR. 2008. The animal in the genome: comparative genomics and evolution. Phil. Trans. R. Soc. B 363, 1453–1461. ( 10.1098/rstb.2007.2235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellegren H. 2008. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 17, 4586–4596. ( 10.1111/j.1365-294X.2008.03954.x) [DOI] [PubMed] [Google Scholar]
- 39.Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35(Suppl 1), D61–D65. ( 10.1093/nar/gkl842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett T, et al. 2007. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 35(Suppl 1), D760–D765. ( 10.1093/nar/gkl887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216. ( 10.1038/nrg2063) [DOI] [PubMed] [Google Scholar]
- 42.Salmanidis M, Pillman K, Goodall G, Bracken C. 2014. Direct transcriptional regulation by nuclear microRNAs. Int. J. Biochem. Cell Biol. 54, 304–311. ( 10.1016/j.biocel.2014.03.010) [DOI] [PubMed] [Google Scholar]
- 43.He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. ( 10.1038/nrg1379) [DOI] [PubMed] [Google Scholar]
- 44.Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. ( 10.1038/nrg3606) [DOI] [PubMed] [Google Scholar]
- 45.Tokita M, Yano W, James HF, Abzhanov A. 2017. Cranial shape evolution in adaptive radiations of birds: comparative morphometrics of Darwin's finches and Hawaiian honeycreepers. Phil. Trans. R. Soc. B 372, 20150481 ( 10.1098/rstb.2015.0481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tucker AS. 2017. Major evolutionary transitions and innovations: the tympanic middle ear. Phil. Trans. R. Soc. B 372, 20150483 ( 10.1098/rstb.2015.0483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llamas B, Willerslev E, Orlando L. 2017. Human evolution: a tale from ancient genomes. Phil. Trans. R. Soc. B 372, 20150484 ( 10.1098/rstb.2015.0484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolpert L. 1992. Gastrulation and the evolution of development. Development 116, 7–13. [PubMed] [Google Scholar]
- 49.Xu H-J, Zhang C-X. 2017. Insulin receptors and wing dimorphism in rice planthoppers. Phil. Trans. R. Soc. B 372, 20150489 ( 10.1098/rstb.2015.0489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corona M, Libbrecht R, Wheeler DE. 2016. Molecular mechanisms of phenotypic plasticity in social insects. Curr. Opin. Insect Sci. 13, 55–60. ( 10.1016/j.cois.2015.12.003) [DOI] [PubMed] [Google Scholar]
- 51.Bush SJ, Chen L, Tovar-Corona JM, Urrutia AO. 2017. Alternative splicing and the evolution of phenotypic novelty. Phil. Trans. R. Soc. B 372, 20150474 ( 10.1098/rstb.2015.0474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavalier-Smith T. 2017. Origin of animal multicellularity: precursors, causes, consequences—the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Phil. Trans. R. Soc. B 372, 20150476 ( 10.1098/rstb.2015.0476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison CJ. 2017. Development and genetics in the evolution of land plant body plans. Phil. Trans. R. Soc. B 372, 20150490 ( 10.1098/rstb.2015.0490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saxena A, Towers M, Cooper KL. 2017. The origins, scaling and loss of tetrapod digits. Phil. Trans. R. Soc. B 372, 20150482 ( 10.1098/rstb.2015.0482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chanderbali AS, Berger BA, Howarth DG, Soltis DE, Soltis PS. 2017. Evolution of floral diversity: genomics, genes and gamma. Phil. Trans. R. Soc. B 372, 20150509 ( 10.1098/rstb.2015.0509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiggins CD, Wallbank RWR, Hanly JJ. 2017. Waiting in the wings: what can we learn about gene co-option from the diversification of butterfly wing patterns? Phil. Trans. R. Soc. B 372, 20150485 ( 10.1098/rstb.2015.0485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan J, Rohner N. 2017. Cavefish and the basis for eye loss. Phil. Trans. R. Soc. B 372, 20150487 ( 10.1098/rstb.2015.0487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peichel CL, Marques DA. 2017. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Phil. Trans. R. Soc. B 372, 20150486 ( 10.1098/rstb.2015.0486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker HG, Harris A, Dreger DL, Davis BW, Ostrander EA. 2017. The bald and the beautiful: hairlessness in domestic dog breeds. Phil. Trans. R. Soc. B 372, 20150488 ( 10.1098/rstb.2015.0488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Tovar-Corona JM, Urrutia AO. 2012. Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int. J. Evol. Biol. 2012, 10 ( 10.1155/2012/596274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tress ML, Abascal F, Valencia A. 2016. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. ( 10.1016/j.tibs.2016.08.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305. ( 10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez-Rios J, et al. 2014. Attenuated sensing of SHH by Ptch1 underlies evolution of bovine limbs. Nature 511, 46–51. ( 10.1038/nature13289) [DOI] [PubMed] [Google Scholar]
- 64.Tschopp P, Sherratt E, Sanger TJ, Groner AC, Aspiras AC, Hu JK, Pourquié O, Gros J, Tabin CJ. 2014. A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature 516, 391–394. ( 10.1038/nature13819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halder G, Callaerts P, Gehring WJ. 1995. Induction of ectopic eyes by targeting expression of the eyeless gene in Drosophila. Science 267, 1788 ( 10.1126/science.7892602) [DOI] [PubMed] [Google Scholar]
- 66.Reilly SK, Noonan JP. 2016. Evolution of gene regulation in humans. Annu. Rev. Genomics Hum. Genet. 17, 45–67. ( 10.1146/annurev-genom-090314-045935) [DOI] [PubMed] [Google Scholar]
- 67.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56. ( 10.1038/nature09941) [DOI] [PubMed] [Google Scholar]
- 68.Pieretti J, Gehrke AR, Schneider I, Adachi N, Nakamura T, Shubin NH. 2015. Organogenesis in deep time: a problem in genomics, development, and paleontology. Proc. Natl Acad. Sci. USA 112, 4871–4876. ( 10.1073/pnas.1403665112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donoghue PC, et al. 2006. Synchrotron X-ray tomographic microscopy of fossil embryos. Nature 442, 680–683. ( 10.1038/nature04890) [DOI] [PubMed] [Google Scholar]
- 70.Gilbert SF, Bosch TC, Ledón-Rettig C. 2015. Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat. Rev. Genet. 16, 611–622. ( 10.1038/nrg3982) [DOI] [PubMed] [Google Scholar]