Abstract
Invasive and endangered species reflect opposite ends of a spectrum of ecological success, yet they experience many similar eco-evolutionary challenges including demographic bottlenecks, hybridization and novel environments. Despite these similarities, important differences exist. Demographic bottlenecks are more transient in invasive species, which (i) maintains ecologically relevant genetic variation, (ii) reduces mutation load, and (iii) increases the efficiency of natural selection relative to genetic drift. Endangered species are less likely to benefit from admixture, which offsets mutation load but also reduces fitness when populations are locally adapted. Invading species generally experience more benign environments with fewer natural enemies, which increases fitness directly and also indirectly by masking inbreeding depression. Adaptive phenotypic plasticity can maintain fitness in novel environments but is more likely to evolve in invasive species encountering variable habitats and to be compromised by demographic factors in endangered species. Placed in an eco-evolutionary context, these differences affect the breadth of the ecological niche, which arises as an emergent property of antagonistic selection and genetic constraints. Comparative studies of invasions and extinctions that apply an eco-evolutionary perspective could provide new insights into the environmental and genetic basis of ecological success in novel environments and improve efforts to preserve global biodiversity.
This article is part of the themed issue ‘Human influences on evolution, and the ecological and societal consequences’.
Keywords: endangered species, niche theory, range limits, evolutionary genetics, plasticity, epigenetics
1. Introduction
Global biodiversity is increasingly under threat from human activity, which has elevated rates of extinction and invasion by several orders of magnitude above historical averages [1,2]. The net result of increasing extinctions and invasions is a homogenization of global biodiversity that may be mitigated by two distinct but complementary goals: (i) suppress long-term viability of invasive populations and (ii) increase population growth rates of endangered native species. Although eradication and enhancement are opposite conservation goals, invasions and extinctions represent two extreme outcomes along a single gradient of ecological success and therefore may be determined by the same core set of ecological and genetic factors. In other words, invasions and extinctions of closely related species may be like reflections in Lewis Carroll's looking glass [3], with similar elements reflecting opposite realities.
Several key environmental and demographic elements that affect population growth and long-term persistence do not differ fundamentally between endangered and invasive species. Both experience strong demographic bottlenecks, hybridization with divergent lineages, and the demands of surviving and reproducing in novel and changing environments. Yet, these common elements can lead to drastically different ‘realities’ or ecological outcomes, with invasive species expanding rapidly and endangered species spiralling towards extinction.
One hypothesis for these contrasting fates is that endangered and invasive species possess distinct sets of developmental and life-history characteristics that are either beneficial or detrimental in human-altered environments. For example, some species may be good invaders, because they have co-evolved with increasing human disturbances [4] or fluctuating environments [5] in their native ranges. However, a meta-analysis of 1813 species did not find evidence that invasive and threatened species possess contrasting traits [6]. It is likely that different characteristics are favoured at particular stages of invasion (i.e. transport, introduction, establishment and spread) [7–9]. But many invasive species are close relatives of taxa that are not invasive [10,11], suggesting that any functional basis for being invasive versus of conservation concern is not often phylogenetically conserved. If development, life history or other phylogenetically conserved traits do not differ consistently between endangered and invasive species, then perhaps more transient ecological and genetic factors are responsible for the varied ecological success of species in nature.
How is it that even related species, having similar growth and life-history traits and encountering similar environmental and demographic challenges, can end up at opposite ends on the spectrum of ecological success? Here, we apply an eco-evolutionary perspective to explore some of the similarities and outline important but often overlooked differences between invasions and extinctions. We focus on three areas of eco-evolutionary theory that reveal important differences between endangered and invasive species likely to affect ecological success: the composition of ecologically relevant genetic variation in natural populations, the genetic basis and evolution of phenotypic plasticity, and evolutionary constraints on the ecological niche. Our overall goal is to examine invasions and extinctions through the same lens of eco-evolutionary theory to suggest common principles for a unified approach to both basic and applied research in these areas.
2. Genetic variation
Human activity exposes invasive and endangered species to new adaptive landscapes in which natural selection is fundamentally different from the ancestral environment [12]. Comparing the evolutionary genetics of invasions and extinctions can shed new light on the fate of natural populations exposed to novel and changing environments (figure 1). In this section, we compare the effects of demographic bottlenecks, inbreeding and admixture on invasive and endangered species. We identify key differences that probably contribute to the very different population dynamics of invasions and extinctions.
Figure 1.
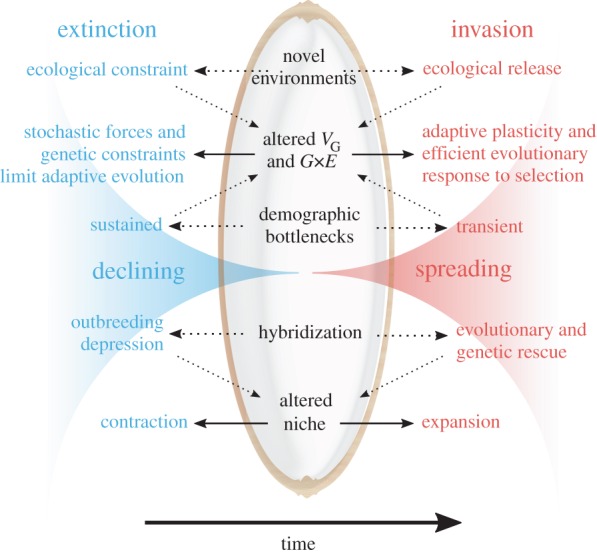
Extinctions and invasions conceptualized ‘Through the Looking Glass’ of evolutionary ecology. Extinctions (left side) represent population decline over time, while biological invasions (right side) represent an increase in abundance. Both invasions and extinctions reflect a common set of elements (central column) because subtle but influential ecological and genetic differences (outer columns) can cause opposite population growth trajectories. (Online version in colour.)
(a). Genetic variation and response to selection
Empirical studies show that moderate to severe population bottlenecks can be relatively common for both endangered and invasive species [13–15]. However, post-bottleneck population dynamics differ in several important ways that translate to large differences in genetic diversity maintained within populations (figure 1). One key difference is the length of the bottleneck period [16], as this directly affects the rate at which genetic diversity is lost [17]. While paired demographic and genetic data are rare for early stage invasions, the magnitude of genetic bottlenecks estimated from neutral markers shows that, on average, invasive populations suffer a detectable but relatively minor loss of genetic diversity [13]. This result suggests that most introduced populations remain small for only a relatively short period before increasing again, because most introductions begin with a strong demographic bottleneck involving a small fraction (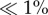 ) of all individuals present in the native range. Endangered species far more commonly experience extended periods of small population size, greatly increasing the loss of genetic diversity. Even if endangered populations do recover in census size, the genetic effective size of the population (Ne) will recover slowly as new mutations introduce variability back to the population.
) of all individuals present in the native range. Endangered species far more commonly experience extended periods of small population size, greatly increasing the loss of genetic diversity. Even if endangered populations do recover in census size, the genetic effective size of the population (Ne) will recover slowly as new mutations introduce variability back to the population.
Another factor mitigating loss of genetic diversity from demographic bottlenecks in invasive species is the high rate of gene flow common among introduced populations, which can promote ‘genetic rescue’ (alleviating inbreeding depression; [18]) of inbred populations and ‘evolutionary rescue’ (introduction of an adaptive variant; [19]) of maladapted populations [20–22]. Such increased gene flow is particularly important within a metapopulation context, because the negative demographic and genetic consequences of small population size and local extinction can be mitigated by connectivity with other populations in the region [15,23,24]. Under this model, gene flow shares diversity among populations so the metapopulation as a whole maintains higher Ne and sustains the invasion even if individual invasive populations are small. By contrast, declining populations of endangered species tend to be few in number and highly isolated, with little connectivity and opportunity for genetic or evolutionary rescue. Efforts to increase population connectivity for endangered species may be particularly beneficial when isolation is prolonged and population sizes are especially small; however, the benefits of increasing gene flow must be evaluated relative to the risk of compromising locally adapted genotypes.
Because the efficacy of selection is directly proportional to Ne, genetic drift has a much stronger effect on evolutionary change in species with limited genetic variability and among small, isolated populations, compared with larger well-connected populations. The reduced ability to respond to selection makes endangered populations particularly susceptible to environmental changes that lead to loss of local adaptation. Here too, the metapopulation context distinguishes biological invasions and provides additional opportunities for these species to respond to selection despite founder effects and genetic drift during establishment. Response to selection is more likely for invasive populations when (i) population sizes are large or increasing, (ii) gene flow from neighbouring populations acts as a genetic or evolutionary rescue, and (iii) local extinction of maladapted populations are followed by recolonization from populations with pre-adapted genotypes. This latter example of inter-demic selection within a meta-population context occurs as a result of non-random extinction and recolonization; this may be an under-appreciated mechanism for rapid evolution of invaders in novel environments [23,25]. In this way, invasions might be useful experimental systems for understanding how population connectivity contributes to the survival of individual subpopulations in variable environments, applying this knowledge in turn to restoration strategies and expectations for native species. Population genetic models that assign source populations to colonists and test for genetic associations between source population characteristics (e.g. density, inbreeding rate and phenotypes) and colonization probability may be a particularly useful approach for investigating the effect of metapopulation context on population persistence [24,26].
It is important to recognize that the genetic consequences of small population size affect variation at neutral marker loci differently than quantitative genetic variation in ecologically important traits, and indeed these two types of variance are often poorly correlated [27]. This is likely a consequence of the broadly polygenic basis of most quantitative traits, as individual loci each have only a small effect on the overall trait variance. Previous studies have shown that substantial quantitative genetic variation (VG) can persist for quantitative traits, even when populations show evidence of bottlenecks at individual loci [28,29]. As a result, rapid population growth of invasive species following even a severe demographic bottleneck will have little effect on quantitative genetic variance [30]. Moreover, given that most introductions fail, repeated founding events create opportunities for natural selection to filter the genetic variants that do establish at the scale of genotypes or even entire populations [31], though direct evidence of this process is lacking to date. By contrast, persistent small population sizes over multiple generations in endangered species will erode quantitative genetic variance through genetic drift, limiting adaptive evolutionary responses to changing selection pressures. Few studies have directly addressed the consequences of invasion or colonization on VG, although one such study showed a loss of VG in younger colonizing populations during (non-invasive) range expansion [32]. A few studies have compared VG between native and introduced populations of invasive species [33,34], but in these cases it is difficult to rule out differences among populations in the strength of stabilizing selection. Increasingly, population genomics is enabling the discovery of genes (or closely linked loci) involved in local adaptation, which may allow a more direct integration of molecular data into conservation strategies. More generally, genomic sequencing can now be combined with ecological surveys and field manipulations to better understand the relationship between genome-wide genetic variation and adaptive trait variation in natural populations, and how both are affected by demographic bottlenecks, gene flow and natural selection.
(b). Genetic load and inbreeding depression
One intriguing difference between endangered and invasive species is the effect of small population size on genetic load and inbreeding depression. Genetic load is a well-established conservation concern for many endangered taxa, as elevated inbreeding in small populations exposes the negative fitness effects of recessive deleterious alleles [17]. These effects directly contribute to increased extinction probability, especially under stressful environments [35]. The genome-wide load of deleterious mutations is sensitive to Ne, which determines how effectively purifying selection can purge genetic load. In endangered populations that are small, declining, or have undergone a bottleneck, avoiding or mitigating inbreeding is critical to maintain genotypes that carry few deleterious alleles (e.g. avoiding Muller's Ratchet). Invaders may also suffer from inbreeding depression if a colonization bottleneck is severe, as exemplified by a multi-species analysis of birds introduced to New Zealand. In this case, species that experienced strong bottlenecks (less than 150 individuals) showed persistent increases in hatching failure compared with less ‘bottlenecked’ species [36]. These effects may be transient in growing populations as some empirical studies have found increases in heterozygosity over time, despite bottlenecks as severe as a single pair of breeding individuals [37,38]. This is consistent with an increase in the efficiency of natural selection to ‘weed out’ homozygous individuals as population sizes increase. However, further studies are needed to determine whether this is a general phenomenon in successful invaders. Invasive species may also accumulate genetic load as a by-product of the range expansion process. Recent theoretical work has shown that small populations at the wavefront of an expanding range face increased probabilities of deleterious mutations drifting to high frequency, termed ‘expansion load’ [39]. The accumulation of expansion load is predicted to lead to legacies of reduced fitness following expansion [40,41]. Long-range dispersal between multiple introductions from distinct genetic sources could partially mitigate this effect by helping to shelter the genetic load in recently expanded invasive populations [42], but to our knowledge this has not been explored in colonization models.
Why do invasive populations not suffer from inbreeding depression more often? First, as described above, if invaders recover from demographic bottlenecks more quickly, they maintain genetic variation and experience less severe genetic drift and inbreeding. Second, as invasive populations grow in Ne, selection should become more effective at reducing the frequency of weakly deleterious alleles, thereby reducing the genetic load [43]. Third, the fitness effects of recessive alleles may be conditional on the environment. Phenotypic effects of deleterious mutations may be conditionally neutral when the environment is benign but amplified by environmental stress, resulting in inbreeding × environment (I × E) interactions [44]. Invasive species tend to experience more benign environments than endangered species, for example, by invading resource-rich or enemy-free environments, and this could mask the expression of deleterious alleles [45]. Consequently, invasive species may avoid the negative fitness effects of inbreeding more often than rare and declining species, even when subject to the same evolutionary dynamics of small population size. An important question is thus how much of the among-population variance in fitness (and underlying allele frequency differentiation) is caused by different degrees of purifying selection acting on weakly deleterious or conditionally neutral (I × E) genetic load? Answering this question could improve insights into the population-level consequences of different amounts of inbreeding in both invasive and endangered species. Spatial comparisons of population demography could be coupled with experimental and/or genomic assessments of genetic load to assess the conditions associated with effective purging. Here, an experimental strength is the high replication potential afforded by many invading populations with different demographic histories.
(c). The pros and cons of genetic admixture
Admixture is a well-established outcome for invasions and arises when multiple genotypes from genetically divergent populations in the native range come into secondary contact during invasion [46–48]. Hybridization and admixture are also a major issue for species at risk, where declining populations may be intentionally admixed during management efforts or may unintentionally hybridize with more abundant species [49,50]. The immediate fitness consequences of admixture can be complex, depending on the degree of divergence of the parental lineages, and can vary among F1 and more advanced recombinant hybrid generations [51,52]. Fitness effects can range from highly beneficial outbreeding to severely deleterious hybrid incompatibilities, decreasing in benefit as parental populations become more locally adapted [53].
A distinct benefit of admixture for both invasive and endangered species is increased heterozygosity at loci containing recessive deleterious mutations. Positive effects of admixture have been observed in invasive populations that show heterozygosity-fitness correlations in the introduced range [46,54]. The fitness benefits of a heterozygous genome are likely to be especially strong for small, declining and inbred populations of endangered species that are unable to purge deleterious mutations [55]. This is frequently seen in zoo or other extremely bottlenecked vertebrate populations. For example, the endangered Florida panther population shrank in size to as few as 20 individuals, with reduced heterozygosity at neutral markers and phenotypic evidence of inbreeding depression, including sperm deformities, kinked tails and reduced survival [56]. Intentional release of eight female Texas pumas into Florida created opportunities for admixture, doubling heterozygosity in the population and alleviating inbreeding depression in many traits.
In many cases, admixture also contributes to increases in standing VG and can broaden the genotypic space available to selection. When adaptive variants from a genetically distinct population introgress, natural selection can act on this enhanced standing variation without waiting for new mutations to arise de novo [57]. For these reasons, and because invasive species frequently experience novel selective environments in their introduced range, admixture has been implicated as a potential factor fuelling rapid evolution and the generation of novel invasive genotypes [33,48,58]. Nevertheless, direct evidence of a link between introgressed variation and the evolution of invasiveness is largely lacking to date. Population genomic studies of admixture/hybrid zones during invasions could test the adaptive introgression hypothesis using methods that identify the signal of differential introgression of positively selected loci against the null expectation based on the degree of admixture across the genomic background (e.g. genomic clines analysis) [59]. The benefit of adaptive introgression should be most pronounced when rates of gene flow and introgression are low relative to Ne and recombination rate, such that selection can efficiently incorporate beneficial variants while eliminating detrimental alleles. Therefore, adaptive introgression of positively selected genomic regions is likely to be much less common in endangered species, where low population sizes and low effective recombination limit the ability of selection to decouple maladaptive from adaptive introgressed alleles.
Populations of endangered species can also benefit from expanded genotypic variation resulting from admixture. Indeed, this is one reason that wildlife corridors are promoted for conservation [60]. The benefit of admixture is reduced when populations are locally adapted and therefore may be less beneficial for native relative to non-native species. Such an influx of genetic variation would be even more detrimental in small populations where natural selection is less efficient at eliminating locally maladapted alleles [61,62]. This genomic swamping of endangered species has become a serious conservation concern when small native populations meet abundant populations of reproductively compatible invasive species. Many native species show relatively low reproductive isolation from introduced species with which they have had no historical contact, and high numbers of introduced genotypes increase the likelihood of hybridization [63,64]. In addition to swamping locally adapted alleles, these native-invasive hybrids can put legal protections of native species in jeopardy as species definitions become questionable [65].
3. Phenotypic plasticity
The developmental, physiological and life-history modifications that have been widely observed in natural populations exposed to altered or novel environments can be due to plastic responses of individuals in a population as well as to selective trait changes [66,67]. To focus on this aspect of diversity, each genotype can be viewed as a repertoire of phenotypes expressed in different environments, or, more technically, a norm of reaction. This perspective makes it possible to evaluate both individual adaptive flexibility and genetic variation as expressed in novel environments. It is important to distinguish adaptive plastic responses, which maintain fitness across a range of environments, from phenotypic responses that arise directly from resource limits or other stresses and may not be adaptive. Reaction norms are products of evolution that, like any phenotypic trait, are inevitably subject to developmental, phylogenetic and genetic constraints [68]. Consequently, individual plasticity is best understood as the result of adaptive and stochastic evolution, rather than as a separate phenomenon [69]. Individual plasticity can play two key roles in the eco-evolutionary dynamics of natural populations. First, the capacity of individuals to express adaptive plasticity in response to novel or changing environments contributes to a population's viability. Second, existing norms of reaction, and genetic variation for these norms, influence the potential for future evolution of adaptive responses to new environments.
(a). Plasticity and tolerance of novel environments
Adaptive plasticity allows individual organisms to survive and reproduce in a variety of environmental conditions. Such plasticity can buffer populations or species from extinction when conditions change rapidly [70–72]. For example, some birds and mammals can advance life-history schedules through plastic responses to seasonal cues, allowing them to keep pace with altered timing of food availability due to rapid climate change (e.g. [73]); the many taxa whose populations lack such plasticity may face an enhanced risk of extinction [74,75]. Adaptive plasticity can include effects of parental environments on offspring phenotypes (transgenerational plasticity). For instance, in the sheepshead minnow, offspring growth rates, body mass and expected fecundity were highest at temperatures previously experienced by parent fish, regardless of whether that temperature was high or low [76]. As a result, warmer water temperatures did not cause negative effects on development and fitness. Adaptive transgenerational effects such as this may be most beneficial to species encountering gradual changes in the environment, including the rise in sea temperatures predicted under current models of global climate change.
Along with promoting species persistence, adaptive plasticity can facilitate the rapid spread of invasive species across diverse new habitats [77–79]. In both animals and plants, the ability to maintain net reproductive rates in contrasting environments promotes invasive spread [80,81]. Additionally, a plastic response to sharply increase fecundity in resource-rich environments may be an important attribute of invasive taxa [82,83] because it increases the ‘propagule pressure’ that fuels colonization [84,85]. Theoretical work indicates that greater adaptive plasticity should be associated with higher rates of colonization of new, and more diverse, environments [86]. However, recent meta-analyses disagree as to whether invasive species consistently show higher plasticity in general than native congeners [82,87]. This inconsistency in part reflects the different plasticity metrics and choice of traits used in various studies. Results of native versus invasive comparisons also depend on environmental variability in the home range that favours high plasticity, and on norm of reaction evolution following a species' introduction (for instance, canalization of a new adaptive phenotype following initially high plasticity) [88].
In addition to maintaining fitness across environments, adaptive plasticity can prevent a decrease in ecological breadth when a genetic bottleneck occurs, as each genotype can accommodate diverse conditions. Such plasticity can mitigate the ecological consequences of a prolonged bottleneck in an endangered species [89] or a short-term bottleneck due to an introduction event [90,91]. In these cases, geographically isolated populations may share similar broad patterns of individual plasticity instead of divergent, locally adapted norms of reaction [92,93]. However, as adaptive plasticity itself has a genetic basis, it can be compromised by the negative consequences of inbreeding and sustained bottlenecks. Moreover, two critical factors will determine the effectiveness of plasticity relative to selective evolution in maintaining the viability of populations exposed to novel environments. First, norms of reaction that evolved under past selection pressures may not encompass the range of phenotypes required to maintain fitness in the new circumstances, and can even produce a disrupted, maladaptive phenotype in response to a novel stress [94]. Second, even with sufficient existing plasticity, adaptive phenotypes will not be produced if changed cues fail to provide accurate information to elicit appropriate and timely responses [69,95]. In such cases, plasticity can promote extinction rather than persistence [29]. This potential adaptive limit may be particularly important in human-altered environments or following introduction to a new continent where abiotic and biotic factors that organisms have evolved to use as plasticity cues may be absent or disrupted.
(b). Plasticity and evolutionary potential
Phenotypic plasticity can allow a population to persist following a change in local conditions or introduction to a new range. If pre-existing reaction norms do not produce sufficiently adaptive phenotypes to maintain a population's viability (e.g. [96]), then further adaptive evolution of plasticity is an essential step to either avoid extinction or permit establishment (modelled by Chevin & Lande [97]; see [98–100] on selective evolution of reaction norms). In invasive species, rapid plasticity evolution can promote subsequent spread into new habitats. For instance, cane toads (Rhinella marina) that have spread to colder regions in Australia have evolved higher metabolic plasticity [101]; invasive freshwater populations of the marine-native copepod Eurytemora affinis have evolved increased ion-transport plasticity [96]; and a shrubby South African Senecio introduced into Spain has evolved greater reproductive output in wet conditions without any loss of fitness in its ancestral dry habitat [102].
Expanded repertoires of adaptive plasticity can result from the environmental heterogeneity encountered within and among sites in the new range, rather than from a changed directional selection pressure compared to the native range [103]. This finding is consistent with theoretical predictions that increased adaptive plasticity is selectively favoured in populations and metapopulations that encounter variable environments ([68,86,104–108] and references therein). This particular selective property may lead to a positive evolutionary feedback for greater invasiveness in non-native taxa that have sufficiently broad norms of reaction to survive their initial introduction. Because successful non-natives tend to (i) colonize disturbed, variable habitats and (ii) have high dispersal capabilities that would cause them to encounter diverse sites, they may be especially likely to evolve greater adaptive plasticity post-introduction [109]. Evolution of increasingly generalist norms of reaction will in turn promote an even broader ecological distribution across habitats ([67] and references therein), expanding the invasion front and possibly creating more contiguous populations that will accelerate colonization of new sites [85,110]. Conversely, if endangered taxa initially have less-plastic norms of reaction that confine them to a narrow range of conditions, they may not encounter the environmental variability that promotes evolution of broader plasticity.
Although a cost of plasticity could in theory limit this type of evolution, evidence for such a cost is weak [107,111], and recent models assume that any plasticity costs are outweighed by benefits (e.g. [97]). Indeed, the previous examples show that increasingly broad adaptive norms of reaction can evolve, at least in certain taxa, without fitness trade-offs that would entail a loss of adaptive responses to ancestral environments. In any given case, the evolution of greater plasticity—like that of any adaptive trait—depends on whether or not genetic or developmental constraints are present that limit the potential for selective change. How widespread among invasive taxa is the potential to evolve ‘jack of all trades’ genotypes? Is the lag phase between introduction and invasion commonly characterized by the evolution of greater adaptive plasticity? These questions can be explored through resurrection experiments designed to compare environmental response patterns of genotypes sampled from introduced taxa across time (e.g. [103]).
As with any phenotypic trait, evolution of plasticity requires genetic variation—in this case, genetic differences in reaction norms measured as genotype × environment (G × E) variance in a statistical model ([67,68,112,113] and references therein). Like other aspects of genetic diversity, populations and taxa will differ in G × E variation due to previous mutation, drift and selection. A defining feature of G × E variation is that a given set of genotypes may express different phenotypes in certain environments, but converge on similar phenotypes in others: in other words, genetic variance itself differs from one environment to another [114,115]. A novel environment such as a new range or altered habitat can lead to rapid adaptive evolution if genotypes in a population express different phenotypes [116]. By contrast, evolutionary response to natural selection is buffered in a new environment where similar phenotypes occur [69,95,117]. Consequently, differences in patterns of G × E variation will influence the ability of populations to evolve new adaptive responses following introduction or in situ environmental challenges. In addition, a better understanding of the genetic architectures underlying reaction norms, such as the effects of modularity versus pleiotropy of regulatory pathways [118], will better inform models of constraint on plasticity evolution in natural populations [67,119].
In general, it is not known whether phenotypes in natural populations are more likely to differ or converge in predicted future environments such as high CO2 and higher temperatures [120,121]. The question of adaptive evolutionary potential is of particular concern with respect to species that may face extinction in the absence of altered plasticity patterns. A case in point is reptiles with temperature-dependent sex determination, which are predicted to produce female-biased sex ratios in warmer future climates [122]. In a study of the leopard gecko Eublepharis macularius [123], populations were found to contain G × E variation for the proportion of males produced at different likely incubation temperatures. Such variation could fuel the selective evolution of new temperature thresholds for sex determination in future populations, promoting the species' persistence. Similarly, certain European populations of great tits (Parus major) contain genetic variation for temperature-based plasticity of reproductive timing, providing the potential for adaptive evolution of life-history plasticity in response to altered seasonal timing of food availability [124]. Examining norm of reaction diversity in populations of various organisms, including its expression under predicted future conditions, is a crucial step to assess (i) the potential for plasticity evolution to prevent extinction and (ii) the critical differences in evolutionary potential between endangered and invasive species.
4. Ecological niche
Invasive and endangered species exist at opposite ends of ecological spectra in geographical range size, niche breadth and population density [125–127] (figure 1). Geographical range size correlates positively with both local abundance [128,129] and niche breadth [130]. Therefore, understanding eco-evolutionary constraints on the ecological niche and local abundance could improve our understanding of why some species remain rare while others become invasive [127]. We use the term ‘ecological niche’ in the broad sense, as the range of environmental conditions and resources that influence the viability of local populations, either in the absence (fundamental niche) or in the presence (realized niche) of biotic interactions [131]. In this section, we apply the eco-evolutionary framework to the ecological niche concept and ask how differences in genetic variation and plasticity might explain niche differences in invasive and endangered species.
Some invasive species have evolved rapidly along environmental gradients during range expansion, increasing genetic variation for ecologically important traits and potentially expanding the niche well beyond that of the founder population(s). For example, clinal genetic variation in traits such as size and phenology have been documented in a number of widespread invasive species across geographical gradients, including latitude (e.g. [132]), elevation (e.g. [133]) and continentality (e.g. [134]). Interestingly, a species' climatic niche breadth in its non-native range often does not exceed its niche breadth in the native range—in other words, climatic niches are frequently conserved between ranges [135]. This raises a biological conundrum: if introduced populations evolve and plasticity allows persistence in a range of environments, why are similar climatic limits re-established in introduced populations [136]?
One explanation is the presence of constraints on niche breadth along environmental gradients that are also conserved across ranges [137]. Genetic constraints on niche breadth could include inbreeding depression, maladaptive gene flow or low heritable genetic variability in single traits, and also along multivariate trait axes (i.e. core trade-offs; reviewed by [138,139]). Core trade-offs provide perhaps the most convincing explanation for why niche limits should be conserved across disparate geographical ranges, despite the evolution of local adaptation [136]. For example, natural selection favours early flowering time and larger size at flowering across the native and introduced ranges of Lythrum salicaria, but evolution of these traits is constrained by a trade-off [132,140]. This trade-off limits the reproductive output of earlier flowering plants in high-latitude populations, helping to set the northern range limit in both the non-native and native ranges. Therefore, while adaptive evolution during invasion can contribute to the niche expansion of founder populations, genetic architecture can limit the extent of adaptive evolution and niche breath. As outlined in previous sections, several genetic attributes of non-native populations promote adaptive evolution of niche breadth. By contrast, high genetic drift, genetic load and small population will limit adaptive responses and could contribute to narrow niche breadth in endangered species.
In addition to selection from abiotic factors, biotic interactions can influence range margins [141], and are likely to differ in importance between native and introduced ranges as well as between invasive and endangered species. There are several mechanisms by which negative interactions (e.g. competition or predation) could contribute to evolutionary constraints on niche breadth (figure 2). First, if selection imposed by species interactions is antagonistic to abiotic selection pressures, this can limit the fitness of local populations and ultimately restrict niche breadth. This would occur if either the same trait was under antagonistic selection from abiotic and biotic factors, or if multiple genetically correlated traits were under antagonistic selection. For example, the evolution of increased competitive ability (EICA) hypothesis predicts a trade-off between herbivore defence and competitive ability in plants [142]. Non-native species that escape regulation by natural enemies, particularly specialists, would therefore experience relaxed selection on traits associated with defence, allowing a response to selection for increased competitive ability. While there is support for the EICA hypothesis in some species [143], the consequences for niche breadth in the non-native range of invasive species have, to our knowledge, not been investigated. Second, resource competition can constrain niche evolution by imposing strong selection against resource switching (i.e. stabilizing selection) even as resource availability drops to unsustainable levels [144,145]. Third, competition that reduces fitness of a focal species can suppress the expression of genetic variation for ecologically important traits, limiting the potential for selective changes in niche breadth. In an elegant experiment across a depth gradient in Californian vernal pools, release from competition enabled the annual plant Lasthenia fremontii to expand its niche breadth, and exposed additive genetic variation in the fundamental niche for which there was positive selection [146]. These examples suggest that a relaxation in evolutionary constraints caused by biotic interactions is likely to facilitate niche expansion, and could contribute to the success of invasive species that experience ecological release from native-range competitors or natural enemies. By contrast, biotic interactions might impose particularly strong evolutionary constraints on endangered species with narrow niche breath, compounded by low genetic variation or plasticity in traits affecting the outcome of interactions.
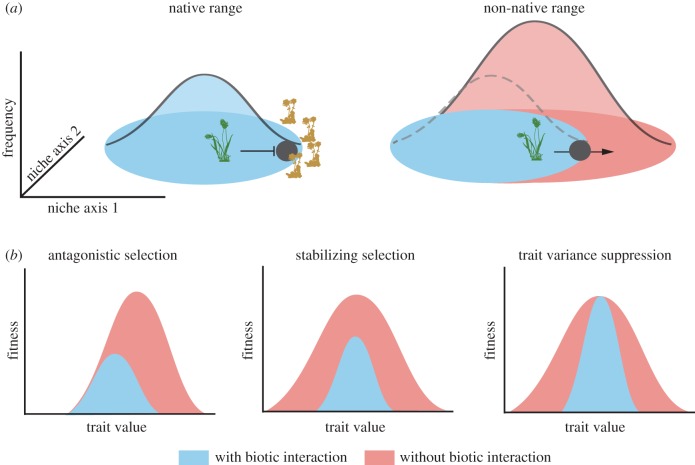
Figure 2.
Eco-evolutionary model of the ecological niche as an emergent property of genetic constraints and spatial variation in abiotic and biotic sources of natural selection. (a) The number of individuals in a population (y-axis) depends on environmental variables that vary along geographical gradients (niche axes 1 and 2), for example, moisture and temperature. Additional constraints on the niche of the focal species (darker green plant) are imposed by negative biotic interactions, such as a competitor (lighter yellow plants). Escape from negative biotic interaction represents an ecological release, which increases population vital rates across a range of environments and thereby expands the ecological niche. (b) Three evolutionary consequences of relaxed biotic interactions for an individual population, measured near the edge of the range of the focal plants (black circles in (a)). Selection at this location is measured either in the presence (lighter blue curve) or in the absence (darker red curve) of competition: antagonistic selection—ecological release relaxes antagonistic selection on a trait that is also under abiotic selection (e.g. phenology) or on a genetically correlated trait (e.g. size at reproduction); stabilizing selection—ecological release relaxes the strength of stabilizing selection across a range of trait values; trait variance suppression—competition suppresses the expression of heritable genetic variation, which increases following ecological release. In the first two cases, ecological release is also accompanied by an increase in absolute fitness as negative selection pressures are removed.
Ecological release is a common explanation for why some invasive species attain higher abundance in their new range [147], and release from negative interactions with competitors and pathogens even appears to have allowed some non-native species to expand their realized niches [148–150]. Indeed, there are examples of species that are considered endangered in their native range, sometimes restricted to a few populations, yet can attain broad non-native ranges (e.g. plants used in horticulture or forestry such as Pinus radiata, Lotus maculatus, Melaleuca quinquenervia) [149,151,152]. Niche expansion following invasion is usually interpreted as a purely ecological response to an altered biotic environment; the possible contribution and relative importance of evolution and phenotypic plasticity, though acknowledged, is rarely tested [149]. Hill et al. [153] showed that niche shifts are associated with the evolution of thermal tolerance in the mite, Halotydeus destructor, in Australia. Evolution has also been associated with the invasion of a narrowly endemic Canary Island shrub across large parts of California [34]. Another form of niche expansion is host shifting in herbivorous insects, including those introduced for biocontrol. For example, non-native populations of the beetle Ophraella communa in Japan have evolved to use Ambrosia trifida as a host, even though this plant is not used by O. communa in its native range. This host shift is partly explained by relaxed herbivore defences in non-native populations of A. trifida, after having escaped natural enemies for approximately 50 generations [154]. However, the contribution of relaxed selection to niche expansion and the role of genetic constraints are still unresolved.
Overall, endangered species can be predicted to experience stronger constraints on niche breadth than invasive taxa for several reasons. First, endangered species might experience greater genetic constraints than invasive species for reasons outlined in the previous sections (e.g. genetic bottlenecks and inbreeding). Second, endangered species might be characterized by especially strong negative biotic interactions. Indeed, invasive and endangered species can be discriminated by the strength of negative interactions with pathogens [155] and competitors [156]. Nonetheless, while rapid evolution associated with environmental gradients is recognized as being important for the persistence of endangered species in changing environments [157] and for the dynamics of species invasions [158], we know much less about the contribution of changing biotic interactions to niche evolution in invasive and endangered species [159]. Understanding whether negative biotic interactions impose constraints on niche evolution, in addition to a purely ecological restriction of niche breadth, is important for accurately predicting evolutionary responses of endangered and invasive species experiencing novel and changing environments.
5. Conclusion and future directions
Like Lewis Carroll's looking glass, endangered and invasive species appear similar at first glance, as both often (i) experience strong demographic bottlenecks, (ii) are subject to hybridization and introgression from other species or divergent populations, and (iii) encounter fitness challenges due to novel and changing environments. Yet these apparently similar reflections differ in key elements that lead to very different eco-evolutionary realities. Specifically, the potential for adaptive evolution of phenotypes and broad individual plasticity is predicted to be higher in invading species as a result of (i) rapid population growth following transient demographic bottlenecks, (ii) lower genetic load, (iii) greater environmental variability, and (iv) relaxed selection from natural enemies and competition. These evolutionary differences directly affect the breadth of the realized ecological niche and ultimately determine the abundance and distribution of species in nature.
Our discussion identifies key elements of what we believe could be a robust framework for exploring eco-evolutionary dynamics using comparative studies of invasive and endangered species. Contrasting invasive and rare or endangered species is not a novel concept, but has tended to focus on interspecific comparisons of functional traits rather than on the local ecological and demographic conditions that directly influence population dynamics. Understanding limits to adaptive evolution (including appropriate plasticity patterns) at the population level may be a more promising avenue of research. We believe these kinds of comparisons between endangered and invasive species represent an under-exploited opportunity to better understand and manage the abundance and distribution of species at both ends of the ecological spectrum. Experimental manipulations could compare phenotypic selection, reaction norms, neutral and functional genetic diversity of closely related endangered and invasive species in several different environments to better understand evolutionary potential and constraints. Endangered (or at least rare native) species that are invasive or spreading elsewhere would be particularly useful study systems to investigate in this way.
One potential obstacle to developing such a framework is the fact that researchers investigating invasive and endangered species rarely collaborate—indeed the authors of this paper work primarily on invasive species and as a result our discussion is more heavily weighted towards this area. Nevertheless, we hope that this review will encourage better communication between these two fields. After all, the goals of conserving endangered species and preventing invasions both require a comprehensive understanding of the evolutionary and ecological factors that affect population persistence in a changing world. Combining knowledge gained from the thousands of published ecological, evolutionary and population genetic studies of invasive and endangered species could lead to more robust tests of theoretical foundations of conservation biology, and to a comprehensive, unified framework for the management of global biodiversity.
Acknowledgements
The authors are grateful to A. Hendry, K. Gotanada and E. Svensson for the invitation to contribute to this special issue and suggestions that improved the manuscript. We are also grateful for conceptual discussions and manuscript suggestions from S. Yakimowski.
Authors' contributions
All authors contributed to the conceptual development, writing and revising of the manuscript. R.I.C. initiated and coordinated the project, and all other authors contributed equally.
Competing interests
We have no competing interests.
Funding
R.I.C. is funded by Canada Research Chair (CRC) and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). K.M.D. is funded by a United States Department of Agriculture (USDA) grant no. 2015-67013-23000. S.R.K. is funded by a USDA Cooperative Agreement project no. 8062-22620-004-17S. S.E.S. is funded by the New Phytologist Trust. J.M.A. is supported by ETH Zurich funding to the Plant Ecology group.
References
- 1.Ricciardi A. 2007. Are modern biological invasions an unprecedented form of global change? Conserv. Biol. 21, 329–336. ( 10.1111/j.1523-1739.2006.00615.x) [DOI] [PubMed] [Google Scholar]
- 2.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752 ( 10.1126/science.1246752) [DOI] [PubMed] [Google Scholar]
- 3.Carroll L. 1871. Through the looking-glass, and what alice found there. London, UK: Macmillan. [Google Scholar]
- 4.Hufbauer RA, Facon B, Ravigné V, Turgeon J, Foucaud J, Lee CE, Rey O, Estoup A. 2012. Anthropogenically induced adaptation to invade (AIAI): contemporary adaptation to human-altered habitats within the native range can promote invasions. Evol. Appl. 5, 89–101. ( 10.1111/j.1752-4571.2011.00211.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CE, Gelembiuk GW. 2008. Evolutionary origins of invasive populations. Evol. Appl. 1, 427–448. ( 10.1111/j.1752-4571.2008.00039.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeschke JM, Strayer DL. 2008. Are threat status and invasion success two sides of the same coin? Ecography 31, 124–130. ( 10.1111/j.2007.0906-7590.05343.x) [DOI] [Google Scholar]
- 7.Colautti RI, Grigorovich IA, MacIsaac HJ. 2006. Propagule pressure: a null model for biological invasions. Biol. Invasions 8, 1023–1037. ( 10.1007/s10530-005-3735-y) [DOI] [Google Scholar]
- 8.van Kleunen M, Weber E, Fischer M. 2010. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 13, 235–245. ( 10.1111/j.1461-0248.2009.01418.x) [DOI] [PubMed] [Google Scholar]
- 9.van Kleunen M, Dawson W, Maurel N. 2015. Characteristics of successful alien plants. Mol. Ecol. 24, 1954–1968. ( 10.1111/mec.13013) [DOI] [PubMed] [Google Scholar]
- 10.Agrawal AA, Kotanen PM, Mitchell CE, Power AG, Godsoe W, Klironomos J. 2005. Enemy release? An experiment with congeneric plant pairs and diverse above- and belowground enemies. Ecology 86, 2979–2989. ( 10.1890/05-0219) [DOI] [Google Scholar]
- 11.Ordonez A. 2014. Functional and phylogenetic similarity of alien plants to co-occurring natives. Ecology 95, 1191–1202. ( 10.1890/13-1002.1) [DOI] [PubMed] [Google Scholar]
- 12.Hendry AP, Gotanda KM, Svensson EI. 2017. Human influences on evolution, and the ecological and societal consequences. Phil. Trans. R. Soc. B 372, 20160028 ( 10.1098/rstb.2016.0028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. ( 10.1111/j.1365-294X.2007.03538.x) [DOI] [PubMed] [Google Scholar]
- 14.Amos W, Balmford A. 2001. When does conservation genetics matter? Heredity 87, 257–265. ( 10.1046/j.1365-2540.2001.00940.x) [DOI] [PubMed] [Google Scholar]
- 15.Hedrick PW. 2001. Conservation genetics: where are we now? Trends Ecol. Evol. 16, 629–636. ( 10.1016/S0169-5347(01)02282-0) [DOI] [Google Scholar]
- 16.Reznick DN, Ghalambor CK. 2001. The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica 112–113, 183–198. ( 10.1023/A:1013352109042) [DOI] [PubMed] [Google Scholar]
- 17.Frankham R. 2005. Genetics and extinction. Biol. Conserv. 126, 131–140. ( 10.1016/j.biocon.2005.05.002) [DOI] [Google Scholar]
- 18.Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA. 2015. Genetic rescue to the rescue. Trends Ecol. Evol. 30, 42–49. ( 10.1016/j.tree.2014.10.009) [DOI] [PubMed] [Google Scholar]
- 19.Carlson SM, Cunningham CJ, Westley PAH. 2014. Evolutionary rescue in a changing world. Trends Ecol. Evol. 29, 521–530. ( 10.1016/j.tree.2014.06.005) [DOI] [PubMed] [Google Scholar]
- 20.Frankham R. 2005. Resolving the genetic paradox in invasive species. Heredity 94, 385 ( 10.1038/sj.hdy.6800634) [DOI] [PubMed] [Google Scholar]
- 21.Hufbauer RA. 2008. Biological invasions: paradox lost and paradise gained. Curr. Biol. 18, R246–R247. ( 10.1016/j.cub.2008.01.038) [DOI] [PubMed] [Google Scholar]
- 22.Richards CM. 2000. Inbreeding depression and genetic rescue in a plant metapopulation. Am. Nat. 155, 383–394. ( 10.1086/303324) [DOI] [PubMed] [Google Scholar]
- 23.Pannell JR, Fields PD. 2014. Evolution in subdivided plant populations: concepts, recent advances and future directions. New Phytol. 201, 417–432. ( 10.1111/nph.12495) [DOI] [PubMed] [Google Scholar]
- 24.Fields PD, Taylor DR. 2014. Determinants of genetic structure in a nonequilibrium metapopulation of the plant Silene latifolia. PLoS ONE 9, e104575 ( 10.1371/journal.pone.0104575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade MJ, Goodnight CJ, Night JG. 1998. The theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution 52, 1537–1553. ( 10.2307/2411328) [DOI] [PubMed] [Google Scholar]
- 26.Faubet P, Gaggiotti OE. 2008. A new Bayesian method to identify the environmental factors that influence recent migration. Genetics 178, 1491–1504. ( 10.1534/genetics.107.082560) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed DH, Frankham R. 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. ( 10.1046/j.1523-1739.2003.01236.x) [DOI] [Google Scholar]
- 28.Taft HR, Roff DA. 2011. Do bottlenecks increase additive genetic variance? Conserv. Genet. 13, 333–342. ( 10.1007/s10592-011-0285-y) [DOI] [Google Scholar]
- 29.Reed DH. 2010. Albatrosses, eagles and newts, Oh My!: exceptions to the prevailing paradigm concerning genetic diversity and population viability? Anim. Conserv 13, 448–457. ( 10.1111/j.1469-1795.2010.00353.x) [DOI] [Google Scholar]
- 30.Nei M, Maruyama T, Chakraborty R. 1975. The bottleneck effect and genetic variability in populations. Evolution 29, 1–10. ( 10.2307/2407137) [DOI] [PubMed] [Google Scholar]
- 31.Lombaert E, Guillemaud T, Cornuet JM, Malausa T, Facon B, Estoup A. 2010. Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PLoS ONE 5, e9743 ( 10.1371/journal.pone.0009743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pujol B, Pannell JR. 2008. Reduced responses to selection after species range expansion. Science 321, 96 ( 10.1126/science.1157570) [DOI] [PubMed] [Google Scholar]
- 33.Lavergne S, Molofsky J. 2007. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. USA 104, 3883–3888. ( 10.1073/pnas.0607324104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dlugosch KM, Parker IM. 2008. Invading populations of an ornamental shrub show rapid life history evolution despite genetic bottlenecks. Ecol. Lett. 11, 701–709. ( 10.1111/j.1461-0248.2008.01181.x) [DOI] [PubMed] [Google Scholar]
- 35.Bijlsma R, Bundgaard J, Boerema AC. 2000. Does inbreeding affect the extinction risk of small populations? Predictions from Drosophila. J. Evol. Biol. 13, 502–514. ( 10.1046/j.1420-9101.2000.00177.x) [DOI] [Google Scholar]
- 36.Briskie JV, Mackintosh M. 2004. Hatching failure increases with severity of population bottlenecks in birds. Proc. Natl Acad. Sci. USA 101, 558–561. ( 10.1073/pnas.0305103101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaeuffer R, Coltman DW, Chapuis J-L, Pontier D, Réale D. 2007. Unexpected heterozygosity in an island mouflon population founded by a single pair of individuals. Proc. R. Soc. B 274, 527–533. ( 10.1098/rspb.2006.3743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labonne J, Kaeuffer R, Guéraud F, Zhou M, Manicki A, Hendry AP. 2016. From the bare minimum: genetics and selection in populations founded by only a few parents. Evol. Ecol. Res. 17, 21–34. [Google Scholar]
- 39.Klopfstein S, Currat M, Excoffier L. 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490. ( 10.1093/molbev/msj057) [DOI] [PubMed] [Google Scholar]
- 40.Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. 2013. On the accumulation of deleterious mutations during range expansions. Mol. Ecol. 22, 5972–5982. ( 10.1111/mec.12524) [DOI] [PubMed] [Google Scholar]
- 41.Flaxman SM. 2013. Surfing downhill: when should population range expansion be characterized by reductions in fitness? Mol. Ecol. 22, 5963–5965. ( 10.1111/mec.12564) [DOI] [PubMed] [Google Scholar]
- 42.Marchini GL, Sherlock NC, Ramakrishnan AP, Rosenthal DM, Cruzan MB. 2015. Rapid purging of genetic load in a metapopulation and consequences for range expansion in an invasive plant. Biol. Invasions 18, 183–196. ( 10.1007/s10530-015-1001-5) [DOI] [Google Scholar]
- 43.Otto SP, Whitlock MC. 1997. The probability of fixation in populations of changing size. Genetics 146, 723–733. ( 10.1534/genetics.104.040089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheptou P-O, Donohue K. 2011. Environment-dependent inbreeding depression: its ecological and evolutionary significance. New Phytol. 189, 395–407. ( 10.1111/j.1469-8137.2010.03541.x) [DOI] [PubMed] [Google Scholar]
- 45.Schrieber K, Lachmuth S. 2016. The Genetic Paradox of Invasions revisited: the potential role of inbreeding×environment interactions in invasion success. Biol. Rev. ( 10.1111/brv.12263) [DOI] [PubMed] [Google Scholar]
- 46.Keller SR, Taylor DR. 2010. Genomic admixture increases fitness during a biological invasion. J. Evol. Biol. 23, 1720–1731. ( 10.1111/j.1420-9101.2010.02037.x) [DOI] [PubMed] [Google Scholar]
- 47.Kolbe JJ, Glor RE, Rodríguez Schettino L, Lara AC, Larson A, Losos JB. 2004. Genetic variation increases during biological invasion by a Cuban lizard. Nature 431, 177–181. ( 10.1038/nature02807) [DOI] [PubMed] [Google Scholar]
- 48.Dlugosch KM, Anderson SR, Braasch J, Cang FA, Gillette HD. 2015. The devil is in the details: genetic variation in introduced populations and its contributions to invasion. Mol. Ecol. 24, 2095–2111. ( 10.1111/mec.13183) [DOI] [PubMed] [Google Scholar]
- 49.Levin DA, Francisco-Ortega J, Jansen RK. 1996. Hybridization and the extinction of rare plant species. Conserv. Biol. 10, 10–16. ( 10.1046/j.1523-1739.1996.10010010.x) [DOI] [Google Scholar]
- 50.Wolf DE, Takebayashi N, Rieseberg LH. 2001. Predicting the risk of extinction through hybridization. Conserv. Biol. 15, 1039–1053. ( 10.1046/j.1523-1739.2001.0150041039.x) [DOI] [Google Scholar]
- 51.Lynch M. 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45, 622–629. ( 10.2307/2409915) [DOI] [PubMed] [Google Scholar]
- 52.Barton NH. 2001. The role of hybridization in evolution. Mol. Ecol. 10, 551–568. ( 10.1046/j.1365-294x.2001.01216.x) [DOI] [PubMed] [Google Scholar]
- 53.Garant D, Forde SE, Hendry AP. 2007. The multifarious effects of dispersal and gene flow on contemporary adaptation. Funct. Ecol. 21, 434–443. ( 10.1111/j.1365-2435.2006.01228.x) [DOI] [Google Scholar]
- 54.Keller SR, Fields PD, Berardi AE, Taylor DR. 2014. Recent admixture generates heterozygosity-fitness correlations during the range expansion of an invading species. J. Evol. Biol. 27, 616–627. ( 10.1111/jeb.12330) [DOI] [PubMed] [Google Scholar]
- 55.Hedrick PW, Kalinowski ST. 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31, 139–162. ( 10.1146/annurev.ecolsys.31.1.139) [DOI] [Google Scholar]
- 56.Johnson WE, et al. 2010. Genetic restoration of the Florida panther. Science 329, 1641–1645. ( 10.1126/science.1192891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gompert Z, Lucas LK, Nice CC, Fordyce JA, Forister ML, Buerkle CA. 2012. Genomic regions with a history of divergent selection affect fitness of hybrids between two butterfly species. Evolution 66, 2167–2181. ( 10.1111/j.1558-5646.2012.01587.x) [DOI] [PubMed] [Google Scholar]
- 58.Facon B, Pointier JP, Jarne P, Sarda V, David P. 2008. High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr. Biol. 18, 363–367. ( 10.1016/j.cub.2008.01.063) [DOI] [PubMed] [Google Scholar]
- 59.Gompert Z, Alex BC. 2011. Bayesian estimation of genomic clines. Mol. Ecol. 20, 2111–2127. ( 10.1111/j.1365-294X.2011.05074.x) [DOI] [PubMed] [Google Scholar]
- 60.Beier P, Gregory AJ. 2012. Desperately seeking stable 50-year-old landscapes with patches and long, wide corridors. PLoS Biol. 10, 31001253 ( 10.1371/journal.pbio.1001253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prentis PJ, White EM, Radford IJ, Lowe AJ, Clarke AR. 2007. Can hybridization cause local extinction: a case for demographic swamping of the Australian native Senecio pinnatifolius by the invasive Senecio madagascariensis? New Phytol. 176, 902–912. ( 10.1111/j.1469-8137.2007.02217.x) [DOI] [PubMed] [Google Scholar]
- 62.Verhoeven KJF, Macel M, Wolfe LM, Biere A. 2011. Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc. R. Soc. B 278, 2–8. ( 10.1098/rspb.2010.1272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonald DB, Parchman TL, Bower MR, Hubert WA, Rahel FJ. 2008. An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proc. Natl Acad. Sci. USA 105, 10 837–10 842. ( 10.1073/pnas.0712002105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovach RP, Luikart G, Lowe WH, Boyer MC, Muhlfeld CC. 2016. Risk and efficacy of human-enabled interspecific hybridization for climate-change adaptation: response to Hamilton and Miller (2016). Conserv. Biol. 30, 428–430. ( 10.1111/cobi.12678) [DOI] [PubMed] [Google Scholar]
- 65.Allendorf FW, Leary RF, Hitt NP, Knudsen KL, Boyer MC, Spruell P. 2005. Cutthroat trout hybridization and the U.S. Endangered Species Act: one species, two policies. Conserv. Biol. 19, 1326–1328. ( 10.1111/j.1523-1739.2005.00223.x) [DOI] [Google Scholar]
- 66.Hendry AP, Farrugia TJ, Kinnison MT. 2008. Human influences on rates of phenotypic change in wild animal populations. Mol. Ecol. 17, 20–29. ( 10.1111/j.1365-294X.2007.03428.x) [DOI] [PubMed] [Google Scholar]
- 67.Chevin L-M, Collins S, Lefèvre F. 2013. Phenotypic plasticity and evolutionary demographic responses to climate change: taking theory out to the field. Funct. Ecol. 27, 967–979. ( 10.1111/j.1365-2435.2012.02043.x) [DOI] [Google Scholar]
- 68.Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68. ( 10.2307/2097172) [DOI] [Google Scholar]
- 69.Sultan SE. 2015. Organism and environment: ecological development, niche construction, and adaptation. New York, NY: Oxford University Press. [Google Scholar]
- 70.Carroll SP, Hendry AP, Reznick DN, Fox CW. 2007. Evolution on ecological time-scales. Funct. Ecol. 21, 387–393. ( 10.1111/j.1365-2435.2007.01289.x) [DOI] [Google Scholar]
- 71.Visser ME. 2008. Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc. R. Soc. B 275, 649–659. ( 10.1098/rspb.2007.0997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicotra AB, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. ( 10.1016/j.tplants.2010.09.008) [DOI] [PubMed] [Google Scholar]
- 73.Berteaux D, Réale D, McAdam AG, Boutin S, Reale D. 2004. Keeping pace with fast climate change: can arctic life count on evolution? Integr. Comp. Biol. 44, 140–151. ( 10.1093/icb/44.2.140) [DOI] [PubMed] [Google Scholar]
- 74.Visser ME, Both C. 2005. Shifts in phenology due to global climate change: the need for a yardstick. Proc. R. Soc. B 272, 2561–2569. ( 10.1098/rspb.2005.3356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willis CG, Ruhfel B, Primack RB, Miller-Rushing AJ, Davis CC. 2008. Phylogenetic patterns of species loss in Thoreau's woods are driven by climate change. Proc. Natl Acad. Sci. USA 105, 17 029–17 033. ( 10.1073/pnas.0806446105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salinas S, Munch SB. 2012. Thermal legacies: transgenerational effects of temperature on growth in a vertebrate. Ecol. Lett. 15, 159–163. ( 10.1111/j.1461-0248.2011.01721.x) [DOI] [PubMed] [Google Scholar]
- 77.Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA. 2000. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710. ( 10.1890/1051-0761(2000)010%5B0689:BICEGC%5D2.0.CO;2) [DOI] [Google Scholar]
- 78.Sultan SE. 2004. Promising directions in plant phenotypic plasticity. Perspect. Plant Ecol. Evol. Syst. 6, 227–233. ( 10.1078/1433-8319-00082) [DOI] [Google Scholar]
- 79.Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. 2006. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 9, 981–993. ( 10.1111/j.1461-0248.2006.00950.x) [DOI] [PubMed] [Google Scholar]
- 80.Lockwood JL, Cassey P, Blackburn T. 2005. The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 20, 223–228. ( 10.1016/j.tree.2005.02.004) [DOI] [PubMed] [Google Scholar]
- 81.Theoharides KA, Dukes JS. 2007. Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytol. 176, 256–273. ( 10.1111/j.1469-8137.2007.02207.x) [DOI] [PubMed] [Google Scholar]
- 82.Davidson AM, Jennions M, Nicotra AB. 2011. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecol. Lett. 14, 419–431. ( 10.1111/j.1461-0248.2011.01596.x) [DOI] [PubMed] [Google Scholar]
- 83.Budy P, Thiede GP, Lobón-Cerviá J, Fernandez GG, Mchugh P, Mcintosh A, Voøllestad LA, Becares E, Jellyman P. 2013. Limitation and facilitation of one of the world's most invasive fish: an intercontinental comparison. Ecology 94, 356–367. ( 10.1890/12-0628.1) [DOI] [PubMed] [Google Scholar]
- 84.Simberloff D. 2009. The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102. ( 10.1146/annurev.ecolsys.110308.120304) [DOI] [Google Scholar]
- 85.Matesanz S, Sultan SE. 2013. High-performance genotypes in an introduced plant: insights to future invasiveness. Ecology 94, 2464–2474. ( 10.1890/12-1359.1) [DOI] [PubMed] [Google Scholar]
- 86.Thibert-Plante X, Hendry AP. 2011. The consequences of phenotypic plasticity for ecological speciation. J. Evol. Biol. 24, 326–342. ( 10.1111/j.1420-9101.2010.02169.x) [DOI] [PubMed] [Google Scholar]
- 87.Palacio-López K, Gianoli E. 2011. Invasive plants do not display greater phenotypic plasticity than their native or non-invasive counterparts: a meta-analysis. Oikos 120, 1393–1401. ( 10.1111/j.1600-0706.2010.19114.x) [DOI] [Google Scholar]
- 88.Lande R. 2015. Evolution of phenotypic plasticity in colonizing species. Mol. Ecol. 24, 2038–2045. ( 10.1111/mec.13037) [DOI] [PubMed] [Google Scholar]
- 89.Noel F, Machon N, Porcher E. 2007. No genetic diversity at molecular markers and strong phenotypic plasticity in populations of Ranunculus nodiflorus, an endangered plant species in France. Ann. Bot. 99, 1203–1212. ( 10.1093/aob/mcm067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Williams DG, Mack RN, Black RA. 1995. Ecophysiology of introduced Pennisetum setaceum on Hawaii: the role of phenotypic plasticity. Ecology 76, 1569–1580. ( 10.2307/1938158) [DOI] [Google Scholar]
- 91.Geng YP, Pan XY, Xu CY, Zhang WJ, Li B, Chen JK, Lu BR, Song ZP. 2007. Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol. Invasions 9, 245–256. ( 10.1007/s10530-006-9029-1) [DOI] [Google Scholar]
- 92.Matesanz S, Horgan-Kobelski T, Sultan SE. 2012. Phenotypic plasticity and population differentiation in an ongoing species invasion. PLoS ONE 7, e44955 ( 10.1371/journal.pone.0044955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sassenhagen I, Wilken S, Godhe A, Rengefors K. 2015. Phenotypic plasticity and differentiation in an invasive freshwater microalga. Harmful Algae 41, 38–45. ( 10.1016/j.hal.2014.11.001) [DOI] [Google Scholar]
- 94.Ghalambor CK, McKay JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407. ( 10.1111/j.1365-2435.2007.01283.x) [DOI] [Google Scholar]
- 95.Sultan SE. 2007. Development in context: the timely emergence of eco-devo. Trends Ecol. Evol. 22, 575–582. ( 10.1016/j.tree.2007.06.014) [DOI] [PubMed] [Google Scholar]
- 96.Lee CE. 2016. Evolutionary mechanisms of habitat invasions, using the copepod Eurytemora affinis as a model system. Evol. Appl. 9, 248–270. ( 10.1111/eva.12334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chevin LM, Lande R. 2010. When do adaptive plasticity and genetic evolution prevent extinction of a density-regulated population? Evolution 64, 1143–1150. ( 10.1111/j.1558-5646.2009.00875.x) [DOI] [PubMed] [Google Scholar]
- 98.West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York, NY: Oxford University Press.
- 99.Badyaev AV. 2009. Evolutionary significance of phenotypic accommodation in novel environments: an empirical test of the Baldwin effect. Phil. Trans. R. Soc. B 364, 1125–1141. ( 10.1098/rstb.2008.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moczek AP, et al. 2011. The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B 278, 2705–2713. ( 10.1098/rspb.2011.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Winwood-Smith HS, Alton LA, Franklin CE, White CR. 2015. Does greater thermal plasticity facilitate range expansion of an invasive terrestrial anuran into higher latitudes? Conserv. Physiol. 3, 1–11. ( 10.1093/conphys/cov010.Introduction) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caño L, Escarré J, Fleck I, Blanco-Moreno JM, Sans FX. 2008. Increased fitness and plasticity of an invasive species in its introduced range: a study using Senecio pterophorus. J. Ecol. 96, 468–476. ( 10.1111/j.1365-2745.2008.01363.x) [DOI] [Google Scholar]
- 103.Sultan SE, Horgan-Kobelski T, Nichols LM, Riggs CE, Waples RK. 2013. A resurrection study reveals rapid adaptive evolution within populations of an invasive plant. Evol. Appl. 6, 266–278. ( 10.1111/j.1752-4571.2012.00287.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moran NA. 1992. The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971 ( 10.1086/285369) [DOI] [Google Scholar]
- 105.Sultan SE, Spencer HG. 2002. Metapopulation structure favors plasticity over local adaptation. Am. Nat. 160, 271–283. ( 10.1086/341015) [DOI] [PubMed] [Google Scholar]
- 106.Berrigan D, Scheiner SM. 2004. Modeling the evolution of phenotypic plasticity. In Phenotypic plasticity: functional and conceptual approaches (eds DeWitt TM, Scheiner SM), pp. 82–97. New York, NY: Oxford University Press. [Google Scholar]
- 107.Scheiner SM, Holt RD. 2012. The genetics of phenotypic plasticity. X. Variation versus uncertainty. Ecol. Evol. 2, 751–767. ( 10.1002/ece3.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baythavong BS. 2011. Linking the spatial scale of environmental variation and the evolution of phenotypic plasticity: selection favors adaptive plasticity in fine-grained environments. Am. Nat. 178, 75–87. ( 10.1086/660281) [DOI] [PubMed] [Google Scholar]
- 109.Sultan SE, Matesanz S. 2015. An ideal weed: plasticity and invasiveness in Polygonum cespitosum. Ann. N. Y. Acad. Sci. 1360, 101–119. ( 10.1111/nyas.12946) [DOI] [PubMed] [Google Scholar]
- 110.Matesanz S, Horgan-Kobelski T, Sultan SE. 2015. Evidence for rapid ecological range expansion in a newly invasive plant. AoB Plants 7, plv038. ( 10.1093/aobpla/plv038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Van Buskirk J, Steiner UK. 2009. The fitness costs of developmental canalization and plasticity. J. Evol. Biol. 22, 852–860. ( 10.1111/j.1420-9101.2009.01685.x) [DOI] [PubMed] [Google Scholar]
- 112.Via S, Lande R. 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522. ( 10.2307/2408649) [DOI] [PubMed] [Google Scholar]
- 113.Scheiner SM. 2006. Genotype-environment interactions and evolution. In Evolutionary genetics: concepts and case studies (eds Fox CW, Wolf CM), pp. 326–338. London, UK: Oxford University Press. [Google Scholar]
- 114.Conner JK, Hartl DL. 2004. A primer of ecological genetics. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 115.Kruuk LEB, Slate J, Wilson AJ. 2008. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 39, 525–548. ( 10.1146/annurev.ecolsys.39.110707.173542) [DOI] [Google Scholar]
- 116.Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Evol. Biol. 22, 1435–1446. ( 10.1111/j.1420-9101.2009.01754.x) [DOI] [PubMed] [Google Scholar]
- 117.Merilä J, Sheldon BC, Kruuk LEB. 2001. Explaining stasis: microevolutionary studies in natural populations. Genetica 112–113, 199–222. ( 10.1023/A:1013391806317) [DOI] [PubMed] [Google Scholar]
- 118.Papakostas S, Vøllestad LA, Bruneaux M, Aykanat T, Vanoverbeke J, Ning M, Primmer CR, Leder EH. 2014. Gene pleiotropy constrains gene expression changes in fish adapted to different thermal conditions. Nat. Commun. 5, 4071 ( 10.1038/ncomms5071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anderson JT, Wagner MR, Rushworth CA, Prasad KVSK, Mitchell-Olds T. 2014. The evolution of quantitative traits in complex environments. Heredity (Edinb). 112, 4–12. ( 10.1038/hdy.2013.33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rice KJ, Emery NC. 2003. Managing microevolution: restoration in the face of global change. Front. Ecol. Environ. 1, 469–478. ( 10.1890/1540-9295(2003)001%5B0469:MMRITF%5D2.0.CO;2) [DOI] [Google Scholar]
- 121.Merilä J, Hendry AP. 2014. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl. 7, 1–14. ( 10.1111/eva.12137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hulin V, Delmas V, Girondot M, Godfrey MH, Guillon JM. 2009. Temperature-dependent sex determination and global change: are some species at greater risk? Oecologia 160, 493–506. ( 10.1007/s00442-009-1313-1) [DOI] [PubMed] [Google Scholar]
- 123.Janes DE, Wayne ML. 2006. Evidence for a genotype×enviroment interaction in sex-determining response to incubation temperature in the leopard gecko, Eublepharis macularius. Herpetologica 62, 56–62. ( 10.1655/04-104.1) [DOI] [Google Scholar]
- 124.Nussey DH, Postma E, Gienapp P, Visser ME. 2005. Selection on heritable phenotypic plasticity in a wild bird population. Science 310, 304–306. ( 10.1126/science.1117004) [DOI] [PubMed] [Google Scholar]
- 125.Goodwin BJ, McAllister AJ, Fahrig L. 1999. Predicting invasiveness of plant species based on biological information. Conserv. Biol. 13, 422–426. ( 10.1046/j.1523-1739.1999.013002422.x) [DOI] [Google Scholar]
- 126.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. 2000. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B 267, 1947–1952. ( 10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bradshaw CJA, Giam X, Tan HTW, Brook BW, Sodhi NS. 2008. Threat or invasive status in legumes is related to opposite extremes of the same ecological and life-history attributes. J. Ecol. 96, 869–883. ( 10.1111/j.1365-2745.2008.01408.x) [DOI] [Google Scholar]
- 128.Gaston KJ, Blackburn TM, Lawton JH. 1997. Interspecific abundance-range size relationships: an appraisal of mechanisms. J. Anim. Ecol. 66, 579–601. ( 10.2307/5951) [DOI] [Google Scholar]
- 129.Holt RD, Lawton JH, Gaston KJ, Blackburn TM. 1997. On the relationship between range size and local abundance: back to basics. Oikos 78, 183–190. ( 10.2307/3545815) [DOI] [Google Scholar]
- 130.Slatyer RA, Hirst M, Sexton JP. 2013. Niche breadth predicts geographical range size: a general ecological pattern. Ecol. Lett. 16, 1104–1114. ( 10.1111/ele.12140) [DOI] [PubMed] [Google Scholar]
- 131.Soberón J. 2007. Grinnellian and Eltonian niches and geographic distributions of species. Ecol. Lett. 10, 1115–1123. ( 10.1111/j.1461-0248.2007.01107.x) [DOI] [PubMed] [Google Scholar]
- 132.Colautti RI, Barrett SCH. 2013. Rapid adaptation to climate facilitates range expansion of an invasive plant. Science 342, 364–366. ( 10.1126/science.1242121) [DOI] [PubMed] [Google Scholar]
- 133.Monty A, Mahy G. 2009. Clinal differentiation during invasion: Senecio inaequidens (Asteraceae) along altitudinal gradients in Europe. Oecologia 159, 305–315. ( 10.1007/s00442-008-1228-2) [DOI] [PubMed] [Google Scholar]
- 134.Leger EA, Rice KJ. 2007. Assessing the speed and predictability of local adaptation in invasive California poppies (Eschscholzia californica). J. Evol. Biol. 20, 1090–1103. ( 10.1111/j.1420-9101.2006.01292.x) [DOI] [PubMed] [Google Scholar]
- 135.Petitpierre B, Kueffer C, Broennimann O, Randin C, Daehler C, Guisan A. 2012. Climatic niche shifts are rare among terrestrial plant invaders. Science 335, 1344–1348. ( 10.1126/science.1215933) [DOI] [PubMed] [Google Scholar]
- 136.Alexander JM, Edwards PJ. 2010. Limits to the niche and range margins of alien species. Oikos 119, 1377–1386. ( 10.1111/j.1600-0706.2009.17977.x) [DOI] [Google Scholar]
- 137.Alexander JM. 2013. Evolution under changing climates: climatic niche stasis despite rapid evolution in a non-native plant. Proc. R. Soc. B 280, 20131446 ( 10.1098/rspb.2013.1446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Blows MW, Hoffmann AA. 2005. A reassessment of genetic limits to evolutionary change. Ecology. 86, 1371–1384. ( 10.1890/04-1209) [DOI] [Google Scholar]
- 139.Sexton JP, McIntyre PJ, Angert AL, Rice KJ. 2009. Evolution and ecology of species range limits. Annu. Rev. Ecol. Evol. Syst. 40, 415–436. ( 10.1146/annurev.ecolsys.110308.120317) [DOI] [Google Scholar]
- 140.Colautti RI, Eckert CG, Barrett SCH. 2010. Evolutionary constraints on adaptive evolution during range expansion in an invasive plant. Proc. R. Soc. B 277, 1799–1806. ( 10.1098/rspb.2009.2231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Case TJ, Holt RD, McPeek MA, Keitt TH. 2005. The community context of species’ borders: ecological and evolutionary perspectives. Oikos 108, 28–46. ( 10.1111/j.0030-1299.2005.13148.x) [DOI] [Google Scholar]
- 142.Blossey B, Nötzold R. 1995. Evolution of increased competitive ability in plants: a hypothesis. J. Ecol. 83, 887–889. ( 10.2307/2261425) [DOI] [Google Scholar]
- 143.Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Prati D. 2005. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11. ( 10.1007/s00442-005-0070-z) [DOI] [PubMed] [Google Scholar]
- 144.Price TD, Kirkpatrick M. 2009. Evolutionarily stable range limits set by interspecific competition. Proc. R. Soc. B 276, 1429–1434. ( 10.1098/rspb.2008.1199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dlugosch KM, Cang FA, Barker BS, Andonian K, Swope SM, Rieseberg LH. 2015. Evolution of invasiveness through increased resource use in a vacant niche. Nat. Plants 1, 15066 ( 10.1038/nplants.2015.66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Emery NC, Ackerly DD. 2014. Ecological release exposes genetically based niche variation. Ecol. Lett. 17, 1149–1157. ( 10.1111/ele.12321) [DOI] [PubMed] [Google Scholar]
- 147.Keane RM, Crawley MJ. 2002. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 17, 164–170. ( 10.1016/S0169-5347(02)02499-0) [DOI] [Google Scholar]
- 148.DeWalt SJ, Denslow JS, Ickes K. 2004. Natural-enemy release facilitates habitat expansion of the invasive tropical shrub Clidemia hirta. Ecology 85, 471–483. ( 10.1890/02-0728) [DOI] [Google Scholar]
- 149.Early R, Sax DF. 2014. Climatic niche shifts between species’ native and naturalized ranges raise concern for ecological forecasts during invasions and climate change. Glob. Ecol. Biogeogr. 23, 1356–1365. ( 10.1111/geb.12208) [DOI] [Google Scholar]
- 150.Tingley R, Vallinoto M, Sequeira F, Kearney MR. 2014. Realized niche shift during a global biological invasion. Proc. Natl Acad. Sci. USA 111, 10 233–10 238. ( 10.1073/pnas.1405766111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Farjon A. 2013. Pinus cembra. IUCN Red List Threat. Species. ( 10.2305/IUCN.UK.2013-1.RLTS.T42349A2974490.en) [DOI] [Google Scholar]
- 152.Marrero Gómez MV, Coello M.2011. Lotus maculatus . IUCN Red List Threat. Species . ( ) [DOI]
- 153.Hill MP, Chown SL, Hoffmann AA. 2013. A predicted niche shift corresponds with increased thermal resistance in an invasive mite, Halotydeus destructor. Glob. Ecol. Biogeogr. 22, 942–951. ( 10.1111/geb.12059) [DOI] [Google Scholar]
- 154.Fukano Y, Doi H, Thomas CE, Takata M, Koyama S, Satoh T. 2016. Contemporary evolution of host plant range expansion in an introduced herbivorous beetle Ophraella communa. J. Evol. Biol. 29, 757–765. ( 10.1111/jeb.12824) [DOI] [PubMed] [Google Scholar]
- 155.Klironomos JN. 2002. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70. ( 10.1038/417067a) [DOI] [PubMed] [Google Scholar]
- 156.Dawson W, Fischer M, van Kleunen M. 2012. Common and rare plant species respond differently to fertilisation and competition, whether they are alien or native. Ecol. Lett. 15, 873–880. ( 10.1111/j.1461-0248.2012.01811.x) [DOI] [PubMed] [Google Scholar]
- 157.Jump AS, Peñuelas J. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecol. Lett. 8, 1010–1020. ( 10.1111/j.1461-0248.2005.00796.x) [DOI] [PubMed] [Google Scholar]
- 158.Sakai AK, et al. 2001. The population biology of invasive species. Annu. Rev. Ecol. Syst. 32, 305–332. ( 10.1146/annurev.ecolsys.32.081501.114037) [DOI] [Google Scholar]
- 159.Urban MC, De Meester L, Vellend M, Stoks R, Vanoverbeke J. 2012. A crucial step toward realism: responses to climate change from an evolving metacommunity perspective. Evol. Appl. 5, 154–167. ( 10.1111/j.1752-4571.2011.00208.x) [DOI] [PMC free article] [PubMed] [Google Scholar]