Abstract
Communication among cells was paramount to the evolutionary increase in cell type diversity and, ultimately, the origin of large body size. Across the diversity of Metazoa, there are only few conserved cell signalling pathways known to orchestrate the complex cell and tissue interactions regulating development; thus, modification to these few pathways has been responsible for generating diversity during the evolution of animals. Here, we summarize evidence for the origin and putative function of the intracellular, membrane-bound and secreted components of seven metazoan cell signalling pathways with a special focus on early branching metazoans (ctenophores, poriferans, placozoans and cnidarians) and basal unikonts (amoebozoans, fungi, filastereans and choanoflagellates). We highlight the modular incorporation of intra- and extracellular components in each signalling pathway and suggest that increases in the complexity of the extracellular matrix may have further promoted the modulation of cell signalling during metazoan evolution. Most importantly, this updated view of metazoan signalling pathways highlights the need for explicit study of canonical signalling pathway components in taxa that do not operate a complete signalling pathway. Studies like these are critical for developing a deeper understanding of the evolution of cell signalling.
This article is part of the themed issue ‘Evo-devo in the genomics era, and the origins of morphological diversity’.
Keywords: metazoa, evolution, cell signalling, ctenophore, complexity
1. Introduction
Multicellularity has evolved numerous times [1], but has only resulted in the expansive diversification of complex organisms in six lineages: basidiomycete and ascomycete fungi, red algae, brown algae, plants and animals. Many factors influenced the transition from unicellular to multicellular life; important among them were the evolution of adhesion that permitted daughter cells to remain in physical contact following replication [2], and the division of labour that enabled the spatial segregation of incompatible/diverse cell functions and enabled the evolution of specialized somatic cell types [3]. Adherent colonies of cells have arisen numerous times and, indeed, classical cell–cell adhesion proteins evolved long before the origin of Metazoa [4], implying that adhesion alone is insufficient to drive the evolution of diversity in metazoans. The origin of cell–cell communication through intercellular signalling pathways enabled cells to communicate their identity to their neighbours, and was essential for stimulating the rise of multicellularity. Without the ability to communicate information among individual cells, a colonial organism can only be a cluster of autonomous units; thus, the evolution of the basement membrane played a critical role in facilitating communication among cells as it provided a scaffold for the retention and transmission of extracellular ligands and inhibitors. The combined evolution of intercellular communication and an extracellular means to transmit signals permitted the evolution of long-range policing of cell identity, promoting the evolution of truly multicellular life where only aggregates had succeeded before. Here, we review recent advancements in our understanding of the evolution of major cell signalling pathways in non-bilaterian unikonts (see glossary) and the origin of the basement membrane as a means to facilitate long-range signal propagation. This review highlights the modular evolution of intracellular, membrane-bound and secreted components of these conserved pathways and points to the evolution of secreted inhibitory molecules as key regulators of pathway diversity.
2. Cell–cell communication and the evolution of signal transduction pathways
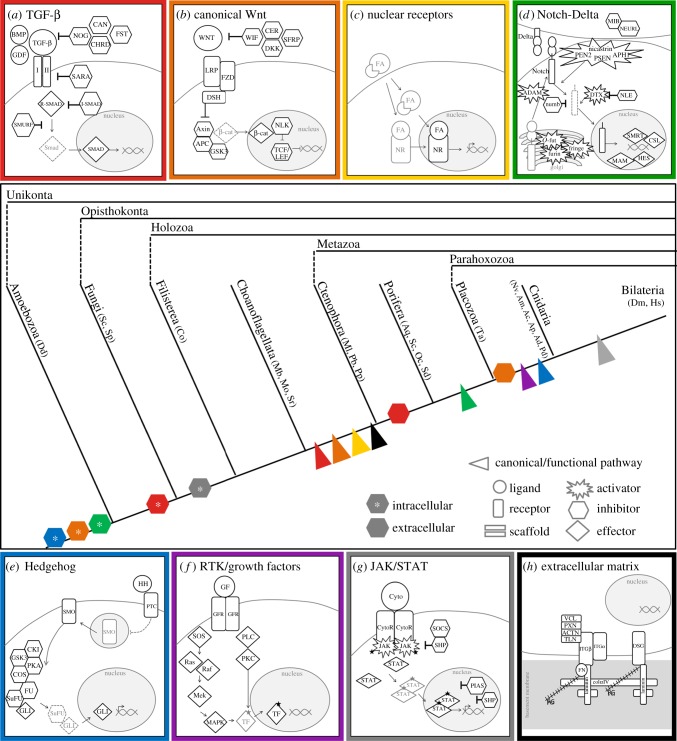
Across Metazoa, seven signalling pathways appear to play similar and essential roles in regulating animal development: the transforming growth factor β (TGF-β) pathway, canonical WNT (cWNT) signalling, nuclear receptors (NRs), receptor tyrosine kinase (RTK) signalling, the Notch/Delta pathway, Hedgehog signalling and the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway. These seven highly conserved signalling pathways transduce extracellular signalling into transcriptional regulation of target genes and are responsible for facilitating the specification of cells differentiating from a proliferative progenitor during embryogenesis. Thus, these signalling pathways may be representative of the ancestral mechanisms permitting the division of labour that resulted in the origin of novel cell types and the evolution of complex multicellular life. However, understanding how the origin of these seven pathways may have influenced the diversification of Metazoa requires an understanding of the functions of these pathways in diverse metazoan lineages. In particular, understanding the developmental context in which they acted to influence the cell biology of ancestral taxa is critical for reconstructing the events that led to the rise of the canonical/functional pathways and the origin of complex multicellular life.
Recent efforts to combine molecular (e.g. transcriptomics) and developmental techniques [5] have provided an opportunity to use expression data as a first attempt to imply function rather than relying on only presence/absence data from genomic studies. Despite having only limited data from several critical taxa, we take a first step towards integrating genomic and developmental studies from diverse non-bilaterian metazoans to attempt to reconstruct ancestral functions of several key signal transduction cascades. First, we summarize the presence/absence of pathway components in cnidarians, placozoans, sponges and ctenophores as well as in the unicellular lineages that preceded the origin of metazoans (choanoflagellates, filastereans, fungi and amoebozoans, where possible). We then summarize available data regarding the developmental role of these genes to explore their ancestral functions. There have been several previous reviews of cell signalling in non-bilaterians [6–9], and we refer the reader to these and additional resources throughout. With this review, we aim to comprehensively integrate genomic and developmental data from ctenophores and other emerging non-bilaterian models with previous observations of signal pathway evolution to build a richer model of metazoan evolution.
3. Using ctenophores to reconstruct the cell biology of the ancestral metazoan
Inferences about the factors that promoted the evolutionary diversification of metazoans are anchored in the phylogeny used to describe the relationships among metazoans. The sequencing of the sponge genome [10] enabled new predictions about the nature of cell biology in what was considered at the time to be the earliest diverging metazoan. Several studies now support ctenophores as sister to the rest of Metazoa [11–13], necessitating a re-review of the evolution of cell signalling pathways in non-bilaterians. Although much progress has been made in understanding the developmental biology of ctenophores in the past decade, there is still much to be learned from increased sampling of this lineage, and we hope this review stimulates renewed interest in characterizing the developmental biology of ctenophores. Notably absent from our developmental surveys below are data from placozoans. As there has been little advancement in efforts to spawn and successfully rear placozoan embryos through development [14], little is known about the spatial distribution of gene expression in these animals, and there is little opportunity to test hypotheses raised by the recent sequencing of the genome and proteome of Trichoplax adhaerens [15,16]. Conversely, there have been numerous recent advancements in our understanding of the evolution and development of diverse sponge taxa [17–19]. Considering sponges to be sister to the rest of Bilateria, Muller [20] suggested that the ancestral metazoan likely had cell adhesion molecules with intracellular signalling capabilities, gradient-forming growth factors, and immune and nervous systems. Given that ctenophores are now largely accepted to be the earliest diverging lineage of metazoans (figure 1), we ask: how does this list of putative ancestral metazoan characteristics change if the ancestral metazoan is reconstructed under the new phylogenetic framework?
Figure 1.
Schematic of the major components of the seven conserved metazoan signalling pathways: TGF-β (red), canonical Wnt (orange), nuclear receptor (yellow), Notch/Delta (green), Hedgehog (blue), RTK/growth factor (violet) and JAK/STAT (grey), and the components that comprise the basement membrane and adhesome complex (black). The origin of the canonical/functional pathway and the origin of secreted/extracellular and intracellular inhibitors are indicated on the cladogram. Abbreviations for taxa represented by each lineage are indicated in the glossary. The evolutionary origin of the canonical/functional version of each pathway is indicated at the node where conserved function has been identified experimentally.
4. Pathways that unite metazoans
(a). Transforming growth factor β signalling
The mechanism underlying TGF-β signalling has been reviewed in detail [21,22] as has the bone morphogenetic protein (BMP) pathway explicitly [23]. Briefly, this pathway becomes activated when a ligand (TGF-β type or BMP type) binds to a receptor complex (heterodimer of type I and type II receptors) to translocate a SMAD-based intracellular cascade into the nucleus activating or repressing transcription (figure 1a). Now known to be unique to Metazoa [24–26], a full complement of TGF-β signalling components (TGF-β and BMP ligands, their receptors, and numerous SMAD cofactors) has been found in cnidarians [27–29], placozoans [15], sponges [30] and, most recently, the ctenophores Mnemiopsis leidyi [31] and Pleurobrachia bachei (table 1). Non-metazoan unikonts lack not only the ligands, receptors and SMAD cofactors [17,24–26], but also most of the inhibitors. This pathway is a clear example of a metazoan innovation [25], and the presence of all elements together in ctenophores suggests that this system was actively transducing extracellular signals into transcriptional responses in the ancestral metazoan. Notably, both ctenophores and sponges appear to lack most of the secreted antagonists to this pathway, including chordin (CHRD), follistatin (FST), noggin (NOG; but see [17]), and members of the CAN family (cerberus, DAN, gremlin), as well as the intracellular cofactors that inactivate SMAD signalling downstream of receptor activation (SARA and SKI/SNO). This suggests that modulation of TGF-β signalling through selective inhibition in specific cells or at specific stages of development may have evolved as a means to diversify the output of this signalling pathway after its origin.
Table 1.
TGF-β signalling pathway components across unikonts. EC, extracellular; IC, intracellular; MB, membrane bound.
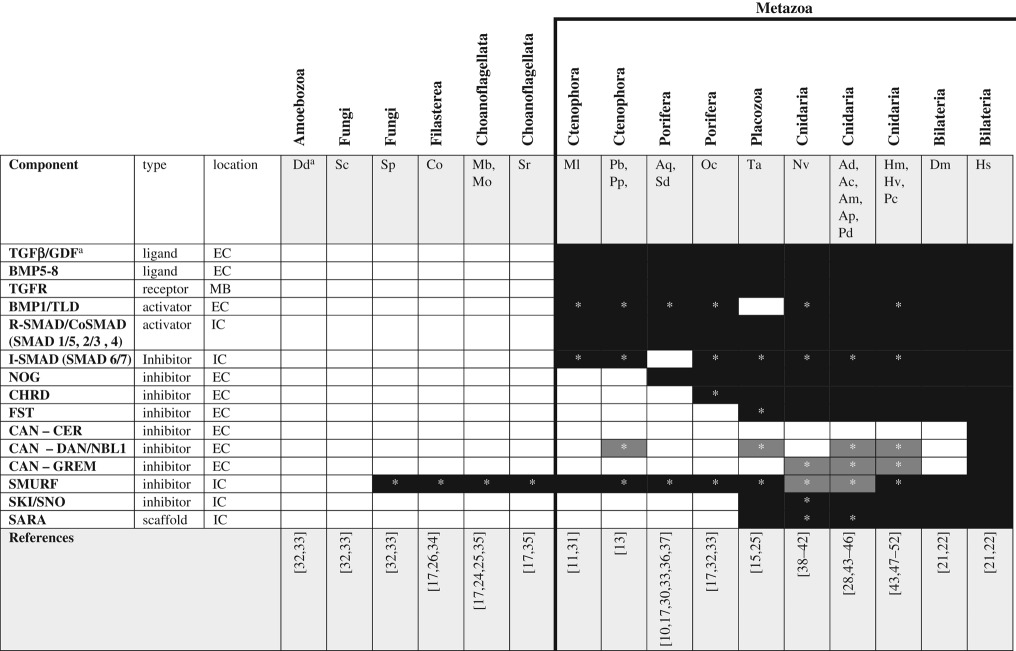 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional pathway is indicated by the bold line.
In bilaterians, TGF-β signalling plays important roles in the promotion of cell and tissue fate, immunity and, embryonically, this pathway is known to be an essential regulator or dorsal/ventral axis specification [25]. The BMP arm of the TGF-β signalling is asymmetrically expressed along the directive axis (orthogonal to the anterior/posterior axis) of the sea anemone Nematostella vectensis [38,53,54] and has been implicated in defining the bilateral axis of symmetry in cnidarians [39,43]. Furthermore, ectopic expression of NvSMAD (a SMAD homologue isolated from Nematostella vectensis) in Xenopus during development has demonstrated that the Nv homologue of this gene is sufficient to recapitulate the ventralized phenotype generated by vertebrate homologues in similar studies [40]. Similarly, using in situ hybridization, Adamska et al. [30] identified asymmetrical expression of TGF-β along the larval swimming axis in sponge embryos and Pang et al. [31] revealed that TGF-β pathway genes are expressed asymmetrically along all three axes in M. leidyi. The non-overlapping expression patterns of TGF-β ligands in early ctenophores suggest these genes may be under the control of maternally deposited RNAs/proteins. Together, these developmental/functional data provide strong evidence for a conserved role for TGF-β signalling in axis specification throughout Metazoa.
In addition to this role in axis specification, Matus et al. [41] propose that the co-expression of transcripts from multiple TGF-β signalling components in the early endoderm of N. vectensis suggests an important role for this pathway in patterning germ layer identity. Indeed, studies of spatial expression of TGF-β components across basal metazoans have uncovered multiple roles for TGF-β signalling in the development and maintenance of differentiated cell types, including: regenerating head cells, tentacles, the oral nerve ring and adult endo/gastrodermal cells in cnidarians [47,48,55,56] and the embryonic pigment ring in sponges [30]. Finally, pharmacological studies in the facultatively symbiotic sea anemone Aiptasia pallida suggest a potential role for TGF-β in facilitating the interaction between the cnidarian host and algal symbiont [57]. Together, these patterns suggest that this pathway may have had an original function in transducing maternally derived molecules into developmental signals regulating axis specification, but clearly this pathway was been co-opted, spatially and temporally, to regulate several aspects of larval and adult tissue identity.
(b). Canonical WNT signalling
The WNT signalling pathways are known to play diverse roles in the cell/tissue biology of metazoans (reviewed by [58]). cWNT signalling is initiated by binding of the extracellular WNT ligand to its receptor complex composed of frizzled (FZD) and lipoprotein receptor-related protein 5/6 (LRP5/6) (figure 1b). This activates an intracellular signal transduction cascade through which dishevelled (DSH) inhibits glycogen synthase kinase 3 (GSK-3) leading to the translocation of β-catenin (β-cat) to the nucleus where it complexes with the transcription factors T-cell-specific transcription factor/lymphoid enhancer binding factor (TCF/LEF) to regulate gene expression. Like TGF-β signalling, cWNT signalling appears to be a metazoan innovation as the ligands, receptors and intracellular transducers are largely excluded from pre-metazoan taxa (table 2). While the genome of the amoebozoan Dictyostelium discoideum encodes a putative orthologue of β-cat [59], several FZD-like receptors [60] and an orthologue of GSK-3 [71], with the exception of GSK-3, no cWNT pathway genes are encoded in the genomes of other unikonts. This suggests either the convergent evolution of FZD-like proteins in amoebozoans and metazoans or that these components were lost after the divergence of amoebozoans and re-evolved at the base of Metazoa. Cnidarians have not only a complete cWNT pathway, but also have a surprising diversity of WNT ligands [65,72]; by contrast, sponges and placozoans have orthologues of FZD, LRP5/6, GSK-3 and β-cat but only few WNT ligands (4 and 3, respectively) [10,15,30]. Pang et al. [73] found a nearly complete set of cWNT pathway genes present in the ctenophore M. leidyi but, like sponges and placozoans, ctenophores have only few (4) WNT ligands [62,73]. As was seen for the TGF-β pathway, cWNT signalling in ctenophores, sponges and placozoans appears to lack inhibitory control—conspicuously absent from ctenophore, sponge and placozoan genomes are WNT inhibitory factor (WIF), dickkopft (DKK) genes and cerberus (CER), which are important inhibitors of cWNT signalling in cnidarians and bilaterians, and axin, which is part of the β-cat degradation complex, is missing from both published ctenophore genomes. Finally, both sponges and ctenophores have fewer ligands and receptors for the cWNT pathway than do cnidarians and bilaterians and the sponge paralogues appear to be the result of independent duplication of an ancestral gene in the sponge lineage [10]. Thus, sponges appear to have undergone their own WNT ligand expansion (albeit, a more limited one) sometime after they diverged from the rest of Metazoa.
Table 2.
Canonical WNT signalling pathway components across unikonts. EC, extracellular; IC, intracellular; MB, membrane bound.
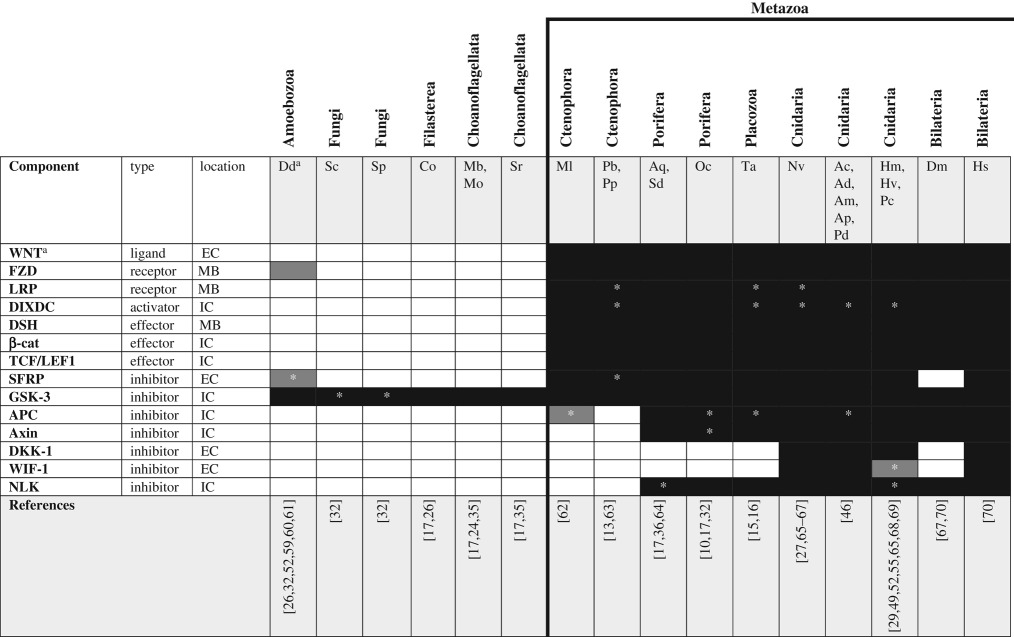 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional pathway is indicated by the bold line.
cWNT signalling plays diverse roles in cell/tissue development and homeostasis. Although this pathway has been implicated in patterning of the primary axis in some bilaterians [63], the earliest developmental role of cWNT appears to be defining the site of gastrulation (the blastopore) and establishing this tissue as an embryonic organizing centre [74–76]. Among cnidarians, cWNT signalling has been shown to facilitate the establishment of embryonic [65,77,78] and adult [29,72] organizing centres in both anthozoan and hydrozoan model systems. Although functional studies in sponges are lacking, studies of WNT gene expression in situ suggest that this pathway is also asymmetrically expressed along the larval swimming axis and during development of the adult axis (defined by the aquiferous system) [36,79,80]. Treatment with pharmacological activators of cWNT signalling (e.g. LiCl) in sponges also point to a possible role for this pathway in specifying the epithelia in adult sponges [63], though clearly functional studies of cWNT signalling in sponges are much needed. Together, these data suggest that cWNT-regulated specification of the blastopore and the subsequent defining of the anterior/posterior axis were early features of metazoan development but that this pathway may have been co-opted to play a role in specification of larval and adult tissues in some lineages. Although gastrulation has not been observed in placozoans [14] and the only identifiable axis in these animals appears to be a dorsal/ventral axis, the genome of T. adhaerens encodes a full complement of WNT/β-cat signalling components [15]. It is intriguing to consider what organizing centre or potential axis may be revealed if analyses of spatial gene expression are one day performed in these animals.
Using whole mount in situ hybridization, Pang et al. [31] described the expression of cWNT pathway genes in M. leidyi. Surprisingly, WNT genes were detected only late in development, after the onset of gastrulation. With the use of RNAseq, WNT transcripts were detected much earlier in the development of P. bachei [13]. No functional studies of cWNT signalling have been performed in ctenophores, so it is unclear if these early expressed WNTs play a homologous role in the specification of the ctenophore blastopore. Interestingly, pharmacological treatment with cWNT activators (e.g. LiCl and alsterpaullone) has little effect on early development in M. leidyi (K Pang and MQ Martindale, personal observations). Alternatively, the asymmetrical division of maternally loaded proteins and/or the ratio of nuclear material to cytoplasm have been suggested to be important drivers of early embryonic cell fate in ctenophores [81]. How the WNT/β-cat pathway interacts with these cytological determinants (e.g. nuclear : cytoplasm ratio) is not known but studies of this type could reveal an ancestral mechanism for the activation of the cWNT pathway in ctenophores. It has been suggested that the overlapping expression domains of three different WNT pathways—cWNT, WNT/planar cell polarity (PCP) and WNT/Ca2+—in cnidarians suggests these pathways work synergistically to pattern the primary axis in this lineage. Curiously, ctenophores lack the essential components of the PCP pathway [11,82], suggesting this synergistic effect of multiple WNT signalling pathways on axis specification must have arisen after ctenophores split from the lineage giving rise to sponges and parahoxozoans.
(c). Nuclear receptor signalling
NRs are transcription factors that translate ligand binding directly into activation or repression of transcription. NRs can be activated by binding a hydrophobic ligand (e.g. fatty acid and steroid), complexing with other proteins, or undergoing post-translational modification, after which they bind DNA directly to regulate (activate or repress) the expression of target genes (figure 1c). There are two conserved functional domains diagnostic of NRs: the DNA binding domain responsible for transcriptional regulation and the ligand binding domain to which hydrophobic ligands bind. NRs are thought to have evolved from ancestral molecules that did not require activation by a ligand [83,84]; thus, the expansive diversity in this group of proteins may have evolved with the independent origins of numerous diverse ligand binding capabilities [85]. Unlike the other cell signalling mechanisms discussed here, NRs act as both the receptor and the effector molecule, providing the most direct link between the extracellular signal and the transcriptional response. Pathways of this type may be the easiest to evolve because they rely on only few signalling components; the massive expansion of the NR gene family in nematodes provides support for this hypothesis [86]. Conversely, the small number of nodes that comprise this pathway means there are fewer opportunities for co-option/modification of this pathway once it evolves.
Like the TGF-β and cWNT pathways, NRs evolved in the stem metazoan. These receptors have been described from cnidarians [87], placozoans [15,88], sponges [89,90] and ctenophores [91], but are missing from choanoflagellates [24], filastereans [26,35], fungi [92] and amoebozoans (table 3). Over 17 NRs have been identified among cnidarians, many of which lack clear orthology with bilaterian NRs but have clear orthologues in other cnidarians, suggesting that this gene family expanded through several rounds of cnidarian-specific duplication events [87,91]. Srivastava et al. [15] identified four NRs from placozoans, orthologues of: hepatocyte nuclear factor 4 (HNF4), retinoid X receptor (RXR), an NR2 family member, and one that appears to lack orthology with other available sequences. Baker [88] also described an oestrogen-related receptor (family NR3) from T. adhaerens but indicated that it contained a hormone binding pocket that was too small to accommodate a hormone, suggesting it may not function the way NR3 family receptors do in bilaterians. Among sponges, two NRs have been identified from Amphimedon queenslandica, Suberites domuncula and Oscarella carmela—one, NR2, appears to be an orthologue of HNF4 and is found only in A. queenslandica and O. carmela; the second, apparently sponge-specific, is found in A. queenslandica, S. domuncula and O. carmela [85,91]. Three NR orthologues have been identified from ctenophores (two from M. leidyi and one from P. bachei), all of which appear to be orthologues of the NR2 family but none of which has a canonical DNA binding domain. This suggests that the DNA binding domain (and, likely, the ability to regulate transcription) may have arisen as a feature of NRs only after ctenophores diverged from the rest of Metazoa [91].
Table 3.
Nuclear receptor signalling pathway components across unikonts. IC, intracellular.
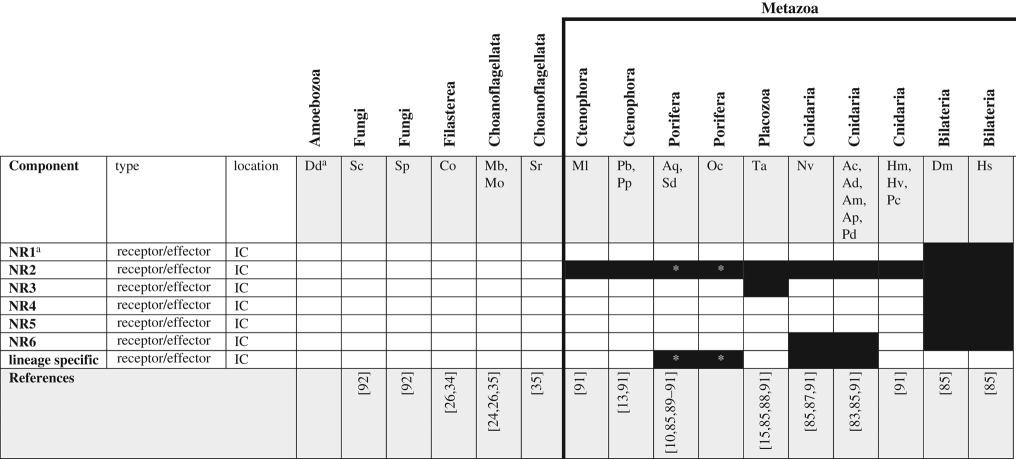 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the Amphimedon queenslandica NR database; accession numbers available in electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional NR pathway is considered to be coincident with the origin of metazoan NR homologs and is indicated by the bold line.
Three classes of NRs have been identified, based on their mode of activation; one group becomes active (and activates transcription) upon binding a hydrophobic ligand, another requires no ligand and is constitutively active, and a third exclusively acts to repress the function of other NRs [85]. All three classes of NRs are represented among bilaterians, and the functional output of these signalling molecules is accordingly diverse. Perhaps most famously, this group of signalling molecules includes the retinoic acid receptors, which are known to play a critical role in establishing morphogen gradients and regulating cell identity during development of bilaterians [93]. Only few studies of NR function have been performed in non-bilaterian animals. Notably, signalling through the RXR receptor has been examined in several species of cnidarian; exogenous application of retinoic acid has been shown to play a role in promoting metamorphosis in the jellyfish Aurelia aurita [94] and regulating the development of the eyes in the box jelly Tripedalia cystophora [95]. Further, the COUP-TF transcription factor (NR2 family) is an important regulator of neurogenesis in hydrozoans [96,97]. While Reitzel & Tarrant [87] demonstrated differential expression of NRs through developmental time in N. vectensis, without spatial analysis of gene expression it is difficult to hypothesize what role these diverse receptors may play in embryogenesis of the sea anemone. The two NRs identified from the demosponge A. queenslandica also appear to be functional during development. The HNF4 homologue is expressed in the ciliated cells of the ectoderm before they are specified to become choanocytes [90] and functional studies suggest that both sponge NRs bind fatty acids but the outcome of this interaction differs. While NR1 activates transcription in response to ligand binding, NR2 appears to repress transcription [85]. Thus, at least two different mechanisms of NR-mediated cell signalling appear to have been present before the divergence of sponges. Although NR signalling has not been characterized in the ctenophores, whole mount in situ hybridization in M. leidyi suggests that transcripts for these receptors are expressed only after the onset of gastrulation [91]. Thus, it seems NRs may not play a prominent role in the earliest specification of cell identity in ctenophores.
5. Parahoxozoan pathways
(a). Notch/Delta signalling
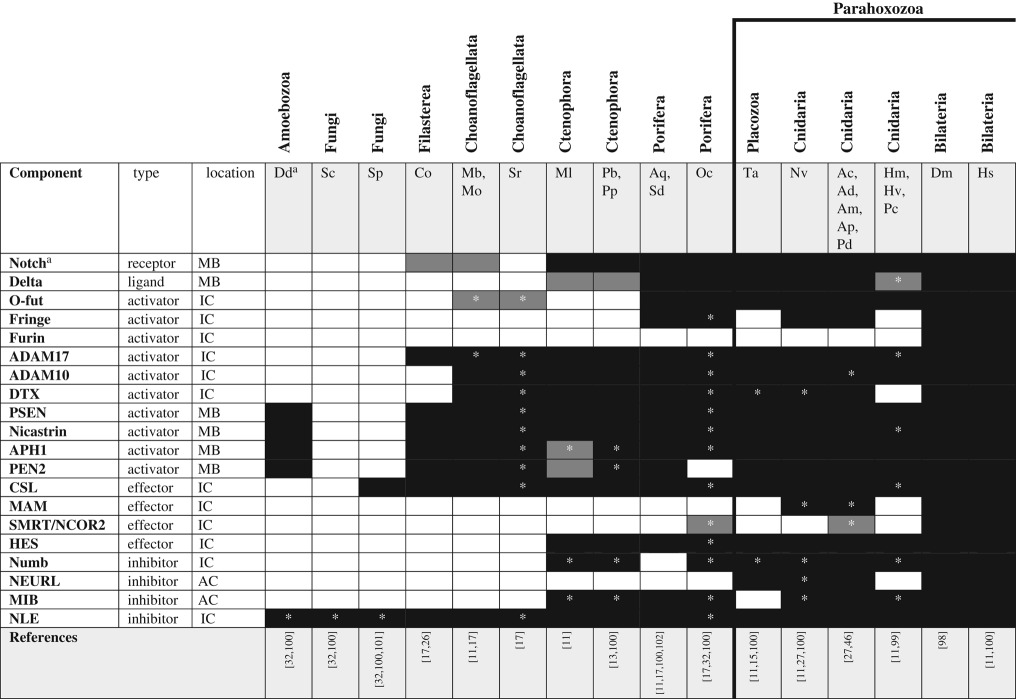
The Notch/Delta signalling cascade is unlike other signalling mechanisms described herein inasmuch as both Notch and its canonical ligand (Delta) are membrane-bound proteins; thus, this system behaves as a juxtacrine signalling system, transducing signals between adjacent cells (see [98] for a thorough review of Notch/Delta signalling in bilaterians). Binding of the ligand (Delta) catalyses cleavage of the Notch intracellular domain that translocates to the nucleus to effect transcriptional change (figure 1d). Beyond the ligand (Delta) and receptor (Notch), the proteases that process the Notch preprotein (furin, O-fut and fringe) and liberate the intracellular domain from the extracellular domain (ADAM-type metalloproteases and the γ-secretase complex) are critically important for effective signalling. Outside of Bilateria, Notch homologues have been found in cnidarians [27,99], placozoans [11,15], sponges [17] and ctenophores [11,13]. Notch-like proteins have also been described from the choanoflagellate Monosiga brevicollis and the filasterean Capsaspora owczarzaki, though these proteins lack key functional domains including the Delta interaction domain (DSL) in the former and the epidermal growth factor (EGF) domains in both taxa [26]. While Delta has been described from each of these taxa as well, analysis of the Delta-like molecule found in M. brevicollis and C. owczarzaki suggests this protein may interact with other EGF domain-containing proteins but likely does not interact with the Notch-like proteins in these taxa [26]. Similarly, the Delta protein described in ctenophores may not function equivalently to the canonical Delta ligand from other taxa because it is missing key diagnostic domains [11]. Only few of the numerous molecules involved in Notch/Delta signalling are metazoan-specific (including both Notch and Delta; table 4). Specifically, many of the enzymes associated with this pathway (e.g. ubiquitin ligases and ADAM-type proteases) were present before the origin of Metazoa, suggesting Notch/Delta signalling may have evolved by co-option of an existing intracellular cascade [100].
Table 4.
Notch/Delta signalling pathway components across unikonts. AC, adjacent cell; IC, intracellular; MB, membrane bound.
 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional pathway is indicated by the bold line.
Notch/Delta signalling functions primarily in promoting differential cell identity in subpopulations of adjacent cells. Two main developmental roles have been described for this pathway: lateral inhibition promoting one of two possible fates and boundary mechanisms that induce a third cell fate at the interface between two other cell fates [100]. The mechanism of Notch/Delta signalling appears to depend on stochastic variation in the abundance of both the ligand and receptor [103]. Notch/Delta signalling pathways are known to be functional in cnidarians and bilaterians but the roles of these pathways may be quite different in these two lineages. Pharmacological studies using the γ-secretase inhibitor DAPT have identified a role for Notch/Delta signalling in neural specification across cnidarians [99,104,105], but knockdown experiments in the sea anemone N. vectensis suggest that cnidarians use the non-canonical Notch pathway, independent of the hairy/enhancer of split (HES) transcription factors, in this process [106]. Curiously, cnidarians have orthologues of HES genes (indeed, HES genes are found throughout Metazoa), suggesting that the canonical Notch/HES signalling pathway may be used in a context other than neural specification in non-bilaterian metazoans. In sponges, Notch and Delta are expressed in adjacent tissues during embryonic development and the downstream transcription factors have been shown to have neurogenic effects when expressed during development in Xenopus and Drosophila [107]. In situ, Notch/Delta signalling seems to regulate cell identity in globular cells of A. queenslandica [107]. Thus, in both cnidarians and sponges, Notch/Delta signalling promotes the differentiation of specialized cell fate during early embryogenesis. The putative function of this pathway has not been examined extensively in ctenophores but preliminary pharmacological assays using the γ-secretase inhibitor DAPT resulted in no detectible effects on development in M. leidyi (K Pang and MQ Martindale, personal observations). Although ctenophores lack a complete Delta ligand [11], abundant research from bilaterians supports a functional role for Delta-like ligands in tissue specification during development. What this pathway may regulate in ctenophores is unknown, but the fact that ctenophores seem to be insensitive to γ-secretase is intriguing and leads us to conclude that the canonical/functional Notch/Delta pathway evolved after ctenophores diverged from other metazoans.
6. Pathways restricted to Cnidaria + Bilateria
(a). Hedgehog signalling
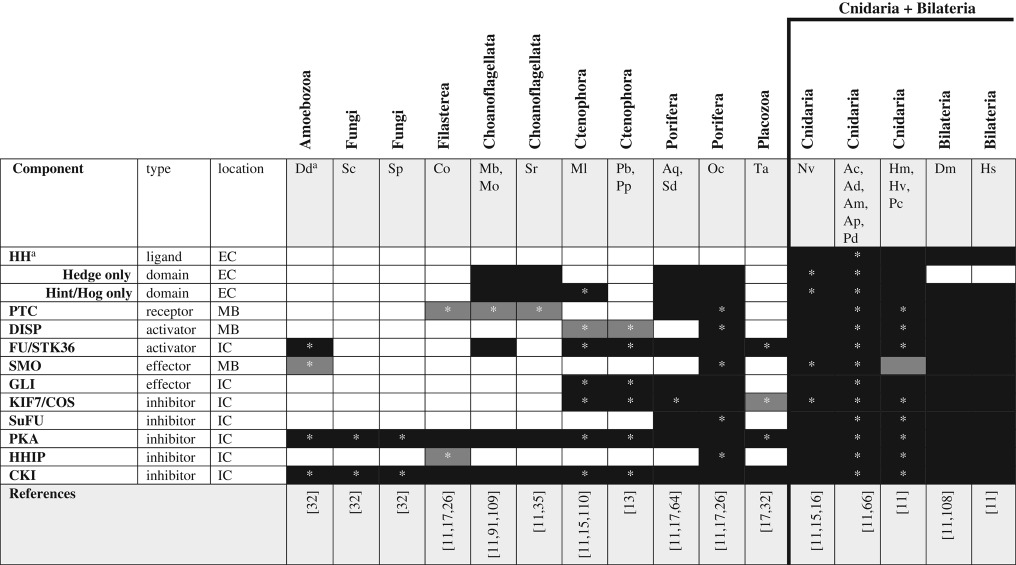
The hedgehog (HH) pathway regulates many aspects of cell and tissue identity during bilaterian development (see [108] for review). Upon binding of the HH ligand to the patched (PTC) receptor, constitutive inhibition of the smoothened (SMO) receptor is relaxed, leading to the translocation of SMO to the cell membrane (figure 1e). Insertion of SMO into the plasma membrane enhances the interaction between SMO and the intracellular HH signalling components and results in the release of the transcription factor GLI from the signalling complex and leads to its translocation into the nucleus to modulate transcription. HH signalling is well known to operate through the establishment of a ligand concentration gradient; thus, cells responding to HH far from the source of the ligand respond differently from those near the source. The HH pathway is an interesting model for the modular evolution of cell signalling pathways; while a complete set of HH signalling components (ligand, dispatched transmembrane transporter, both PTC and SMO receptors, and the GLI transcription factor) is restricted to the group containing cnidarians and bilaterians (table 5), there is clear evidence for the origin of individual pathway components well before this. Both the ‘Hedge’ and ‘Hog’ domains that define the ligand arose before the origin of the complete HH gene [111]. Indeed, choanoflagellates, sponges and cnidarians have proteins with Hedge domains that lack the associated Hog domain characteristic of the true HH ligand [11]. Even without a Hog domain, these so-called Hedgling genes contain many functional domains (e.g. von willebrand factor, cadherin and EGF domains) suggesting they are capable of interacting with many other signalling components [30].
Table 5.
Hedgehog signalling pathway components across unikonts. EC, extracellular; IC, intracellular; MB, membrane bound.
 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional pathway is indicated by the bold line.
Ctenophores lack the orthologues of several HH pathway components. Orthologues of both HH and Hedgling ligands are missing from the genomes of M. leidyi and P. bachei, as are the PTC and SMO receptors [11]. The phylogenetic patterns represented in table 5 suggest that the lack of PTC and Hedgling orthologues may represent secondary loss in ctenophores, indicating that these molecules may have experienced relaxed selection associated with loss of a specific pathway or function during the divergence of ctenophores from rest of Metazoa. Understanding how PTC and Hedgling orthologues function in filastereans and choanoflagellates is critical to understanding the evolution of the canonical pathway from these early precursors. Furthermore, while M. leidyi does have an orthologue of the GLI transcription factor, this gene encodes only four of the five diagnostic zinc finger domains [110], thus it is unknown whether this transcription factor is functional. Interestingly, placozoans do not have orthologues of any of the essential components of HH signalling [11,15] but given their position between sponges and cnidarians, this likely represents a lineage-specific loss of the pathway.
The role of HH genes in bilaterian development has been well characterized as they are known to play a crucial role in establishing tissue-specific organizers (e.g. the limb ZPA and neural floor plate) during embryogenesis [112]. Outside of Bilateria, cnidarians are the only metazoans to have true HH orthologues and their role in development has been studied only by whole mount in situ hybridization in the sea anemone N. vectensis [66]. The overlapping expression of the PTC receptor and GLI transcription factor in the endodermal mesenteries and their proximity to the expression of both HH genes suggests this pathway may be regulating the development of endodermal cell identity in cnidarians. Although they are not true HH orthologues, Hedgling genes are also expressed during development in cnidarians and sponges [30]. Indeed, the overlapping/abutting expression of TGF-β, WNT and Hedgling revealed by in situ hybridization during formation of the pigment ring in A. queenslandica and the endodermal mesenteries in N. vectensis [19,30] is reminiscent of the overlapping expression of TGF-β/WNT/HH expression during bilaterian organ specification and suggests that Hedgling and HH may play similar regulatory roles in non-bilaterians. Understanding the putative differences between the roles of HH and Hedgling in cnidarians and the function of Hedglings in sponges and choanoflagellates would shed much-needed light on the origin/evolution of this protein family and functional studies would be particularly useful in this context. Ctenophores encode a Hog domain, but there is no evidence of Hedge domains in this lineage [11]. Neither the spatial/temporal expression of this Hog-domain containing transcript nor the putative function of the protein it encodes has been studied in ctenophores; thus, the putative role that HH pathway genes play in the development of ctenophores remains elusive.
(b). Receptor tyrosine kinase/growth factor signalling
Protein tyrosine kinases (PTKs) function to phosphorylate other proteins, enabling them to manipulate (activate/inhibit) the function of other proteins. RTKs are one large class of PTKs that have evolved a transmembrane domain and transduce the binding of extracellular signals (e.g. growth factors, cytokines and amino acids) into altered phosphorylation states of their targets. Binding of the ligand induces dimerization and stabilization of the receptor in the cell membrane. This stabilization of the RTK activates an intracellular signalling pathway, typically the Ras/Raf/MAPK protein kinase pathway, resulting in phosphorylation of a target transcription factor that can then move into the nucleus to promote or repress transcription (figure 1f). Certain members of the RTK family signal through alternative means, relying instead on the activation of phospholipase C (PLC) and phosphorylation of target genes by protein kinase C (PKC) or even through the JAK/STAT pathway (described below). The diversity of RTKs is matched by the equally diverse mechanisms by which they transduce signals and the numerous roles they play in cell biology [113]. Similar to the expansive repertoire of NR paralogues described above, this may suggest the evolutionary expansion of RTKs resulted from duplication followed by modifications to the ligand binding domain that allowed them to respond to diverse extracellular cues. Accordingly, the intracellular kinase domain of RTKs exhibits greater conservation than the extracellular ligand-binding domains.
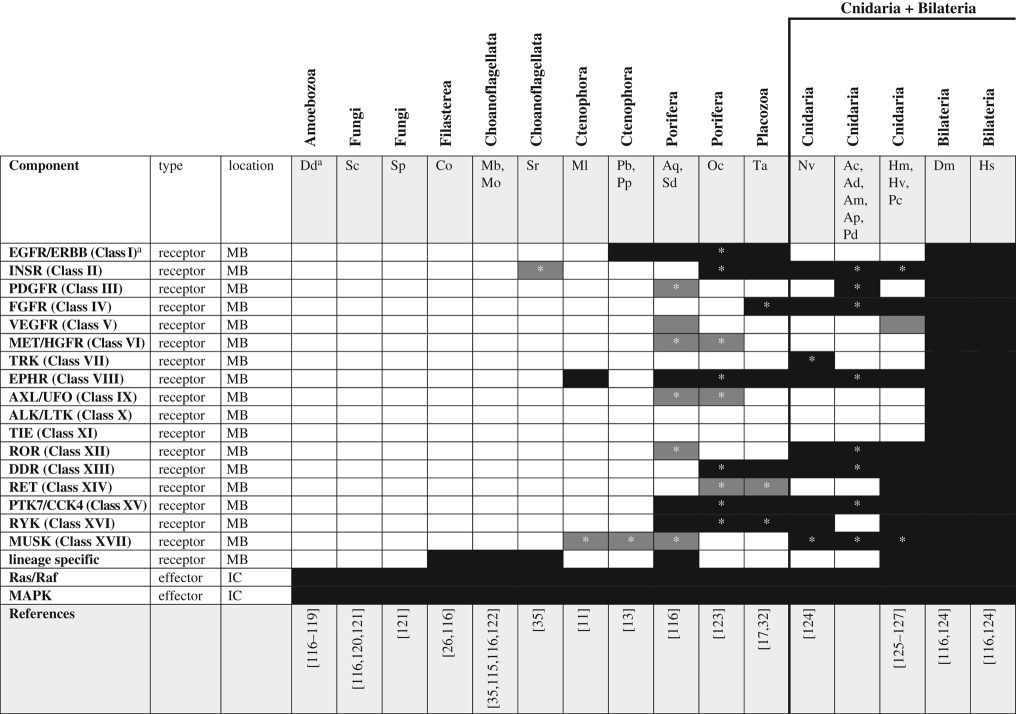
Bilaterians have dozens of family members, but the origin of RTKs predates the dawn of Metazoa as numerous RTKs have been described from choanoflagellates [114,115] and filastereans [116]. The genomes of amoebozoans and fungi encode PTKs but not RTKs (table 6) [117]. A comparison of the protein domains in RTKs from filastereans, choanoflagellates and metazoans suggests that the pre-metazoan RTKs lack homologues in other lineages, suggesting RTKs underwent expansive diversification independently in each lineage before the origin of Metazoa [116]. Clear homologues of bilaterian RTKs first appear in the Ctenophora with the emergence of an EPHR in M. leidyi [11] and an EGFR/ERBB receptor in P. bachei [13] and expanded considerably in sponges and cnidarians. Indeed, analysis of the A. queenslandica genome identified approximately 150 RTKs from sponges [10], only some of which appear to have ligands that are orthologous to other metazoan RTK ligands (e.g. EGFR, EPHR and INSR). Thus, the origin of the bilaterian family of RTKs did not preclude further evolution of lineage-specific RTKs [123,125] and it will be interesting to learn whether ctenophores have their own lineage-specific RTKs. A novel RTK family—venus kinase receptor (VKR)—was recently identified in N. vectensis as well as several bilaterian lineages but is absent from sponges and Hydra. A search of the M. leidyi genome for the diagnostic VFT domain of VKR-related proteins failed to find any significant matches in this ctenophore, suggesting VKRs also evolved after ctenophores and sponges diverged from the cnidarian/bilaterian ancestor [126]. Also absent from ctenophores and sponges is the FGFR family of RTKs [128], which is known to have many important roles governing the development of cell and tissue identity in Cnidarians and Bilaterians. Finally, despite the lack of conserved RTK orthologues among basal unikonts, the intracellular effectors of RTK signalling (i.e. Ras/Raf/MAPK) were present in amoebozoans. Again, this signalling pathway seems to have evolved by co-option of an existing intracellular signalling mechanism after the origin of the ligand/receptor.
Table 6.
RTK signalling pathway components across unikonts. IC, intracellular; MB, membrane bound.
 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in the electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/functional pathway is indicated by the bold line.
While the bilaterian RTK family is expansive [124] and well known for promoting cell differentiation during tissue patterning, the role of RTKs in directing the development of non-bilaterians is poorly understood. Fibroblast growth factor (FGF) orthologues first emerged before cnidarians diverged from bilaterians. In N. vectensis, FGF signalling was assayed using a combination of descriptive and functional techniques and was shown to regulate the development of the aboral sensory organ (apical tuft) in planula larvae [129–131]. Recent studies of Hydra have identified a putative role for FGF in maintenance of the interstitial stem cell lineage and for defining the termini of the animal, whereas another growth factor (vascular endothelial growth factor, VEGF) was shown to be involved in specification of neurons [132,133]. Lemon, an orthologue of protein tyrosine kinase-like 7 (PTK7/CCK4) family RTKs from Hydra vulgaris, is expressed in interstitial stem cells and upregulated during times of gametogenesis [127]. Combined, these data suggest that RTKs play numerous diverse roles in regulating cell specification and tissue morphogenesis in cnidarians, supporting the origin of functional RTK signalling before the cnidarian/bilaterian split. In addition to bona fide FGFs, FGF-like proteins have been identified from choanoflagellates, ctenophores, sponges, placozoans and cnidarians [128]. These proteins are characterized by a β-trefoil fold and group more closely with bona fide FGFs than with any other protein family, but the potential role of these proteins in the developmental biology of these taxa is unknown. FGFR-like proteins were recently characterized in cnidarians and chordates and appear to be expressed in early development [134], suggesting this putative FGF-like pathway may have a role in directing cell/tissue specification or aspects of morphogenesis. The presence of putative RTK homologues in ctenophores, sponges and placozoans suggests that these pathways may operate (canonically or not) in these taxa as well, but this remains to be empirically tested.
7. Bilaterian-specific signalling pathways
(a). Janus kinase/signal transducer and activator of transcription signalling
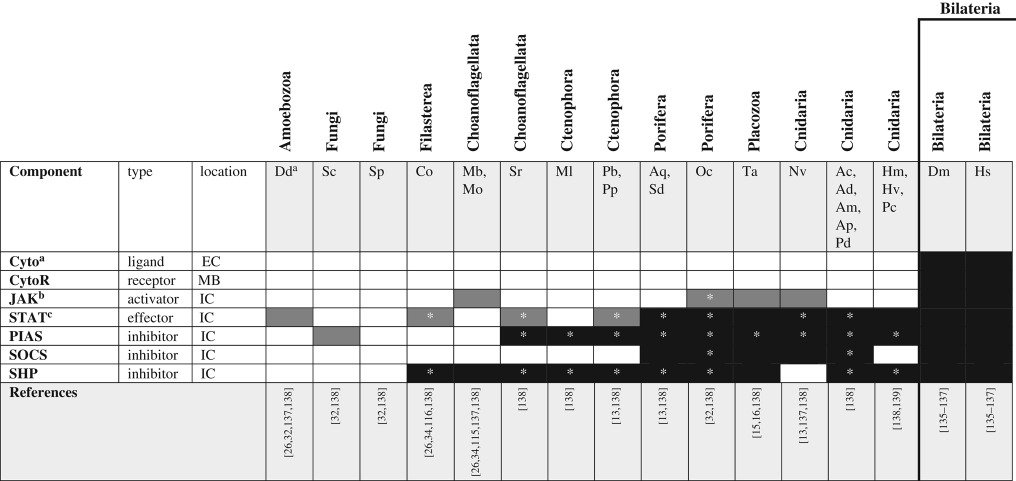
The JAK/STAT signalling cascade plays an essential role in cell signalling in bilaterians by transducing secreted peptide signals (typically cytokines) into transcriptional activation responses (see [135,136] for recent reviews). Intracellular JAK proteins become activated by complexing with transmembrane receptors in the cytokine receptor class I/II families. This interaction leads to the phosphorylation and dimerization of STAT transcription factors in the cytoplasm that then undergo translocation to the nucleus, where they activate transcription of target genes (figure 1g). Inactivation of the pathway is accomplished through dephosphorylation by SH2-domain containing protein tyrosine phosphatases (SHPs) or through negative regulation by molecules including suppressor of cytokine signalling (SOCS) and protein inhibitor of activated STAT (PIAS). Studies in Drosophila revealed a non-canonical JAK signalling pathway responsible for modification of heterochromatin architecture, revealing the potential for this pathway to affect many genes beyond the specific targets of STAT activation [135].
The complete repertoire of proteins comprising the canonical signalling pathway (cytokines, cytokine receptors, JAK, STAT, SHP and SOCS) is restricted to the Bilateria as there are no clear orthologues of JAK in other Unikonts. As was the case with hedgehog signalling, however, many of the intracellular components of JAK/STAT signalling were present before the evolution of the canonical/functional pathway (table 7). A proto-JAK first appeared before the origin of Porifera, but JAK-like proteins in sponges and cnidarians both lack a pseudo PTK domain [137]. The evolutionary history of STAT proteins may be slightly older than that of JAK. STAT-like transcription factors have been reported from all major lineages of unikonts; however, most of these molecules lack the C-terminal transactivation (TAD) domain. Despite this, the cnidarian STAT-like protein appears to complex with nuclear factor kappa-B (NFκB) to serve as part of the rudimentary immune system [140]. This suggests that STAT-like proteins may have engaged in transcriptional regulation before becoming associated with the JAK signalling cascade. Clearly, the involvement of ancestral STAT-like proteins and their partners (including proto-JAKs) in cell–cell signalling has not been sufficiently studied in any of the non-bilaterians. This pathway, however, appears to be one of the few/unusual examples whereby the negative regulator evolved before the full pathway as SOCS, PIAS and SHPs all evolved long before the origin of canonical JAKs and STATs. Understanding what other functions these inhibitory molecules play in organisms that lack the full JAK/STAT pathway may indeed reveal novel protein–protein interactions and point to previously uncharacterized signalling pathways.
Table 7.
JAK/STAT signalling pathway components across unikonts. EC, extracellular; IC, intracellular; MB, membrane bound.
 |
aSee glossary for gene and species abbreviations.
bJAK homologues were identified by the presence of the following domains: FERM, SH2 and at least one PTK.
cNon-bilaterian STAT homologues were identified by the presence of the following domains: N-terminal, alpha coil, DNA binding, and SH2; the TAD domain is clearly a bilaterian novelty [137].
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in the electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present.
8. Tying the room together: the role of the extracellular matrix in cell–cell communication
The evolution of the extracellular matrix (ECM) was pivotal in permitting the evolution of large complex organisms as it allowed the directed transmission of extracellular signals among cells. The ECM is now known to play a critical role in cell–cell signalling [141], and much progress has been made in characterizing the evolutionary history of the ECM and animal epithelia [142–144]. Epithelia are not unique to metazoans; indeed, the fruiting body of the amoebozoan D. discoideum forms a polarized epithelium in its multicellular stage, using a catenin-based system of cell–cell adhesion [145,146]. Although cell–cell adhesion evolved before the origin of Holozoa, the rise of multicellularity did not occur until much later. The origin of the ECM and its ability to facilitate the development of diffusion gradients likely provided a significant selection advantage at the origin of complex life. Here, we focus specifically on two aspects of ECM: the evolution of the basement membrane, which provided the infrastructure for the development of signal gradients, and the evolution of the adhesome complex (integrin and its associated molecules), which provided a means to keep cells attached to the ECM.
Essential components of the basement membrane include type IV collagen (ColαIV), the multiple subunits of laminin (LAMA, LAMB1 and LAMC), and both membrane-bound and free/secreted heparan sulfate proteoglycans (HSPGs; e.g. glypican and perlecan; figure 1h). Several components of the ECM are known to facilitate growth factor gradient formation. In particular, fibronectin (FN) facilitates contact between the ECM and cell surface molecules (including cytokine receptors, HSPGs, integrins and dystroglycans), providing a functional link among these proteins and making it an essential component of extracellular signal transduction in bilaterians. Furthermore, perlecan is known to stabilize secreted peptide ligands (cytokines, FGFs and VEGFs), increasing the efficacy of FGF [141] and hedgehog signalling [112] in bilaterians. The canonical/complete basement membrane is a metazoan-specific novelty, as clear ColαIV and laminin orthologues first appear in the Ctenophora but the adhesome complex (integrin and its intracellular partners vinculin, α-actinin, talin and paxilin) dates back to the origin of Holozoa as both α and β integrins have been identified from filastereans (table 8). This suggests that integrins may have played a role in intracellular signalling long before they became associated with the ECM and, further, that the ECM may have evolved scaffolding properties first (through the integration of structural proteins such as laminin and collagen) and developed the ability to act as a signalling centre only after the origin of proteoglycans and linker proteins such as fibronectin.
Table 8.
ECM components across unikonts. EC, extracellular; IC, intracellular; MB, membrane bound.
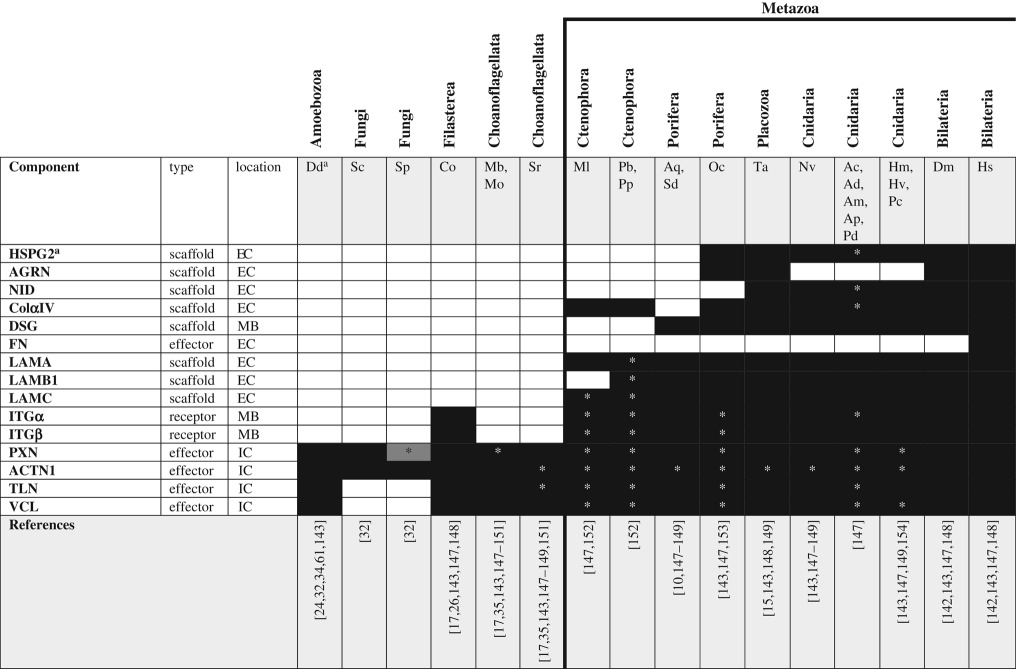 |
aSee glossary for gene and species abbreviations.
*Homologues identified by reciprocal BLAST against the human NR database; accession numbers available in the electronic supplementary material. White boxes—no homologue reported in the literature or detected by BLAST. Grey boxes—putative/degenerate homologue. Black boxes—homologue present. The origin of the canonical/complete basement membrane is indicated by the bold line.
Combinations of in vivo and in vitro assays have revealed the numerous important roles cnidarian basement membrane plays in directing cell differentiation, cell migration and tissue regeneration (see [155] for a review of these processes in Hydra). However, because the focus of these studies has largely been on hydrozoans, little is known about the development of the basement membrane in other cnidarians. Likewise, studies of the spatial expression and/or function of basement membrane transcripts in sponges are lacking. The situation with sponges is a particularly unusual one as homoscleromorphs appear to be the only sponges to secrete a basement membrane [156]. Considering that basement membrane-specific HSPGs also arose before the divergence of sponges and parahoxozoans, future studies aimed specifically at assessing the signalling capacity of the basement membrane in homoscleromorph sponges would be extremely informative. Basement membrane is known to underlie both epithelia in ctenophores [157] but again, the development of this structure has not been studied. Assaying stage-specific transcriptomes from M. leidyi throughout early development suggests that the expression of both ColαIV and laminins peaks just after gastrulation, coincident with the development of the mesoglea (LS Babonis and MQ Martindale, personal observations). Neither FN nor any of the basement membrane-specific HSPGs are encoded in the genome of M. leidyi, suggesting that the basement membrane of ctenophores may play only a structural role, rather than a role in cell signalling. In Drosophila melanogaster, DPP (the homologue of BMP 2/4) is known to bind directly to ColαIV in the basement membrane without the aid of HSPGs [158]. Both BMPs and ColαIV are present in ctenophores just after gastrulation. It would be interesting to learn if this direct relationship between ColαIV and secreted ligands for the WNT/TGF-β pathways also functions in ctenophores, suggesting it was present in the stem metazoan.
Using in situ hybridization and quantitative PCR, integrins have been shown to be expressed throughout all life stages in the coral Acropora millepora and the hydrozoan Podocoryne carnea and specifically upregulated in the endoderm during the invagination movements of gastrulation in the former [154,159]. Furthermore, integrin expression was maintained throughout the dynamic morphogenic processes associated with transdifferentiation and cell migration in P. carnea [154] but in N. vectensis, integrins seem to be specifically upregulated during regeneration [160]. Thus, studies of quantitative and spatial expression suggest that integrins play many diverse roles in the cell and tissue biology of cnidarians. Transduction of integrin-mediated signals into upregulated DNA synthesis was also demonstrated in the sponge S. domuncula [161], confirming that the inward transmission of signals from ECM to nucleus was functional before the origin of Porifera. Integrin signalling during development of ctenophores has not been studied but it is reasonable to hypothesize that intracellular signalling through integrins is likely operational in these animals as they possess the complete set of intracellular binding partners. Whether ctenophore integrins also play a role in facilitating embryonic morphogenesis or regeneration is not known. Studies explicitly examining the effect of perturbed integrin expression during ctenophore development are necessary to evaluate these ideas.
9. Commonalities in the evolution of novel proteins and novel pathways
Although changes in the spatial and temporal expression of existing genes are sufficient to induce novel expression patterns and signal new outcomes, the de novo evolution of new/novel molecules is also a critical driver of novel cell functions. Duplication and divergence may be the primary source of novel genes [162], but domain/exon shuffling is a well-established means for linking historically disparate functional domains into a single molecule [163]. Exon shuffling and gene fusion are two prominent ways in which the pathways described here have evolved. In the case of each pathway, functional domains necessary for protein–protein interactions arose long before the origin of the molecule itself. Indeed, the hedgehog pathway is an excellent example of this—hedge and hog domains are present in the lineages leading up to the Metazoa but the complete hedgehog gene did not arise until the stem lineage giving rise to Cnidaria + Bilateria [109]. Likewise, there is very good evidence for the piecewise assembly of genes in the JAK/STAT pathway [137], Notch/Delta pathway [100] and the evolution of the ECM [142]. Another striking pattern to be drawn from this summary is the timing of evolution of the components of each pathway. Figure 1 shows clearly that intracellular pathway components tend to arise before the membrane-bound and secreted components. Furthermore, it seems that the evolution of a ‘canonical pathway’ is largely coincident with the origin of the ligand, although evidence of individual pathway components before the evolution of a canonical pathway underscores the need for additional functional studies in diverse unikont systems. Together, these data support previous assertions that cell–cell communication evolved by co-option of existing intracellular pathways by novel extracellular and membrane-bound modulators [10,116].
10. Summary/outlook
To conclude, we return to Müller's [20] reconstruction of the ancestral metazoan but consider the new data gathered from ctenophores. Like Müller, we think it is reasonable to assume the ancestral metazoan had cell adhesion molecules, intracellular and intercellular signalling capabilities, and immune and nervous systems. In contrast, we suggest that the ability to form growth factor gradients evolved after ctenophores split from the rest of Metazoa. Perhaps more importantly, this review highlights the need for more explicit study of the ancestral components of canonical cell signalling pathways in ctenophores and other non-bilaterian taxa. Studies aimed at understanding complexity often start with a hypothesis derived from observations of bilaterians; thus, these studies focus on understanding when the parts of the bilaterian cell signalling systems first arose and when they were assembled into the canonical pathways represented by modern bilaterians. If, however, we are to understand how evolution produced these signalling pathways, we need explicit studies of the functions of pathway components in basal taxa before they acquired the role they play in bilaterians. Although challenging, these studies have the potential to reveal novel biochemical features of pathway components and the mechanisms by which gene functional domains evolve. These studies are essential for understanding the evolution of complex multi-component signalling pathways and the evolution of novel cell communication systems in general. Studying these interactions may well reveal biochemical properties of proteins that have gone overlooked as a result of our focus on bilaterian biology.
11. Methods
In cases where pathway components were not available in the literature, we used BLAST to search for homologues. Query sequences were collected from the human NR protein database (NCBI accession numbers provided in electronic supplementary material, file S1). To identify putative homologues in other taxa, we performed reciprocal BLAST searches against the NR protein databases (NCBI) for most of the taxa listed in tables 1–8. To identify homologues from M. leidyi, we queried human sequences against the Protein Models 2.2 database at NHGRI (https://research.nhgri.nih.gov/mnemiopsis/blast/), for P. bachei, we queried against the Unfiltered_Gene_Models database at NeuroBase (http://neurobase.rc.ufl.edu/pleurobrachia/blast?view=blast), and for O. carmela, we queried the OCAR_T-PEP_130911 database at Compagen (http://www.compagen.org/blast.html). Alignments were inspected by eye and putative homologues (with low support) are indicated separately from clear homologues in tables 1–8. In cases where our BLAST results conflicted with previous observations, the results of our BLAST searches are provided.
Supplementary Material
Acknowledgements
The authors thank Dr Joseph Ryan (University of Florida) for helpful discussions of ctenophore phylogenetics and BLAST analysis.
Authors' contributions
L.S.B. wrote the manuscript and performed BLAST searches. M.Q.M. evaluated and edited the manuscript for content. Both authors contributed to conceptualization of the study and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This work was supported by the University of Florida and by the National Aeronautics and Space Administration (NNX14AG70G).
Glossary
- Unikonta
the monophyletic group composed of amoebozoans, fungi, filastereans, choanoflagellates and metazoans.
- Opisthokonta
the monophyletic group composed of fungi, filastereans, choanoflagellates and metazoans.
- Holozoa
the monophyletic group composed of filastereans, choanoflagellates and metazoans.
- Metazoa
the monophyletic group composed of ctenophores, sponges, placozoans, cnidarians and bilaterians.
- Parahoxozoa
the monophyletic group composed of placozoans, cnidarians and bilaterians.
Gene abbreviations
- ADAM
a disintegrin and metalloproteinase domain
- ACTN1
α-actinin
- AGRN
agrin
- ALK/LTK
anaplastic lymphoma tyrosine kinase, leukocyte tyrosine kinase
- APC
adenomatous polyposis coli
- APH-1
anterior pharynx defective homolog 1, γ-secretase subunit
- AXL/UFO
AXL/unidentified function
- β-cat
β-catenin
- BMP
bone morphogenic protein
- CAN
cerberus/DAN/gremlin family
- CER
cerberus
- CHRD
chordin
- CKI
casein kinase 1
- ColαIV
type IV α-collagen
- COS
costal
- CSL
CBF1, suppressor of hairless, LAG-1
- Cyto
cytokine
- CytoR
cytokine receptor
- DAN/NBL1
neuroblastoma suppressor of tumorigenicity 1 isoform 1
- DDR
Discoidin domain receptor
- DAPT
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester; γ-secretase inhibitor
- DISP
dispatched receptor
- DIXDC
dixin domain containing
- DKK
dickkopft
- DPP
decapentaplegic, Drosophila homologue of BMP
- DSG
dystroglycan
- DSL
delta interaction domain
- DSH
disheveled
- DTX
deltex
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EGFR/ERBB
EGF receptor, erythroblastic leukemia viral oncogene
- EPHR
ephrin receptor
- FA
fatty acid
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- FN
fibronectin
- FST
follistatin
- FU
fused
- FZD
frizzled
- GDF
growth/development factor
- GF
growth factor
- GFR
growth factor receptor
- GLI
glioma-associated oncogene
- GREM
gremlin
- GSK-3
glycogen synthase kinase 3
- HES
hairy and enhancer of split
- HH
hedgehog
- HHIP
hedgehog interacting protein
- HNF4
hepatocyte nuclear factor 4
- HSPG2
heparin sulfate proteoglycan 2/perlecan
- INSR
insulin receptor
- ITG
integrin α/β
- JAK
janus kinase
- LAMA
laminin subunit α
- LAMB1
laminin subunit β
- LAMC
laminin subunit γ
- LRP
low density lipoprotein receptor-related protein
- MAM
mastermind
- MAPK
mitogen activated protein kinase
- MEK
MAPK/ERK kinase
- MET/HGFR
met proto-oncogene, hepatocyte growth factor receptor
- MIB
mindbomb
- MUSK
muscle-specific kinase
- NEURL
neuralized
- NFκβ
nuclear factor κ-β
- NLE
Notchless
- NLK
nemo-like kinase
- NOG
noggin
- NR
nuclear receptor
- NvSMAD
a SMAD homologue isolated from Nematostella vectensis
- O-fut
o-fucosyltransferase
- PCP
planar cell polarity
- PDGFR
platelet-derived growth factor receptor
- PEN2
presenilin enchancer 2, γ-secretase subunit
- PG
proteoglycan
- PIAS
protein inhibitor of activated STAT
- PKC
protein kinase C
- PKA
phosphokinase A
- PLC
phospholipase C
- PSEN
presenilin-1, γ-secretase subunit
- PTC
patched receptor
- PTKs
protein tyrosine kinases
- PTK7/CCK4
protein tyrosine kinase 7, colon carcinoma kinase 4
- PXN
paxillin
- Ras/Raf
proto-oncogene serine/threonine kinase
- RET
rearranged during transfection proto-oncogene
- ROR
receptor tyrosine kinase-like orphan receptor
- RTK
receptor tyrosine kinase
- RXR
retinoid X receptor
- RYK
related to receptor tyrosine kinase
- SARA/ZFYVE9
SMAD anchor for receptor activation
- SFRP
secreted frizzled receptor protein
- SH2
SRC homology 2 domain
- SHP
SH2 domain protein tyrosine phosphatase
- SKI/SNO
SKI proto-oncogene/SKI-related gene
- SMAD
mothers against decapentaplegic homologue
- SMO
smoothened receptor
- SMRT/NCOR2
nuclear receptor co-repressor
- SMURF
SMAD specific E3 ubiquitin protein ligase
- SOCS
suppressor of cytokine signalling
- SOS
SOS Ras/Rac guanine nucleotide exchange factor 1
- STAT
signal transducer and activator of transcription
- STK36
serine/threonine kinase 36 (homologue of fused)
- SuFU
suppressor of Fused
- TAD
C-terminal transactivation domain
- TCF/LEF
transcription factor/lymphoid enhancer binding factor
- TF
transcription factor
- TGF-β
transforming growth factor β
- TGFR
TGF-β receptor
- TIE
tyrosine kinase with immunoglobulin-like and EGF-like domains
- TLD
tolloid/BMP-1
- TLN
talin
- TRK
tropomyosin receptor kinase
- VCL
vinculin
- VEGF
vascular endothelial growth factor
- VEGFR
VEGF receptor
- VFT
venus fly trap domain
- VKR
venus kinase receptor
- WIF
WNT inhibitory factor
- WNT
wingless-type MMTV integration site
- ZPA
zone of polarizing activity
Species abbreviations
- Dd
Dictyostelium discoideum
- Sc
Saccharomyces cerevisiae
- Sp
Schizosaccharomyces pombe
- Co
Capsaspora owczarzaki
- Mb
Monosiga brevicollis
- Mo
Monosiga ovata
- Sr
Salpingoeca rosetta
- Ml
Mnemiopsis leidyi
- Pb
Pleurobrachia bachei
- Pp
Pleurobrachia pileus
- Aq
Amphimedon queenslandica
- Oc
Oscarella carmela
- Sd
Suberites domuncula
- Ta
Trichoplax adhaerens
- Nv
Nematostella vectensis
- Ac
Acropora cervicornis
- Ap
Acropora palmata
- Ad
Acropora digitifera
- Am
Acropora millepora
- Pd
Pocillopora damicornis
- Hm
Hydra magnipapillata
- Hv
Hydra vulgaris
- Pc
Podocoryne carnea
- Dm
Drosophila melanogaster
- Hs
Homo sapiens
References
- 1.Grosberg RK, Strathmann RR. 2007. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654. ( 10.1146/annurev.ecolsys.36.102403.114735) [DOI] [Google Scholar]
- 2.Abedin M, King N.. 2010. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 20, 734–742. ( 10.1016/j.tcb.2010.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ispolatov I, Ackermann M, Doebeli M.. 2012. Division of labour and the evolution of multicellularity. Proc. R. Soc. B 279, 1768–1776. ( 10.1098/rspb.2011.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray PS, Zaidel-Bar R. 2014. Pre-metazoan origins and evolution of the cadherin adhesome. Biol. Open 3, 1183–1195. ( 10.1242/bio.20149761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin M, et al. 2016. The mid-developmental transition and the evolution of animal body plans. Nature 531, 637–641. ( 10.1038/nature16994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter DJ, King N. 2013. The genomic and cellular foundations of animal origins. Annu. Rev. Genet. 47, 509–537. ( 10.1146/annurev-genet-111212-133456) [DOI] [PubMed] [Google Scholar]
- 7.Pires-daSilva A, Sommer RJ. 2003. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39–49. ( 10.1038/nrg977) [DOI] [PubMed] [Google Scholar]
- 8.Rokas A. 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu. Rev. Genet. 42, 235–251. ( 10.1146/annurev.genet.42.110807.091513) [DOI] [PubMed] [Google Scholar]
- 9.Richards GS, Degnan BM. 2009. The dawn of developmental signaling in the metazoa. Cold Spring Harb. Symp. Quant. Biol. 74, 81–90. ( 10.1101/sqb.2009.74.028) [DOI] [PubMed] [Google Scholar]
- 10.Srivastava M, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. ( 10.1038/nature09201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 ( 10.1126/science.1242592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270. ( 10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. ( 10.1038/nature13400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitel M, Guidi L, Hadrys H, Balsamo M, Schierwater B. 2011. New insights into placozoan sexual reproduction and development. PLoS ONE 6, e19639 ( 10.1371/journal.pone.0019639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava M, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. ( 10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- 16.Ringrose JH, van den Toorn HW, Eitel M, Post H, Neerincx P, Schierwater B, Altelaar AF, Heck AJ. 2013. Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat. Commun. 4, 1408 ( 10.1038/ncomms2424) [DOI] [PubMed] [Google Scholar]
- 17.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102–1120. ( 10.1093/molbev/msu057) [DOI] [PubMed] [Google Scholar]
- 18.Worheide G, Dohrmann M, Erpenbeck D, Larroux C, Maldonado M, Voigt O, Borchiellini C, Lavrov DV. 2012. Deep phylogeny and evolution of sponges (phylum Porifera). Adv. Mar. Biol. 61, 1–78. ( 10.1016/B978-0-12-387787-1.00007-6) [DOI] [PubMed] [Google Scholar]
- 19.Adamska M, Degnan BM, Green K, Zwafink C.. 2011. What sponges can tell us about the evolution of developmental processes. Zoology 114, 1–10. ( 10.1016/j.zool.2010.10.003) [DOI] [PubMed] [Google Scholar]
- 20.Müller WEG. 2001. Review: how was metazoan threshold crossed? The hypothetical Urmetazoa. Comp. Biochem. Physiol. A 129, 433–460. ( 10.1016/S1095-6433(00)00360-3) [DOI] [PubMed] [Google Scholar]
- 21.Massague J. 2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. ( 10.1038/nrm3434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss A, Attisano L.. 2013. The TGFbeta superfamily signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2, 47–63. ( 10.1002/wdev.86) [DOI] [PubMed] [Google Scholar]
- 23.Blitz IL, Cho KW. 2009. Finding partners: how BMPs select their targets. Dev. Dyn. 238, 1321–1331. ( 10.1002/dvdy.21984) [DOI] [PubMed] [Google Scholar]
- 24.King N, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788. ( 10.1038/nature06617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. 2009. Emergence, development and diversification of the TGF-beta signalling pathway within the animal kingdom. BMC Evol. Biol. 9, 28 ( 10.1186/1471-2148-9-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4, 2325 ( 10.1038/ncomms3325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Technau U, et al. 2005. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633–639. ( 10.1016/j.tig.2005.09.007) [DOI] [PubMed] [Google Scholar]
- 28.Samuel G, Miller D, Saint R.. 2001. Conservation of a DPP/BMP signaling pathway in the nonbilateral cnidarian Acropora millepora. Evol. Dev. 3, 241–250. ( 10.1046/j.1525-142x.2001.003004241.x) [DOI] [PubMed] [Google Scholar]
- 29.Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. 2000. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186–189. ( 10.1038/35025063) [DOI] [PubMed] [Google Scholar]
- 30.Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. 2007. Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE 2, e1031 ( 10.1371/journal.pone.0001031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang K, Ryan JF, Baxevanis AD, Martindale MQ. 2011. Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS ONE 6, e24152 ( 10.1371/journal.pone.0024152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols SA, Dirks W, Pearse JS, King N.. 2006. Early evolution of animal cell signaling and adhesion genes. Proc. Natl Acad. Sci. USA 103, 12 451–12 456. ( 10.1073/pnas.0604065103) [DOI] [Google Scholar]
- 33.Roch GJ, Sherwood NM. 2014. Glycoprotein hormones and their receptors emerged at the origin of metazoans. Genome Biol. Evol. 6, 1466–1479. ( 10.1093/gbe/evu118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. 2010. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl Acad. Sci. USA 107, 10 142–10 147. ( 10.1073/pnas.1002257107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairclough SR, et al. 2013. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14, R15 ( 10.1186/gb-2013-14-2-r15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leininger S, et al. 2014. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat. Commun. 5, 3905 ( 10.1038/ncomms4905) [DOI] [PubMed] [Google Scholar]
- 37.Müller WEG, Korzhev M, Le Pennec G, Müller IM, Schröder HC. 2003. Origin of metazoan stem cell system in sponges: first approach to establish the model (Suberites domuncula). Biomol. Eng. 20, 369–379. ( 10.1016/s1389-0344(03)00055-8) [DOI] [PubMed] [Google Scholar]
- 38.Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. 2004. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304, 1335–1337. ( 10.1126/science.1091946) [DOI] [PubMed] [Google Scholar]
- 39.Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. 2006. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc. Natl Acad. Sci. USA 103, 11 195–11 200. ( 10.1073/pnas.0601257103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorrentino GM, Gillis WQ, Oomen-Hajagos J, Thomsen GH. 2012. Conservation and evolutionary divergence in the activity of receptor-regulated sMADS. EvoDevo 3, 22 ( 10.1186/2041-9139-3-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matus DQ, Thomsen GH, Martindale MQ. 2006. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during Cnidarian gastrulation. Curr. Biol. 16, 499–505. ( 10.1016/j.cub.2006.01.052) [DOI] [PubMed] [Google Scholar]
- 42.Rentzsch F, Anton R, Saina M, Hammerschmidt M, Holstein TW, Technau U.. 2006. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev. Biol. 296, 375–387. ( 10.1016/j.ydbio.2006.06.003) [DOI] [PubMed] [Google Scholar]
- 43.Watanabe H, Schmidt HA, Kuhn A, Hoger SK, Kocagoz Y, Laumann-Lipp N, Ozbek S, Holstein TW. 2014. Nodal signalling determines biradial asymmetry in Hydra. Nature 515, 112–115. ( 10.1038/nature13666) [DOI] [PubMed] [Google Scholar]
- 44.Hayward DC, Grasso LC, Saint R, Miller DJ, Ball EE. 2015. The organizer in evolution—gastrulation and organizer gene expression highlight the importance of Brachyury during development of the coral, Acropora millepora. Dev. Biol. 399, 337–347. ( 10.1016/j.ydbio.2015.01.006) [DOI] [PubMed] [Google Scholar]
- 45.Hayward DC, Samuel G, Pontynen PC, Catmull J, Saint R, Miller DJ, Ball EE. 2002. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc. Natl Acad. Sci. USA 99, 8106–8111. ( 10.1073/pnas.112021499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemond EM, Kaluziak ST, Vollmer SV. 2014. The genetics of colony form and function in Caribbean Acropora corals. BMC Genomics 15, 1133 ( 10.1186/1471-2164-15-1133) [DOI] [PMC free article] [PubMed]
- 47.Reinhardt B, Broun M, Blitz IL, Bode HR. 2004. HyBMP5-8b, a BMP5-8 orthologue, acts during axial patterning and tentacle formation in hydra. Dev. Biol. 267, 43–59. ( 10.1016/j.ydbio.2003.10.031) [DOI] [PubMed] [Google Scholar]
- 48.Rentzsch F, Guder C, Vocke D, Hobmayer B, Holstein TW. 2007. An ancient chordin-like gene in organizer formation of Hydra. Proc. Natl Acad. Sci. USA 104, 3249–3254. ( 10.1073/pnas.0604501104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464, 592–596. ( 10.1038/nature08830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hobmayer B, Rentzsch F, Holstein TW. 2001. Identification and expression of HySmad1, a member of the R-Smad family of TGFβ signal transducers, in the diploblastic metazoan Hydra. Dev. Genes Evol. 211, 597–602. ( 10.1007/s00427-001-0198-8) [DOI] [PubMed] [Google Scholar]
- 51.Reber-Müller S, Streitwolf-Engel R, Yanze N, Schmid V, Stierwald M, Erb M, Seipel K.. 2006. BMP2/4 and BMP5-8 in jellyfish development and transdifferentiation. Int. J. Dev. Biol. 50, 377–384. ( 10.1387/ijdb.052085sr) [DOI] [PubMed] [Google Scholar]
- 52.Petersen HO, et al. 2015. A comprehensive transcriptomic and proteomic analysis of Hydra head regeneration. Mol. Biol. Evol. 32, 1928–1947. ( 10.1093/molbev/msv079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saina M, Genikhovich G, Renfer E, Technau U.. 2009. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl Acad. Sci. USA 106, 18 592–18 597. ( 10.1073/pnas.0900151106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genikhovich G, Fried P, Prunster MM, Schinko JB, Gilles AF, Fredman D, Meier K, Iber D, Technau U. 2015. Axis patterning by BMPs: cnidarian network reveals evolutionary constraints. Cell Rep. ( 10.1016/j.celrep.2015.02.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe H, Kuhn A, Fushiki M, Agata K, Ozbek S, Fujisawa T, Holstein TW. 2014. Sequential actions of β-catenin and Bmp pattern the oral nerve net in Nematostella vectensis. Nat. Commun. 5, 5536 ( 10.1038/ncomms6536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saina M, Technau U.. 2009. Characterization of myostatin/gdf8/11 in the starlet sea anemone Nematostella vectensis. J. Exp. Zool. B Mol. Dev. Evol. 312, 780–788. ( 10.1002/jez.b.21304) [DOI] [PubMed] [Google Scholar]
- 57.Detournay O, Schnitzler CE, Poole A, Weis VM. 2012. Regulation of cnidarian–dinoflagellate mutualisms: evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev. Comp. Immunol. 38, 525–537. ( 10.1016/j.dci.2012.08.008) [DOI] [PubMed] [Google Scholar]
- 58.Holstein TW. 2012. The evolution of the Wnt pathway. Cold Spring Harb. Perspect. Biol. 4, a007922 ( 10.1101/cshperspect.a007922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ. 2000. Adherens junctions and β-catenin-mediated cell signalling in a non-metazoan organism. Nature 408, 727–731. ( 10.1038/35047099) [DOI] [PubMed] [Google Scholar]
- 60.Prabhu Y, Eichinger L.. 2006. The Dictyostelium repertoire of seven transmembrane domain receptors. Eur. J. Cell Biol. 85, 937–946. ( 10.1016/j.ejcb.2006.04.003) [DOI] [PubMed] [Google Scholar]
- 61.Eichinger L, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57. ( 10.1038/nature03481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jager M, Dayraud C, Mialot A, Queinnec E, le Guyader H, Manuel M. 2013. Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neuro-sensory system in an adult ctenophore. PLoS ONE 8, e84363 ( 10.1371/journal.pone.0084363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersen CP, Reddien PW. 2009. Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068. ( 10.1016/j.cell.2009.11.035) [DOI] [PubMed] [Google Scholar]
- 64.Lapebie P, Gazave E, Ereskovsky A, Derelle R, Bezac C, Renard E, Houliston E, Borchiellini C. 2009. WNT/β-catenin signalling and epithelial patterning in the homoscleromorph sponge Oscarella. PLoS ONE 4, e5823 ( 10.1371/journal.pone.0005823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kusserow A, et al. 2005. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160. ( 10.1038/nature03158) [DOI] [PubMed] [Google Scholar]
- 66.Matus DQ, Magie CR, Pang K, Martindale MQ, Thomsen GH. 2008. The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev. Biol. 313, 501–518. ( 10.1016/j.ydbio.2007.09.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guder C, Philipp I, Lengfeld T, Watanabe H, Hobmayer B, Holstein TW. 2006. The Wnt code: cnidarians signal the way. Oncogene 25, 7450–7460. ( 10.1038/sj.onc.1210052) [DOI] [PubMed] [Google Scholar]
- 68.Rentzsch F, Hobmayer B, Holstein TW. 2005. Glycogen synthase kinase 3 has a proapoptotic function in Hydra gametogenesis. Dev. Biol. 278, 1–12. ( 10.1016/j.ydbio.2004.10.007) [DOI] [PubMed] [Google Scholar]
- 69.Minobe S, Fei KY, Yan L, Sarras MP, Werle MJ. 2000. Identification and characterization of the epithelial polarity receptor “Frizzled” in Hydra vulgaris. Dev. Genes Evol. 210, 258–262. ( 10.1007/s004270050312) [DOI] [PubMed] [Google Scholar]
- 70.Logan CY, Nusse R.. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell. Dev. Biol. 20, 781–810. ( 10.1146/annurev.cellbio.20.010403.113126) [DOI] [PubMed] [Google Scholar]
- 71.Schilde C, Araki T, Williams H, Harwood A, Williams JG. 2004. GSK3 is a multifunctional regulator of Dictyostelium development. Development 131, 4555–4565. ( 10.1242/dev.01330) [DOI] [PubMed] [Google Scholar]
- 72.Lengfeld T, et al. 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev. Biol. 330, 186–199. ( 10.1016/j.ydbio.2009.02.004) [DOI] [PubMed] [Google Scholar]
- 73.Pang K, Ryan JF, Program NCS, Mullikin JC, Baxevanis AD, Martindale MQ. 2010. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. EvoDevo 1, 10 ( 10.1186/2041-9139-1-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arkell RM, Fossat N, Tam PP. 2013. Wnt signalling in mouse gastrulation and anterior development: new players in the pathway and signal output. Curr. Opin. Genet. Dev. 23, 454–460. ( 10.1016/j.gde.2013.03.001) [DOI] [PubMed] [Google Scholar]
- 75.Martindale MQ. 2005. The evolution of metazoan axial properties. Nat. Rev. Genet. 6, 917–927. ( 10.1038/nrg1725) [DOI] [PubMed] [Google Scholar]
- 76.Martindale MQ, Lee PN. 2013. The development of form: causes and consequences of developmental reprogramming associated with rapid body plan evolution in the Bilaterian radiation. Biol. Theory 8, 253–264. ( 10.1007/s13752-013-0117-z) [DOI] [Google Scholar]
- 77.Lee PN, Kumburegama S, Martindale MQ, Wikramanayake AH. 2006. Disheveled is a localized maternal determinant mediating egg polarity and germ layer segregation in the cnidarian, Nematostella vectensis. Integr. Comp. Biol. 46, E221. [Google Scholar]
- 78.Kraus Y, Aman A, Technau U, Genikhovich G. 2016. Pre-bilaterian origin of the blastoporal axial organizer. Nat. Commun. 7, 11694 ( 10.1038/ncomms11694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Windsor PJ, Leys SP. 2010. Wnt signaling and induction in the sponge aquiferous system: evidence for an ancient origin of the organizer. Evol. Dev. 12, 484–493. ( 10.1111/j.1525-142X.2010.00434.x) [DOI] [PubMed] [Google Scholar]
- 80.Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM. 2010. Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol. Dev. 12, 494–518. ( 10.1111/j.1525-142X.2010.00435.x) [DOI] [PubMed] [Google Scholar]
- 81.Fischer AHL, Pang K, Henry JQ, Martindale MQ. 2014. A cleavage clock regulates features of lineage-specific differentiation in the development of a basal branching metazoan, the ctenophore Mnemiopsis leidyi. EvoDevo 5, 4 ( 10.1186/2041-9139-5-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schenkelaars Q, Fierro-Constain L, Renard E, Borchiellini C. 2016. Retracing the path of planar cell polarity. BMC Evol. Biol. 16, 69 ( 10.1186/s12862-016-0641-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grasso LC, Hayward DC, Trueman JW, Hardie KM, Janssens PA, Ball EE. 2001. The evolution of nuclear receptors: evidence from the coral Acropora. Mol. Phylogenet. Evol. 21, 93–102. ( 10.1006/mpev.2001.0994) [DOI] [PubMed] [Google Scholar]
- 84.Laudet V. 1997. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 19, 207–226. ( 10.1677/jme.0.0190207) [DOI] [PubMed] [Google Scholar]
- 85.Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier ME, Ortlund EA, Degnan BM, Thornton JW. 2010. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8, e1000497 ( 10.1371/journal.pbio.1000497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taubert S, Ward JD, Yamamoto KR. 2011. Nuclear hormone receptors in nematodes: evolution and function. Mol. Cell. Endocrinol. 334, 49–55. ( 10.1016/j.mce.2010.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reitzel AM, Tarrant AM. 2009. Nuclear receptor complement of the cnidarian Nematostella vectensis: phylogenetic relationships and developmental expression patterns. BMC Evol. Biol. 9, 230 ( 10.1186/1471-2148-9-230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baker ME. 2008. Trichoplax, the simplest known animal, contains an estrogen-related receptor but no estrogen receptor: implications for estrogen receptor evolution. Biochem. Biophys. Res. Commun. 375, 623–627. ( 10.1016/j.bbrc.2008.08.047) [DOI] [PubMed] [Google Scholar]
- 89.Wiens M. 2003. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J. Exp. Biol. 206, 3261–3271. ( 10.1242/jeb.00541) [DOI] [PubMed] [Google Scholar]
- 90.Larroux C, et al. 2006. Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol. Dev. 8, 150–173. ( 10.1111/j.1525-142X.2006.00086.x) [DOI] [PubMed] [Google Scholar]
- 91.Reitzel AM, Pang K, Ryan JF, Mullikin JC, Martindale MQ, Baxevanis AD, Tarrant AM. 2011. Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily? EvoDevo 2, 3 ( 10.1186/2041-9139-2-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Escriva H, Langlois MC, Mendonca RL, Pierce R, Laudet V.. 1998. Evolution and diversification of the nuclear receptor superfamily. Ann. N.Y. Acad. Sci. 839, 143–146. ( 10.1111/j.1749-6632.1998.tb10747.x) [DOI] [PubMed] [Google Scholar]
- 93.Rhinn M, Dolle P.. 2012. Retinoic acid signalling during development. Development 139, 843–858. ( 10.1242/dev.065938) [DOI] [PubMed] [Google Scholar]
- 94.Fuchs B, et al. 2014. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr. Biol. 24, 263–273. ( 10.1016/j.cub.2013.12.003) [DOI] [PubMed] [Google Scholar]
- 95.Kostrouch Z, Kostrouchova M, Love W, Jannini E, Piatigorsky J, Rall JE. 1998. Retinoic acid X receptor in the diploblast, Tripedalia cystophora. Proc. Natl Acad. Sci. USA 95, 13 442–13 447. ( 10.1073/pnas.95.23.13442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miljkovic-Licina M, Gauchat D, Galliot B.. 2004. Neuronal evolution: analysis of regulatory genes in a first-evolved nervous system, the hydra nervous system. Biosystems 76, 75–87. ( 10.1016/j.biosystems.2004.05.030) [DOI] [PubMed] [Google Scholar]
- 97.Duffy DJ, Frank U. 2011. Modulation of COUP-TF expression in a cnidarian by ectopic Wnt signalling and allorecognition. PLoS ONE 6, e19443 ( 10.1371/journal.pone.0019443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andersson ER, Sandberg R, Lendahl U.. 2011. Notch signaling: simplicity in design, versatility in function. Development 138, 3593–3612. ( 10.1242/dev.063610) [DOI] [PubMed] [Google Scholar]
- 99.Kasbauer T, Towb P, Alexandrova O, David CN, Dall'armi E, Staudigl A, Stiening B, Bottger A.. 2007. The Notch signaling pathway in the cnidarian Hydra. Dev. Biol. 303, 376–390. ( 10.1016/j.ydbio.2006.11.022) [DOI] [PubMed] [Google Scholar]
- 100.Gazave E, Lapebie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. 2009. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol. Biol. 9, 249 ( 10.1186/1471-2148-9-249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prevorovsky M, Grousl T, Stanurova J, Rynes J, Nellen W, Puta F, Folk P.. 2009. Cbf11 and Cbf12, the fission yeast CSL proteins, play opposing roles in cell adhesion and coordination of cell and nuclear division. Exp. Cell Res. 315, 1533–1547. ( 10.1016/j.yexcr.2008.12.001) [DOI] [PubMed] [Google Scholar]
- 102.Richards GS, Degnan BM. 2012. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. EvoDevo 3, 15 ( 10.1186/2041-9139-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rudel D, Sommer RJ. 2003. The evolution of developmental mechanisms. Dev. Biol. 264, 15–37. ( 10.1016/s0012-1606(03)00353-1) [DOI] [PubMed] [Google Scholar]
- 104.Marlow H, Roettinger E, Boekhout M, Martindale MQ. 2012. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev. Biol. 362, 295–308. ( 10.1016/j.ydbio.2011.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kelava I, Rentzsch F, Technau U. 2015. Evolution of eumetazoan nervous systems: insights from cnidarians. Phil. Trans. R. Soc. B 370, 20150065 ( 10.1098/rstb.2015.0065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Layden MJ, Martindale MQ. 2014. Non-canonical Notch signaling represents an ancestral mechanism to regulate neural differentiation. EvoDevo 5, 30 ( 10.1186/2041-9139-5-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. 2008. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 18, 1156–1161. ( 10.1016/j.cub.2008.06.074) [DOI] [PubMed] [Google Scholar]
- 108.Ingham PW, Nakano Y, Seger C.. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406. ( 10.1038/nrg2984) [DOI] [PubMed] [Google Scholar]
- 109.Snell EA, Brooke NM, Taylor WR, Casane D, Philippe H, Holland PW. 2006. An unusual choanoflagellate protein released by Hedgehog autocatalytic processing. Proc. R. Soc. B 273, 401–407. ( 10.1098/rspb.2005.3263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Layden MJ, Meyer NP, Pang K, Seaver EC, Martindale MQ. 2010. Expression and phylogenetic analysis of the zic gene family in the evolution and development of metazoans. EvoDevo 1, 12 ( 10.1186/2041-9139-1-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Burglin TR. 2008. The Hedgehog protein family. Genome Biol. 9, 241 ( 10.1186/gb-2008-9-11-241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Briscoe J, Therond PP. 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429. ( 10.1038/nrm3598) [DOI] [PubMed] [Google Scholar]
- 113.Lemmon MA, Schlessinger J.. 2010. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134. ( 10.1016/j.cell.2010.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suga H, Sasaki G, Kuma K, Nishiyori H, Hirose N, Su ZH, Iwabe N, Miyata T.. 2008. Ancient divergence of animal protein tyrosine kinase genes demonstrated by a gene family tree including choanoflagellate genes. FEBS Lett. 582, 815–818. ( 10.1016/j.febslet.2008.02.002) [DOI] [PubMed] [Google Scholar]
- 115.Manning G, Young SL, Miller WT, Zhai Y.. 2008. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc. Natl Acad. Sci. USA 105, 9674–9679. ( 10.1073/pnas.0801314105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suga H, Dacre M, de Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. 2012. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci. Signal. 5, ra35. ( 10.1126/scisignal.2002733) [DOI] [PubMed] [Google Scholar]
- 117.Tan JL, Spudich JA. 1990. Developmentally regulated protein-tyrosine kinase genes in Dictyostelium discoideum. Mol. Cell. Biol. 10, 3578–3583. ( 10.1128/MCB.10.7.3578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kortholt A, Kataria R, Keizer-Gunnink I, Van Egmond WN, Khanna A, Van Haastert PJ. 2011. Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 12, 1273–1279. ( 10.1038/embor.2011.210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwebs DJ, Hadwiger JA. 2015. The Dictyostelium MAPK ERK1 is phosphorylated in a secondary response to early developmental signaling. Cell Signal. 27, 147–155. ( 10.1016/j.cellsig.2014.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao X, Mehrabi R, Xu JR. 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6, 1701–1714. ( 10.1128/EC.00216-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao Z, Jin Q, Xu JR, Liu H. 2014. Identification of a fungi-specific lineage of protein kinases closely related to tyrosine kinases. PLoS ONE 9, e89813 ( 10.1371/journal.pone.0089813) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.King N, Carroll SB. 2001. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc. Natl Acad. Sci. USA 98, 15 032–15 037. ( 10.1073/pnas.261477698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Suga H, Katoh K, Miyata T.. 2001. Sponge homologs of vertebrate protein tyrosine kinases and frequent domain shufflings in the early evolution of animals before the parazoan–eumetazoan split. Gene 280, 195–201. ( 10.1016/S0378-1119(01)00784-3) [DOI] [PubMed] [Google Scholar]
- 124.D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jimenez-Delgado S, Bertrand S, Garcia-Fernandez J. 2008. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol. Biol. Evol. 25, 1841–1854. ( 10.1093/molbev/msn132) [DOI] [PubMed] [Google Scholar]
- 125.Reddy PC, Bidaye SS, Ghaskadbi S.. 2011. Genome-wide screening reveals the emergence and divergence of RTK homologues in basal Metazoan Hydra magnipapillata. J. Biosci. 36, 289–296. ( 10.1007/s12038-011-9065-6) [DOI] [PubMed] [Google Scholar]
- 126.Vanderstraete M, Gouignard N, Ahier A, Morel M, Vicogne J, Dissous C. 2013. The venus kinase receptor (VKR) family: structure and evolution. BMC Genomics 14, 361 ( 10.1186/1471-2164-14-361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller MA, Steele RE. 2000. Lemon encodes an unusual receptor protein-tyrosine kinase expressed during gametogenesis in Hydra. Dev. Biol. 224, 286–298. ( 10.1006/dbio.2000.9786) [DOI] [PubMed] [Google Scholar]
- 128.Bertrand S, Iwema T, Escriva H.. 2014. FGF signaling emerged concomitantly with the origin of Eumetazoans. Mol. Biol. Evol. 31, 310–318. ( 10.1093/molbev/mst222) [DOI] [PubMed] [Google Scholar]
- 129.Matus DQ, Thomsen GH, Martindale MQ. 2007. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev. Genes Evol. 217, 137–148. ( 10.1007/s00427-006-0122-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U.. 2008. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135, 1761–1769. ( 10.1242/dev.020784) [DOI] [PubMed] [Google Scholar]
- 131.Sinigaglia C, Busengdal H, Leclere L, Technau U, Rentzsch F. 2013. The bilaterian head patterning gene six3/6 controls aboral domain development in a cnidarian. PLoS Biol. 11, e1001488 ( 10.1371/journal.pbio.1001488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Krishnapati LS, Ghaskadbi S.. 2013. Identification and characterization of VEGF and FGF from Hydra. Int. J. Dev. Biol. 57, 897–906. ( 10.1387/ijdb.130077sg) [DOI] [PubMed] [Google Scholar]
- 133.Lange E, Bertrand S, Holz O, Rebscher N, Hassel M.. 2014. Dynamic expression of a Hydra FGF at boundaries and termini. Dev. Genes Evol. 224, 235–244. ( 10.1007/s00427-014-0480-1) [DOI] [PubMed] [Google Scholar]
- 134.Bertrand S, Somorjai I, Garcia-Fernandez J, Lamonerie T, Escriva H. 2009. FGFRL1 is a neglected putative actor of the FGF signalling pathway present in all major metazoan phyla. BMC Evol. Biol. 9, 226 ( 10.1186/1471-2148-9-226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li WX. 2008. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 18, 545–551. ( 10.1016/j.tcb.2008.08.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liongue C, O'Sullivan LA, Trengove MC, Ward AC. 2012. Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS ONE 7, e32777 ( 10.1371/journal.pone.0032777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liongue C, Ward AC. 2013. Evolution of the JAK-STAT pathway. JAKSTAT 2, e22756 ( 10.4161/jkst.22756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liongue C, Sertori R, Ward AC. 2016. Evolution of cytokine receptor signaling. J. Immunol. 197, 11–18. ( 10.4049/jimmunol.1600372) [DOI] [PubMed] [Google Scholar]
- 139.Wenger Y, Buzgariu W, Reiter S, Galliot B. 2014. Injury-induced immune responses in Hydra. Semin. Immunol. 26, 277–294. ( 10.1016/j.smim.2014.06.004) [DOI] [PubMed] [Google Scholar]
- 140.Sinkovics JG. 2015. The cnidarian origin of the proto-oncogenes NF-kappaB/STAT and WNT-like oncogenic pathway drives the ctenophores (Review). Int. J. Oncol. 47, 1211–1229. ( 10.3892/ijo.2015.3102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim SH, Turnbull J, Guimond S.. 2011. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 209, 139–151. ( 10.1530/JOE-10-0377) [DOI] [PubMed] [Google Scholar]
- 142.Hynes RO. 2012. The evolution of metazoan extracellular matrix. J. Cell Biol. 196, 671–679. ( 10.1083/jcb.201109041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ozbek S, Balasubramanian PG, Chiquet-Ehrismann R, Tucker RP, Adams JC. 2010. The evolution of extracellular matrix. Mol. Biol. Cell 21, 4300–4305. ( 10.1091/mbc.E10-03-0251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fahey B, Degnan BM. 2010. Origin of animal epithelia: insights from the sponge genome. Evol. Dev. 12, 601–617. ( 10.1111/j.1525-142X.2010.00445.x) [DOI] [PubMed] [Google Scholar]
- 145.Dickinson DJ, Nelson WJ, Weis WI. 2011. A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331, 1336–1339. ( 10.1126/science.1199633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dickinson DJ, Nelson WJ, Weis WI. 2012. An epithelial tissue in Dictyostelium challenges the traditional origin of metazoan multicellularity. Bioessays 34, 833–840. ( 10.1002/bies.201100187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Adams JC, Brancaccio A.. 2015. The evolution of the dystroglycan complex, a major mediator of muscle integrity. Biol. Open 4, 1163–1179. ( 10.1242/bio.012468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. 2011. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol. Biol. Evol. 28, 1241–1254. ( 10.1093/molbev/msq309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Adams JC. 2013. Extracellular matrix evolution: an overview. 1–25. ( 10.1007/978-3-642-36002-2_1) [DOI] [Google Scholar]
- 150.Exposito JY, Larroux C, Cluzel C, Valcourt U, Lethias C, Degnan BM. 2008. Demosponge and sea anemone fibrillar collagen diversity reveals the early emergence of A/C clades and the maintenance of the modular structure of type V/XI collagens from sponge to human. J. Biol. Chem. 283, 28 226–28 235. ( 10.1074/jbc.M804573200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Williams F, Tew HA, Paul CE, Adams JC. 2014. The predicted secretomes of Monosiga brevicollis and Capsaspora owczarzaki, close unicellular relatives of metazoans, reveal new insights into the evolution of the metazoan extracellular matrix. Matrix Biol. 37, 60–68. ( 10.1016/j.matbio.2014.02.002) [DOI] [PubMed] [Google Scholar]
- 152.Warren CR, Kassir E, Spurlin J, Martinez J, Putnam NH, Farach-Carson MC. 2015. Evolution of the perlecan/HSPG2 gene and its activation in regenerating Nematostella vectensis. PLoS ONE 10, e0124578 ( 10.1371/journal.pone.0124578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ereskovsky AV, Borchiellini C, Gazave E, Ivanisevic J, Lapebie P, Perez T, Renard E, Vacelet J.. 2009. The Homoscleromorph sponge Oscarella lobularis, a promising sponge model in evolutionary and developmental biology: model sponge Oscarella lobularis. Bioessays 31, 89–97. ( 10.1002/bies.080058) [DOI] [PubMed] [Google Scholar]
- 154.Reber-Müller S, Studer R, Müller P, Yanze N, Schmid V.. 2001. Integrin and talin in the jellyfish Podocoryne carnea. Cell Biol. Int. 25, 753–769. ( 10.1006/cbir.2000.0708) [DOI] [PubMed] [Google Scholar]
- 155.Sarras MP. 2012. Components, structure, biogenesis and function of the Hydra extracellular matrix in regeneration, pattern formation and cell differentiation. Int. J. Dev. Biol. 56, 567–576. ( 10.1387/ijdb.113445ms) [DOI] [PubMed] [Google Scholar]
- 156.Boute N, Exposito JY, BouryEsnault N, Vacelet J, Noro N, Miyazaki K, Yoshizato K, Garrone R.. 1996. Type IV collagen in sponges, the missing link in basement membrane ubiquity. Biol. Cell 88, 37–44. ( 10.1016/S0248-4900(97)86829-3) [DOI] [PubMed] [Google Scholar]
- 157.Hernandez-Nicaise ML. 1991. Ctenophora. In Microscopic anatomy of invertebrates, placozoa, porifera, cnidaria, and ctenophora (eds Harrison FW, Westfall JA), pp. 359–418. London, UK: Wiley. [Google Scholar]
- 158.Wang XM, Harris RE, Bayston LJ, Ashe HL, 2008. Type IV collagens regulate BMP signalling in Drosophila. Nature 455, 72–77. ( 10.1038/nature07214) [DOI] [PubMed] [Google Scholar]
- 159.Knack BA, Iguchi A, Shinzato C, Hayward DC, Ball EE, Miller DJ. 2008. Unexpected diversity of cnidarian integrins: expression during coral gastrulation. BMC Evol. Biol. 8, 136 ( 10.1186/1471-2148-8-136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gong QZ, Garvey K, Qian CH, Yin I, Wong G, Tucker RP. 2014. Integrins of the starlet sea anemone Nematostella vectensis. Biol. Bull. 227, 211–220. ( 10.1086/BBLv227n3p211) [DOI] [PubMed] [Google Scholar]
- 161.Wimmer W, Perovic S, Kruse M, Schroder HC, Krasko A, Batel R, Müller WEG. 1999. Origin of the integrin-mediated signal transduction - functional studies with cell cultures from the sponge Suberites domuncula. Eur. J. Biochem. 260, 156–165. ( 10.1046/j.1432-1327.1999.00146.x) [DOI] [PubMed] [Google Scholar]
- 162.Ding Y, Zhou Q, Wang W.. 2012. Origins of new genes and evolution of their novel functions. Annu. Rev. Ecol. Evol. Syst. 43, 345–363. ( 10.1146/annurev-ecolsys-110411-160513) [DOI] [Google Scholar]
- 163.Cheatle Jarvela AM, Hinman VF. 2015. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. EvoDevo 6, 3 ( 10.1186/2041-9139-6-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.