Abstract
Purpose
To investigate the influence of transcytolemmal water exchange on estimates of tissue microstructural parameters derived from diffusion MRI using conventional PGSE and IMPULSED methods.
Methods
Computer simulations were performed to incorporate a broad range of intracellular water life times τin (50 – ∞ ms), cell diameters d (5 – 15 μm), and intrinsic diffusion coefficient Din (0.6 – 2 μm2/ms) for different values of signal to noise ratio (10 to 50). For experiments, murine erythroleukemia (MEL) cancer cells were cultured and treated with saponin to selectively change cell membrane permeability. All fitted microstructural parameters from simulations and experiments in vitro were compared with ground-truth values.
Results
Simulations showed that, for both PGSE and IMPULSED methods, cell diameter d can be reliably fit with sufficient SNR (≥ 50), while intracellular volume fraction fin is intrinsically underestimated due to transcytolemmal water exchange. Din can be reliably fit only with sufficient SNR and using the IMPULSED method with short diffusion times. These results were confirmed with those obtained in the cell culture experiments in vitro.
Conclusion
For the sequences and models considered in this study, transcytolemmal water exchange has minor impacts on fittings of d and Din with physiologically relevant membrane permeabilities if SNR is sufficient (> 50), but fin is intrinsically underestimated.
Keywords: diffusion, transcytolemmal water exchange, cell size, intracellular, diffusion time, OGSE, MRI
Introduction
Diffusion-weighted MRI provides a non-invasive means to probe biological tissue microstructure, and has been widely used for the diagnosis of neurodegenerative diseases and cancer (1). Apparent diffusion coefficients (ADC) obtained from conventional diffusion MRI measurements report the overall averaged diffusion properties of biological tissues. ADC has been found to be sensitive to variations of a variety of tissue properties, including but not limited to cell size (2), cell membrane permeability (3), intra- and extracellular diffusion coefficients (4), and intracellular volume fraction (5). When diffusion times are short enough (such as those obtained using oscillating gradient spin echo (OGSE) sequences at moderately high gradient frequencies(6)), ADC can be sensitive to intracellular microstructural variations such as the sizes of nuclei (7) and organelles (8). Therefore, ADC provides a powerful approach to probe a variety of tissue features from sub- to supra-cellular scales if appropriate diffusion times are used. However, specific microstructural changes usually cannot be directly determined from ADC measurements alone. In many cases, the detection of specific microstructural parameters such as cell size and intracellular volume fraction may provide additional and sometimes more useful diagnostic information (9). Hence, there is a need to develop quantitative diffusion MRI methods to derive more specific microstructural changes from experimental measurements.
To date, there are various diffusion MRI methods developed for quantitative characterization of tissue microstructure. Q-space imaging provides a means to measure apparent compartment sizes (2), but tends to bias cell size because of the inability of distinguishing contributions from intra- and extracellular spaces (10) and the violation of the short gradient pulse condition (11). If the transcytolemmal water exchange between intra- and extracellular spaces is slow enough to be ignored, the overall diffusion signals can be assumed to represent simply the sum of signals from each space. This provides an opportunity to decouple intra- and extracellular signals, and thereby to measure specific microstructural parameters. The AxCaliber method, based on the CHARMED model that assumes cylindrical compartments with hindered diffusion in the extracellular space (with a constant diffusion coefficient), has been implemented to measure axon size distributions in animals in vitro (12) and in vivo (13), and humans in vivo (14). The ActiveAx method was developed to measure indices of mean axon diameter instead of a diameter distribution for better clinical feasibility (15,16). In addition to these conventional pulse gradient spin echo (PGSE) based methods, double-PGSE (17) and OGSE (18) sequences have also been developed for quantitative characterization of cell size and intracellular volume fraction. Recently, a temporal diffusion spectroscopy (TDS) based approach (19) has been developed to combine PGSE (long diffusion times) and OGSE (short diffusion times) acquisitions to cover a broader range of effective diffusion times for better quantitative characterization of microstructure at different length scales simultaneously, i.e. from intracellular diffusion effects up to the impact of cell size and spacing. We term this new experimental approach as the Imaging Microstructural Parameters Using Limited Spectrally Edited Diffusion (IMPULSED) method.
All the models used to interpret data from these quantitative diffusion methods assume that transcytolemmal water exchange between intra- and extracellular spaces can be ignored. This assumption is usually believed true in neural tissues because the typical diffusion time tD in PGSE measurements is < 100 ms, smaller than the intracellular water lifetime τin of brain tissues in vivo reported as 622±29 ms (20) or ∼ 550 ms (21). However, some studies have suggested that the “slow” water exchange still significantly affects MRI measurements of nerves in vitro (22) and in vivo (23). Our recent study indicates that the influence of transcytolemmal water exchange on ADC measurements can be ignored only when tD is at least one order of magnitude smaller than τin (24). More complicatedly, transcytolemmal water exchange typically increases significantly in e.g. lesions caused by Parkinson's (25) and Alzheimer diseases (26), and tumors (27) especially in apoptotic regions (28). This raises concerns if the estimates of microstructural parameters are biased by transcytolemmal water exchange. Previous computer simulations have suggested that the estimated intracellular volume fraction may be underestimated in case of short exchange times (29,30). However, comprehensive studies have not been reported up to date to investigate the influence of transcytolemmal water exchange on the accuracy of fitted microstructural parameters obtained using quantitative diffusion MRI methods.
The present work aims to address the concerns mentioned above. Diffusion MRI sequences using different diffusion times were investigated. Computer simulations and cell culture studies in vitro were performed, and the cell membrane permeability was selectively changed without affecting other cellular microstructure in order to evaluate the precise impact of transcytolemmal water exchange on fitted microstructural parameters. In addition, the influences of signal-to-noise (SNR) and the choices of pre-assigned Din used in the fittings were also studied to evaluate their impacts on fitted parameters.
Methods
Theory
Diffusion MRI signals were modeled as the sum of signals from intra- and extracellular spaces without water exchange between them, namely,
| [1] |
where fin is the water fraction of intracellular space, Sin and Sex are the signal magnitudes from intra- and extracellular spaces, respectively. Note that T2 relaxation is not considered here. For simplicity, tissues were modeled as tightly-packed spherical cells with an effect mean cell diameter d. The analytical expressions to predict diffusion signals of intracellular water restricted by spheres have been reported previously (31). The expression for extracellular water depends on the effective diffusion time, as detailed below. Note that Eq.[1] is widely-used for various diffusion methods (15,18,32). After the diffusion signals were obtained from simulations or experiments, all diffusion MRI signals were fit to Eq.[1]. By comparing fitted values and ground truth, the impact of transcytolemmal water exchange on quantitative characterization of tissue microstructure can be evaluated. Three diffusion methods PGSE_I, PGSE_II and IMPULSED, were investigated.
PGSE_I and PGSE_II methods
The analytical expression for Sin using PGSE acquisitions based on the Gaussian phase approximation is given by
| [2] |
Where S0,in is T2-weighted intracellular signal, γ is the gyromagnetic ratio, δ is the gradient duration, Δ is the gradient separation, g is the gradient strength, Din is the intracellular free diffusion coefficient, λk are Bk are cell diameter d dependent parameters (19,31).
Previous analyses of PGSE data (12–16) usually assume the extracellular diffusion is hindered, i.e. extracellular diffusion coefficient is a constant independent of diffusion time, and hence
| [3] |
Note that the accuracy of this approximation has been previously questioned (33,34). Nevertheless, Eq.[3] was used to describe extracellular diffusion to follow the current widely-used approaches.
Altogether, four independent parameters may be fit using this method: d, fin, Din, and Dex. We term this combination of measurement and analysis as the PGSE_I method. It has been reported that PGSE acquisitions with relatively long diffusion times have low sensitivity to intracellular features (6,7), and hence previous studies have assigned Din to an empirical constant value in fittings to reduce the number of free parameters (9,13,15,16). In this case, three free parameters may be fit: d, fin, and Dex, and we term this as the PGSE_II method. Note that the influence of different choices of Din has not been fully investigated before, so we studied this influence on fitted microstructural parameters in the simulations in the present work.
IMPULSED method
We previously proposed a new experimental IMPULSED method. In this method, PGSE acquisitions are used to obtain data with a long diffusion time tD (e.g. 52 ms) and acquisitions using oscillating gradients with two or more frequencies are used to sample other regions of the diffusion spectrum with shorter diffusion times (where the oscillating frequency f is related to the effective diffusion time as tD = 1/(4f) for cosine-modulated waveforms). By such as means, it covers a broader range of diffusion times and therefore provides a more comprehensive sensitivity to different length scales (19). The analytic expressions for intracellular cosine-modulated OGSE signals have been derived (31) and validated with phantoms (35) previously as
| [4] |
where f is diffusion gradient frequency.
When the experimental diffusion times are relatively short, the extracellular diffusion usually cannot be assumed as a tD–independent constant. Due to the complexity of extracellular structure, it is challenging to derive a general analytical equation to describe the diffusion spectra arising from the extracellular space. An empirical model for extracellular diffusion has been recently proposed to describe packed cylindrical axons (36). However, previous studies have found that if only a narrow range of relatively low gradient frequencies is used (such as 0, 40 and 80 Hz used in the current work), the extracellular diffusion coefficient can be approximated as a linear function of gradient frequency (18,33,34,37). Therefore, extracellular diffusion signals can be simplified as
| [5] |
where Dex0 is the apparent diffusion coefficient of extra-cellular space measured at very long diffusion times (f → 0) and βex is the slope of Dex with respect to gradient frequency f. Dex0 is mainly determined by the extracellular tortuosity and βex indirectly reflects microstructural information (38). PGSE measurements with a single long diffusion time are used in IMPULSED method and treated as 0 frequency. Note that this only affects the modeling of extracellular diffusion, while intracellular diffusion of PGSE measurements is diffusion time dependent determined by Eq. [2]. Therefore, five independent parameters are fit using the IMPULSED method: d, fin, Din, βex and Dex0. The IMPULSED method has been successfully implemented to measure mean axon diameter (18) and cell size (19), both with validations using light microscopy.
Computer simulations
A finite difference method was used to simulate diffusion signals obtained using different sequences as shown in (19). The tissue was modeled as tightly-packed spherical cells on a face-centered-cubic lattice (Figure 2 in ref. (31)) with fin = 61.8%, Din = 1 μm2/ms, Dex = 2 μm2/ms, and homogenous relaxation times everywhere for simplicity. 11 different values of cell diameter d evenly distributed from 5 to 20 μm were simulated to emulate typical cancer cell sizes, and each d incorporated 16 different intracellular water lifetimes τin's: 50, 60, 70, 80, 90, 100, 125, 150, 200, 250, 300, 350, 400, 500, and ∞ ms. Note that τin of rat brain in vivo has been reported as ∼ 550 ms (21). The transmembrane water apparent exchange rate has been reported as 2.9 ± 0.8 s-1 (τin ∼ 500 ms) for viable human brain tumor (27) and tumor cell exchange rate is expected to increase during treatment-induced apoptosis (28). The finite difference simulations actually incorporated cell membrane permeability Pm instead of τin as described previously (39), where Pm is related to τin as (40)
| [6] |
All the other pulse sequence parameters in the simulations were the same as those used in the MR experiments (see below). All simulations (176 combinations of 11 cell diameters and 16 intracellular water lifetimes) took ∼18.2 hours using Matlab R2015a running on a 64bit Linux machine with a Xeon 3.30 GHz CPU.
Three studies were performed based on simulation-generated datasets:
[Study I] To compare the accuracy of fitted d, fin, and Din using IMPULSED, PGSE_I, and PGSE_II methods. Note the PGSE_II method does not provide fitted Din. The percentage differences of fitted and ground truth values were evaluated with a broad range of τin 50 - ∞ ms and d 5 – 15 μm.
[Study II] To investigate the different choices of pre-defined Din on the accuracy of fitted d and fin using the PGSE_II method. Only cells with d = 10 μm were simulated here. Five Din values were investigated: 0.6, 0.8, 1.0 (true value), 1.5, and 2.0 μm2/ms. Fitted results with different choices of Din were compared with ground truth values.
[Study III] To investigate the influence of SNR on the contrast and stability of the fitted parameters obtained using the PGSE_II and IMPULSED methods. Three SNR's were investigated: 10, 20, and 50, and each with four τin's: 50, 100, 250, and 500 ms. Random noise with targeted SNR's based on the non-diffusion-weighted signals were added to simulated noise-free signals according to the approach described in (41), and then microstructural parameters were fit based on signals with noise. This process was repeated 100 times for each of the 12 combinations of SNR and τin, and the mean of the 100 fitted values of each microstructural parameter (d, or fin, or Din) was calculated and compared with the ground truth values.
Cell sample preparation
Murine erythroleukemia (MEL) cancer cells purchased from American Type Culture Collection (Manassas, Virginia, USA) grew under standard culture conditions described previously (24). Large scale of MEL cells were cultured in multiples 150 mm dishes, after spun down and washed with PBS, the cells were fixed with 4% paraformaldehyde in PBS for about 2 hours. After fixation, cells were washed and divided into four groups at a cell density 4.2×107 cell/ml, each treated with 0%, 0.01%, 0.025% and 0.05% (w/v) -saponin at room temperature for 30 minutes to induce various degree of cell membrane permeability. Note that saponin does not cross cell membranes at low concentration, so that it is unlikely to change other cell microstructure e.g. nuclear envelope. For each concentration of saponin treatment, cells were divided approximately evenly into six 0.65ml microtubes (each tube contains about 120 million cells). After centrifuge at 6000g for 2 minutes, top fluid was carefully removed and the cell pellet samples were then used for MR experiments. Small aliquoted sample was spotted on glass slides and imaged directly under phase contrast microscopy for cell diameter estimation.
MR measurements
All MR diffusion measurements were performed on a Varian 4.7T MRI spectrometer. The sample temperature was maintained at ∼ 17°C using a cooling water circulation system. The acquisition time was ∼7 min for measurements of τin, and 9 min for PGSE and IMPULSED experiments.
Measurements of τin
The intracellular exchange lifetime τin was estimated using constant gradient (CG) experiments (20,40). With a stimulated echo (STEAM) sequence, diffusion weighting was achieved by keeping δ = 10 ms and varying the gradient separation Δ in 30 increments. The minimum Δ was 20 ms and the maximum Δ was 426, 426, 223, 121.5 ms for cell samples with saponin concentrations of 0, 0.01%, 0.025% and 0.05%, respectively. Diffusion gradients were applied simultaneously on three axes with gradient strength g = 5 G/cm. The experiment was repeated with a lower gradient strength g = 0.5 G/cm to normalize the higher gradient dataset in order to correct for T1 relaxation effect during the mixing time. The ADC of the slowly decaying component (DB) was determined by linear regression using the last 10 b-value points. The τin was then estimated as (20,40,42)
| [7] |
Note that Eq. [7] is derived from Karger model based on several assumptions and its validity is supported by computer simulations (40).
PGSE experiments
Diffusion signals were measured with PGSE sequences at four different diffusion times (δ = 4 ms, and Δ = 10, 25, 35, 50 ms). Diffusion gradients were applied simultaneously on three axes with 11 gradient strengths varying linearly from 0 to 20 G/cm. Other parameters included: repetition time TR = 3.5 sec; number of excitation = 2; number of dummy scans = 2; receiver bandwidth = 50 kHz; spectral resolution = 390.625 Hz; echo time TE = 60 ms.
IMPULSED experiments
The IMPULSED measurements used both PGSE and OGSE sequences. For the PGSE measurements, δ = 4 ms, and Δ = 52 ms. For OGSE experiments, cosine-modulated waveforms were applied with δ/Δ = 25/30 ms at two oscillation frequencies (f = 40 and 80 Hz). TR = 3.5 sec and TE = 60 ms for both PGSE and OGSE measurements. Nine b values evenly distributed between 0 and 2 ms/μm2 were used in both measurements. The maximum gradient strength used in this study was 30 G/cm, which is achievable on e.g. the cutting-the-edge human brain gradient coil (43).
Data analysis
Diffusion signals obtained in PGSE acquisitions were fit to Eqs.[1-3], and those from IMPULSED experiments were fit to Eqs.[1], [2], and [4-5]. Four parameters were fit using the PGSE_I method: d, fin, Din, and Dex, three using PGSE_II by assuming Din = 1 μm2/ms: d, fin, and Dex, and five using the IMPULSED method: d, fin, Din, βex, and Dex0. The lsqcurvefit function in Matlab was used in the optimization with constraints of parameters limited to biophysically meaningful values: 0 ≤ d ≤ 30 μm, 0 ≤ fin ≤ 1, 0.1 ≤ Din ≤ 3 μm2/ms, 0.1 ≤ Dex, Dex0 ≤ 3 μm2/ms, and 0 ≤ βex ≤ 30 μm2. To avoid local minima, each fitting was repeated 100 times with randomly-generated initial conditions, and the analyses corresponding to the smallest fitting residuals were chosen as the final results.
Results
Simulated influence of fitted parameters on τin and d
Figure 1 shows the simulated fitting errors of fitted parameters compared with ground truth values dependent on d and τin for all three methods. Both IMPULSED and PGSE_II provide accurate estimates of d with error < 5% for τin > 150 ms and d > 7 μm, while the PGSE_I can provide ≤ 5% errors only when τin > 350 ms and d was between 10 and 16 μm. Consistent with previous computer simulation results (29,30), all three methods significantly underestimated fin, except for IMPULSED and PGSE_II methods when τin is infinitely large. All fitted fin decreases rapidly with smaller τin, indicating transcytolemmal water exchange has a strong influence on fitted fin. The IMPULSED method provides reasonable fits of Din with errors ≤ 10% for most τin and d values, although the fits with errors ≤ 5% occurred only when d was between 7 – 13 μm. The PGSE_I could provide fits of Din with errors < 10% only when d is large (≥ 12 μm) and τin is long (≥ 400 ms), and those with ≤ 5% errors only occur at a few combinations of τin and d. We need to reemphasize that Din was empirically pre-defined in the PGSE_II method, so it cannot be obtained in the fittings. Note that some plots in Figure 1 appear to be noisy, the main reason is that some fitted parameters reached the boundaries of loose fitting constraints.
Figure 1.
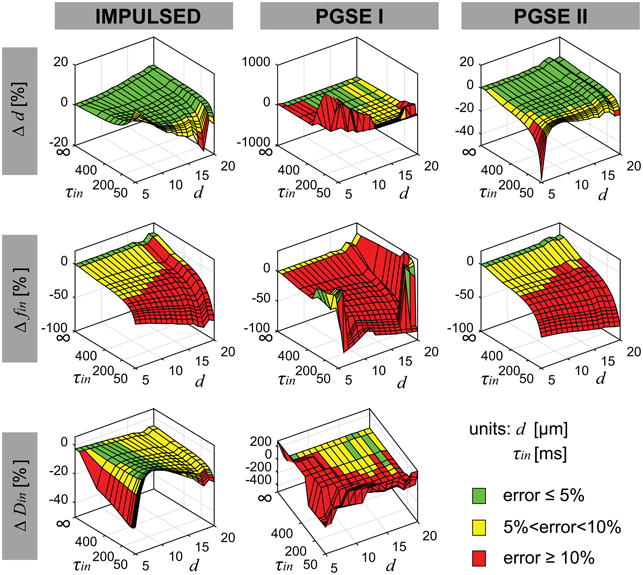
Simulated dependence of the fitting errors compared with ground truth values of fitted parameters on d and τin for all three methods.
Simulated influence of pre-defined Din on PGSE_II fitting
Figure 2 shows the simulated dependence of the fitted PGSE_II parameters on the choice of pre-defined Din used in the fittings. Note that only d = 10 μm was simulated here. Except for fast transcytolemmal water exchange (τin < 100 ms), the choices of Din had minor effects on the fittings of PGSE_II. For example, the fitted d were all within 5% difference when the pre-defined Din was in the range of 0.6 – 2.0 μm2/ms when τin > 100 ms, indicating the accuracy of fitted d is not influenced by the choice of Din used in the fittings. fin is consistently underestimated across all τin values, while the discrepancy decreases with longer τin. The ground truth extracellular diffusion coefficient is provided in Figure 2, which is much higher than fitted Dex which is the extracellular diffusion coefficient at very long diffusion times, i.e., mainly determined by the extracellular tortuosity. Note that both the fitted fin and Dex had small dependences on pre-defined Din: < 10% changes occur when Din changes from 0.6 to 2.0 μm2/ms. These results are encouraging. In most cases, Din is unknown. If the influence of Din on the fitted parameters of the PGSE_II method is minor, an empirical value of Din can be implemented and the fitting accuracy is preserved.
Figure 2.
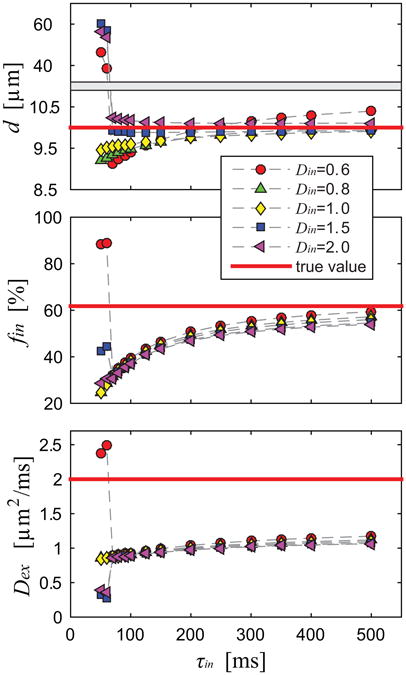
Simulated parameters obtained using the PGSE_II method dependent on different choices of pre-defined Din used in the fittings.
Simulated influence of SNR
The influence of SNR on fitted d using the IMPULSED and PGSE_II methods is shown in Figure 3. For each real d value, the fittings were repeated 100 times and each with different noises at the same SNR. Hence, 100 fitted d values were obtained (green +) for each real d, and the mean of the repetitions (black ×) are also shown in Figure 3 to indicate the fitting accuracy. For both IMPULSED and PGSE_II methods at low SNR = 20, there was a broad range of fitted d values for each real d, indicating the precision was significantly reduced by noise. By contrast, at SNR = 50, d could be reliably fit using the IMPULSED or PGSE_II methods despite different τin. The mean values of all fitted results indicate the accuracy of fittings. At SNR = 20, the PGSE_II method provided relatively more accurate fittings compared with the IMPULSED method, while the latter overestimated d for all ground-truth d and τin values. Note that this is at least partially due to the less data points (28 b values) acquired using the IMPULSED method compared with the 45 b values used in the PGSE_II method. By contrast, both diffusion methods provided more reliable fittings at SNR = 50. The IMPULSED method provided more accurate and precise fittings compared with the PGSE_II method especially for d 5 – 10 μm. The fitted results showed slightly less variations with higher τin and SNR = 50, indicating transcytolemmal water exchange has a minor influence on the fittings of d when SNR is sufficient. For SNR = 20, the influence of τin on d fittings was more significant, especially for the IMPULSED method. Based on these findings, we can conclude that when SNR is low (e.g. 20 in the current simulation), the transcytolemmal water exchange does affect fitted d, and higher τin (slower water exchange) and higher SNR would lead to more accurate estimates of d.
Figure 3.
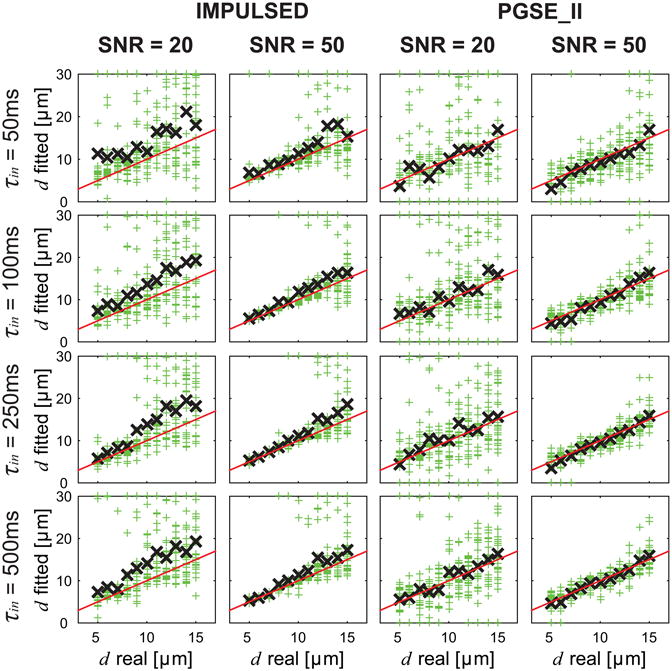
Simulated influence of SNR on fitted d using the IMPULSED and PGSE_II methods. For each real d, the fittings were repeated 100 times (green +) each with different noises but at the same SNR level, and the mean of the repetitions (black ×) are also shown. The red lines represent the identity lines.
Figure 4 shows the simulated influence of SNR and τin on fitted Din values using the IMPULSED method. SNR significantly affects the accuracy of fitted Din values. For SNR = ∞, Din can be fit well with < 10% errors except when d ≤ 6 μm. This is consistent with previous findings that Din cannot be reliably fit when the restriction dimension is too small and the highest gradient frequency used is limited (35). For SNR = 10, Din tends to be overestimated with large variations. For SNR = 20, better estimates of Din were achieved when τin > 250 ms and real d ≥ 10 μm. For SNR = 50, it seems Din were all fit reasonably well with most errors < 20% except for τin = 50 ms. This suggests that the accuracy and precision of Din fitting using the IMPULSED approach were mainly limited by SNR when transcytolemmal water exchange is not too fast (e.g. τin > 100 ms). Recall that the PGSE_II method is incapable of providing any Din information, while the PGSE_I method estimates Din poorly even with the noise-free signals (see Figure 1). This suggests an advantage of using the IMPULSED method to quantitatively characterize tissues in practice.
Figure 4.
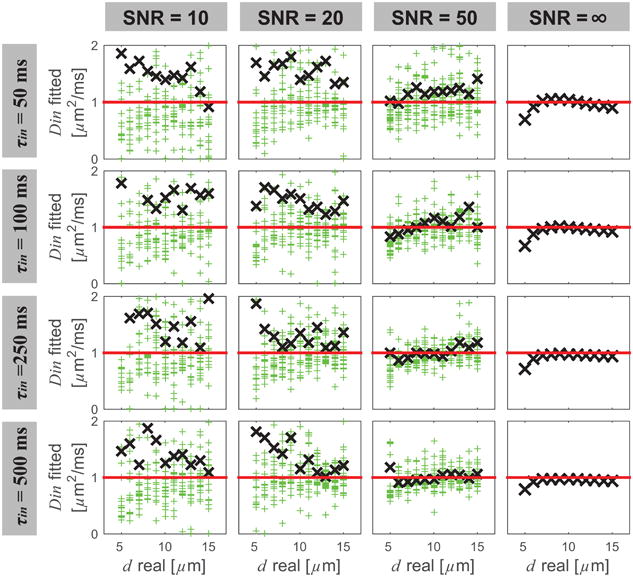
Simulated influence of SNR and τin on fitted Din values using the IMPULSED method. Meanings of symbols and lines are the same as in Figure 3.
Experimental results of in vitro studies
Cell size and τin
Based on the light microscopy experiments and the analysis approach described previously (19), the MEL cell diameter was measured as 11.34±1.68 μm, consistent with previous reported 11.74±1.30 μm (19). The volume weighted cell diameter d was then calculated as 12.11 μm used as ground truth value in Figure 5. The CG-experiments provided τin values of 161.8±9.4, 157.8±8.9, 106.6±4.3, and 59.4±3.7 ms for 0, 0.01, 0.025, and 0.05% concentration of saponin, respectively.
Figure 5.
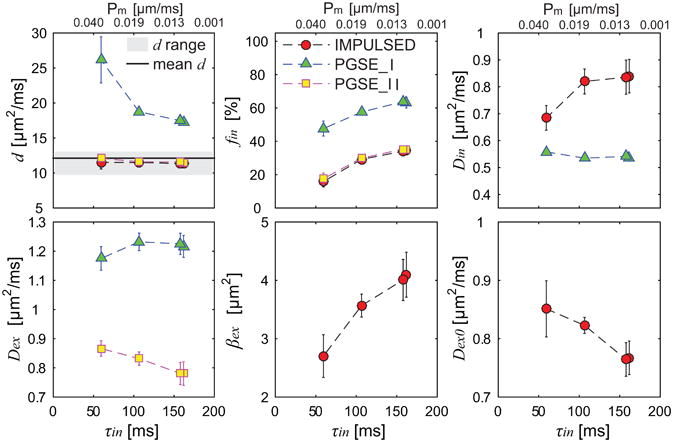
Summary of fitted microstructural parameters vs τin using three diffusion methods. Error bars in each sub-figure denote across-sample STD. d range means histology-derived mean cell diameter ± STD of all cells, and mean d is volume-weighted cell diameter.
Microstructural parameters of cell samples
Figure 5 summarizes all fitted microstructural parameters of cultured MEL cells with four different cell membrane permeabilities. The IMPULSED and PGSE_II methods provide accurate fittings of d independent of permeability over a broad range of τin (59.40 – 161.80 ms), while fitted fin decreased rapidly with τin. This is consistent with simulated results shown in Figure 1 and Figure 3 that, if SNR is not too low (e.g. > 50) and τin is not too small (e.g. > 50 ms), transcytolemmal water exchange has minor influence on the estimations of d over a broad range of τin values, while estimations of fin could be significantly biased by fast transcytolemmal water exchange i.e. higher τin values. Din estimated by the IMPULSED method showed approximately no dependence over the τin range of 106.58 to 161.80 ms, but decreased to 0.69 μm2/ms when τin decreased to 59.40 ms. This suggests that although Din is slightly more susceptible to transcytolemmal water exchange compared with d, Din still can be reliably estimated if τin is not too small (e.g. > 100 ms). Din fitted from the PGSE method were approximately constant over the τin range of 106.58 to 161.80 ms. However, as suggested by the simulations shown in Figure 1, the PGSE method intrinsically underestimates Din even with noise-free signals, and hence Din obtained using the PGSE method may not be reliable. All other fitted parameters, Dex from the PGSE_I and PGSE_II methods, kex and Dex0 from the IMPULSED method, showed dependence on τin.
Discussion
The present work investigated the influence of transcytolemmal water exchange on quantitative characterization of tissue microstructure using diffusion MRI methods. Despite the significant variations of τin present in simulations and in vitro experiments, all diffusion signal data sets were fit to currently widely-used models that assume no water exchange between intra- and extracellular spaces. The results suggest that fitted d is relatively insensitive to τin with reasonable SNR in both PGSE_II and IMPULSED experiments, while fin is significantly underestimated for all diffusion methods tested. Din can be reliably fit only using IMPULSED experiments when SNR is sufficient (e.g. 50). These findings may assist better interpretation of quantitative characterization of tissue microstructure in tumors, in which the transcytolemmal water exchange is often much faster than normal tissue and cannot be ignored (27,28). For example, a previous study using the IMPULSED method provided d and fin maps of an MDA-MBA-231 triple-negative breast cancer xenograft in vivo (44). Although d was fit reasonably well and confirmed with histology, fin was significantly underestimated (< 50%) in most regions. Based on the findings of the present work, the fitting accuracy of these in vivo findings can be explained by the enhanced transcytolemmal water exchange in tumors. To prove this, the transcytolemmal water exchange needs to be mapped using a method such as filter exchange imaging (FEXI) (27,45) in addition to quantitative diffusion MRI measurements.
It is possible to incorporate transcytolemmal water exchange into quantitative diffusion models, and hence fit all microstructural parameters including τin simultaneously. The two-exchanging-component-system describing diffusion measurements has been thoroughly investigated by Karger et al. by assuming two freely-diffusing components (46), and was later improved to include restrictions (i.e. d and Din) on the intracellular water component (40,42,47). However, this method requires very high b values e.g. 580 ms/μm2, as was used in rat brain in vivo (20). Stanisz et al. performed comprehensive fittings to measure cell size, volume fraction, membrane permeability simultaneously in bovine optic nerves ex vivo (48). However, high b values ≈ 40 ms/μm2, high gradient strength 140 G/cm and rise time ∼ 500 μs were used, which are not achievable in practical imaging. Therefore, one major disadvantage of combining transcytolemmal water exchange in quantitative diffusion models is that these methods would become complicated and demand very high SNR and long scanning time. This will significantly limit the feasibility of translating these methods into clinical practice. Diffusion MRI usually suffers low SNR and long scanning time in practice and hence it is desirable to keep quantitative models simple with fewer free parameters. Partially due to this reason, the PGSE_II method was able to provide more accurate fits of d compared with the PGSE_I method because PGSE_II has less free parameters.
The minor influence of pre-defined Din on the fitting accuracy of the PGSE_II method is encouraging. Din is usually unknown in practice. Previous studies assigned Din empirical values in the fittings (9,13,15,16), but the exact influence of different choices of empirical Din values on fitted results has not been fully investigated. The current study suggests that the influence of Din can be ignored if assigned Din is in a reasonable range (between -40% and +100% difference in the present work) when τin is not too short (> 100 ms shown in Figure 2). Since this influence is minor on fitted results, it is recommended to use the PGSE_II method other than the PGSE_I method due to its better accuracy and stability (see Figure 1). Note that neither PGSE_I nor PGSE_II methods used in the present work can reliably fit Din, presumably because of the relatively long diffusion times used. By incorporating PGSE acquisitions with a long diffusion time (Δ = 50 ms) and OGSE acquisitions with short diffusion times (40 and 80 Hz, corresponding to tD 6.25 and 3.125 ms), the IMPULSED method is capable of providing a more comprehensive means to fit both intracellular Din and cellular dimension d simultaneously. Note that the IMPULSED method used in the current work incorporates three different tD values, and fits five parameters, while the PGSE_II method incorporates four tD values and fits three parameters. The fitted d using the IMPULSED method with less data points is more prone to noise than the PGSE_II method shown in Figure 3. We need to emphasize that it is possible to include more acquisitions with additional diffusion times in the IMPULSED method to enhance the sensitivity to d, but this in turn will increase the total scanning time. Therefore, it is desirable to optimize the IMPULSED method for specific applications in vivo.
Two relatively low frequencies (40 and 80 Hz) were used in the current work. The two low frequencies correspond to tD 6.25 and 3.125 ms, and root-mean-square displacements of 3.5 and 2.5 μm if diffusion coefficient is 1 μm2/ms. This is already much smaller than typical cancer cell size (e.g. 11.34 μm in the present work), which enables the feasibility to probe Din. Note that the fitted Din represents an averaged intracellular diffusion properties, and even smaller intracellular microstructure such as nuclear size and organelles may not be probed by the frequencies used in the present work (8). To be sensitive to smaller sizes (∼ 2 μm), much higher frequencies (∼ 250 Hz) would be required (18,38).
To ensure translatability of our results, maximum gradient strengths were limited < 30 G/cm which is achievable on the cutting-the-edge human gradient coils (43). Note that even with typical maximum gradient strength 8 G/cm, OGSE has been implemented on human scanners and shows promising potential to probe microstructure of human brain in vivo (49,50) and may assist interpret ADC data in stroke patients (51). However, the lowest frequency achievable is limited by echo time, so OGSE acquisitions have reduced sensitivity to larger scale dimensions. Combined with PGSE acquisitions with long diffusion times, sensitivity to both intra- and supracellular scales can be achieved. In return, both d and Din can be fit which provides more microstructural information than d alone as provided by the PGSE_II method. Note that higher gradient performance can improve the estimates of the fitting results (52). This is particular true for the IMPULSED method which uses OGSE acquisitions (53).
Conclusions
Both computer simulations and in vitro cell studies suggest that: for PGSE methods with relatively long diffusion times, empirically assigning Din as a constant in fittings significantly increases the accuracy of other fit parameters. Both IMPULSED and PGSE_II methods provide accurate fits of mean cell diameter d independent of transcytolemmal water exchange with sufficient SNR (e.g. 50), while intracellular volume fraction fin was intrinsically underestimated in both methods and decreased rapidly with shorter τin. The IMPULSED method is capable of estimating intracellular diffusion coefficient Din when appropriate ranges of d (7 – 15 μm), τin > 100 ms and sufficient SNR e.g. 50 were satisfied, while Din cannot be reliably estimated using the PGSE methods even for noise-free data. These results can assist better interpreting diffusion data for quantitative characterization of e.g. tumors, where transcytolemmal water exchange cannot be ignored.
Acknowledgments
This work was funded by NIH grants NIH K25CA168936; R01CA109106; R01CA173593.
Grant Support: NIH K25CA168936; R01CA109106; R01CA173593.
References
- 1.Gillard JH, Waldman AD, Barker PB. Clinical MR Neuroimaging: Diffusion, Perfusion and Spectroscopy. 1. Cambridge University Press; Dec 27, 2004. 2004. [Google Scholar]
- 2.Cory DG, Garroway AN. Measurement of translational displacement probabilities by NMR: an indicator of compartmentation. Magn Reson Med. 1990;14:435–444. doi: 10.1002/mrm.1910140303. [DOI] [PubMed] [Google Scholar]
- 3.Tanner JE. Transient diffusion in a system partitioned by permeable barriers - application to NMR measurements with a pulsed field gradient. J Chem Phys. 1978;69:1748–1754. [Google Scholar]
- 4.Tanner J. Intracellular diffusion of water. Arch Biochem Biophys. 1983;224:416–428. doi: 10.1016/0003-9861(83)90228-x. [DOI] [PubMed] [Google Scholar]
- 5.van der Toorn A, Syková E, Dijkhuizen RM, Vorísek I, Vargová L, Skobisová E, van Lookeren Campagne M, Reese T, Nicolay K. Dynamic changes in water ADC, energy metabolism, extracellular space volume, and tortuosity in neonatal rat brain during global ischemia. Magn Reson Med. 1996;36:52–60. doi: 10.1002/mrm.1910360110. [DOI] [PubMed] [Google Scholar]
- 6.Gore JC, Xu J, Colvin DC, Yankeelov TE, Parsons EC, Does MD. Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. NMR Biomed. 2010;23:745–756. doi: 10.1002/nbm.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: Effects of nuclear size. Magn Reson Med. 2009;61:828–833. doi: 10.1002/mrm.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colvin DC, Jourquin J, Xu J, Does MD, Estrada L, Gore JC. Effects of intracellular organelles on the apparent diffusion coefficient of water molecules in cultured human embryonic kidney cells. Magn Reson Med. 2011;65:796–801. doi: 10.1002/mrm.22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res. 2014;74:1902–12. doi: 10.1158/0008-5472.CAN-13-2511. [DOI] [PubMed] [Google Scholar]
- 10.Ong HH, Wright AC, Wehrli SL, Souza A, Schwartz ED, Hwang SN, Wehrli FW. Indirect measurement of regional axon diameter in excised mouse spinal cord with q-space imaging: simulation and experimental studies. Neuroimage. 2008;40:1619–32. doi: 10.1016/j.neuroimage.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lätt J, Nilsson M, Malmborg C, Rosquist H, Wirestam R, Ståhlberg F, Topgaard D, Brockstedt S. Accuracy of q-space related parameters in MRI: Simulations and phantom measurements. IEEE Trans Med Imaging. 2007;26:1437–1447. doi: 10.1109/TMI.2007.907278. [DOI] [PubMed] [Google Scholar]
- 12.Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med. 2008;59:1347–1354. doi: 10.1002/mrm.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barazany D, Basser PJ, Assaf Y. In vivo measurement of axon diameter distribution in the corpus callosum of rat brain. Brain. 2009;132:1210–1220. doi: 10.1093/brain/awp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz A, Barazany D, Tavor I, Bernstein M, Yovel G, Assaf Y. In vivo correlation between axon diameter and conduction velocity in the human brain. Brain Struct Funct. 2015;220:1777–1788. doi: 10.1007/s00429-014-0871-0. [DOI] [PubMed] [Google Scholar]
- 15.Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJM, Dyrby TB. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage. 2010;52:1374–89. doi: 10.1016/j.neuroimage.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 16.Dyrby TB, Søgaard LV, Hall MG, Ptito M, Alexander DC. Contrast and stability of the axon diameter index from microstructure imaging with diffusion MRI. Magn Reson Med. 2013;70:711–721. doi: 10.1002/mrm.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shemesh N, Özarslan E, Basser PJ, Cohen Y. Accurate noninvasive measurement of cell size and compartment shape anisotropy in yeast cells using double-pulsed field gradient MR. NMR Biomed. 2012;25:236–46. doi: 10.1002/nbm.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Li H, Harkins KD, Jiang X, Xie J, Kang H, Does MD, Gore JC. Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. Neuroimage. 2014;103:10–19. doi: 10.1016/j.neuroimage.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med. 2015 doi: 10.1002/mrm.25684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier C, Dreher W, Leibfritz D. Diffusion in compartmental systems. II. Diffusion-weighted measurements of rat brain tissue in vivo and postmortem at very large b-values. Magn Reson Med. 2003;50:510–4. doi: 10.1002/mrm.10558. [DOI] [PubMed] [Google Scholar]
- 21.Quirk JD, Bretthorst GL, Duong TQ, Snyder AZ, Springer CS, Ackerman JJH, Neil JJ. Equilibrium water exchange between the intra- and extracellular spaces of mammalian brain. Magn Reson Med. 2003;50:493–9. doi: 10.1002/mrm.10565. [DOI] [PubMed] [Google Scholar]
- 22.Dula AN, Gochberg DF, Valentine HL, Valentine WM, Does MD. Multiexponential T2, magnetization transfer, and quantitative histology in white matter tracts of rat spinal cord. Magn Reson Med. 2010;63:902–9. doi: 10.1002/mrm.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harkins KD, Dula AN, Does MD. Effect of intercompartmental water exchange on the apparent myelin water fraction in multiexponential T2 measurements of rat spinal cord. Magn Reson Med. 2012;67:793–800. doi: 10.1002/mrm.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Jiang X, Xie J, McIntyre JO, Gore JC, Xu J. Time-dependent influence of cell membrane permeability on MR diffusion measurements. Magn Reson Med. 2015 doi: 10.1002/mrm.25724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volles MJ, Lansbury PT. Vesicle permeabilization by protofibrillar α-synuclein is sensitive to Parkinson's disease-linked mutations and occurs by a pore-like mechanism. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 26.Moftakhar P, Lynch MD, Pomakian JL, Vinters HV. Aquaporin expression in the brains of patients with or without cerebral amyloid angiopathy. J Neuropathol Exp Neurol. 2010;69:1201–9. doi: 10.1097/NEN.0b013e3181fd252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson M, Lätt J, van Westen D, Brockstedt S, Lasič S, Ståhlberg F, Topgaard D. Noninvasive mapping of water diffusional exchange in the human brain using filter-exchange imaging. Magn Reson Med. 2013;69:1573–81. doi: 10.1002/mrm.24395. [DOI] [PubMed] [Google Scholar]
- 28.Bailey C, Moosvi F, Stanisz GJ. Mapping water exchange rates in rat tumor xenografts using the late-stage uptake following bolus injections of contrast agent. Magn Reson Med. 2013;71:1874–1887. doi: 10.1002/mrm.24847. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson M, Lätt J, Nordh E, Wirestam R, Ståhlberg F, Brockstedt S. On the effects of a varied diffusion time in vivo: is the diffusion in white matter restricted? Magn Reson Imaging. 2009;27:176–187. doi: 10.1016/j.mri.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson M, Alerstam E, Wirestam R, Ståhlberg F, Brockstedt S, Lätt J. Evaluating the accuracy and precision of a two-compartment Kärger model using Monte Carlo simulations. J Magn Reson. 2010;206:59–67. doi: 10.1016/j.jmr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Does MD, Gore JC. Quantitative characterization of tissue microstructure with temporal diffusion spectroscopy. J Magn Reson. 2009;200:189–197. doi: 10.1016/j.jmr.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Assaf Y, Freidlin RZ, Rohde GK, Basser PJ. New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magn Reson Med. 2004;52:965–978. doi: 10.1002/mrm.20274. [DOI] [PubMed] [Google Scholar]
- 33.Burcaw LM, Fieremans E, Novikov DS. Mesoscopic structure of neuronal tracts from time-dependent diffusion. Neuroimage. 2015;114:18–37. doi: 10.1016/j.neuroimage.2015.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novikov DS, Jensen JH, Helpern JA, Fieremans E. Revealing mesoscopic structural universality with diffusion. Proc Natl Acad Sci. 2014;111:5088–5093. doi: 10.1073/pnas.1316944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Gore JC, Xu J. Fast and robust measurement of microstructural dimensions using temporal diffusion spectroscopy. J Magn Reson. 2014;242:4–9. doi: 10.1016/j.jmr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam WW, Jbabdi S, Miller KL. A model for extra-axonal diffusion spectra with frequency-dependent restriction. Magn Reson Med. 2014 doi: 10.1002/mrm.25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aggarwal M, Jones MV, Calabresi PA, Mori S, Zhang J. Probing mouse brain microstructure using oscillating gradient diffusion MRI. Magn Reson Med. 2012;67:98–109. doi: 10.1002/mrm.22981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Jiang X, Wang F, Xu J, Gore JC. Structural information revealed by the dispersion of ADC with frequency. Magn Reson Imaging. 2015;33:1083–1090. doi: 10.1016/j.mri.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang SN, Chin CL, Wehrli FW, Hackney DB. An image-based finite difference model for simulating restricted diffusion. Magn Reson Med. 2003;50:373–382. doi: 10.1002/mrm.10536. [DOI] [PubMed] [Google Scholar]
- 40.Meier C, Dreher W, Leibfritz D. Diffusion in compartmental systems. I. A comparison of an analytical model with simulations. Magn Reson Med. 2003;50:500–9. doi: 10.1002/mrm.10557. [DOI] [PubMed] [Google Scholar]
- 41.Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- 42.Pfeuffer J, Flögel U, Dreher W, Leibfritz D. Restricted diffusion and exchange of intracellular water: theoretical modelling and diffusion time dependence of 1H NMR measurements on perfused glial cells. NMR Biomed. 1998;11:19–31. doi: 10.1002/(sici)1099-1492(199802)11:1<19::aid-nbm499>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 43.Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, et al. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage. 2013;80:220–33. doi: 10.1016/j.neuroimage.2013.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, Li H, Zhao P, Xie J, Gore JC, Xu J. Quantification of mean cell size and intracellular volume fraction using temporal diffusion spectroscopy. Proceedings of the 23rd Annual Meeting ISMRM. 2015:615. [Google Scholar]
- 45.Lasič S, Nilsson M, Lätt J, Ståhlberg F, Topgaard D. Apparent exchange rate mapping with diffusion MRI. Magn Reson Med. 2011;66:356–365. doi: 10.1002/mrm.22782. [DOI] [PubMed] [Google Scholar]
- 46.Karger J, Pfeifer H, Heink W. Principles and applications of self-diffusion measurements by nuclear magnetic resonance. In: Waugh JS, editor. Advances in Magnetic Resonance. Vol. 12. 1250 Sixth Avenue San Diego, California 92101: Academic Press Inc.; 1988. pp. 1–89. [Google Scholar]
- 47.Price W, Barzykin A, Hayamizu K, Tachiya M. A model for diffusive transport through a spherical interface probed by pulsed-field gradient NMR. Biophys J. 1998;74:2259–2271. doi: 10.1016/S0006-3495(98)77935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanisz GJ, Szafer A, Wright GA, Henkelman RM. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med. 1997;37:103–111. doi: 10.1002/mrm.1910370115. [DOI] [PubMed] [Google Scholar]
- 49.Van AT, Holdsworth SJ, Bammer R. In vivo investigation of restricted diffusion in the human brain with optimized oscillating diffusion gradient encoding. Magn Reson Med. 2014;71:83–94. doi: 10.1002/mrm.24632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baron CA, Beaulieu C. Oscillating Gradient Spin-Echo (OGSE) diffusion tensor imaging of the human brain. Magn Reson Med. 2014;72:726–736. doi: 10.1002/mrm.24987. [DOI] [PubMed] [Google Scholar]
- 51.Allan Baron C, Kate M, Gioia L, Butcher K, Emery D, Budde M, Beaulieu C. Reduction of Diffusion-Weighted Imaging Contrast of Acute Ischemic Stroke at Short Diffusion Times. Stroke. 2015 doi: 10.1161/STROKEAHA.115.008815. [DOI] [PubMed] [Google Scholar]
- 52.Huang SY, Nummenmaa A, Witzel T, Duval T, Cohen-Adad J, Wald LL, McNab JA. The impact of gradient strength on in vivo diffusion MRI estimates of axon diameter. Neuroimage. 2015;106:464–472. doi: 10.1016/j.neuroimage.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drobnjak I, Zhang H, Ianuş A, Kaden E, Alexander DC. PGSE, OGSE, and sensitivity to axon diameter in diffusion MRI: Insight from a simulation study. Magn Reson Med. 2015;00:n/a–n/a. doi: 10.1002/mrm.25631. [DOI] [PMC free article] [PubMed] [Google Scholar]


