Abstract
The principle of homology is central to conceptualizing the comparative aspects of morphological evolution. The distinctions between homologous or non-homologous structures have become blurred, however, as modern evolutionary developmental biology (evo-devo) has shown that novel features often result from modification of pre-existing developmental modules, rather than arising completely de novo. With this realization in mind, the term ‘deep homology’ was coined, in recognition of the remarkably conserved gene expression during the development of certain animal structures that would not be considered homologous by previous strict definitions. At its core, it can help to formulate an understanding of deeper layers of ontogenetic conservation for anatomical features that lack any clear phylogenetic continuity. Here, we review deep homology and related concepts in the context of a gene expression-based homology discussion. We then focus on how these conceptual frameworks have profited from the recent rise of high-throughput next-generation sequencing. These techniques have greatly expanded the range of organisms amenable to such studies. Moreover, they helped to elevate the traditional gene-by-gene comparison to a transcriptome-wide level. We will end with an outlook on the next challenges in the field and how technological advances might provide exciting new strategies to tackle these questions.
This article is part of the themed issue ‘Evo-devo in the genomics era, and the origins of morphological diversity’.
Keywords: morphological novelty, regulatory evolution, transcriptomics, gene regulatory networks, enhancers, genome structure
1. Introduction
Understanding the origins of the vast array of morphological features displayed by creatures in the natural world is a core problem in evolutionary biology. Much of the variety we can observe in living species arises through changes to structures present in ancestral species, a process neatly explained by Darwin's concept of descent with modification. It has been more problematic to explain the appearance of seemingly novel anatomic structures. The principle of homology was developed to distinguish between these situations, referring to the existence of shared ancestry between a pair of structures. However, how to best define homology has been contentious [1,2]. As others have previously pointed out, this has to some extent been a problem of the hierarchical level at which homology is considered [3–5]. For example, while all vertebrate forelimbs can be considered homologous as structural units, this structural entity has been modified evolutionarily to various different ‘character states’, facilitating distinct, yet in some cases analogous modes of locomotion, such as e.g. wings used for flying. The skeletal elements inside the wings of birds, bats or pterodactyls clearly imply a common basic pattern. This can be traced back to a common forelimb ground state that has subsequently been modified in each of the three lineages. Their inferred historical continuity therefore allows us to identify them as homologous as forelimbs. However, although all three are used for flying, they have to be considered functionally analogous as wings, because flight has evolved independently in these clades. The structures are thus homologous at the level of forelimbs but not at the level of wings, i.e. whether traits are classified as homologous or not becomes a hierarchy issue, dependent on the level at which homology is being discussed.
Conflicting semantics aside, at the core of the homology concept is the notion of ‘sameness’ and some sort of ‘historical continuity.’ This has also led to the inclusion of the level of genes and proteins within the general realm of homology, followed by the rise of modern molecular biology [6]. A similar case can be made at yet another level of organizational hierarchy, when considering homology among different cell types, rather than entire organs. Again, phylogenetic as well as functional or structural criteria have historically been used to assess homology of these entities [7].
In this context, it is perhaps best to frame a discussion of homology around its original definition. As first defined by Sir Richard Owen, homology refers to ‘the same organ in different animals under every variety of form and function’ [8]. His pre-Darwinian definition of homology was thus based very much on underlying structural similarities that could not simply be explained by functional constraints. With the advent of Darwinism, Owen's concept of homology became linked to the historical continuity of morphological structures (e.g. [9]), thus implying descent with modification from an ‘archetype’ of a common ancestor. This type of homology is therefore intricately linked to phylogenetics and systematics, and how certain morphological characters are distributed over the evolutionary tree. It is often referred to as ‘historical homology’ [5].
With the advent of comparative ‘evo-devo’ biology, and a better understanding of the molecular mechanisms driving embryogenesis in classical model organisms, some have argued that considering ontogeny would sometimes be more informative when evaluating homology [10–12]. In particular, rather than following strict genealogical criteria, mechanistic constraints on the development of morphological features should be taken into account. This was largely inspired by the realization that distantly related species use a remarkably conserved gene toolkit during embryogenesis, for example for patterning their main body axes [13,14]. For the evo-devo field, this represented a unique opportunity to reframe the homology concept with a particular focus on developmental constraints. One of the prevailing criticisms of historical homology to this point was that anatomical structures, unlike genes, are not directly copied and inherited, but rather are generated de novo during the embryonic development of each generation [15,16]. As individualized parts of a species’ phenotype, these structures can change independently owing to adaptive processes. Their development, however, is usually internally constrained by the underlying genetic blueprint as well as morphogenetic processes that can be inherently self-regulatory. There, the ‘biological homology’ concept, as it has been referred to, is defined for anatomical structures that have a shared set of developmental constraints for their individualization [15,17]. As such, this form of homology mainly concerns phenotypes that result from complex regulatory interactions, rather than single-gene traits, such as colour variants [18,19]. Moreover, it also insinuates a certain degree of modularity, within which self-contained developmental units can undergo evolutionary change, for example at the level of gene regulation.
2. Homology and gene expression: kernels, character identity networks and deep homology
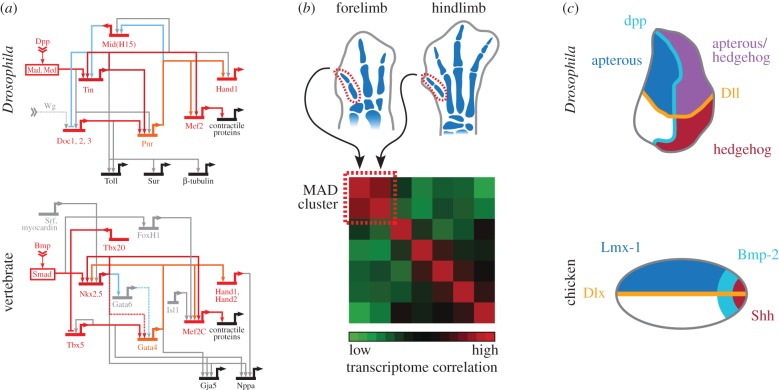
Consideration of hierarchy in the regulation of genes led to the formulation of the ‘kernel’ concept [20]. Fundamental to this approach, the genome is treated as a regulatory blueprint for embryogenesis, layered in both its functional impact on developmental patterning as well as its evolutionary age (with newer modules superimposed upon older ones). At the top of this regulatory hierarchy lie the so-called kernels, sub-units of gene regulatory networks (GRN) that are central to bodyplan patterning, exhibit deep evolutionary conservation and are refractory to regulatory rewiring. Their static behaviour, and importance in defining fundamental embryonic patterns, have been argued to underlie the stability exhibited by different animal bodyplans since the Cambrian explosion [21]. At the base of this GRN hierarchy, so-called gene differentiation batteries direct terminal cell or organ differentiation. These are assemblies of effector genes that underlie functional specification, but lack regulatory information. Their deployment is controlled by so-called intermediate plug-ins, or I/O-switches, that transmit kernel-contained patterning information down to its final differentiation output. From an evolutionary perspective, regulatory modifications are most likely to occur at this ‘plug-in’ level, to ultimately result in structural novelty. Therefore, a hierarchy of regulatory homology can, to some extent, be inferred from the re-deployment of these switches. There are a number of examples of extraordinarily deep evolutionary GRN conservation that display ‘kernel’-like behaviour: these include endomesoderm specification in echinoderms, hindbrain regionalization in chordates or, most spectacularly, the specification of heart development in clades as distant as arthopods and chordates (see references in [20]). Albeit structurally very distinct, a core set of regulatory interactions is equally important in directing heart development in these two distantly related phyla. Sub-circuit formations as well as downstream effector genes are remarkably conserved, implying a common regulatory blueprint that traces back to a primitive circulatory organ at the base of the Bilateria [20,22,23] (figure 1a). However, one of the potential shortcomings of the kernel concept in assessing homology is its focus on transcription factors and their associated cis-regulatory sequences, with much less attention given to signalling pathways. Moreover, central to its definition is the deep evolutionary conservation that GRN parts have to display in order to be considered a kernel.
Figure 1.
Gene expression-based homology assessment. (a) The kernel concept. Deep evolutionary conservation of the core gene regulatory network underlying bilaterian heart development. Parts of the gene regulatory diagrams for Drosophila (top) and mouse (bottom) heart development are depicted. Genes and regulatory interactions conserved between the two distantly related species, i.e. the kernel, are highlighted in red. Interactions conserved via intermediate relays are highlighted in blue. Modified after [20,23]. (b) The character identity network concept. During their development, the most anterior digits (MAD) of both chicken forelimbs and hindlimbs cluster together based on high transcriptome correlation, identifying them as homologous with regards to their underlying gene regulatory signature. Modified after [24]. (c) The deep homology concept. Molecular patterning of developing bilaterian body appendages, in the Drosophila wing disc (top) and the chicken limb bud (bottom). Although these structures do not share any historical homology, they have their embryonic axes defined by remarkably conserved genetic circuitries, and hence display deep homology. Modified after [25].
A slightly more flexible approach is provided by the character identity network (ChIN) concept [26]. Again, historical continuity of character-defining GRN is key to its definition. However, and unlike kernels, these do not have to be evolutionary ancient (i.e. phylum or sub-phylum level for kernels, down to species level for ChINs). Central to the applicability of ChINs in discussing homology is the inherent modularity of developmental systems. Different body parts and organs develop, and are patterned, in a semi-autonomous fashion, a fact known since the early days of experimental embryology [27]. The division into discrete developmental sub-systems allows for their individualized evolution, yet shared ChINs underlying their formation in different organisms help us to identify the resulting anatomical features as homologous. By introducing ChINs, a historical continuity is inferred by means of their repetitive re-deployment during the embryogenesis of each following generation, while modifying their output results in varying character states across species [26]. Such reasoning can help to disentangle conflicting results coming from various lines of research, such as embryology versus palaeontology, when trying to establish homologies between morphological characters. This has been demonstrated in the assessment of digit identity in extant avian wings, where a pentadactyl ground state has been reduced to a three-digit formula. While the most anterior wing digit develops from an embryonic position usually associated with digit II, the palaeontological record of theropod digit loss suggests the remaining most anterior digit to be digit I ([28,29]; see also Saxena et al. [30]). Using comparative RNA-sequencing revealed a strong transcriptional signature uniting the most anterior digits (MAD) of the forelimbs and hindlimbs (figure 1b). This implies, at the ChIN level, that the most anterior digit of the avian wing shares a common developmental blueprint with its hindlimb counterpart, and hence the forelimb digit formula should be considered I, II, III, regardless of the anatomical position from which they develop [24,31]. These findings were recently corroborated by studies using gene expression patterns to identify homeotic identities of digit primordia in species that actually have lost digit I [32]. In the meantime, ChIN-based approaches of homology have also been expanded to address the evolution of novel cell types [33].
Both kernel and ChIN arguments for homology are continuous, at least in part, with the concept of ‘deep homology’ [25], while also mechanistically refining it. The term deep homology was originally coined to describe the repeated use of highly conserved genetic circuits in the development of anatomical features that do not share homology in a strict historical or developmental sense. For example, although evolutionary separated since the Cambrian, and morphologically and developmentally highly divergent, the development of insect and vertebrate appendages share striking similarities in specifying their embryonic axes [25,34] (figure 1c). Such a degree of conservation in developmental patterning, it was argued, would be most parsimoniously explained by a common ancestor that possessed a primitive body-wall outgrowth programme [34,35]. The genetic blueprint for this outgrowth programme would then have been co-opted and re-deployed for the independent evolution of body appendages in different animal phyla, as well as being reactivated in different anatomical locations within a developing organism to give rise to serial homologues (e.g. tetrapod forelimbs and hindlimbs). Modification of this deeply conserved genetic programme would thus represent the molecular framework within which the morphological diversifications of animal appendages would have to be considered. Their development thus shares historical continuity at the level of the gene regulatory circuits, and is said to display deep homology (figure 1c). Likewise, the use of similar cellular building blocks, such as the deployment of an ancestral photosensitive cell in the generation of animal eyes, led to the dependence on homologous gene regulatory interactions and thus resulted in morphological structures that display deep homology [34]. As such, the structural entity itself (in this example, the eye) is termed ‘deeply homologous’, based on the re-deployment of genetic circuitries and/or developmental mechanism that themselves display true homology, i.e. a common historical origin. Ultimately, determining whether the expression of similar genetic cassettes underlying the development of two historically non-homologous structures represents deep homology or convergent, and potentially coincidental, deployment of related genes in two independent settings, depends on assessing the number of genes used in common and, more importantly, their epistatic relationships. Accordingly, the ability to confidently identify deep homology is already enhanced through the use of more comprehensive approaches to determine gene expression similarities.
3. Deep homology goes global: from transcripts to transcriptomes
The obvious advantage of assessing deep homology at a genome-wide level is strength in numbers, each probed gene adding robustness to formulate meaningful predictions. The emergence of hybridization-based array-techniques first opened the possibility of measuring the expression of a large set of genes in a single experiment. Moreover, compared with in situ hybridization techniques, array-based experiments also yield information about quantitative differences in gene expression (although often at the expense of spatial information). Indeed, several pioneering evo-devo studies exploited the potential of microarrays for comparative gene expression studies across multiple species. Variation in hybridization efficiencies, due to species-inherent sequence differences in the targeted parts of mRNAs, and other technical issues made direct interpretations difficult. A number of normalization and analytics procedures helped to bypass these problems [36,37]. However, most of these shortcomings can now easily be avoided, thanks to the development of high-throughput next-generation sequencing (NGS) techniques [38]. The advantages of NGS over array-based methods of gene expression measurements are manifold. Massively paralleled sequencing of cDNAs, known as RNA-seq, now allows for genome-wide interrogation of expression status, as well as the global description of transcript structures [39]. RNA-seq experiments also yield a better quantitative assessment of gene expression differences, thanks to a higher dynamic range as compared to hybridization-based methods [40]. Moreover, RNA-seq opens the possibility to compare a broad range of species that traditionally would have been considered ‘non-model organisms’, including those previously excluded by a lack of an available genome sequence [41,42]. As a consequence, comparative large-scale transcriptome studies, covering different species and tissue types, have emerged in recent years as a powerful approach to address questions of morphological evolution and homology [43]. These approaches have been used across several taxonomic levels, from comparing transcriptomes of closely related species in a given genus, to spanning the entire metazoan kingdom [44–46]. Such studies are a powerful proof of how one can now go far beyond the standard realm of model organisms, eliminating the need to focus on just a select few taxa [47,48]. Moreover, NGS enables the global assessment of quantitative gene expression differences, a parameter known to influence a variety of phenotypes [49–51]. There are, however, also analytical challenges in normalizing and comparing these datasets, especially when working with evolutionarily distant organisms [51–54]. While many of the early trans-species RNA-seq studies have mainly focused on adult tissue samples, increasingly this approach is being expanded to developmental time-series, across species and embryonic stages. Heterochrony, i.e. species-specific differences in the relative order in which certain morphological structures appear during embryogenesis, may potentially confound such transcriptome comparisons [54,55]. However, focusing on the temporal dynamics of transcriptome evolution holds the greatest potential to inform us about putative developmental homologies of different anatomical features [24,52]. NGS-based global and quantitative assessment of transcriptome dynamics, across a range of species and developmental time-points, is therefore likely to reveal more cases of deep homology in the near future.
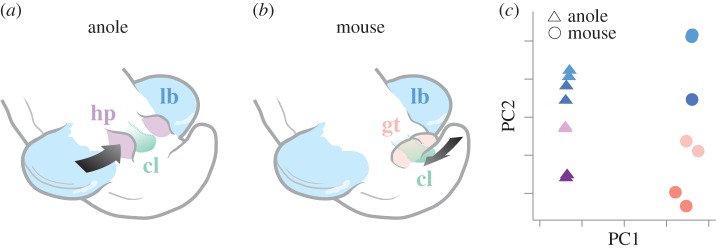
Combining next-generation transcriptomic analyses with comparative embryological and functional experiments can inform us about the developmental mechanisms that lead to the appearance of deep homology. For example, such was the motivation in studying the evolution of genital bud transcriptomes in amniotes, an embryonic structure that was known to share substantial gene expression similarities with developing limbs [56,57]. Comparative lineage tracing experiments uncovered considerable variation in the embryonic origins of amniote external genitalia [52]. In the case of squamates (lizards and snakes), early limb and genital buds share a common cellular source that results in high transcriptome similarity between the two tissues (figure 2a,c). In its most radical interpretation, squamate external genitalia development could therefore be considered serially homologous to hindlimbs. By contrast, in mammals, the genital bud originates from the tailbud mesenchyme. As the genitalia of squamates and amniotes form from distinct embryonic tissues, they are not historically homologous. How then did these critical, functionally analogous, structures evolve? The answer comes from the realization that this shift in tissue of origin in the two clades is accompanied by a relative repositioning of the genitalia-inducing signalling centre, the embryonic cloaca. Once positioned next to a different tissue source, the cloaca elicited similar downstream transcriptional responses in a different mesenchymal cell population. This indicates that the mammalian tail bud is competent to respond in a manner reminiscent of the squamate lateral plate tissue. Given the lack of a well-documented fossil record for developing amniote external genitalia, it is difficult to argue for either the squamate or mammalian situation to be the ancestral condition. However, the still recognizable similarities in gene regulatory programmes underlying both amniote limb and genital development suggest an ancestral limb-like embryonic origin for amniote external genitalia as the more likely scenario [52]. In this manner, mammalian genitalia development maintained a limb-like regulatory blueprint similar to the squamates, despite its now distinct embryonic origin (figure 2b,c). Thanks to this regulatory co-option, owing to the common inductive signals originating from the cloaca, and a shared transcriptional response to it, squamate and mammalian external genitalia display a deep homology in their development [52]. These transcriptional similarities are likely to be mechanistically linked at the cis-regulatory level, as suggested by comparative enhancer studies [58]. Intriguingly, a similar co-option of an ancestral gene regulatory network seems to have paved the way for the emergence of morphological novelties in the external genitalia of several members of the Drosophila melanogaster clade [59]. Although shared developmental trajectories seem unlikely in this case of regulatory co-option, the Drosophila example further underscores the power of re-utilizing pre-existing genetic cassettes, especially in rapidly evolving structures such as external genitalia [60,61].
Figure 2.
Deep homology of amniote external genitalia. (a) Stylized scheme of external genitalia development in a squamate, the anole lizard. For hemipenis bud initiation, the cloacal signalling centre recruits cells with a hindlimb-like developmental origin (black arrow). (b) In mammals, by contrast, the cloaca is positioned closer to the posterior end, and attracts cells from the tailbud for genital tubercle outgrowth (black arrow). (c) Representative principal component analysis (PCA) depiction of limb bud and genitalia transcriptomes in anole lizard and mouse. While PC1 is dominated by a species signal, PC2 reveals tissue-type similarities between the two species, anoles (triangles) and mice (circles). Anole limb and genitalia transcriptomes show a higher degree of relatedness than in mouse, owing to a shared embryonic origin. The development of both lizard and mouse genitalia, however, still shows high PC2 tissue-type similarity, and hence displays deep homology at the transcriptome level. lb, hindlimb; hp, hemipenis; gt, genital tubercle; cl, cloaca. Modified after [52].
4. Homology assessment: gene expression and regulatory strategies
A pressing question when evaluating homology based on gene expression similarities is whether the observed common patterns of gene activity are caused by the same underlying regulatory strategies, or are rather the result of convergent evolution. Even though the evolution of gene expression seems much more constrained than originally assumed [62], it still displays a considerable amount of plasticity and can rapidly diverge in response to altered selective pressures [63]. Consequently, it has been suggested that studying underlying regulatory strategies, rather than gene expression patterns alone, is more informative when evaluating potential synapomorphies of anatomical structures [64]. Likewise, a much stronger argument for deep homology can be made if the cis-regulatory circuitries causing the observed gene expression similarities are also conserved. In the case of deep homology, however, pre-existing gene regulatory modules that can easily be co-opted by the gain of expression of single ‘gatekeeper’ transcription factors also need to be assessed. Probing gene expression at a transcriptome-wide level, thanks to NGS approaches, has opened new experimental avenues to address these questions. By restricting the analysis of whole-transcriptome data to functional sub-classes of genes, GRN inputs can be approximated from the expression status of signalling molecules and/or transcription factors [24,52]. Given the propensity of the latter to bind to DNA, some similarity in GRN regulatory input can be expected in tissue types that show a high degree of correlation for their transcription factor expression profiles. Eventually, though, dedicated experimental data are required to arrive at a more molecular understanding of the regulatory mechanism causing any observed gene expression similarities. Again, several NGS-based approaches pave the way for such epigenomic annotation of regulatory elements, in a variety of tissue types and species. These include chromatin immunoprecipitation followed by sequencing (ChIP-seq) of histone modifications associated with enhancer function [65], as well as methods to assess local and/or global chromatin structure [66]. Of particular interest are transcriptional enhancers, cis-regulatory elements that can activate target genes over hundreds of kilobases away. These elements can function in a highly tissue-specific and temporally resolved fashion, making them potent drivers of morphological evolution. At the same time, by changing only the regulation of a gene, rather than its coding sequence, pleiotropic effects can be avoided [67]. Hence, potential evolutionary modifications of enhancer activities have been the subject of intense investigation in the field of regulatory evolution. For example, using ChIP-seq for histone H3 lysine 27 acetylation (H3K27Ac), an active enhancer mark, the evolutionary and developmental dynamics of enhancer activities have been mapped in various different organs and across several taxonomic ranks [68–70]. Ideally, such enhancer activity maps will be complemented by the binding profiles of transcription factors known to be important for the development or function of the tissues in question [71,72]. At the level of chromatin structure, local DNA accessibility as well as higher-order folding can inform us of potential enhancer sequences and regulatory strategies at a given locus. DNA accessibility, as defined by nucleosome-sparse regions, can be probed with a variety of NGS-based assays, such as DnaseI-, FAIRE- or ATAC-seq [73–75]. As a result, potential transcription factor binding sites can be defined bioinformatically, using motif search algorithms. Advantages of these techniques include the considerably smaller cell numbers that they require as input, compared with transcription factor ChIP-seq experiments. These methods have been successfully employed to compare global DNA accessibility maps for different species and organs, in adult tissues or across developmental time-points [76,77].
The importance of three-dimensional folding of the DNA strand itself for correct execution of gene regulatory programmes is also becoming increasingly appreciated. Such DNA looping can range from enhancer–promoter interactions over several hundred kilobases, all the way to supra-structural territories on the mega-base scale called topological associated domains, or TADs [78]. Inside of these TADs, regulatory ‘promiscuity’ of enhancer–promoter contacts can occur to some degree. However, regulatory interactions across TAD boundaries seem inhibited, underlining their importance in maintaining proper genome organization and gene regulation [79,80]. Cross-species comparisons of TADs have revealed both deeply conserved, as well as step-wise evolutionary dynamic assembly of such regulatory domains [81,82]. TAD conservation could thus reveal deep homology of entire regulatory landscapes, just as deep conservation of enhancer activities might inform us about evolutionary relationships of different morphological structures that they help to pattern [76]. Clearly, however, the most instructive insights at the gene regulatory level would come from the integration of different NGS technologies to first describe the regulatory architecture at loci of interest, and then test them in reciprocal, cross-species enhancer reporter experiments [83–85].
5. Concluding remarks and outlook
NGS has already significantly advanced our ability to address questions of homology, both experimentally as well as conceptually, and will probably continue to do so. Overall, it's fair to assume that the trend of incorporating technological advances from different fields of biology, and indeed the natural sciences in general, will continue in the field of evolutionary and developmental biology, to make it a more inclusive science [86,87]. Already, the next revolution in transcriptome sequencing is on its way, with the recent ability to use single cells as input material in high-throughput experiments [88]. For the field of homology assessment, this holds special importance at several levels. First, it will allow us to study the molecular mechanisms driving cell type differentiation at the relevant resolution, and learn about the evolution and putative homologous counterparts in different species, using comparative approaches [7]. Moreover, at the organ level, it enables for the cellular deconstruction of morphological character development, which will help to resolve confounding organ cell heterogeneity that might differ from species to species [54]. Spatial transcriptome information lost during organ dissociation can then be recovered in silico, down to cellular resolution, by re-mapping single-cell RNA-seq data onto grids of known marker gene in situ hybridization patterns [89,90]. Such high-throughput in vivo approaches will benefit from complementary cell culture experiments, where the controlled parameters of an in vitro environment can be exploited. Various cell and organ development pathways can already be re-capitulated in vitro, thereby helping to define their minimal differentiation requirements [91,92]. Expanding such rationale to a comparative level, between cell lines and organoids from different organisms, will allow for a reductionist approach to character identity development. Moreover, cell culture-based assays for large-scale comparative studies of epigenomic states and enhancer activities can function as invaluable proxies to delineate regulatory logic across species boundaries [93,94].
A more comprehensive inclusion of published datasets, for example through large-scale consortia, has the potential to increase the predictive power of more targeted comparative studies on gene regulatory strategies, across species, organs and developmental time-points [95–97]. The creation of public repositories will certainly help this endeavour (see resources in [98]), especially when created with an explicit evo-devo approach in mind [99]. Undoubtedly, though, this will present new challenges in terms of data analysis and integration. Dedicated bioinformatics efforts will thus be required to tackle the problems inherent to cross-species comparisons, especially when considering large evolutionary distances [51,53,100]. Ideally, the future of evo-devo will gravitate toward such an integrative approach, where comparative embryology, transcriptomics and epigenomics, bioinformatics, as well as functional in vivo and in vitro work will be incorporated to study these questions at the systems level [86,87,101]. Including comparative embryological and gene regulatory data at the micro-evolutionary level, in addition to the macro-evolutionary perspective of classical evo-devo, will allow for the integration of ecological constraints, as well as population genetics data [102].
Ultimately, a definite answer to address questions of homology among morphological features will only result from a highly integrative approach, taking into account a well-curated palaeontological record, studies on the developmental dynamics underlying the ontogeny of these structures in extant species, as well as a molecular and genetic understanding of their underlying mechanism. At the very least, exploiting the power of NGS technologies to investigate molecular mechanisms during development might help to disentangle conflicting results from the two former fields, i.e. palaeontology and embryology (see e.g. digit identities in bird wings [31]) Moreover, building on such qualitative and quantitative molecular data during embryogenesis, modelling approaches could then move the field forward and help to consider deep homology beyond a simple comparative description of gene expression. In particular, a combination of experimental data and modelling approaches might help to delineate a ‘configuration space’, in which different individualized developmental and gene regulatory systems were free to evolve [87,103]. Following such synthesis, evo-devo research in general might indeed gain certain predictive powers about possible evolutionary trajectories, by defining the range of possible ontogenetic roadmaps [47,48,87]. The concept of deep homology, with its emphasis on how gene transcription can be co-opted in an evolutionary novel context, is likely to prove particularly powerful in delineating the gene regulatory dimension of such configuration spaces.
Acknowledgements
The authors wish to apologize to colleagues whose work could not be cited due to space limitations.
Authors' contributions
P.T. and C.J.T. wrote the paper.
Competing interests
The authors declare no competing interests.
Funding
P.T. is supported by an Advanced Postdoc. Mobility fellowship (P300P3_158525) from the Swiss National Science Foundation. Work in the Tabin laboratory is funded by grant HD032443 from the NIH.
References
- 1.Brigandt I. 2003. Homology in comparative, molecular, and evolutionary developmental biology: the radiation of a concept. J. Exp. Zool. 299, 9–17. ( 10.1002/jez.b.36) [DOI] [PubMed] [Google Scholar]
- 2.Roux J, Robinson-Rechavi M. 2010. An ontology to clarify homology-related concepts. Trends Genet. 26, 99–102. ( 10.1016/j.tig.2009.12.012) [DOI] [PubMed] [Google Scholar]
- 3.Roth VL. 1991. Homology and hierarchies: problems solved and unresolved. J. Evol. Biol. 4, 167–194. ( 10.1046/j.1420-9101.1991.4020167.x) [DOI] [Google Scholar]
- 4.Wake DB. 1994. Comparative terminology. Science 265, 268–269. ( 10.1126/science.265.5169.268) [DOI] [PubMed] [Google Scholar]
- 5.Butler AB, Saidel WM. 2000. Defining sameness: historical, biological, and generative homology. Bioessays 22, 846–853. ( 10.1002/1521-1878(200009)22:9%3C846::AID-BIES10%3E3.0.CO;2-R) [DOI] [PubMed] [Google Scholar]
- 6.Patterson C. 1988. Homology in classical and molecular biology. Mol. Biol. Evol. 5, 603–625. [DOI] [PubMed] [Google Scholar]
- 7.Arendt D. 2008. The evolution of cell types in animals: emerging principles from molecular studies. Nat. Rev. Genet. 9, 868–882. ( 10.1038/nrg2416) [DOI] [PubMed] [Google Scholar]
- 8.Owen R. 1843. Lectures on the comparative anatomy and physiology of the invertebrate animals, delivered at the Royal College of Surgeons in 1843. London, UK: Longman, Brown, Green & Longmans.
- 9.Lankester ER. 1870. On the use of the term homology in modern zoology, and the distinction between homogenetic and homoplastic agreements. Ann. Mag. Nat. Hist. Zool. Bot. Geol. 6, 34–43. [Google Scholar]
- 10.Goodwin BC. 1982. Development and evolution. J. Theor. Biol. 97, 43–55. ( 10.1016/0022-5193(82)90275-2) [DOI] [PubMed] [Google Scholar]
- 11.Mindell DP, Meyer A. 2001. Homology evolving. Trends Ecol. Evol. 16, 434–440. ( 10.1016/S0169-5347(01)02206-6) [DOI] [Google Scholar]
- 12.Arthur W. 2002. The emerging conceptual framework of evolutionary developmental biology. Nature 415, 757–764. ( 10.1038/415757a) [DOI] [PubMed] [Google Scholar]
- 13.McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. 1984. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37, 403–408. ( 10.1016/0092-8674(84)90370-2) [DOI] [PubMed] [Google Scholar]
- 14.Krumlauf R. 1994. Hox genes in vertebrate development. Cell 78, 191–201. ( 10.1016/0092-8674(94)90290-9) [DOI] [PubMed] [Google Scholar]
- 15.Wagner G. 1989. The biological homology concept. Annu. Rev. Ecol. Syst. 20, 51–69. ( 10.1146/annurev.ecolsys.20.1.51) [DOI] [Google Scholar]
- 16.Müller GB. 2007. Evo-devo: extending the evolutionary synthesis. Nat. Rev. Genet. 8, 943–949. ( 10.1038/nrg2219) [DOI] [PubMed] [Google Scholar]
- 17.Roth VL. 1984. On homology. Biol. J. Linn. Soc. 22, 13–29. ( 10.1111/j.1095-8312.1984.tb00796.x) [DOI] [Google Scholar]
- 18.Nachman MW, Hoekstra HE, D'Agostino SL. 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. USA 100, 5268–5273. ( 10.1073/pnas.0431157100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittkopp PJ, Carroll SB, Kopp A. 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19, 495–504. ( 10.1016/S0168-9525(03)00194-X) [DOI] [PubMed] [Google Scholar]
- 20.Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800. ( 10.1126/science.1113832) [DOI] [PubMed] [Google Scholar]
- 21.Davidson EH, et al. 2002. A genomic regulatory network for development. Science 295, 1669–1678. ( 10.1126/science.1069883) [DOI] [PubMed] [Google Scholar]
- 22.Reim I, Frasch M. 2005. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132, 4911–4925. ( 10.1242/dev.02077) [DOI] [PubMed] [Google Scholar]
- 23.Bruneau BG. 2013. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 5, a008292 ( 10.1101/cshperspect.a008292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Young RL, Xue H, Wagner GP. 2011. Transcriptomic analysis of avian digits reveals conserved and derived digit identities in birds. Nature 477, 583–586. ( 10.1038/nature10391) [DOI] [PubMed] [Google Scholar]
- 25.Shubin N, Tabin C, Carroll S. 1997. Fossils, genes and the evolution of animal limbs. Nature 388, 639–648. ( 10.1038/41710) [DOI] [PubMed] [Google Scholar]
- 26.Wagner GP. 2007. The developmental genetics of homology. Nat. Rev. Genet. 8, 473–479. ( 10.1038/nrg2099) [DOI] [PubMed] [Google Scholar]
- 27.von Dassow G, Munro E. 1999. Modularity in animal development and evolution: elements of a conceptual framework for EvoDevo. J. Exp. Zool. 285, 307–325. ( 10.1002/(SICI)1097-010X(19991215)285:4%3C307::AID-JEZ2%3E3.0.CO;2-V) [DOI] [PubMed] [Google Scholar]
- 28.Burke AC, Feduccia A. 1997. Developmental patterns and the identification of homologies in the avian hand. Science 278, 666–668. ( 10.1126/science.278.5338.666) [DOI] [Google Scholar]
- 29.Vargas AO, Fallon JF. 2005. The digits of the wing of birds are 1, 2, and 3. A review. J. Exp. Zool. 304B, 206–219. ( 10.1002/jez.b.21051) [DOI] [PubMed] [Google Scholar]
- 30.Saxena A, Towers M, Cooper KL. 2017 doi: 10.1098/rstb.2015.0482. The origins, scaling and loss of tetrapod digits. Phil. Trans. R. Soc. B 372 , 20150482. ( ) [DOI] [Google Scholar]
- 31.Young RL, Wagner GP. 2011. Why ontogenetic homology criteria can be misleading: lessons from digit identity transformations. J. Exp. Zool. 316B, 165–170. ( 10.1002/jez.b.21396) [DOI] [PubMed] [Google Scholar]
- 32.Salinas-Saavedra M, Gonzalez-Cabrera C, Ossa-Fuentes L, Botelho JF, Ruiz-Flores M, Vargas AO. 2014. New developmental evidence supports a homeotic frameshift of digit identity in the evolution of the bird wing. Front. Zool. 11, 33 ( 10.1186/1742-9994-11-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kin K, Nnamani MC, Lynch VJ, Michaelides E, Wagner GP. 2015. Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep. 10, 1398–1409. ( 10.1016/j.celrep.2015.01.062) [DOI] [PubMed] [Google Scholar]
- 34.Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823. ( 10.1038/nature07891) [DOI] [PubMed] [Google Scholar]
- 35.Panganiban G, et al. 1997. The origin and evolution of animal appendages. Proc. Natl Acad. Sci. USA 94, 5162–5166. ( 10.1073/pnas.94.10.5162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Huggins P, Bar-Joseph Z. 2009. Cross species analysis of microarray expression data. Bioinformatics 25, 1476–1483. ( 10.1093/bioinformatics/btp247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilad Y, Rifkin SA, Bertone P, Gerstein M, White KP. 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15, 674–680. ( 10.1101/gr.3335705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzker ML. 2010. Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46. ( 10.1038/nrg2626) [DOI] [PubMed] [Google Scholar]
- 39.Ozsolak F, Milos PM. 2011. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98. ( 10.1038/nrg2934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517. ( 10.1101/gr.079558.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cook CE, Chenevert J, Larsson TA, Arendt D, Houliston E, Lénárt P. 2016. Old knowledge and new technologies allow rapid development of model organisms. Mol. Biol. Cell 27, 882–887. ( 10.1091/mbc.E15-10-0682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romero IG, Ruvinsky I, Gilad Y. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13, 505–516. ( 10.1038/nrg3229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Assis R, Zhou Q, Bachtrog D. 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 4, 1189–1200. ( 10.1093/gbe/evs093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. ( 10.1038/nature12943) [DOI] [PubMed] [Google Scholar]
- 46.Levin M, et al. 2016. The mid-developmental transition and the evolution of animal body plans. Nature 531, 1–18. ( 10.1038/nature16994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duboule D. 2010. The evo-devo comet. EMBO Rep. 11, 489 ( 10.1038/embor.2010.94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaeger J, Laubichler M, Callebaut W. 2015. The comet cometh: evolving developmental systems. Biol. Theory 10, 36–49. ( 10.1007/s13752-015-0203-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montavon T, Le Garrec JF, Kerszberg M, Duboule D. 2008. Modeling Hox gene regulation in digits: reverse collinearity and the molecular origin of thumbness. Genes Dev. 22, 346–359. ( 10.1101/gad.1631708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Navas LF, Reed H, Akam M, Barrio R, Alonso CR, Sánchez-Herrero E. 2011. Integration of RNA processing and expression level control modulates the function of the Drosophila Hox gene Ultrabithorax during adult development. Development 138, 107–116. ( 10.1242/dev.051409) [DOI] [PubMed] [Google Scholar]
- 51.Brawand D, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348. ( 10.1038/nature10532) [DOI] [PubMed] [Google Scholar]
- 52.Tschopp P, Sherratt E, Sanger TJ, Groner AC, Aspiras AC, Hu JK, Pourquié O, Gros J, Tabin CJ. 2014. A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature 516, 391–394. ( 10.1038/nature13819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musser JM, Wagner GP. 2015. Character trees from transcriptome data: origin and individuation of morphological characters and the so-called ‘species signal’. J. Exp. Zool. 324, 588–604. ( 10.1002/jez.b.22636) [DOI] [PubMed] [Google Scholar]
- 54.Roux J, Rosikiewicz M, Robinson-Rechavi M. 2015. What to compare and how: comparative transcriptomics for Evo-Devo. J. Exp. Zool. 324, 372–382. ( 10.1002/jez.b.22618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anavy L, Levin M, Khair S, Nakanishi N, Fernandez-Valverde SL, Degnan BM, Yanai I. 2014. BLIND ordering of large-scale transcriptomic developmental timecourses. Development 141, 1161–1166. ( 10.1242/dev.105288) [DOI] [PubMed] [Google Scholar]
- 56.Lin C, Yin Y, Bell SM, Veith GM, Chen H, Huh S-H, Ornitz DM, Ma L. 2013. Delineating a conserved genetic cassette promoting outgrowth of body appendages. PLoS Genet. 9, e1003231 ( 10.1371/journal.pgen.1003231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada G, et al. 2006. Molecular genetic cascades for external genitalia formation: an emerging organogenesis program. Dev. Dyn. 235, 1738–1752. ( 10.1002/dvdy.20807) [DOI] [PubMed] [Google Scholar]
- 58.Infante CR, Mihala AG, Park S, Wang JS, Johnson KK, Lauderdale JD, Menke DB. 2015. Shared enhancer activity in the limbs and phallus and functional divergence of a limb-genital cis-regulatory element in snakes. Dev. Cell 35, 107–119. ( 10.1016/j.devcel.2015.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glassford WJ, Johnson WC, Dall NR, Smith SJ, Liu Y, Boll W, Noll M, Rebeiz M. 2015. Co-option of an ancestral Hox-regulated network underlies a recently evolved morphological novelty. Dev. Cell 34, 520–531. ( 10.1016/j.devcel.2015.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosken DJ, Stockley P. 2004. Sexual selection and genital evolution. Trends Ecol. Evol. 19, 87–93. ( 10.1016/j.tree.2003.11.012) [DOI] [PubMed] [Google Scholar]
- 61.Eberhard WG. 2010. Evolution of genitalia: theories, evidence, and new directions. Genetica 138, 5–18. ( 10.1007/s10709-009-9358-y) [DOI] [PubMed] [Google Scholar]
- 62.Necsulea A, Kaessmann H. 2014. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 15, 734–748. ( 10.1038/nrg3802) [DOI] [PubMed] [Google Scholar]
- 63.Ghalambor CK, Hoke KL, Ruell EW, Fischer EK, Reznick DN, Hughes KA. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525, 372–375. ( 10.1038/nature15256) [DOI] [PubMed] [Google Scholar]
- 64.Woltering JM, Duboule D. 2010. The origin of digits: expression patterns versus regulatory mechanisms. Dev. Cell 18, 526–532. ( 10.1016/j.devcel.2010.04.002) [DOI] [PubMed] [Google Scholar]
- 65.Harmston N, Lenhard B. 2013. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 41, 7185–7199. ( 10.1093/nar/gkt499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nora EP, Dekker J, Heard E. 2013. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 35, 818–828. ( 10.1002/bies.201300040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levine M. 2010. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763. ( 10.1016/j.cub.2010.06.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cotney J, Leng J, Yin J, Reilly SK, DeMare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. 2013. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154, 185–196. ( 10.1016/j.cell.2013.05.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. 2015. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159. ( 10.1126/science.1260943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Villar D, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160, 554–566. ( 10.1016/j.cell.2015.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626. ( 10.1038/nrg3207) [DOI] [PubMed] [Google Scholar]
- 72.Villar D, Flicek P, Odom DT. 2014. Evolution of transcription factor binding in metazoans—mechanisms and functional implications. Nat. Rev. Genet. 15, 221–233. ( 10.1038/nrg3481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.John S, et al. 2013. Genome-scale mapping of DNase I hypersensitivity. Curr. Protoc. Mol. Biol. Ch. 27, Unit 21.27 ( 10.1002/0471142727.mb2127s103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885. ( 10.1101/gr.5533506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Meth. 10, 1213–1218. ( 10.1038/nmeth.2688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gehrke AR, et al. 2015. Deep conservation of wrist and digit enhancers in fish. Proc. Natl Acad. Sci. USA 112, 803–808. ( 10.1073/pnas.1420208112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vierstra J, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012. ( 10.1126/science.1246426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Remeseiro S, Hörnblad A, Spitz F. 2016. Gene regulation during development in the light of topologically associating domains. Wiley Interdiscip. Rev. Dev. Biol. 5, 169–185. ( 10.1002/wdev.218) [DOI] [PubMed] [Google Scholar]
- 79.Lupiáñez DG, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025. ( 10.1016/j.cell.2015.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsujimura T, Klein FA, Langenfeld K, Glaser J, Huber W, Spitz F. 2015. A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes. PLoS Genet. 11, e1004897 ( 10.1371/journal.pgen.1004897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gómez-Marín C, et al. 2015. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl Acad. Sci. USA 112, 7542–7547. ( 10.1073/pnas.1505463112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Acemel RD, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 48, 336–341. ( 10.1038/ng.3497) [DOI] [PubMed] [Google Scholar]
- 83.Woltering JM, Noordermeer D, Leleu M, Duboule D. 2014. Conservation and divergence of regulatory strategies at Hox loci and the origin of tetrapod digits. PLoS Biol. 12, e1001773 ( 10.1371/journal.pbio.1001773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parker HJ, Bronner ME, Krumlauf R. 2014. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 514, 490–493. ( 10.1038/nature13723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao Y, et al. 2016. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nat. Genet. 48, 575–580. ( 10.1038/ng.3542) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wagner GP. 2012. Next gen devo-evo. J. Exp. Zool. 318, 519–520. ( 10.1002/jez.b.22463) [DOI] [PubMed] [Google Scholar]
- 87.Soyer OS, O'Malley MA. 2013. Evolutionary systems biology: what it is and why it matters. Bioessays 35, 696–705. ( 10.1002/bies.201300029) [DOI] [PubMed] [Google Scholar]
- 88.Saliba A-E, Westermann AJ, Gorski SA, Vogel J. 2014. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 42, 8845–8860. ( 10.1093/nar/gku555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. ( 10.1038/nbt.3192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Achim K, Pettit J-B, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. 2015. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat. Biotechnol. 33, 503–509. ( 10.1038/nbt.3209) [DOI] [PubMed] [Google Scholar]
- 91.Sánchez Alvarado A, Yamanaka S. 2014. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 157, 110–119. ( 10.1016/j.cell.2014.02.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato T, Clevers H. 2015. SnapShot: growing organoids from stem cells. Cell 161, 1700–1700.e1. ( 10.1016/j.cell.2015.06.028) [DOI] [PubMed] [Google Scholar]
- 93.Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat. Genet. 46, 685–692. ( 10.1038/ng.3009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J. 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163, 68–83. ( 10.1016/j.cell.2015.08.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerstein MB, et al. 2014. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448. ( 10.1038/nature13424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ho JWK, et al. 2014. Comparative analysis of metazoan chromatin organization. Nature 512, 449–452. ( 10.1038/nature13415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stergachis AB, et al. 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370. ( 10.1038/nature13972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rung J, Brazma A. 2012. Reuse of public genome-wide gene expression data. Nat. Rev. Genet. 14, 89–99. ( 10.1038/nrg3394) [DOI] [PubMed] [Google Scholar]
- 99.Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. 2008. Bgee: integrating and comparing heterogeneous transcriptome data among species. In Data integration in the life sciences, pp. 124–131. Berlin, Germany: Springer. [Google Scholar]
- 100.Piasecka B, Kutalik Z, Roux J, Bergmann S, Robinson-Rechavi M. 2012. Comparative modular analysis of gene expression in vertebrate organs. BMC Genomics 13, 124 ( 10.1186/1471-2164-13-124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thompson D, Regev A, Roy S. 2015. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu. Rev. Cell Dev. Biol. 31, 399–428. ( 10.1146/annurev-cellbio-100913-012908) [DOI] [PubMed] [Google Scholar]
- 102.Nunes MDS, Arif S, Schlötterer C, McGregor AP. 2013. A perspective on micro-evo-devo: progress and potential. Genetics 195, 625–634. ( 10.1534/genetics.113.156463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crombach A, Hogeweg P. 2008. Evolution of evolvability in gene regulatory networks. PLoS Comput. Biol. 4, e1000112 ( 10.1371/journal.pcbi.1000112) [DOI] [PMC free article] [PubMed] [Google Scholar]