Abstract
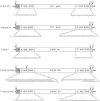
Influenza A and B viruses have not been shown to form reassortants. It had been assumed that the lack of genotypic mixing between influenza virus types reflected differences in polymerase and packaging specificity. In this study, we show that an influenza A virus polymerase transcribes and replicates a chloramphenicol acetyltransferase (CAT) gene flanked by the nontranslated sequences of an influenza B virus gene. Although the transcription level of this CAT gene was several times lower than that of a CAT gene flanked by the homologous nontranslated sequences of an influenza A virus, we proceeded to construct a chimeric type A/B influenza virus. Using recombinant DNA techniques, a chimeric neuraminidase gene was introduced into the genome of influenza A/WSN/33 virus. The hybrid influenza A/B virus gene contained the coding region of the A/WSN neuraminidase and the 3' and 5' nontranslated sequences of the nonstructural gene of influenza B/Lee virus. The resulting chimeric virus formed plaques in Madin-Darby bovine kidney cells but replicated more slowly and achieved lower titers than wild-type influenza A/WSN/33 virus. The chimeric virus was attenuated for mice as indicated by a 400-fold increase in its LD50. Interestingly, the virus was greatly restricted in replication in the upper respiratory tract and partially restricted in the lungs. Animals infected with the transfectant virus were highly resistant to influenza virus challenge. It appears that this chimeric virus has many of the properties desirable for a live attenuated virus vaccine.
Full text
PDF
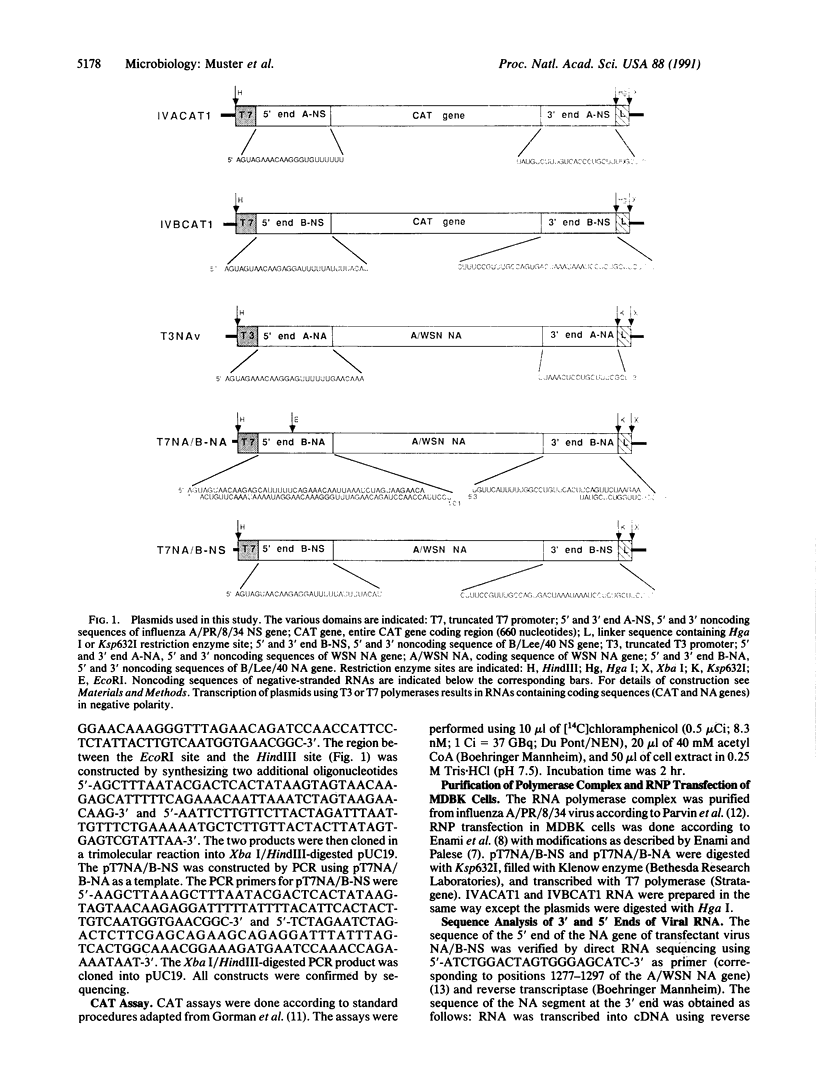


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Enami M., Luytjes W., Krystal M., Palese P. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci U S A. 1990 May;87(10):3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M., Palese P. High-efficiency formation of influenza virus transfectants. J Virol. 1991 May;65(5):2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenkina D. B., Ghendon Y. Z. Recombination and complementation between orthomyxoviruses under conditions of abortive infection. Acta Virol. 1979 Mar;23(2):97–106. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliburton I. W., Honess R. W., Killington R. A. Virulence is not conserved in recombinants between herpes simplex virus types 1 and 2. J Gen Virol. 1987 May;68(Pt 5):1435–1440. doi: 10.1099/0022-1317-68-5-1435. [DOI] [PubMed] [Google Scholar]
- Hiti A. L., Nayak D. P. Complete nucleotide sequence of the neuraminidase gene of human influenza virus A/WSN/33. J Virol. 1982 Feb;41(2):730–734. doi: 10.1128/jvi.41.2.730-734.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson V. H., Semler B. L. Defined recombinants of poliovirus and coxsackievirus: sequence-specific deletions and functional substitutions in the 5'-noncoding regions of viral RNAs. Virology. 1988 Jan;162(1):47–57. doi: 10.1016/0042-6822(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Luytjes W., Krystal M., Enami M., Parvin J. D., Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989 Dec 22;59(6):1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- Murphy B. R., Phelan M. A., Nelson D. L., Yarchoan R., Tierney E. L., Alling D. W., Chanock R. M. Hemagglutinin-specific enzyme-linked immunosorbent assay for antibodies to influenza A and B viruses. J Clin Microbiol. 1981 Mar;13(3):554–560. doi: 10.1128/jcm.13.3.554-560.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. The genes of influenza virus. Cell. 1977 Jan;10(1):1–10. doi: 10.1016/0092-8674(77)90133-7. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., Palese P., Honda A., Ishihama A., Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989 Dec;63(12):5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D. D., Murphy B. R. The association of the temperature-sensitive phenotype with viral attenuation in animals and humans: implications for the development and use of live virus vaccines. Rev Infect Dis. 1979 May-Jun;1(3):413–433. doi: 10.1093/clinids/1.3.413. [DOI] [PubMed] [Google Scholar]
- Schulman J. L., Palese P. Virulence factors of influenza A viruses: WSN virus neuraminidase required for plaque production in MDBK cells. J Virol. 1977 Oct;24(1):170–176. doi: 10.1128/jvi.24.1.170-176.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. H., Betts R. F., DeBorde D., Tierney E. L., Clements M. L., Herrington D., Sears S. D., Dolin R., Maassab H. F., Murphy B. R. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988 Feb;62(2):488–495. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. H., Buckler-White A. J., London W. T., Tierney E. L., Murphy B. R. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987 Sep;61(9):2857–2863. doi: 10.1128/jvi.61.9.2857-2863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle M. Y., Shaw M. W., Choppin P. W. Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs. Proc Natl Acad Sci U S A. 1987 May;84(9):2703–2707. doi: 10.1073/pnas.84.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A., Tobita K., Kilbourne E. D. Isolation and preliminary characterization of temperature-sensitive mutants of influenza virus. J Virol. 1972 Oct;10(4):639–647. doi: 10.1128/jvi.10.4.639-647.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. L., Rogers S. K., Zerhusen M. A. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology. 1989 Oct;172(2):435–450. doi: 10.1016/0042-6822(89)90186-4. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Krystal M., Fitch W. M., Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988 Mar;163(1):112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- Yilma T., Zee Y. C., Osebold J. W. Immunofluorescence determination of the pathogenesis of infection with influenza virus in mice following exposure to aerosolized virus. J Infect Dis. 1979 Apr;139(4):458–464. doi: 10.1093/infdis/139.4.458. [DOI] [PubMed] [Google Scholar]