Abstract
The density and diversity of post-translational modifications (PTMs) observed in histone proteins typically limits their purification to homogeneity from biological sources. Access to quantities of uniformly modified histones is, however, critical for investigating the downstream effects of histone PTMs on chromatin-templated processes. Therefore, a number of semisynthetic methodologies have been developed to generate histones bearing precisely defined PTMs or close analogs thereof. In this chapter, we present two optimized and rapid strategies for generating functional analogs of site-specifically acetylated and sumoylated histones. First, we describe a convergent strategy to site-specifically attach the small ubiquitin-like modifier-3 (SUMO-3) protein to the site of Lys12 in histone H4 by means of a disulfide linkage. We then describe the generation of thialysine analogs of histone H3 acetylated at Lys 14 or Lys 56, using thiol-ene coupling chemistry. Both strategies afford multi-milligram quantities of uniformly modified histones that are easily incorporated into mononucleosomes and nucleosome arrays for biophysical and biochemical investigations. These methods are readily extendable to any desired sites in the four core nucleosomal histones and their variant forms.
Keywords: chromatin, histone, acetylation, SUMO, sumoylation semisynthesis, disulfide
1. Introduction
Post-translational modifications (PTMs) of amino acid side-chains in eukaryotic histones add epigenetic diversity to the chromatin landscape.1,2 Histone PTMs can directly influence chromatin structure and/or mediate protein-protein interactions that drive the transcription, replication and repair of genetic material.3 Histone PTMs range from small chemical moieties such as methyl, acetyl, phosphoryl and glycosyl groups to entire proteins such as ubiquitin and the small ubiquitin-like modifier protein (SUMO).4 Given their central role in regulating gene function and repair, the misregulation or misinterpretation of histone PTMs is closely associated with human diseases such as cancers, ataxias and muscular dystrophy.5
Early genomic approaches with modification-specific antibodies identified distinct sets of histone PTMs at specific chromatin loci and correlated these with transcriptionally activity of the associated genes. These observations inspired the histone code hypothesis for gene regulation, which postulates that different histone PTMs may act individually or in combination to regulate chromatin-templated functions.6 Advances in mass spectrometric techniques have led to the identification of about 15 chemically unique PTMs in histones, and combinations of these modifications lead to hundreds of uniquely identifiable histone states.7 Therefore, synthetic and semisynthetic techniques that yield rapid access to various modified histones are crucial when testing elements of the histone code hypothesis.
Peterson and co-workers first demonstrated the power of protein semisynthesis in interrogating the effects of acetylation at lysine 16 in H4 (H4 K16ac) on chromatin structure.8 In their protocol, a 22-amino acid long histone H4 N-terminal peptide containing acetylated lysine 16 and a C-terminal α-thioester was produced by solid-phase peptide synthesis. Full-length acetylated H4 was then generated by native chemical ligation (NCL)9 of the acetylated α-thioester peptide with a heterologously expressed C-terminal fragment of histone H4 (residues 23–102) containing an arginine-to-cysteine mutation at the N-terminus. The authors successfully incorporated acetylated H4 protein into nucleosome arrays and observed that H4 K16ac significantly impairs chromatin condensation and higher-order fiber formation. Despite its utility, the synthesis of long peptide thioesters required for NCL continues to be technically challenging and the semisynthesis of acetylated histones by NCL has remained limited to specialized laboratories.10
In contrast with acetylation, the modification of histone lysines by conjugation with the proteins ubiquitin and SUMO leads to dramatic changes in the overall physical and chemical properties of nucleosomes. However, the fact that ubiquitylation and sumoylation (modification by Ub and SUMO, respectively) are transient modifications that mark a small fraction of core histones in cells (typically < 5%, with ubiquitylated H2A being the exception at ~10%), has limited the isolation of quantities of ubiquitylated and sumoylated histones for in vitro mechanistic studies.11,12 Recent progress toward understanding the roles for these modifications has been made through the efforts of several research groups in developing novel chemical handles and ligation auxiliaries that permit NCL at lysine side-chains.13,14 However, the technical challenges in generating wild-type ubiquitylated and sumoylated histones go beyond those encountered in generating semisynthetic acetylated histones.
A key turning point in facile semisynthetic access to post-translationally modified histones was the discovery that thialysine analogs of methylated lysine side-chains in histones are reasonable substrates for chromatin-modifying enzymes.15 Thialysine analogs are readily generated by alkylating genetically encoded cysteine residues in full-length histones with N-methylated forms of 2-aminoethyl halides, thereby obviating the need for preparing peptide thioesters for NCL. Several thiol-directed strategies provide rapid access to close functional analogs of wild-type PTMs of lysines and arginines.16 In this chapter, we provide detailed methods for chemical strategies that yield analogs of acetylated and sumoylated histones.17 A high-yielding disulfide forming reaction18 is used to generate H4 site-specifically sumoylated at Lys 12 (suH4ss) and radical-mediated thiol-ene coupling chemistry is used to introduce acetylated thialysine in histone H3 at positions 14 and 56.19 Both strategies provide versatile and straightforward approaches to generate multi-milligram quantities of modified histones suitable for a wide range of biophysical and biochemical studies. Furthermore, we describe the incorporation of these modified histones into octamers, which are applied to reconstitute both mononucleosomes and chromatin-like nucleosome arrays.
2. Materials and Methods
2.1 General materials and methods
All commonly used chemical reagents and solvents were purchased from either Sigma-Aldrich Chemical Company (Milwaukee, WI) or Fischer Scientific (Pittsburgh, PA). 2xYT medium was reconstituted by mixing 16 g Bacto Tryptone, 10 g Bacto Yeast Extract, and 5 g NaCl per liter of water. A Superdex S-200 10/300 GL size-exclusion column was purchased from GE Healthcare (Waukesha, WI). Chemically competent DH5αand BL21(DE3) cells were purchased from Novagen (Madison, WI). T4 DNA ligase and restriction enzymes were purchased from New England BioLabs (Ipswitch, MA). DNA primer synthesis and gene sequencing were performed by Integrated DNA Technologies (Coralville, IA) and Genewiz (South Plainfeld, NJ), respectively. Site-directed mutagenesis was performed with a QuikChange Site-Directed Mutagenesis kit from Agilent Technologies (Santa Clara, CA). Criterion 5% TBE gels were purchased from Bio-Rad (Hercules, CA). Centrifugal filtration units were from Sartorius (Goettingen, Germany) and Slide-A-Lyzer dialysis cassettes and MINI dialysis units were from Pierce (Rockford, IL). PCR purification and gel extraction kits were purchased from Qiagen (Valencia, CA). Size-exclusion chromatography was performed on an AKTA FPLC system (GE Healthcare) equipped with a P-920 pump and UPC-900 monitor. Analytical reversed-phase HPLC (RP-HPLC) was performed on a Varian ProStar instrument with Vydac C18 or C4 columns (5 micron, 4 × 150 mm), employing 0.1% trifluoroacetic acid (TFA) in water (HPLC buffer A), and 90% acetonitrile, 0.1% TFA in water (HPLC buffer B) as the mobile phases. Typical analytical gradients were 30–70% buffer B over 30 min at a flow rate of 1 mL/min. Preparative scale purifications were conducted on Vydac C18 or C4 preparative columns (15–20 micron, 50 × 250 mm) over 60 min at a flow rate of 9 mL/min. Electrospray ionization mass spectrometry (ESI-MS) analysis was performed on a Bruker Esquire LC-ion trap spectrometer (Bruker Daltonics, Billerica, Massachusetts). All protein starting materials and ligation products were analyzed by both C18 or C4 analytical RP-HPLC and ESI-MS.
3. Semisynthesis of Sumoylated Histone H4
3.1 Overall design of the semisynthesis
Histones that are heterologously produced in Escherichia coli are ideal candidates for installing cysteine-derived PTM analogs. E. coli does not have any known histone modifying enzymes, which ensures that histones expressed in this organism are devoid of PTMs. Further, of the four core histones, H3 is the only histone in higher eukaryotes with a cysteine at position 110.20 This residue can be mutated to an alanine without discernible effects on nucleosome structure or function. Working in this mutant background ensures that cysteine residues introduced by mutagenesis at desired positions in the core histones are the only thiols present, thereby allowing site-selective chemical manipulations.
Histone sumoylation is a well-documented but poorly understood modification.12,17,21 Similar to ubiquitylation, sumoylation has been observed on all four core histones as well as the linker histone H1. Although proteomic studies have identified Lys12 in the H4 tail as a site of modification,22,23 the consequences on chromatin structure and function remain unknown. The first reported semisynthesis of a sumoylated histone H2B C-terminal peptide employed a photocleavable NCL auxiliary group appended to the side-chain of a lysine in the target peptide.24 Ligation between the peptide and a C-terminally truncated SUMO α-thioester followed by UV irradiation resulted in a site-specifically sumoylated peptide. While this method affords a native isopeptide linkage, the complex synthetic route poses challenges for obtaining bulk quantities of sumoylated proteins.
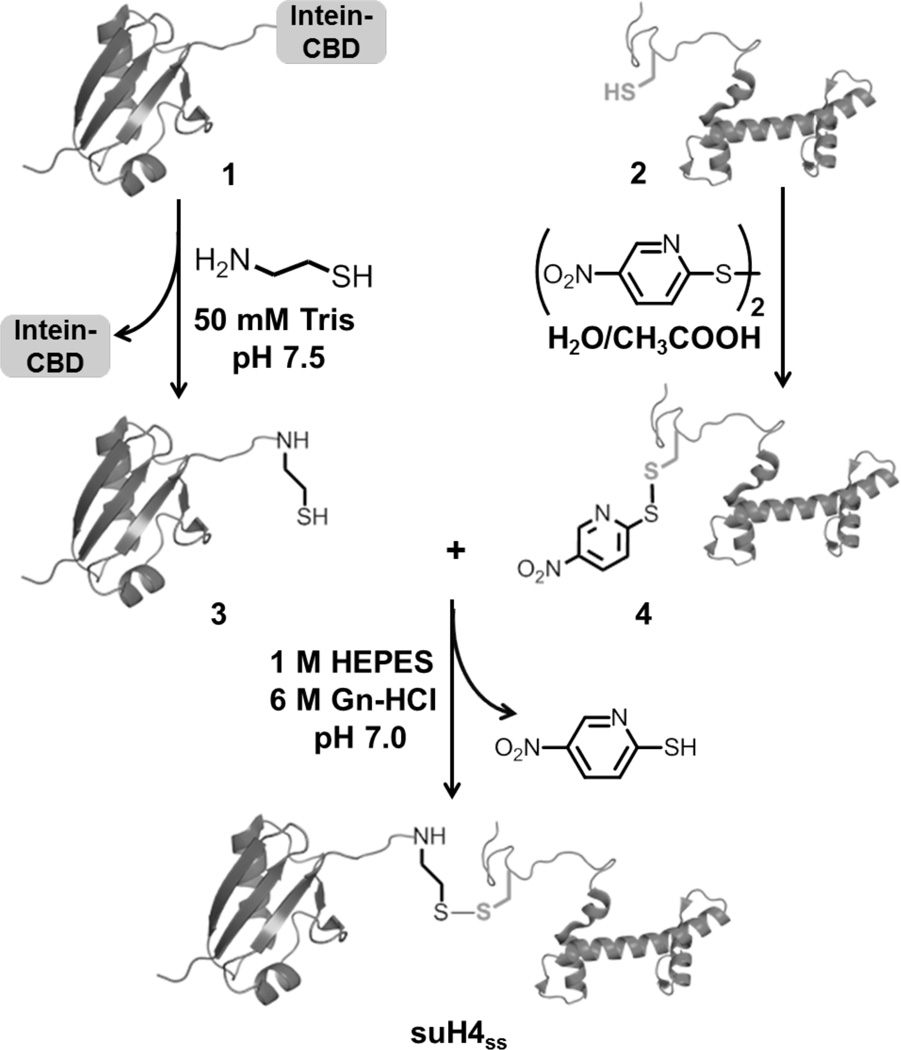
In order to address the gap in our understanding of histone sumoylation, we employed a disulfide-directed approach to generate histone H4 uniformly sumoylated at Lys12 (suH4ss) in milligram quantities. The semisynthesis is achieved in two parts (Figure 1). First, the target Lys 12 in H4 is replaced with a cysteine by site-directed mutagenesis. After affinity-tag enrichment and purification of H4 K12C, the sulfhydryl group of the lone cysteine is activated using a small molecule disulfide. This generates a reactive asymmetric disulfide on H4, poised for nucleophilic attack by an external thiol. Additionally, a free nucleophilic thiol is introduced at the C-terminus of SUMO-3 C47S by intein-mediated chemistry.25 In the second step, mixing the SUMO-3 C47S with activated H4 K12C leads to the desired product in near-quantitative conversion over the course of 2–3 hours.
Figure. 1. Semisynthesis of disulfide-linked sumoylated histone H4 (suH4ss).
Synthetic scheme for the generation of SUMO-3 C47S-aminoethanethiol, 3, using intein-mediated thiolysis and its subsequent reaction with the activated H4 K12C mutant, 4, to generate suH4ss.
3.2 Preparation of recombinant histone H4 K12C
Site-directed mutagenesis is employed to generate the mutant H4 K12C from human histone H4 cloned between the NdeI and BamHI restriction sites in a pET15b vector. A Tobacco Etch Virus (TEV) protease cleavage sequence is introduced between the His6-tag and N-terminus of H4. The following primers are used with the QuikChange Site-Directed mutagenesis kit to introduce the K12C mutations: 5’- GGT AAA GGT GGT AAA GGT CTG GGT TGC GGT GGT GCT AAA CGT CAC CGT AAA -3’ and 5’- TTT ACG GTG ACG TTT AGC ACC ACC GCA ACC CAG ACC TTT ACC ACC TTT ACC -3’. The resulting plasmid is verified by gene sequencing. E. coli BL21(DE3) cells are transformed with the mutant plasmids and grown at 37 °C in 6 L 2x YT medium until OD600 ~0.7. Protein expression is induced by the addition of 0.3 mM IPTG for 3 h at 37 °C. Cells are harvested by centrifugation at 7000×g, resuspended in 150 mM NaCl, 50 mM Tris at pH 7.5 and lysed by sonication. The lysate is centrifuged at 20,000×g for 20 min and insoluble histones are recovered from inclusion bodies with an extraction buffer consisting of 6 M Gn-HCl, 100 mM NaCl, 50 mM Tris at pH 7.5. The re-solubilized histones are purified by Ni2+-affinity chromatography. Briefly, proteins are bound to the Ni2+-column overnight at 4 °C with gentle shaking and the column subsequently washed with 3 volumes of extraction buffer containing 25 mM Imidazole. Immobilized proteins are eluted using 500 mM Imidazole in the extraction buffer. Histone-containing fractions are identified by 15% SDS-PAGE, combined and dialyzed into 1 mM DTT overnight at 4 °C. Purified His6-TEV protease is then added in a 1:10 molar ratio (enzyme:substrate) and the His6-tag is cleaved overnight in a buffer containing 1 mM EDTA, 10 mM DTT, 10 mM Cys, 50 mM Tris, pH 6.9. The cleavage products are dialyzed into extraction buffer and re-applied to a Ni2+-column. This leads to retention of the cleaved His6-tag, His6-TEV and any uncleaved histone on the column. The eluted His6-tag-free H4 K12C is further purified to homogeneity by preparative RP-HPLC and characterized by ESI-MS. Purified H4 K12C (2) is then dissolved in a 1:3 mixture of water:acetic acid (v/v) and reacted with an excess of 2,2'-dithiobis(5-nitropyridine) (DTNP) for 2 h at 25 °C with vigorous shaking. The desired product H4 K12C-Npys (4) is purified away from unreacted starting materials by RP-HPLC and characterized by ESI-MS.
3.3 Preparation of recombinant SUMO-3-aminoethanethiol
Site-directed mutagenesis is employed to generate the SUMO-3 C47S mutant from human SUMO-3 cloned in the pTXB1 vector. The following primers are used with the QuikChange Site-Directed mutagenesis kit: 5’- AGC AAG CTG ATG AAG GCC TAC TCT GAG AGG CAG GGC TTG TCA ATG -3’ and 5’- CAT TGA CAA GCC CTG CCT CTC AGA GTA GGC CTT CAT CAG CTT GCT -3’. The resulting plasmid is verified by gene sequencing. E. coli BL21(DE3) cells are transformed with SUMO-3 C47S plasmids and grown to an OD600 of ~0.5 at 37 °C followed by the addition of 0.3 mM IPTG. The cells are grown for another 4 h at 25 °C, harvested by centrifugation at 7,000×g, resuspended in lysis buffer containing 200 mM NaCl, 20 mM Tris, pH 7.5 and lysed by sonication. The lysate is centrifuged at 20,000×g for 20 min and the soluble supernatant containing SUMO-3 C47S-intein-chitin binding domain fusion protein (1) is bound to chitin beads at 4 °C overnight. The column is subsequently washed with 3 column volumes of lysis buffer and intein-mediated production of SUMO-3 C47S-aminoethanethiol (3) is initiated by incubating the column with lysis buffer containing 100 mM cysteamine, pH 7.5, for 48 h at 4 °C. After this period, the desired adducts are eluted from the column with lysis buffer, further purified by preparative RP-HPLC and characterized by ESI-MS.
3.3 Generation of sumoylated histone H4
One equivalent of H4 K12C-Npys (4) and two equivalents of SUMO-3 C47S-aminoethanethiol (3) are dissolved in a reaction buffer consisting of 6 M Gn-HCl, 1 M HEPES, pH 6.9 and allowed to react for 1 h at 25 °C with continuous shaking. The desired product, suH4ss is purified away from starting materials by RP-HPLC and characterized by ESI-MS.
4. Preparation of acetylated histone H3 analogs
4.1 Overall design of the semisynthesis
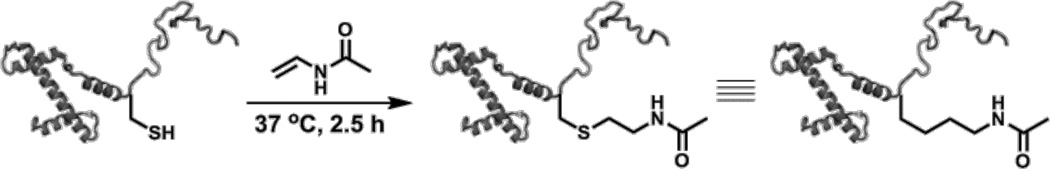
The incorporation of acetyllysine into histones can be achieved by protein semisynthesis8 or amber suppression,26 but these approaches can be low yielding and complex. In contrast, a method reported by Liu and coworkers in 2011 is uniquely well-suited for generating mimics of acetyllysine in a single step on recombinant histone proteins, which can be purified in large quantities.19 The acetyllysine mimic is installed via a thiol-ene reaction between the sulfhydryl group of Cys and N-vinylacetamide, thus the target protein must contain a Cys residue only at the desired site of Lys acetylation (Figure 2). The radical thiol-ene click reaction is fast, site-specific and gives high product yields. Further, we observe that initiation with heat rather than UV light further reduces the possibility of undesired side reactions.17 In addition, multiple sites may be simultaneously and efficiently modified. The resulting N-acetyl thialysine, an isostere for acetyllysine at the side-chain γ position, is functionally similar to the natural modification, as evidenced by its identical effect on nucleosome array compaction, α-acetyllysine antibody reactivity, and susceptibility to enzymatic deacetylation.19 As an example of site-specific incorporation of acetyllysine mimics, we describe the generation of H3 C110A bearing this modification at position 14 or 56, each a site of functionally important H3 acetylation.27
Figure 2. Thiol-ene coupling chemistry at Cys with N-vinylacetamide to generate acetylated thialysine.
.
4.2 Preparation of recombinant histone H3
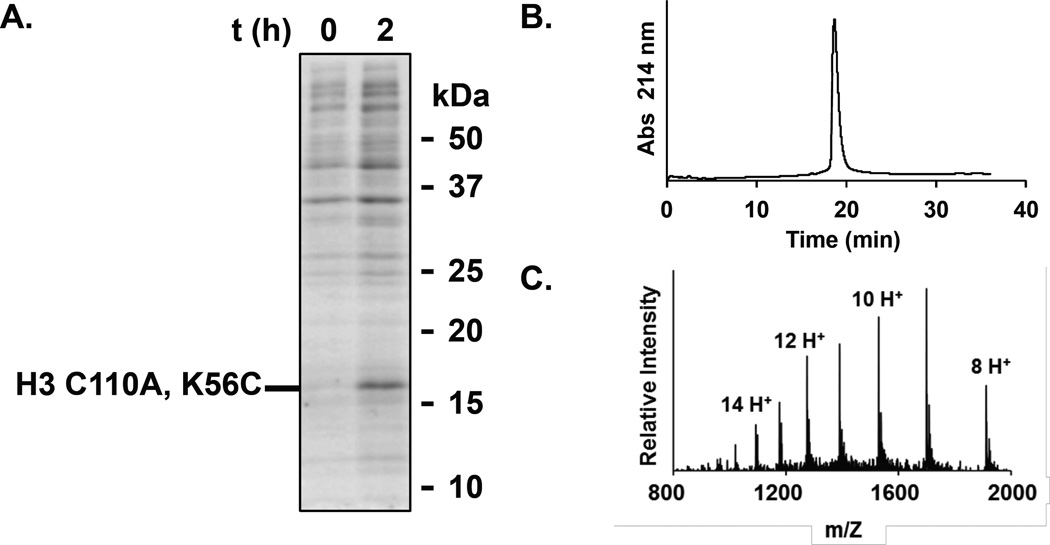
We have reported a pET3a vector containing the gene for human histone variant H3.2, HIST2H3C, bearing a C110A point mutation.17 This plasmid is subjected to QuikChange Mutagenesis (Qiagen) to incorporate the K14C or K56C point mutation utilizing the primers: For K14C, 5’- CAG ACG GCT CGG AAA TCC ACC GGC GGT TGC GCG CCA CGC AAG CAG CTG GCT ACC AAG -3’ and 5’- CTT GGT AGC CAG CTG CTT GCG TGG CGC GCA ACC GCC GGT GGA TTT CCG AGC CGT CTG -3’; and for K56C, 5’- GCT CTG CGC GAG ATC CGC CGC TAC CAA TGC TCG ACC GAG TTG CTG ATT CGG AAG CTG -3’ and 5’- CAG CTT CCG AAT CAG CAA CTC GGT CGA GCA TTG GTA GCG GCG GAT CTC GCG CAG AGC -3’. The resulting plasmids are verified by gene sequencing. E. coli BL21(DE3) cells transformed with the pET3a-H3 C110A, K14C or pET3a-H3 C110A, K56C vector are grown in 2 L of 2x YT broth supplemented with 100 µg/mL ampicillin at 37 °C and with shaking at 250 r.p.m. until the OD600 reaches 0.6–0.8. Overexpression is then induced by the addition of IPTG from a stock solution (1 M in H2O) to a concentration of 0.3 mM, and growth is continued for no longer than 2 h at 37 °C. Protein overexpression is confirmed by SDS-PAGE analysis of the lysed cells at the end of growth (Figure 3A). The cells are harvested by centrifugation at 7,000×g for 15 min, then resuspended in 4 M Gn-HCl, 20 mM Tris, 1 mM DTT, pH 7.5 (pH adjusted for use at 4 °C). The cells are kept on ice and lysed by sonication. The presence of Gn-HCl causes significant frothing of the buffer, and longer sonication times may be required. If the cell suspension becomes warm, it can be allowed to rest on ice before further sonication to prevent overheating. After lysis, the cells are centrifuged at 20,000×g for 15 min. The lysate supernatant is passed through a 0.45 µm filter, then applied to a 500 mL Superdex S-200 size-exclusion column equilibrated with lysis buffer eluting at 1 mL/min at 4 °C. Fractions are collected and evaluated for the presence of the desired protein by SDS-PAGE. Note that Gn-HCl in the buffer precipitates in the presence of SDS and causes gels to run irregularly. To avoid this, samples can be centrifuged at 20,000×g for 5 min after boiling with loading dye, and the supernatant loaded on the gel. Fractions containing a disulfide of the product may be present. This can be evaluated by adding 10 mM DTT to the loading dye. Fractions containing product are pooled and dialyzed against 4 L of H2O containing 1 mM DTT, pH 7.2, at 4 °C for 6 h. The sample is then lyophilized and re-dissolved in 6 M Gn-HCl, 100 mM Na2HPO4, 100 mM TCEP, pH 7.0–7.5 and incubated at 4 °C for 30 min. The reduced product is then purified by RP-HPLC and characterized by ESI-MS. ESI-MS for H3 C110A, K14C and H3 C110A, K56C. Calculated m/z [M+H]+ 15,200.7 Da, observed m/z [M+H]+ 15,204.3 ± 4.4 and 15,203.8 ± 2.9 Da, respectively.
Figure 3. Generation of H3 (C110A)Kc56ac analog.
A. SDS-PAGE of E. coli cultures containing pET3a-H3 C110A, K56C at 0 h and 2.5 h after the induction of overexpression with IPTG. 15% SDS-PAGE gel run at 200 V for 45 min, and stained with Coomassie. B. RP-HPLC chromatogram of pure H3(C110A)KC56ac analog. C. ESI-MS of pure H3 (C110A)KC56ac analog.
4.3 Generation of thialysine analogs of acetylated histone H3
Purified H3 C110A, K14C or H3 C110A, K56C is dissolved at 0.1 mM in 6 M Gn-HCl, 200 mM sodium acetate, pH 5.0. An inert atmosphere is crucial for this reaction, so the buffer is freeze-thaw degassed thrice under N2, and the reaction kept under an atmosphere of N2. To this solution is added 15 mM L-glutathione, 50 mM N-vinylacetamide, 100 mM dimethyl sulfide, and 50 mM of the azo radical initiator VA-044 (2,2’-azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride). The thiol-ene click reaction is initiated by incubation at 37 °C for 2.5 h. Longer incubation times should be avoided as they may lead to alkylation at undesired sites, such as the protein N-terminus. The product is then purified by RP-HPLC and characterized by ESI-MS (Figure 3B and 3C). ESI-MS for H3 (C110A)Kc14Ac and H3 (C110A)Kc56Ac, calculated m/z [M+H]+ 15,284.8 Da, observed m/z [M+H]+ 15,287.3 ± 2.6 and 15,288.1 ± 3.6 Da, respectively.
5. Generation of designer mononucleosomes
5.1 Octamer formation using modified histones
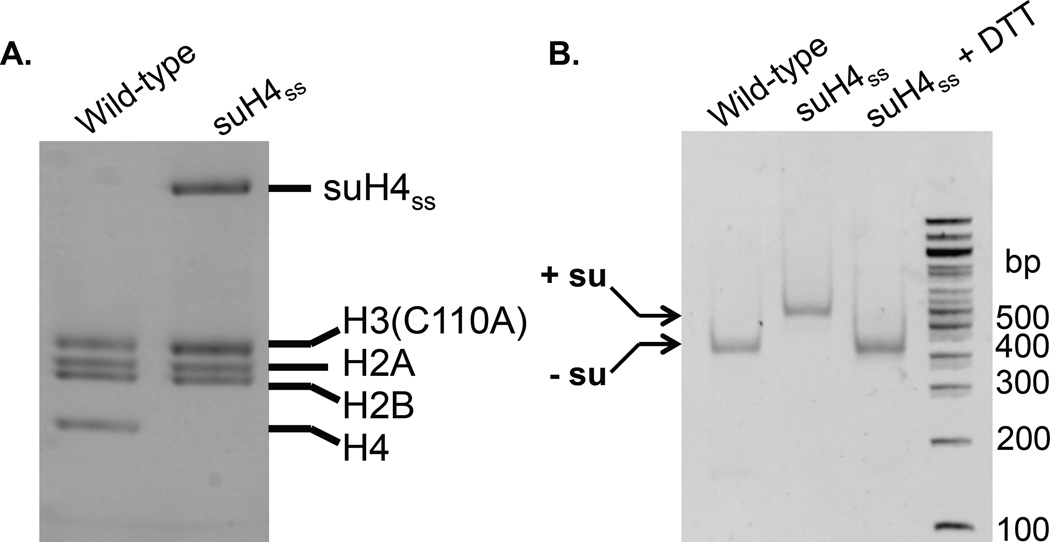
Histone octamers are assembled with minor modification of previous reports with the strict exclusion of reducing agents.28 Briefly, each core histone is dissolved at ~4 mg/mL in an unfolding buffer containing 7 M Gn-HCl, 20 mM Tris, pH 7.5. It is important to use Ultra-pure Gn-HCl at this step to allow accurate protein quantitation by Abs280. The web-based ExPASy Protein Parameters Tool at http://web.expasy.org/protparam/ is used to compute the extinction coefficient for each protein. The histones are mixed in equimolar amounts and the resulting mixture is dialyzed into a refolding buffer (3 × 1 L) consisting of 2 M NaCl, 1 mM EDTA, 10 mM Tris, pH 7.5 at 4 °C. Crude octamers are concentrated by Vivaspin 500 concentrators and purified by size-exclusion on a Superdex S-200 column equilibrated in the octamer-refolding buffer. Fractions containing pure histone octamers are identified by 15% SDS-PAGE stained with Coomassie, combined and concentrated prior to nucleosome assembly (Figure 4A).
Figure 4. Generation of designer mononucleosomes.
A. Wild-type and symmetrically sumoylated octamers visualized by 15% SDS-PAGE gel run at 200 V for 45 min and stained with Coomassie. B. Wild-type and symmetrically sumoylated octamers visualized by 5% TBE gel run at 180 V for 50 min and stained with ethidium bromide. su= SUMO-3 C47S.
5.2 Generation of 147 bp 601 DNA
The 147 bp 601 DNA is amplified from a plasmid containing the 1_147_601 fragment29 by PCR in 5 mL net volume using the forward primer 5'-CTG GAG AAT CCC GGT GCC GAG G-3' and reverse primer 5'- ACA GGA TGT ATA TAT CTG ACA CG-3'. The PCR product is purified using a QIAquick PCR purification kit and eluted in sterile water (pH 7.5). The eluate is lyophilized to concentrate the DNA and then resuspended in sterile water to a final concentration of 20–25 µM. The DNA is visualized for purity by running on a 2% Agarose gel at 150 V for 30 min in TBE buffer and staining with ethidium bromide.
5.3 Reconstitution of mononucleosomes
Following a modified stepwise dilution protocol reported by Workman and co-workers,30 pure histone octamers and 147 bp 601 DNA are combined in 10 µL of a high-salt refolding buffer consisting of 2 M NaCl, 1 mM EDTA, 10 mM Tris, pH 7.5, to a final concentration of 2 µM in octamers and DNA. After incubation at 37 °C for 15 min, 3.3 µL of Dilution Buffer 1 containing 1 mM EDTA, 0.5 mM PMSF, 10 mM HEPES, pH 7.9, is added and the temperature dropped to 30 °C. Further dilutions of 6.7, 5, 3.6, 4.7, 6.7, 10, 30, and 20 µL, respectively, are performed every 15 min. A final dilution is undertaken with 100 µL of Dilution Buffer 2 containing 1 mM EDTA, 0.1% (v/v) NP-40, 0.5 mM PMSF, 20% (v/v) glycerol, 10 mM Tris, pH 7.5. After an additional 15 min at 30 °C, the mononucleosomes (MNs) are concentrated and analyzed by separation on a Criterion 5% TBE gel run in 0.5X TBE, followed by staining with ethidium bromide. For sumoylated MNs, reduction with 1 mM DTT for 30 min on ice leads to the removal of SUMO and co-migration of the reduced MNs with unmodified wild-type MNs. This is used to confirm the core histone composition as being identical to that of wild-type MNs (Figure 4B).
5.4 Preparation of 177 bp repeat 601 DNA
A plasmid containing 12 copies of a 177 bp repeat of the 601 nucleosome positioning sequence (12_177_601)31 flanked by EcoRV sites is purified from a 6 L culture of DH5α cells using a Qiafilter Plasmid Giga kit. The 12_177_601 sequence is obtained by preparative-scale digestion of the plasmid with EcoRV. This is followed by selective precipitation of the 12_177_601 fragment with 6% polyethylene glycol (PEG)-6000 on ice and centrifugation at 26,000×g for 30 min at 4 °C. After phenol extraction and ethanol precipitation, the DNA is re-dissolved in TE buffer (1 mM EDTA, 10 mM Tris, pH 7.5) and quantified by Abs260 prior to array formation.
5.5 Generation of 12-mer nucleosome arrays
Pure histone octamers (2 µM) and 12_177_601 DNA (0.17 µM in DNA, 2 µM in octamer binding sites) are combined in 75 µL of reconstitution buffer consisting of 2 M KCl, 0.1 mM EDTA, 10 mM Tris pH 7.8. Additionally, 0.7 µM of a weaker-binding 147 bp fragment of the MMTV A DNA is added to prevent overloading of the array with octamers. Stepwise dialysis is performed at 4°C against reconstitution buffer containing 1.4 M NaCl, 1.2 M NaCl, 1 M NaCl, 0.8 M NaCl, 0.5 M NaCl and 10 mM NaCl for 90 min each, followed by a final dialysis step against reconstitution buffer containing 10 mM NaCl. DNA fragments are removed by selective precipitation of the arrays with MgCl2. After removal of the supernatant, the arrays are resuspended in TEN buffer (10 mM Tris, 1 mM EDTA, 100 mM NaCl, pH 8.0), dialyzed against fresh TEN buffer and quantified by Abs260. Wild-type and modified arrays are visualized on 1% agarose-2% polyacrylamide gel electrophoresis (APAGE) by running gels at 180 V for 20 min at 4 °C followed by staining with ethidium bromide (Figure 5A).
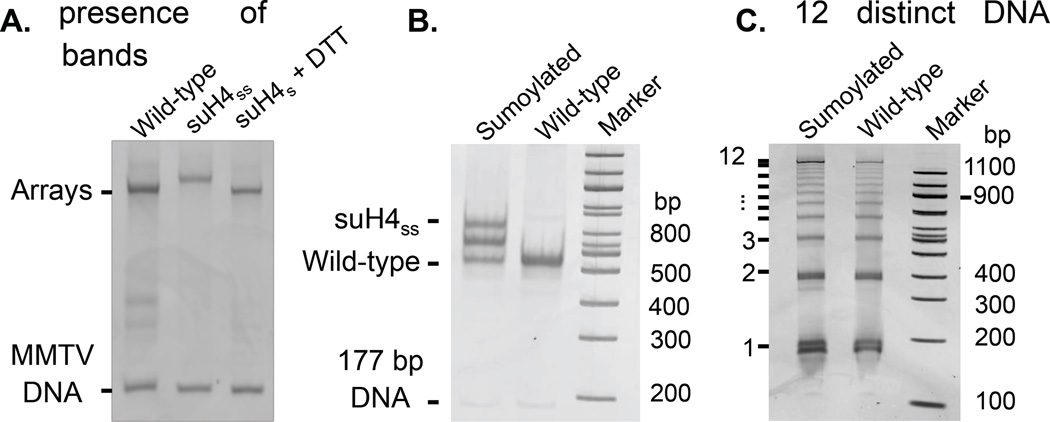
Figure 5. Generation and characterization of designer nucleosome arrays.
A. 1% APAGE gel stained with ethidium bromide showing wild-type and uniformly sumoylated 12-mer arrays and the desumoylation product upon addition of a reducing agent. B. 5% TBE gel stained with ethidium bromide showing ScaI digestion products of wild-type and sumoylated 12-mer arrays. The presence of three bands in the sumoylated sample corresponds to di-, mono-, and non-sumoylated mononucleosomes arising from partial reduction. C. 5% TBE gel stained with ethidium bromide showing digested DNA arising from limited Micrococcal nuclease digestion of wild-type and sumoylated 12-mer arrays. The presence of 12 DNA bands of increasing size is indicated.
The saturation of octamer-binding sites in 12-mer arrays is confirmed by the digestion of 0.17 pmol of arrays with 10 U of ScaI restriction enzyme in NEB buffer 3 (50 mM Tris, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, pH 7.9) at 25 °C for 12 h, followed by separation on a Criterion 5% TBE gel run in 0.5 × TBE buffer and staining with ethidium bromide. The presence of an MN band as well as the absence of free DNA and higher molecular weight species indicates full array occupancy. DTT in the digestion and restriction enzyme buffer leads to partial loss of the SUMO moiety in sumoylated arrays resulting in three distinct MN bands corresponding to the doubly, singly and non-sumoylated species (Figure 5B). The presence of 12 MNs per array is also confirmed by partial digestion with the enzyme Micrococcal nuclease. In a typical assay, 0.17 pmol of 12-mer arrays are digested with 0.2 U of Micrococcal nuclease for 60 s on ice. The reaction was stopped by the addition of 0.2% (w/v) SDS and 20 mM EDTA followed by DNA purification using a QIAquick PCR purification kit (Qiagen). The DNA fragments were separated on a Criterion 5% TBE gel run in 0.5× TBE buffer and visualized after ethidium bromide staining (Figure 5C). The presence of 12 distinct DNA bands indicates the uniform distribution of nucleosomes on the 12_177_601 DNA.
6. Summary and Conclusions
Synthetic access to uniformly and site-specifically modified histones is key when investigating the molecular details of chromatin-mediated mechanisms that drive gene regulation, replication and repair. Entirely synthetic32 and combinations of synthetic and recombinant technologies, embodied by the technique of expressed protein ligation,25 allow exquisite chemical control of both modification site and type in full-length histones. However, these methodologies remain challenging for laboratories not equipped for advanced chemical synthesis. The discovery of multiple functional analogs of modified histones provide convenient alternatives that are accessible by one or two-step transformations on recombinant proteins with commercially available reagents.16 The two methods presented to generate analogs of acetylated and sumoylated histones are readily accomplished in a two-week period, starting from heterologous expression, and provide access to milligram quantities of the modified histones. Although disulfide-linked sumoylated histones cannot be used in the presence of reducing agents, these can easily be excluded from in vitro assays, and may even be tolerated based on the solvent accessibility of the modified residue. Importantly, these methods can be easily adapted toward histone ubiquitylation as well as for generating multiply acetylated histones.33
Acknowledgments
We would like to thank the Department of Chemistry and the Royalty Research Fund at the University of Washington for generous support. CC is supported by NIGMS grant 1R01M110430. C.E.W. gratefully acknowledges support from the NSF GRFP (grant number DGH-1256082) and an ARCS foundation fellowship.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Allis CD, Jenuwein T, Reinberg D, Caparros M-L. Epigenetics. 2007:502. [Google Scholar]
- 3.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell. Biol. 2013;14:211. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhall A, Chatterjee C. Chemical approaches to understand the language of histone modifications. ACS Chem. Biol. 2011;6:987. doi: 10.1021/cb200142c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods. 2007;4:487. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 8.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 9.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 10.Kent SBH. Total chemical synthesis of proteins. Chem. Soc. Rev. 2009;38:338. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 11.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim. Biophys. Acta. 1994;1218:187. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 12.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, Berger SL. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long L, Thelen JP, Furgason M, Haj-Yahya M, Brik A, Cheng D, Peng J, Yao T. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 2014;289(8916) doi: 10.1074/jbc.M114.551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon M, Chu F, Racki L, Delacruz C, Burlingame A, Panning B, Narlikar G, Shokat K. The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones. Cell. 2007;128:1003. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhall A, Chatterjee C. In: Epigenetic Technological Applications. Zheng YG, editor. New York: Academic Press; 2015. p. 149. [Google Scholar]
- 17.Dhall A, Wei S, Fierz B, Woodcock CL, Lee TH, Chatterjee C. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J. Biol. Chem. 2014;289:33827. doi: 10.1074/jbc.M114.591644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabanal F, DeGrado W, Dutton P. Use of 2,2'-dithiobis(5-nitropyridine) for the heterodimerization of cysteine containing peptides. Introduction of the 5-nitro-2-pyridinesulfenyl group. Tet. Lett. 1996;37(1347) [Google Scholar]
- 19.Li F, Allahverdi A, Yang R, Lua GBJ, Zhang X, Cao Y, Korolev N, Nordenskiöld L, Liu C-F. A direct method for site-specific protein acetylation. Angew. Chem. Int. Ed. Eng. 2011;50:9611. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]
- 20.Marino-Ramirez L, Levine KM, Morales M, Zhang S, Moreland RT, Baxevanis AD, Landsman D. The Histone Database: an integrated resource for histones and histone fold-containing proteins. Database. 2011;2011:bar048. doi: 10.1093/database/bar048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shilo Y, Eisenman R. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. SciU.SA. 2003;100:13225. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014;21:927. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galisson F, Mahrouche L, Courcelles M, Bonneil E, Meloche S, Chelbi-Alix MK, Thibault P. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.004796. M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee C, McGinty RK, Pellois J-P, Muir TW. Auxiliary-mediated site-specific peptide ubiquitylation. Angew. Chem. Int. Ed. Eng. 2007;46:2814. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 25.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. SciU.SA. 1998;95:6705. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008;4:232. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 27.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 28.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 29.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 30.Utley RT, Owen-Hughes TA, Juan LJ, Cote J, Adams CC, Workman JL. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods in Enzymology. 1996;274:276. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 31.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 32.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of Fully Synthetic Histone H3 Reveals That Acetyl-Lysine 56 Facilitates Protein Binding Within Nucleosomes. J. Mol. Biol. 2011;408:187. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010;6:267. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]