Abstract
A salient feature of flowering plant diversification is the emergence of a novel suite of floral features coinciding with the origin of the most species-rich lineage, Pentapetalae. Advances in phylogenetics, developmental genetics and genomics, including new analyses presented here, are helping to reconstruct the specific evolutionary steps involved in the evolution of this clade. The enormous floral diversity among Pentapetalae appears to be built on a highly conserved ground plan of five-parted (pentamerous) flowers with whorled phyllotaxis. By contrast, lability in the number and arrangement of component parts of the flower characterize the early-diverging eudicot lineages subtending Pentapetalae. The diversification of Pentapetalae also coincides closely with ancient hexaploidy, referred to as the gamma whole-genome triplication, for which the phylogenetic timing, mechanistic details and molecular evolutionary consequences are as yet not fully resolved. Transcription factors regulating floral development often persist in duplicate or triplicate in gamma-derived genomes, and both individual genes and whole transcriptional programmes exhibit a shift from broadly overlapping to tightly defined expression domains in Pentapetalae flowers. Investigations of these changes associated with the origin of Pentapetalae can lead to a more comprehensive understanding of what is arguably one of the most important evolutionary diversification events within terrestrial plants.
This article is part of the themed issue ‘Evo-devo in the genomics era, and the origins of morphological diversity’.
Keywords: floral evolution, eudicots, gamma palaeohexaploidy, Gunneridae, Pentapetalae
1. Introduction
The flowering plants (angiosperms) constitute the largest and most diverse extant group of the plant kingdom. Approximately 350 000 species of flowering plants, classified in 416 families and 14 559 genera, have been recorded to date, accounting for nearly 90% of all known land plant species [1,2]. The angiosperms are also the youngest of the major green plant lineages, having arisen and radiated long after plants colonized the terrestrial habitat about 500–470 million years ago (Ma) during the Ordovician ([3]; see also Harrison [4]). A diverse assortment of angiosperms appears abruptly in the fossil record of the Early Cretaceous, starting approximately 125 Ma [5], and representatives of all major extant flowering plant lineages can be recognized in Mid-Cretaceous deposits, about 100 Ma [6]. Molecular-based estimates suggest a somewhat older origin of angiosperms, ranging from 180 to 140 Ma, but, consistent with the fossil record, they support a rapid radiation occurring 5–10 Myr after the evolution of the flowering plant lineage [7–9]. The precipitous origin and rapid diversification of flowering plants was famously referred to as an ‘abominable mystery’ by Charles Darwin because their rapid appearance contradicted his gradualist view of evolutionary change [10]. The ‘mystery’ has duly received considerable attention from developmental and evolutionary plant biologists, with the two major fields of enquiry providing plausible solutions.
The most conspicuous key evolutionary innovation of angiosperms is the flower itself, and breakthroughs in floral developmental genetics provided new impetus for studies of floral evolution and development—floral evo-devo—from which have emerged numerous new hypotheses [11,12]. Among these are novel ideas about how flowers evolved from transformed gymnosperm cones [13–16], an ancestral ‘fading borders’ model for flower development [17–20] and floral diversification through ‘sliding boundaries’ of organ identity functions [21–24]. The prototypical flower is composed of four types of organs arranged such that carpels (the female reproductive organs, collectively the ‘gynoecium’) are innermost and surrounded by stamens (the male reproductive organs, collectively ‘androecium’) which are, in turn, surrounded by petals (usually colourful, collectively ‘corolla’) and then sepals (leaf-like, collectively ‘calyx’). The corolla and calyx collectively constitute the perianth. Variations in the number and arrangement of these four primary floral organs account for much of floral diversity, and can now be understood in the context of genetic specification of floral organ identity [25–27] and floral symmetry [28]. Floral evo-devo studies also offer explanations for the origins of stamens and carpels from gymnosperm precursors via ‘mostly male’, ‘out-of-male’ and ‘out-of-female’ mechanisms [14–16], and the origins of petals from stamens (andropetals) or bracts (bracteopetals) during the course of angiosperm diversification [29,30].
Complementing these developments in floral evo-devo, analyses of the burgeoning collection of flowering plant genome sequences have suggested a role for whole-genome duplications (WGDs; i.e. polyploidy) in the origin and subsequent diversification of flowering plants [31–37]. For example, an ancient polyploidy event has been inferred for the common ancestor of all angiosperms [38,39], three sequential polyploidy events in the monocots pre-date the radiation of the grasses [40,41] and ancient hexaploidy characterizes most eudicots [42–45]. Additional WGDs have been identified among many relatively younger branches of the flowering plant evolutionary tree, mostly among the eudicots [46], many of which coincide closely with the Cretaceous/Tertiary (K/T) boundary about 65 Ma [32]. Moreover, genes involved in signalling and transcriptional regulation tend to be preferentially retained in duplicate following WGD, expanding the repertoire of genetic tools with which evolutionary novelties may be constructed [47–50]. Thus, WGDs and their impact on genes directing floral development and other processes may have been especially important factors in the evolution and diversification of angiosperms [51]. By contrast, WGD may have been less important than tandem gene duplication during animal evolution (see Holland et al. [52]).
Here, we review the current understanding of the evolutionary context from which the most diverse group of extant flowering plants, Pentapetalae, emerged. We emphasize the diverse contributions of phylogenetics, genetics and genomics to understanding key evolutionary changes associated with the Pentapetalae radiation and relate new analyses to unresolved questions surrounding enigmatic WGD events that pre-date their origin.
2. Angiosperm phylogeny: emergence and radiation of the Pentapetalae
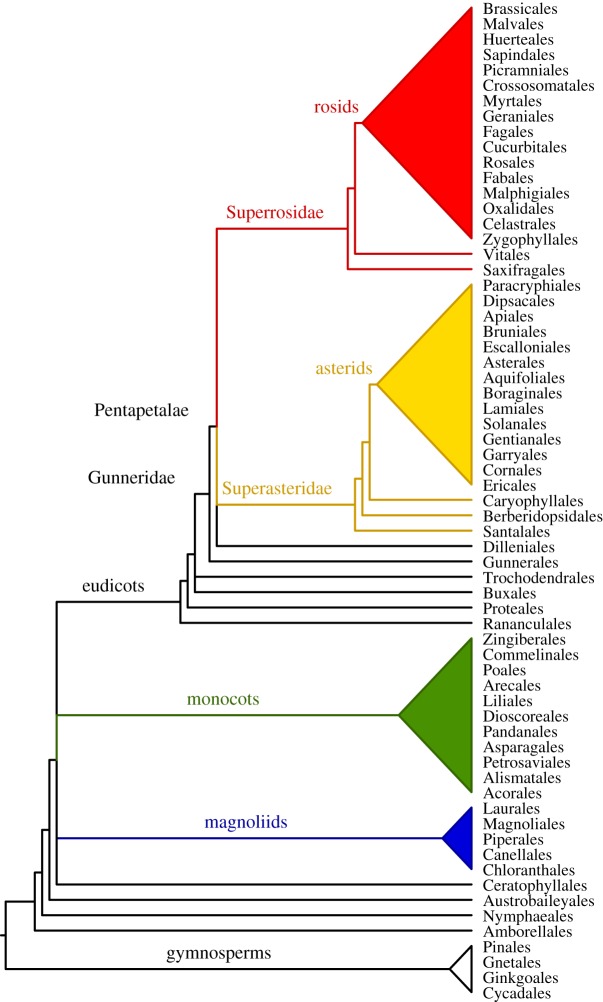
Improved understanding of the relationships among flowering plant lineages has provided an ever-expanding framework for hypothesis testing. Amborella, Nymphaeales (water lilies) and Austrobaileyales are successive sisters to the remaining angiosperms (Mesangiospermae), which comprise three major lineages: magnoliids, monocots and eudicots [53] (figure 1). The eudicots are the largest of the extant angiosperm clades accommodating approximately 75% of angiosperm diversity [2]. The eudicot clade arose early in angiosperm evolution, perhaps within approximately 10 Myr of the initial angiosperm radiation [8,9], and is well supported by biochemical (e.g. production of ellagic and gallic acids), morphological (tricolpate or tricolpate-derived pollen) and a wealth of DNA sequence data [53–56]. Inferences of relationships among the eudicots [2,53,55,57] have so far been derived largely from chloroplast molecular sequence data [53,55,57], but given the rapid diversification of eudicots [8,9,58], as well as multiple WGD events [37], these maternally inherited markers may be revealing only a partial glimpse into the evolutionary history of the clade. Current chloroplast-based estimates indicate with strong support that Ranunculales are sister to all other eudicots; Proteales (including Sabiaceae) diverge next; either Trochodendrales or Buxales are successive sister lineages to the rest, although their relative positions remain uncertain; and Gunnerales are sister to a large clade that has been formally named Pentapetalae [53,59]. Pentapetalae alone accommodates about 70% of extant angiosperm species, and together with Gunnerales constitute the Gunneridae (core eudicots). Thus, the eudicots appear to be represented by relatively species-poor lineages (Ranunculales, Proteales, Trochodendrales, Buxales and Gunnerales) that form a basal grade subtending the Pentapetalae, the largest and most diverse group of extant angiosperms.
Figure 1.
Phylogenetic relationships among the main lineages of flowering plants and their sister group, the extant gymnosperms, based on nuclear, mitochondrial and chloroplast DNA sequence data. Amborellales, Nymphaeales and Austrobaileyales form a basal grade below all other flowering plants (Mesangiospermae). Relationships among the three major clades of Mesangiospermae (magnoliids, monocots and eudicots) and Ceratophyllales are currently unresolved. Among eudicots, Ranunculales diverge first, followed by Proteales before a trichotomy comprising Buxales, Trochodendrales and core eudicots. Among core eudicots, Gunnerales are sister to Pentapetalae, which comprise the large Superrosidae and Superasteridae clades and Dilleniales in an unresolved a trichotomy. Superrosidae includes Saxifragales plus Rosidae (rosids), while Superasteridae comprises a basal grade of Santalales, Berberidopsidales and Caryophyllales subtending the large Asteridae (asterids) clade.
Pentapetalae comprises two major clades, formally named Superrosidae (superrosids) and Superasteridae (superasterids), each accommodating approximately one-third of extant angiosperm species [53,60]. Superrosidae and Superasteridae each include a major subclade that corresponds well with morphology-based classifications (e.g. [61,62]), Rosidae (rosids) and Asteridae (asterids), respectively (figure 1). Readily observable floral features that generally distinguish rosids from asterids (figure 2a–c) include: (i) petals free versus fused and (ii) stamens in two whorls and not fused with petals versus a single whorl of stamens fused with petals. The fusion of petals (sympetaly) into a tubular corolla in most asterids has been recognized as a morphological innovation for centuries [63,64]. Other floral features have evolved repeatedly among rosids and asterids, but tend to be more frequent in one or the other. For example, flowers of rosids tend to be small and simply constructed with radial symmetry, while asterid flowers are often elaborate and complex with bilateral symmetry [65].
Figure 2.
Representatives of eudicot floral diversity. (a) Saxifraga rotundifolia (Saxifragales) displays the five-part flowers typical of the Superrosidae clade of Pentapetalae. (b) The five petals are fused in Petunia sp. (Solanales), as is characteristic of the Asteridae clade of Pentapetalae. (c) Antirrhinum majus (snapdragon; Lamiales), a model species for floral symmetry developmental genetics, has zygomorphic flowers in which dorsal, lateral and ventral petal lobes emerge from the corolla tube. (d) Gunnera (Gunnerales) flowers are minute and densely packed in spicate inflorescences. (e) Trochodendron aralioides (Trochodendrales) flowers are polymerous with numerous stamens and carpels. (f) Inflorescences of Pachysandra procumbens (Buxales) bearing dimerous flowers, each with two pairs of stamens. (g) Grevillea sericea (Proteales) flowers displaying four sepal lobes (dimerous) to which stamens are fused, and an elongated pistil. (h) Nelumbo nucifera (Proteales) flowers are polymerous with numerous petals, stamens and carpels. (i) Eschscholzia californica (Ranunculales) flowers are dimerous with two pairs of decussate petals. Photo credits: (a) ‘Saxifraga rotundifolia’ (CC BY-NC 2.0) by cetp; (b) ‘Petunia’ (CC BY-NC-ND 2.0) by Ava Babili; (c) ‘Antirrhinum majus ‘ (CC BY-NC-SA 2.0) by francesco_43; (d) ‘Gunnera’ (CC BY-NC-ND 2.0) by allisoncake; (e) ‘Trochodendron aralioides ‘ (CC BY-NC-SA 2.0) by dogtooth77; (f) ‘Pachysandra procumbens (Allegheny spurge)’ (CC BY-NC-SA 2.0) by tgpotterfield; (g) ‘Grevillea sericea’ (CC BY-NC-SA 2.0) by Marine Explorer; (h) ‘sacred lotus’ (CC BY-NC-SA 2.0) by faria!; (i) ‘Eschscholzia’ (CC BY-NC 2.0) by Nickiz77.
3. Floral roots of Pentapetalae
The most striking feature of Pentapetalae, reflected in the name of the clade, is the transition to a highly conserved, canonical floral ground plan consisting of: (i) whorled arrangement of organs (whorled phyllotaxis); (ii) a fixed merosity or merism (number of organs per whorl); (iii) an ancestrally five-parted (pentamerous) calyx, corolla and androecium (with transitions to four-parted (tetramery) and other merosities); (iv) alternation of organs in adjacent whorls and (v) a single whorl each of sepals and petals [59,65–67]. This canonical floral ground plan (figure 2a) represents a marked departure from the variable arrangement, merosity and morphology of floral organs in early-diverging eudicot lineages [67–69]. Although some of these characters are occasionally found outside Pentapetalae, all five together are a hallmark of this clade. The distribution of floral features among extant basal eudicots suggests that this suite of characters was established along the immediate stem lineage of Pentapetalae, after their divergence from Gunnerales [66,70].
Gunnerales produce dimerous flowers, with two of each type of floral organ (i.e. two sepals, two petals, two stamens and two carpels), all of which may be greatly reduced (figure 2d), perhaps reflecting a shift to wind pollination in this group [70,71]. Flowers of Tetracentron (Trochodendrales) are dimerous and may show intrafloral switches in merosity, changing from dimerous perianth and androecium to tetramerous gynoecium [72,73], and those of Trochodendron (also of Trochodendrales) are polymerous (figure 2e) [65]. The flowers of Buxales (figure 2f) are predominantly dimerous, but with shifts in merosity involving the inner organs of flowers (e.g. tetramerous androecia and trimerous gynoecia are common), and shifts in phyllotaxis that correlate with flower sex (i.e. female flowers are spiral, while male flowers are whorled) [74]. In Proteales, flowers of Proteaceae are dimerous (figure 2g), those of Platanaceae exhibit a shift to trimery and tetramery, those of Nelumbonaceae are polymerous, with a greatly expanded and variable number of floral organs (figure 2h) and those of Sabiaceae represent another independent derivation of pentamery but with spiral phyllotaxis [65,66,75,76]. Among Ranunculales, the sister lineage of all other eudicots, dimerous, trimerous and pentamerous flowers all occur within Ranunculaceae [24,30,66,77]. As in Sabiaceae, and unlike in Pentapetalae, pentamery in Ranunculaceae is coupled with spiral (rather than whorled) organ initiation, and therefore represents another independent derivation of this kind of flower organization [66,77,78] Thus, the non-Pentapetalae eudicots are predominantly dimerous with a few notable exceptions.
4. Genetic origin of the Pentapetalae flower
The whorled pentamerous flower established in Pentapetalae is potentially a key innovation contributing to the success of the clade [79], but the genetic basis of these traits remains unclear. Of the several transcription factor families that are known to play a role in flower development and morphogenesis (e.g. MADS, TCP, MYB, CUC and YABBY), none is a smoking gun for the transition to a whorled pentamerous flower, although they all have potential functions that may have contributed. Together, these gene families pattern the development of morphological traits, such as organ identity, symmetry, fusion, polarity, elongation and growth.
Phylogenetic analyses suggest that all of the MADS genes that regulate floral organ identity experienced either one or two duplication events prior to the radiation of Pentapetalae [80–83]. As a result, Pentapetalae lineages maintain either two or three paralogous forms of each of these genes. Along with the increase in number of MADS genes in Pentapetalae, the spatial expression of these genes shifted from the broadly overlapping ‘fading borders’ pattern of basal angiosperms to sharply restricted expression domains [18,84–86]. Similar evolutionary changes are reported in comparisons of whole transcriptional programmes in floral organs [20,85,87,88]. Much progress has been made in our understanding of floral developmental genetics in Ranunculales [78,89–94], but the basal eudicot grade has not been representatively studied to date. The available data suggest that genetic programmes for floral organ identity are often more broadly deployed in the flowers of non-Pentapetalae angiosperms than in the flowers of Pentapetalae.
Similar to patterns seen in floral MADS-box genes, increased numbers of floral symmetry genes are also associated with the origin of Pentapetalae. Most Pentapetalae flowers are oriented such that there is a single ventral or abaxial petal, two lateral petals and two dorsal or adaxial petals [95]. In radially symmetrical groups, the five petals are identical in form and equidistant from each other, but there have been frequent transitions to bilateral symmetry in which the petals act as three separate modules (dorsal, lateral and ventral). Floral symmetry genes appear to function in these three modules of the flower independently to produce complex petal arrangements—a phenomenon with multiple, independent derivations [96,97]. The primary genetic regulators of floral symmetry are the CYCLOIDEA (CYC) TCP domain transcription factors and the MYB domain transcription factors DIVARICATA (DIV) and RADIALIS (RAD) [28,96]. Phylogenetic analyses suggest that the CYC, DIV and RAD genes expanded into two or three paralogous lineages prior to the origin of Pentapetalae [98–100] and the three CYC clades may have been established through duplications between the divergence of Proteales and the diversification of Gunneridae [101].
A recurrent feature of MADS and TCP genes is poor phylogenetic resolution among triplicated clades that emerged near the origin of Gunneridae. MADS gene trees all show a polytomy below three core eudicot-wide clades [83], as does the gene tree for TCP genes [101]. This lack of phylogenetic resolution may be due to, in part, or in combination with, several factors, including the nature of the duplication event or events (see §5), the rapid speciation of the eudicots and differential gene evolution [101].
5. Origin of the Pentapetalae genome
Arabidopsis thaliana (Brassicaceae; Brassicales), which is the premier genetic model for plant developmental genetics, and Vitis vinifera (grapevine; Vitaceae; Vitales) have been instrumental in shaping our understanding of genome evolution in Pentapetalae. Early examinations of the Arabidopsis genome revealed three WGD events in its evolutionary history, termed alpha (α), beta (β) and gamma (γ) [102,103]. Subsequent analyses of the Vitis genome sequence revealed three large syntenic gene blocks, representing three ancestral genomes brought together in an anciently hexaploid genome (palaeohexaploidy). Importantly, each of the three Vitis syntenic blocks corresponds to four separate regions in the Arabidopsis genome, suggesting that the two WGD events in the Arabidopsis lineage represent the alpha and beta WGDs, while the shared palaeohexaploidy is the gamma event [42]. Each of the Vitis triplicate regions corresponds to two genomic regions in Populus trichocarpa (poplar) [104], reflecting shared palaeohexaploidy followed by a single additional WGD in the poplar lineage. A one-to-one correspondence between Vitis and Carica papaya syntenic regions also indicated shared palaeohexaploidy, but without further WGDs in Carica [105]. All four of these species belong to the rosid subclade of Pentapetalae, but comparisons involving Solanum lycopersicum, Utricularia gibba, Mimulus guttatus and Coffea canephora indicate that the palaeohexaploidy event is shared with these species of the asterid clade and therefore pre-dates the radiation of Pentapetalae [43,106–109]. Notably, like Carica and Vitis, the Coffea genome has not experienced post-gamma WGDs, and as such there exists a 1 : 1 : 1 correspondence between Vitis–Carica–Coffea syntenic regions [109], underscoring their shared palaeohexaploidy.
The triplicate structure of gamma-derived genomes is particularly well preserved in Vitis [42], facilitating intragenomic analyses that explore the historical nature of this hexaploidy. Importantly, two of the three Vitis subgenomes are more fractionated with respect to one another than to the third, suggesting they co-existed in the same nucleus and experienced differential gene loss for a longer period [110,111]. These observations support a ‘two-step’ model for gamma in which the ancestral palaeohexaploid was formed via a tetraploid intermediate in which fractionation was well advanced by the time the third subgenome was added through a wide cross [108,110,111]. Similar fractionation patterns have been found in Brassica rapa, S. lycopersicum and Capsicum annuum, supporting two-step hexaploidies in Brassica and Solanaceae [109,112]. A two-step process for the gamma hexaploidy is also supported by our understanding of the polyploidization process: unreduced gamete formation results in diploid gametes, not triploid ones, and a hexaploid is formed via crossing between a diploid and a tetraploid and further duplication. Thus, hexaploidy is derived via two successive WGDs as in the formation of bread wheat (Triticum aestivum) through a cross between still extant tetraploid and diploid species approximately 8000 years ago [113,114]. However, unlike hexaploid bread wheat, the antiquity of the hypothesized Brassica, Solanaceae and gamma palaeohexaploidies hinders empirical assessment of the two-step hypothesis, and alternative epigenetic modifications could also account for the observed fractionation patterns [112].
Efforts to elucidate the gamma event further have used synteny-based analyses to determine the origin of gamma-derived genomes, and in the absence of genomic data, phylogenomic analyses have been used to estimate the origins of gamma-derived paralogues. It has been established that gamma is absent in Amborella [39], monocots [42], magnoliids, Ranunculales [44] and Proteales [44,45], effectively narrowing the possibilities to the distal branches of the basal eudicots, possibly just prior to the origin of the Gunneridae [83]. The only study implementing both synteny and phylogenomic analyses for a basal eudicot indicated that the triplicate genome structure of gamma does not exist in Nelumbo nucifera (sacred lotus; Proteales), but, surprisingly, approximately 50% of the gene trees support clades that include Nelumbo genes and gamma-derived Vitis paralogues [45]. Close relationships between putative gamma-derived paralogues and basal eudicot genes were found in earlier phylogenomic studies [44,83], but their significance was not explored.
The apparent conflict between synteny and phylogenomics was seen as potentially consistent with the two-step model for gamma palaeohexaploidy [45]. The lack of a gamma-like structure in the Nelumbo genome coupled with phylogenetic grouping of many Nelumbo genes with gamma paralogues could be explained if (i) the initial tetraploidy event in the two-step model post-dates the divergence of Proteales from other eudicots but pre-dates the diversification of Pentapetalae, and (ii) the donor of the third genome to the tetraploid intermediate is a direct, or even older, ancestor of extant Proteales [45]. This speculative two-step scenario was not supported by synteny-based genome halving analyses of the Nelumbo and Vitis genomes [115], which suggested, instead, that the gamma palaeohexaploidy should be placed after the divergence of Nelumbo from other eudicots, somewhere along the stem lineage leading to Pentapetalae. Therefore, to accommodate a two-step model, a more closely related third genome donor than that of a Nelumbo ancestor must be postulated [115]. The three lineages that occupy branches between the divergence of Nelumbo and the radiation of Pentapetalae (i.e. Trochodendrales, Buxales and Gunnerales) are, therefore, pivotal to understanding gamma palaeohexaploidy, but whole-genome sequences for these taxa are not currently available.
6. Towards an elucidation of the gamma event(s)
Previous studies implementing a phylogenomic approach have relied on clustering algorithms, such as OrthoMCL [116], to circumscribe narrowly defined gene families, or orthogroups, the duplication histories of which can be reconstructed phylogenetically [44,45,83]. Such orthogroups ideally define sets of genes descended from a single ancestral gene in the common ancestor of the taxa under consideration [117], but they can be circumscribed more broadly or narrowly depending on the taxon sampling and algorithm settings employed [114]. Therefore, whether putative sets of gamma-derived paralogues are assigned to the same orthogroup, as is necessary for phylogenomic analyses, is a matter of concern that has to be addressed post hoc. For example, only 123 of approximately 1800 gene trees analysed by Vekemans et al. [83] include putative gamma-derived Vitis paralogues, and Jiao et al. [44] combined orthogroups that would have otherwise kept such paralogues separate. Alternatively, in an approach that has not been attempted to date, the synteny-based orthogroups circumscribed for Vitis [43] may be used as a reference to which genes from other species can be assigned, facilitating phylogenomics within the prescribed context of putative gamma-derived paralogues.
Here, we illustrate the use of both the synteny-based orthogroups of Tang et al. [43] and the cluster-based orthogroups circumscribed by OrthoFinder [117] in our own phylogenomic analyses. We include genes from Amborella, two monocots (Oryza and Sorghum), two magnoliids (Liriodendron and Persea) and eudicots representing Ranunculales, Proteales, Trochodendrales, Buxales, Gunnerales and Pentapetalae (table 1). For each orthogroup, protein alignments were constructed using MAFFT [124], converted into nucleotide alignments using PAL2NAL [125], and trimmed by eliminating spurious sequences and alignment positions using trimAl [126]. The resulting orthogroups were then screened for the presence of Amborella (the designated outgroup), Ranunculales, Proteales, duplicate Vitis genes, and at least Buxales, Trochodendrales or Gunnerales using custom Perl scripts. Orthogroups passing these filtering steps were used to construct gene trees with bootstrap support (BS) values (100 replicates) using RAxML [127]. In order to use the resulting trees to trace duplication events as in a previously described pipeline [41], a phylogenetic tree for the included species is required. Given uncertainty of relationships for critically important Buxales and Trochodendrales, we used the MarkerMiner pipeline [128] to construct phylogenetic data-sets based on single-copy nuclear (SCN) loci. Individual datasets were analysed using RAxML as described above to generate species trees using the ASTRAL coalescent approach [129] as well as a supermatrix of the SCN loci (produced using FASconCAT [130]).
Table 1.
Source of datasets for the 30 species included in this study. 1KP, 1000 Green Plant Transcriptome Project [118]; AAGP, Ancestral Angiosperm Genome Project [119], Lotus-DB [120,121], Phytozome [122,123].
| species | lineage | source | no. genes/unigenes |
|---|---|---|---|
| Akebia trifoliata | Ranunculales | 1KPa | 20 366 |
| Amborella trichopoda | basal angiosperm | Amborella genome projectb | 26 846 |
| Aquilegia coerulea | Ranunculales | Phytozomeb | 24 823 |
| Arabidopsis thaliana | Rosidae | Phytozomeb | 27 416 |
| Buxus sempervirens | Buxales | 1KPa | 20 186 |
| Carica papaya | Rosidae | Phytozomeb | 27 751 |
| Citrus sinensis | Rosidae | Phytozomeb | 25 379 |
| Eschscholzia californica | Ranunculales | 1KPa | 26 317 |
| Euptelea pleiosperma | Ranunculales | 1KPa | 21 659 |
| Glycine max | Rosidae | Phytozomeb | 56 044 |
| Grevillea robusta | Proteales | 1KPa | 16 728 |
| Gunnera manicata | core eudicot | 1KPa | 16 606 |
| Kalanchoe marnieriana | Superrosidae | Phytozomeb | 50 461 |
| Liriodendron tulipifera | magnoliid | AAGPa | 12 067 |
| Meliosma cuneifolia | Proteales | 1KPa | 17 784 |
| Meliosma dillenifolia | Proteales | this studya | 33 175 |
| Mimulus guttatus | Asteridae | Phytozomeb | 28 140 |
| Nandina domestica | Ranunculales | 1KPa | 17 453 |
| Nelumbo nucifera | Proteales | Lotus-DBb | 26 685 |
| Oryza sativa | monocot | Phytozomeb | 39 049 |
| Papaver rhoeas | Ranunculales | 1KPa | 32 741 |
| Papaver somniferum | Ranunculales | 1KPa | 32 169 |
| Persea americana | magnoliid | AAGPa | 19 335 |
| Platanus occidentalis | Proteales | 1KPa | 22 347 |
| Populus trichocarpa | Rosidae | Phytozomeb | 41 335 |
| Sanguinaria canadensis | Ranunculales | 1KPa | 18 993 |
| Solanum lycopersicum | Asteridae | Phytozomeb | 34 727 |
| Sorghum bicolor | monocot | Phytozomeb | 33 032 |
| Trochodendron aralioides | Trochodendrales | 1KPa | 18 636 |
| Vitis vinifera | Superrosidae | Genoscope 8x Releaseb | 30 434 |
aTranscriptome assembly.
bGenome annotation.
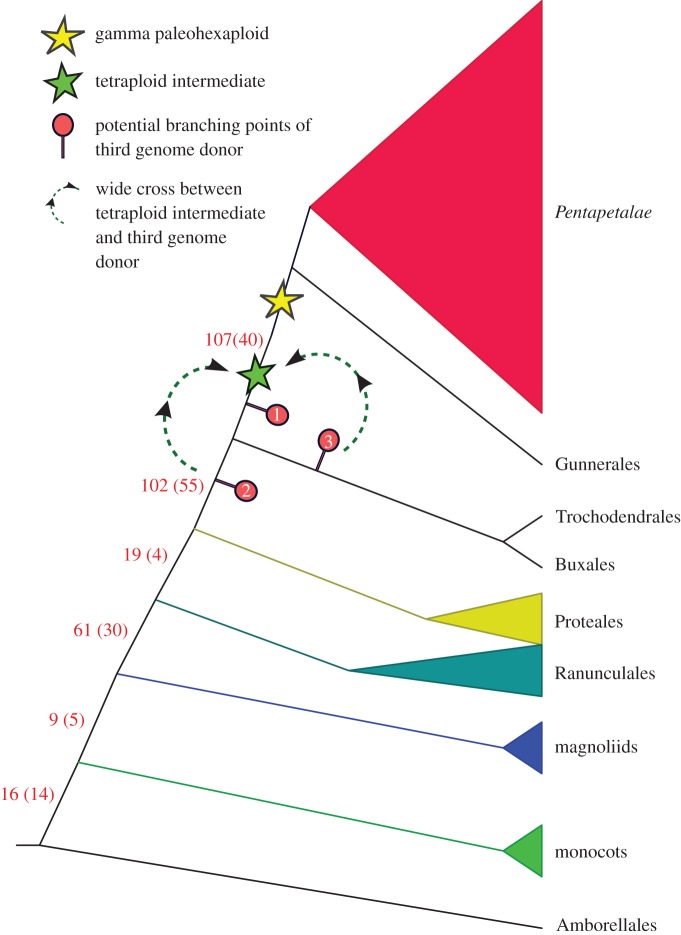
Our results indicate that Buxales and Trochodendrales are sister taxa collectively sister to the core eudicots, and that the origins of gamma-derived Vitis paralogues (palaeologues) are concentrated along two consecutive stem lineages immediately ‘below’ the Gunneridae (table 2 and figure 3). In the analyses of synteny-based orthogroups, 410 pairs of Vitis palaeologues could be assessed phylogenetically. The origins of 107 (40 with 50% BS or more) were placed along the branch that immediately precedes Gunneridae. The second prominent set of gamma duplications, 102 in total (55 with 50% BS or more), was placed along the branch subtending Buxales, Trochodendrales and Gunneridae (post-Proteales). A noteworthy proportion of Vitis palaeologues (61 in total; 30 with 50% BS or more) was estimated to have originated prior to the radiation of all extant eudicots. Similarly, substantial numbers of ‘core eudicot-wide’ duplications were also found in the phylogenomic analyses of Jiao et al. [44], but as noted above (see [115]), they do not appear to be relevant to gamma palaeohexaploidy. Analyses of Orthofinder-circumscribed groups showed a similar distribution of duplication events (table 2).
Table 2.
Phylogenetic origin of Vitis paralogue duplications inferred from orthogroup phylogenies.
| palaeolog origin | synteny-based orthogroups |
OrthoFinder-based orthogroups |
||||
|---|---|---|---|---|---|---|
| BS ≥ 80 | BS ≥ 50 | BS ≥ 0 | BS ≥ 80 | BS ≥ 50 | BS ≥ 0 | |
| Pentapetalae-wide | 0 | 0 | 1 | 0 | 0 | 1 |
| Gunneridae-wide | 8 | 40 | 107 | 6 | 43 | 135 |
| post-Proteales | 21 | 55 | 102 | 19 | 36 | 78 |
| pre-Proteales | 0 | 4 | 19 | 1 | 5 | 21 |
| Eudicot-wide | 12 | 30 | 61 | 11 | 33 | 65 |
| pre-magnoliids | 2 | 5 | 9 | 3 | 5 | 11 |
| pre-monocots | 12 | 14 | 16 | 13 | 13 | 17 |
Figure 3.
Evolutionary origin of gamma-derived Vitis paralogues. The branch labels along the backbone of the phylogenetic tree [total(no. with greater than 50% BS)] indicate the number of Vitis paralogues estimated to have arisen along the respective stem linages assuming strict tetraploidy events. Possible phylogenetic origins of the third subgenome via the two-step model of gamma hexaploidy are indicated by 1, 2 and 3. Scenarios in which a tetraploid is crossed with a close relative that branched off the core eudicot stem lineage (position 1) or off an older stem lineage (position 2) can be reconciled with our gene trees if extensive gene loss and/or extinction is invoked. The scenario of a wide cross between the core eudicot tetraploid and a species that branched off the stem lineage ‘below’ Buxales and Trochodendrales (position 3) is less complex evolutionarily.
These findings are consistent with a two-step model for gamma palaeohexaploidy in which a tetraploidy event occurred in the immediate common ancestor of Gunneridae, followed by donation of the third gamma subgenome from among the ancestors of both Buxales and Trochodendrales. Robustly resolved gene trees with representatives of all three gamma-derived subgenomes were not observed in our dataset, perhaps a consequence of extensive fractionation as previously noted [110]. The two-step scenario is, therefore, largely supported by gene trees that include two duplicate gamma-derived gene lineages: Buxus and/or Trochodendron genes are either (1) sister to duplicate core eudicot gene lineages, or (2) sister to one of two duplicate core eudicot gene lineages. Following the logic outlined by Ming et al. (fig. 3 in [45]), gene tree topologies of type (1) are most parsimoniously interpreted as representing a tetraploidy event along the core eudicot stem branch after the divergence of Buxales and Trochodendrales, but are ambiguous with regard to the origin of the third gamma subgenome. They can be reconciled with loss of any one of the three ancestral genes if the third genome was donated from a branch off the stem lineage of core eudicots below the tetraploidy event (position 1 in figure 3) or loss of a gene donated from a branch off the stem lineage below Buxales, Trochodendrales and core eudicots (position 2 in figure 3). Type (2) gene trees effectively pair a gene lineage that was inherited by core eudicots, Buxales and Trochodendrales, with a core eudicot-specific gene lineage. If the core eudicot-specific gene lineage is one of the tetraploidy-derived duplicates, this topology can be reconciled with organismal phylogeny by postulating a wide cross involving the core eudicot tetraploid and a species that diverged below the common ancestor of Buxales and Trochodendrales (position 3 in figure 3). Alternatively, it is consistent with the donation of the eudicot-specific gene lineage from an extinct line that branched below Buxales, Trochodendrales and core eudicots (at position 2 in figure 3). Thus, barring complex extinction scenarios, our gene trees do not support placing all gamma-associated duplications after the divergence of Buxales and Trochodendrales, nor do they support a WGD in the common ancestor of Buxales, Trochodendrales and core eudicots. Instead, the inclusion of Buxus and/or Trochodendron genes in one of the putative gamma-derived gene lineages is more easily reconciled with the donation of a third subgenome through a wide cross involving an ancestor of Buxales and Trochodendrales, as envisioned in the two-step model (figure 3).
7. Implications of gamma palaeohexaploidy
The close phylogenetic coincidence of the gamma palaeohexaploidy and the origin of pentamerous flowers suggests a causal relationship. As noted, gamma likely arose via a two-step process, with each WGD yielding a set of duplicated genes at each locus. Thus, barring extensive gene loss, we expect a minimum of two or three paralogues for all genes relative to the gene complement present in basal eudicots, monocots and basal angiosperms. In fact, as reviewed in §6, such paralogue diversity is indeed present for many of the key regulators of floral development within Pentapetalae. Especially relevant are transcription factors of the MADS-box, TCP domain and MYB domain gene families, all of which show duplications or triplications prior to the origin of Pentapetalae. For example, multiple duplications in the MADS-box family trace to gamma, and the resulting paralogues of the APETALA1, APETALA3, AGAMOUS and SEPALLATA subfamilies have typically diverged in sequence, expression and function (see [51] for review). Likewise, multiple duplications of TCP genes are also coincident with gamma [101], and paralogous gene lineages have assumed roles in floral symmetry, regulation of vegetative branching and unknown functions in flowers [131–134].
WGD provides the stimulus and genetic raw material for evolutionary novelty [11,37,135]. Although evidence for a causal role of gene duplication in morphological novelty remains limited, data are beginning to accumulate in support of a functional link. For example, differential expression patterns of three paralogues of AP3 coupled with PI control petaloidy in Aquilegia (Ranunculales) and appear to be responsible for the novel features of columbine flowers [90,136]. Duplications of entire genomes allow more complex intergenic interactions, involving multiple paralogues of all genes in the genome, with potentially greater morphological effect than duplications of single genes. Moreover, sequential WGDs, such as those responsible for the palaeohexaploidy recognized as gamma, have even greater potential for novelty than a single WGD.
Narrowing the phylogenetic placement of gamma provides the framework for much more detailed examination of the key features of Pentapetalae. Although we have emphasized the pentamerous, whorled flower of Pentapetalae, other complex floral features, such as bilateral symmetry and highly synorganized flowers (with closely associated floral organs, arising through either fusion or special physical placement of floral parts) also originated within Pentapetalae, perhaps built on the genetic diversity residing in these gamma-derived genomes. Further, because WGD is a common feature of angiosperm evolution, WGDs that both preceded and followed gamma may also have contributed to floral diversity in Pentapetalae. The effects of ancient WGD may not be immediately manifested on a phylogenetic tree; in fact, a phylogenetic ‘lag’ often occurs between WGD and the diversification that may be related to a key innovation [34,137]. Finally, although we focus here on floral traits, we note that other novel features, such as the chemical compound ellagic acid, also trace to Gunneridae or Pentapetalae [49], and further investigation of gamma will have implications for our understanding of many of the key traits that characterize nearly 75% of all angiosperm species.
8. Summary and future prospects
The vast majority of flowering plant diversity can be attributed to the success of a single clade, Pentapetalae, nested within the eudicots. The origin of Pentapetalae coincides with the evolution of a novel suite of floral features (whorled pentamery) and closely follows the gamma genome triplication. These two evolutionary events appear to have had an important impact on flowering plant evolution, but are yet not fully understood. Previous analyses, including the new phylogenomic analysis we present here, have been limited by the available genomic data for three phylogenetically critical lineages: Buxales, Trochodendrales and Gunnerales. These taxa are critical to understanding the timing and nature of gamma palaeohexaploidy, the functional diversification of genes duplicated through this WGD event(s), and relationships between these events and the origin of Pentapetalae. A more fully elucidated evolutionary history of Pentapetalae will, therefore, require the integration of these taxa into several facets of contemporary biological research, including phylogenetics, genomics and functional genetics, which probe the relationships between WGDs, gene duplication, sub- or neofunctionalization, morphological novelty, ecological opportunity and biological radiations.
Acknowledgements
We thank Saravanaraj Ayyampalayam for the scripts used to assign genes to pre-determined orthogroups, Michael McKain for scripts to place gene duplication events on phylogenetic trees and two anonymous reviewers for their useful comments on an earlier version of this manuscript.
Data accessibility
Phylogenomic data, including all alignments and trees analysed here, are available from the Dryad Digital Repository http://dx.doi.org/10.5061/dryad.bc80r.
Authors' contributions
A.S.C., B.A.B., D.G.H., D.E.S. and P.S.S. contributed equally to the conception and design of the study. A.S.C. performed the data analyses and drafted the primary manuscript. Additional text and discussion were provided by B.A.B., D.G.H., D.E.S. and P.S.S. B.A.B. and D.G.H. provided data for Meliosma dillenifolia. All authors approved the final version.
Competing interests
We have no competing interests.
Funding
This work was supported by NSF grants no. IOS-1121301, IOS-0922742, DEB-1455601 and DEB-1457440.
References
- 1.Anon. 2013. The plant list v1.1. See http://www.theplantlist.org/ (accessed 1 January 2016).
- 2.The Angiosperm Phylogeny Group 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. ( 10.1111/boj.12385) [DOI] [Google Scholar]
- 3.Sanderson MJ, Thorne JL, Wikström N, Bremer K. 2004. Molecular evidence on plant divergence times. Am. J. Bot. 91, 1656–1665. ( 10.3732/ajb.91.10.1656) [DOI] [PubMed] [Google Scholar]
- 4.Harrison CJ. 2017. Development and genetics in the evolution of land plant body plans. Phil. Trans. R. Soc. B 372, 20150490 ( 10.1098/rstb.2015.0490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun G, Ji Q, Dilcher DL, Zheng S, Nixon KC, Wang X. 2002. Archaefructaceae, a new basal angiosperm family. Science 296, 899–904. ( 10.1126/science.1069439) [DOI] [PubMed] [Google Scholar]
- 6.Friis EM, Pedersen KR, Crane PR. 2006. Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Palaeogeogr. Palaeoclimatol. Palaeoecol. 232, 251–293. ( 10.1016/j.palaeo.2005.07.006) [DOI] [Google Scholar]
- 7.Soltis DE, Bell CD, Kim S, Soltis PS. 2008. Origin and early evolution of angiosperms. Ann. NY Acad. Sci. 1133, 3–25. ( 10.1196/annals.1438.005) [DOI] [PubMed] [Google Scholar]
- 8.Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97, 1296–1303. ( 10.3732/ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 9.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453. ( 10.1111/nph.13264) [DOI] [PubMed] [Google Scholar]
- 10.Friedman WE. 2009. The meaning of Darwin's ‘abominable mystery’. Am. J. Bot. 96, 5–21. ( 10.3732/ajb.0800150) [DOI] [PubMed] [Google Scholar]
- 11.Soltis PS, Soltis DE. 2014. Flower diversity and angiosperm diversification. Methods Mol. Biol. (Clifton, NJ) 1110, 85–102. ( 10.1007/978-1-4614-9408-9_4) [DOI] [PubMed] [Google Scholar]
- 12.Chanderbali AS, Berger BA, Howarth DG, Soltis PS, Soltis DE. 2016. Evolving ideas on the origin and evolution of flowers: new perspectives in the genomic era. Genetics 202, 1255–1265. ( 10.1534/genetics.115.182964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frohlich MW. 2003. An evolutionary scenario for the origin of flowers. Nat. Rev. Genet. 4, 559–566. ( 10.1038/nrg1114) [DOI] [PubMed] [Google Scholar]
- 14.Baum DA, Hileman LC. 2006. A developmental genetic model for the origin of the flower. In Annual plant reviews 20: flowering and its manipulation (ed. Ainsworth C.), pp. 1–27. Oxford, UK: Blackwell Publishing. [Google Scholar]
- 15.Theissen G, Melzer R. 2007. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 100, 603–619. ( 10.1093/aob/mcm143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frohlich MW. 2006. Recent developments regarding the evolutionary origin of flowers. In Advances in botanical research 44: developmental genetics of the flower (eds Soltis DE, Leebens-Mack JH, Soltis PS), pp. 63–127. San Diego, CA: Elsevier. [Google Scholar]
- 17.Buzgo M, Soltis PS, Soltis DE. 2004. Floral developmental morphology of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 165, 925–947. ( 10.1086/424024) [DOI] [Google Scholar]
- 18.Kim S, Koh J, Yoo M-J, Kong H, Hu Y, Ma H, Soltis PS, Soltis DE. 2005. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. Cell Mol. Biol. 43, 724–744. ( 10.1111/j.1365-313X.2005.02487.x) [DOI] [PubMed] [Google Scholar]
- 19.Soltis PS, Brockington SF, Yoo M-J, Piedrahita A, Latvis M, Moore MJ, Chanderbali AS, Soltis DE. 2009. Floral variation and floral genetics in basal angiosperms. Am. J. Bot. 96, 110–128. ( 10.3732/ajb.0800182) [DOI] [PubMed] [Google Scholar]
- 20.Chanderbali AS, et al. 2010. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc. Natl Acad. Sci. USA 107, 22 570–22 575. ( 10.1073/pnas.1013395108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Tunen AJ, Eikelboom W, Angenent GC. 1993. Floral organogenesis in Tulipa. Flower. Newsl. 16, 33–38. [Google Scholar]
- 22.Bowman JL. 1997. Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J. Biosci. 22, 515–527. ( 10.1007/BF02703197) [DOI] [Google Scholar]
- 23.Albert VA, Gustafsson MHG, Laurenzio LD. 1998. Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal. In Molecular systematics of plants II (eds Soltis DE, Soltis PS, Doyle JJ), pp. 349–374. New York, NY: Springer. [Google Scholar]
- 24.Kramer EM, Di Stilio VS, Schlüter PM. 2003. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164, 1–11. ( 10.1086/344694) [DOI] [Google Scholar]
- 25.Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. ( 10.1105/tpc.1.1.37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coen ES, Meyerowitz EM. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. ( 10.1038/353031a0) [DOI] [PubMed] [Google Scholar]
- 27.Theißen G. 2001. Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. ( 10.1016/S1369-5266(00)00139-4) [DOI] [PubMed] [Google Scholar]
- 28.Corley SB, Carpenter R, Copsey L, Coen E. 2005. Floral asymmetry involves an interplay between TCP and MYB transcription factors in Antirrhinum. Proc. Natl Acad. Sci. USA 102, 5068–5073. ( 10.1073/pnas.0501340102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronse De Craene LP. 2007. Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Ann. Bot. 100, 621–630. ( 10.1093/aob/mcm076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronse De Craene LP, Brockington SF. 2013. Origin and evolution of petals in angiosperms. Plant Ecol. Evol. 146, 5–25. ( 10.5091/plecevo.2013.738) [DOI] [Google Scholar]
- 31.Soltis DE, et al. 2014. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytol. 202, 1105–1117. ( 10.1111/nph.12756) [DOI] [PubMed] [Google Scholar]
- 32.Vanneste K, Maere S, de Peer YV. 2014. Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Phil. Trans. R. Soc. B 369, 20130353 ( 10.1098/rstb.2013.0353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayrose I, Zhan SH, Rothfels CJ, Arrigo N, Barker MS, Rieseberg LH, Otto SP. 2015. Methods for studying polyploid diversification and the dead end hypothesis: a reply to Soltis et al. (2014). New Phytol. 206, 27–35. ( 10.1111/nph.13192) [DOI] [PubMed] [Google Scholar]
- 34.Tank DC, Eastman JM, Pennell MW, Soltis PS, Soltis DE, Hinchliff CE, Brown JW, Sessa EB, Harmon LJ. 2015. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol. 207, 454–467. ( 10.1111/nph.13491) [DOI] [PubMed] [Google Scholar]
- 35.Dodsworth S, Chase MW, Leitch AR. 2016. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Diploidization in polyploid angiosperms. Bot. J. Linn. Soc. 180, 1–5. ( 10.1111/boj.12357) [DOI] [Google Scholar]
- 36.Kellogg EA. 2016. Has the connection between polyploidy and diversification actually been tested? Curr. Opin. Plant Biol. 30, 25–32. ( 10.1016/j.pbi.2016.01.002) [DOI] [PubMed] [Google Scholar]
- 37.Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165. ( 10.1016/j.pbi.2016.03.015) [DOI] [PubMed] [Google Scholar]
- 38.Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. ( 10.1038/nature09916) [DOI] [PubMed] [Google Scholar]
- 39.Amborella Genome Project 2013. The Amborella Genome and the Evolution of Flowering Plants. Science 342, 1241089 ( 10.1126/science.1241089) [DOI] [PubMed] [Google Scholar]
- 40.Tang H, Bowers JE, Wang X, Paterson AH. 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl Acad. Sci. USA. 107, 472–477. ( 10.1073/pnas.0908007107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKain MR, et al. 2016. A phylogenomic assessment of ancient polyploidy and genome evolution across the Poales. Genome Biol. Evol. 8, 1150–1164. ( 10.1093/gbe/evw060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaillon O, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. ( 10.1038/nature06148) [DOI] [PubMed] [Google Scholar]
- 43.Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. 2008. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18, 1944–1954. ( 10.1101/gr.080978.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiao Y, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13, R3 ( 10.1186/gb-2012-13-1-r3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ming R, et al. 2013. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14, R41 ( 10.1186/gb-2013-14-5-r41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnable J, Lyons E. 2015. Plant paleopolyploidy. ( 10.6084/m9.figshare.1538627.v1) [DOI] [Google Scholar]
- 47.Blanc G, Wolfe KH. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. ( 10.1105/tpc.021345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao Y, Paterson AH. 2014. Polyploidy-associated genome modifications during land plant evolution. Phil. Trans. R. Soc. B 369, 20130355 ( 10.1098/rstb.2013.0355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soltis PS, Liu X, Marchant DB, Visger CJ, Soltis DE. 2014. Polyploidy and novelty: Gottlieb's legacy. Phil. Trans. R. Soc. B 369, 20130351 ( 10.1098/rstb.2013.0351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payne JL, Wagner A. 2015. Mechanisms of mutational robustness in transcriptional regulation. Bioinform. Comput. Biol. 6, 322 ( 10.3389/fgene.2015.00322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soltis PS, Burleigh JG, Chanderbali AS, Yoo M-J, Soltis DE. 2010. Gene and genome duplications in plants. In Evolution after gene duplication (eds Dittmar K, Liberles D), pp. 269–298. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 52.Holland PWH, Marlétaz F, Maeso I, Dunwell TL, Paps J. 2017. New genes from old: asymmetric divergence of gene duplicates and the evolution of development. Phil. Trans. R. Soc. B 372, 20150480 ( 10.1098/rstb.2015.0480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soltis DE, et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730. ( 10.3732/ajb.1000404) [DOI] [PubMed] [Google Scholar]
- 54.Soltis DE, Soltis PS, Endress PK, Chase MW. 2005. Phylogeny and evolution of angiosperms. Sunderland, MA: Sinauer. [Google Scholar]
- 55.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14, 23 ( 10.1186/1471-2148-14-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wickett NJ, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA 111, E4859–E4868. ( 10.1073/pnas.1323926111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y, et al. 2016. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol. Phylogenet. Evol. 96, 93–101. ( 10.1016/j.ympev.2015.12.006) [DOI] [PubMed] [Google Scholar]
- 58.Anderson CL, Bremer K, Friis EM. 2005. Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am. J. Bot. 92, 1737–1748. ( 10.3732/ajb.92.10.1737) [DOI] [PubMed] [Google Scholar]
- 59.Cantino PD, Doyle JA, Graham SW, Judd WS, Olmstead RG, Soltis DE, Soltis PS, Donoghue MJ. 2007. Towards a phylogenetic nomenclature of Tracheophyta. Taxon 56, 822–846. ( 10.2307/25065865) [DOI] [Google Scholar]
- 60.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl Acad. Sci. USA 107, 4623–4628. ( 10.1073/pnas.0907801107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takhtajan AL. 1980. Outline of the classification of flowering plants (magnoliophyta). Bot. Rev. 46, 225–359. ( 10.1007/BF02861558) [DOI] [Google Scholar]
- 62.Cronquist A. 1981. An integrated system of classification of flowering plants. New York, NY: Columbia University Press. [Google Scholar]
- 63.de Jussieu AL. 1789. Antonii Laurentii de Jussieu Genera plantarum: secundum ordines naturales disposita, juxta methodum in horto regio parisiensi exaratam. Paris, France: Herissant et Theophilum Barrois. [Google Scholar]
- 64.Reichenbach HGL. 1827. Dr. Joh. Christ. Mössler's Handbuch der Gewächskunde. Altona, Germany: J. F. Hammerich. [Google Scholar]
- 65.Endress PK. 2010. Flower structure and trends of evolution in eudicots and their major subclades. Ann. Mo. Bot. Gard. 97, 541–583. ( 10.3417/2009139) [DOI] [Google Scholar]
- 66.Soltis DE, Senters AE, Zanis MJ, Kim S, Thompson JD, Soltis PS, Ronse De Craene LP, Endress PK, Farris JS. 2003. Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. Am. J. Bot. 90, 461–470. ( 10.3732/ajb.90.3.461) [DOI] [PubMed] [Google Scholar]
- 67.Endress PK. 2006. Angiosperm floral evolution: morphological developmental framework. In Advances in botanical research 44: developmental genetics of the flower (eds Soltis DE, Leebens-Mack JH, Soltis PS), pp. 1–61. San Diego, CA: Elsevier. [Google Scholar]
- 68.Endress PK. 1996. Diversity and evolutionary biology of tropical flowers. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 69.Soltis DE, Soltis PS, Albert VA, Oppenheimer DG, dePamphilis CW, Ma H, Frohlich MW, Theißen G. 2002. Missing links: the genetic architecture of flower and floral diversification. Trends Plant Sci. 7, 22–31. ( 10.1016/S1360-1385(01)02098-2) [DOI] [PubMed] [Google Scholar]
- 70.Wanntorp L, Ronse De Craene LP. 2005. The Gunnera flower: key to eudicot diversification or response to pollination mode? Int. J. Plant Sci. 166, 945–953. ( 10.1086/467474) [DOI] [Google Scholar]
- 71.Ronse De Craene LP, Wanntorp L. 2006. Evolution of floral characters in Gunnera (Gunneraceae). Syst. Bot. 31, 671–688. ( 10.1600/036364406779695951) [DOI] [Google Scholar]
- 72.Endress PK. 1986. Floral structure, systematics, and phylogeny in Trochodendrales. Ann. Mo. Bot. Gard. 73, 297–324. ( 10.2307/2399115) [DOI] [Google Scholar]
- 73.Chen L, Ren Y, Endress PK, Tian XH, Zhang XH. 2007. Floral organogenesis in Tetracentron sinense (Trochodendraceae) and its systematic significance. Plant Syst. Evol. 264, 183–193. ( 10.1007/s00606-006-0505-y) [DOI] [Google Scholar]
- 74.von Balthazar M, Endress PK. 2002. Development of inflorescences and flowers in Buxaceae and the problem of perianth interpretation. Int. J. Plant Sci. 163, 847–876. ( 10.1086/342714) [DOI] [Google Scholar]
- 75.Wanntorp L, Ronse De Craene LP. 2007. Flower development of Meliosma (Sabiaceae): evidence for multiple origins of pentamery in the eudicots. Am. J. Bot. 94, 1828–1836. ( 10.3732/ajb.94.11.1828) [DOI] [PubMed] [Google Scholar]
- 76.Ronse De Craene LP, Quandt D, Wanntorp L. 2015. Floral development of Sabia (Sabiaceae): evidence for the derivation of pentamery from a trimerous ancestry. Am. J. Bot. 102, 336–349. ( 10.3732/ajb.1400388) [DOI] [PubMed] [Google Scholar]
- 77.Damerval C, Nadot S. 2007. Evolution of perianth and stamen characteristics with respect to floral symmetry in Ranunculales. Ann. Bot. 100, 631–640. ( 10.1093/aob/mcm041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang P, Liao H, Zhang W, Yu X, Zhang R, Shan H, Duan X, Yao X, Kong H. 2015. Flexibility in the structure of spiral flowers and its underlying mechanisms. Nat. Plants 2, 15188 ( 10.1038/nplants.2015.188) [DOI] [PubMed] [Google Scholar]
- 79.Ronse De Craene L. 2015. Meristic changes in flowering plants: how flowers play with numbers. Flora Morphol. Distrib. Funct. Ecol. Plants 221, 22–37. ( 10.1016/j.flora.2015.08.005) [DOI] [Google Scholar]
- 80.Litt A, Irish VF. 2003. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165, 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S, Yoo M-J, Albert VA, Farris JS, Soltis PS, Soltis DE. 2004. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91, 2102–2118. ( 10.3732/ajb.91.12.2102) [DOI] [PubMed] [Google Scholar]
- 82.Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H. 2005. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223. ( 10.1534/genetics.104.037770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vekemans D, Proost S, Vanneste K, Coenen H, Viaene T, Ruelens P, Maere S, de Peer YV, Geuten K. 2012. Gamma paleohexaploidy in the stem-lineage of core eudicots: significance for MADS-box gene and species diversification. Mol. Biol. Evol. 29, 3793–3806. ( 10.1093/molbev/mss183) [DOI] [PubMed] [Google Scholar]
- 84.Soltis PS, Soltis DE, Kim S, Chanderbali A, Buzgo M. 2006. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC-function. In Advances in botanical research 44: developmental genetics of the flower (eds Soltis DE, Leebens-Mack JH, Soltis PS), pp. 483–506. San Diego, CA: Elsevier. [Google Scholar]
- 85.Chanderbali AS, Albert VA, Leebens-Mack J, Altman NS, Soltis DE, Soltis PS. 2009. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae). Proc. Natl Acad. Sci. USA 106, 8929–8934. ( 10.1073/pnas.0811476106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoo M-J, Soltis PS, Soltis DE. 2010. Expression of floral MADS-Box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int. J. Plant Sci. 171, 121–146. ( 10.1086/648986) [DOI] [Google Scholar]
- 87.Voelckel C, Borevitz JO, Kramer EM, Hodges SA. 2010. Within and between whorls: comparative transcriptional profiling of Aquilegia and Arabidopsis. PLoS ONE 5, e9735 ( 10.1371/journal.pone.0009735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoo M-J, Chanderbali AS, Altman NS, Soltis PS, Soltis DE. 2010. Evolutionary trends in the floral transcriptome: insights from one of the basalmost angiosperms, the water lily Nuphar advena (Nymphaeaceae). Plant J. 64, 687–698. ( 10.1111/j.1365-313X.2010.04357.x) [DOI] [PubMed] [Google Scholar]
- 89.Kramer EM. 2009. Aquilegia: a new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol. 60, 261–277. ( 10.1146/annurev.arplant.043008.092051) [DOI] [PubMed] [Google Scholar]
- 90.Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM. 2007. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19, 750–766. ( 10.1105/tpc.107.050385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang R, et al. 2013. Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc. Natl Acad. Sci. USA 110, 5074–5079. ( 10.1073/pnas.1219690110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gonçalves B, Nougué O, Jabbour F, Ridel C, Morin H, Laufs P, Manicacci D, Damerval C. 2013. An APETALA3 homolog controls both petal identity and floral meristem patterning in Nigella damascena L. (Ranunculaceae). Plant J. Cell Mol. Biol. 76, 223–235. ( 10.1111/tpj.12284) [DOI] [PubMed] [Google Scholar]
- 93.Bartholmes C, Hidalgo O, Gleissberg S. 2012. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. Stuttg. Ger. 14, 11–23. ( 10.1111/j.1438-8677.2011.00486.x) [DOI] [PubMed] [Google Scholar]
- 94.Hidalgo O, Bartholmes C, Gleissberg S. 2012. Virus-induced gene silencing (VIGS) in Cysticapnos vesicaria, a zygomorphic-flowered Papaveraceae (Ranunculales, basal eudicots). Ann. Bot. 109, 911–920. ( 10.1093/aob/mcs008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donoghue MJ, Ree RH, Baum DA. 1998. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 3, 311–317. ( 10.1016/S1360-1385(98)01278-3) [DOI] [Google Scholar]
- 96.Hileman LC. 2014. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Phil. Trans. R. Soc. B 369, 20130348 ( 10.1098/rstb.2013.0348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Specht CD, Howarth DG. 2015. Adaptation in flower form: a comparative evodevo approach. New Phytol. 206, 74–90. ( 10.1111/nph.13198) [DOI] [PubMed] [Google Scholar]
- 98.Boyden GS, Donoghue MJ, Howarth DG. 2012. Duplications and expression of RADIALIS-like genes in Dipsacales. Int. J. Plant Sci. 173, 971–983. ( 10.1086/667626) [DOI] [Google Scholar]
- 99.Howarth DG, Donoghue MJ. 2006. Phylogenetic analysis of the ‘ECE’ (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl Acad. Sci. USA 103, 9101–9106. ( 10.1073/pnas.0602827103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Howarth DG, Donoghue MJ. 2009. Duplications and expression of DIVARICATA-like genes in Dipsacales. Mol. Biol. Evol. 26, 1245–1258. ( 10.1093/molbev/msp051) [DOI] [PubMed] [Google Scholar]
- 101.Citerne HL, Guilloux ML, Sannier J, Nadot S, Damerval C. 2013. Combining phylogenetic and syntenic analyses for understanding the evolution of TCP ECE genes in eudicots. PLOS ONE 8, e74803 ( 10.1371/journal.pone.0074803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blanc G, Hokamp K, Wolfe KH. 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. ( 10.1101/gr.751803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. ( 10.1038/nature01521) [DOI] [PubMed] [Google Scholar]
- 104.Tuskan GA, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. ( 10.1126/science.1128691) [DOI] [PubMed] [Google Scholar]
- 105.Ming R, et al. 2008. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996. ( 10.1038/nature06856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Consortium TTG. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. ( 10.1038/nature11119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ibarra-Laclette E, et al. 2013. Architecture and evolution of a minute plant genome. Nature 498, 94–98. ( 10.1038/nature12132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tang H, Lyons E, Schnable JC. 2014. Early history of the angiosperms. In Advances in botanical research (ed. Paterson AH.), pp. 195–222. New York, NY: Academic Press. [Google Scholar]
- 109.Denoeud F, et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181–1184. ( 10.1126/science.1255274) [DOI] [PubMed] [Google Scholar]
- 110.Lyons E, Pedersen B, Kane J, Freeling M. 2008. The Value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the Rosids. Trop. Plant Biol. 1, 181–190. ( 10.1007/s12042-008-9017-y) [DOI] [Google Scholar]
- 111.Murat F, Zhang R, Guizard S, Gavranović H, Flores R, Steinbach D, Quesneville H, Tannier E, Salse J. 2015. Karyotype and gene order evolution from reconstructed extinct ancestors highlight contrasts in genome plasticity of modern Rosid crops. Genome Biol. Evol. 7, 735–749. ( 10.1093/gbe/evv014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang H, Woodhouse MR, Cheng F, Schnable JC, Pedersen BS, Conant G, Wang X, Freeling M, Pires JC. 2012. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of Paleohexaploidy. Genetics 190, 1563–1574. ( 10.1534/genetics.111.137349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brenchley R, et al. 2012. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. ( 10.1038/nature11650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.International Wheat Genome Sequencing Consortium (IWGSC) 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 ( 10.1126/science.1251788) [DOI] [PubMed] [Google Scholar]
- 115.Zheng C, Sankoff D. 2014. Practical halving: the Nelumbo nucifera evidence on early eudicot evolution. Comput. Biol. Chem. 50, 75–81. ( 10.1016/j.compbiolchem.2014.01.010) [DOI] [PubMed] [Google Scholar]
- 116.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. ( 10.1101/gr.1224503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 ( 10.1186/s13059-015-0721-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. 1000 Green Plant Transcriptome Project. See http://www.onekp.com .
- 119. Ancestral Angiosperm Genome Project. See http://ancangio.uga.edu .
- 120.Wang K, Deng J, Damaris RN, Yang M, Xu L, Yang P. 2015. LOTUS-DB: an integrative and interactive database for Nelumbo nucifera study. Database 2015, bav023. ( 10.1093/database/bav023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sacred lotus genome annotation project. See http://lotus-db.wbgcas.cn .
- 122.Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. ( 10.1093/nar/gkr944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Phytozome v11.0. See http://www.phytozome.net .
- 124.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. ( 10.1093/nar/gkl315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chamala S, García N, Godden GT, Krishnakumar V, Jordon-Thaden IE, Smet RD, Barbazuk WB, Soltis DE, Soltis PS. 2015. MarkerMiner 1.0: a new application for phylogenetic marker development using angiosperm transcriptomes. Appl. Plant Sci. 3, 1400115 ( 10.3732/apps.1400115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinforma. Oxf. Engl. 31, 44–i52. ( 10.1093/bioinformatics/btv234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kück P, Meusemann K. 2010. FASconCAT: convenient handling of data matrices. Mol. Phylogenet. Evol. 56, 1115–1118. ( 10.1016/j.ympev.2010.04.024) [DOI] [PubMed] [Google Scholar]
- 131.Aguilar-Martínez JA, Poza-Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. ( 10.1105/tpc.106.048934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Finlayson SA. 2007. Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 48, 667–677. ( 10.1093/pcp/pcm044) [DOI] [PubMed] [Google Scholar]
- 133.Martín-Trillo M, et al. 2011. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. Cell Mol. Biol. 67, 701–714. ( 10.1111/j.1365-313X.2011.04629.x) [DOI] [PubMed] [Google Scholar]
- 134.Martín-Trillo M, Cubas P. 2010. TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. ( 10.1016/j.tplants.2009.11.003) [DOI] [PubMed] [Google Scholar]
- 135.Ohno S. 1970. Evolution by gene duplication. New York, NY: Springer. [Google Scholar]
- 136.Rasmussen DA, Kramer EM, Zimmer EA. 2009. One size fits all? Molecular evidence for a commonly inherited petal identity program in Ranunculales. Am. J. Bot. 96, 96–109. ( 10.3732/ajb.0800038) [DOI] [PubMed] [Google Scholar]
- 137.Schranz ME, Mohammadin S, Edger PP. 2012. Ancient whole genome duplications, novelty and diversification: the WGD radiation lag-time model. Curr. Opin. Plant Biol. 15, 147–153. ( 10.1016/j.pbi.2012.03.011) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phylogenomic data, including all alignments and trees analysed here, are available from the Dryad Digital Repository http://dx.doi.org/10.5061/dryad.bc80r.