Abstract
The HIV-1 accessory protein Vpu exhibits high inter- and intra- subtype genetic diversity that may influence Vpu function and possibly contribute to HIV-1 pathogenesis. However, scalable methods to evaluate genotype/phenotype relationships in natural Vpu sequences are limited, particularly those expressing the protein in CD4+ T-cells, the natural target of HIV-1 infection. A major impediment to assay scalability is the extensive genetic diversity within, and immediately upstream of, Vpu's initial 5' coding region, which has necessitated the design of oligonucleotide primers specific for each individual HIV-1 isolate (or subtype). To address this, we developed two universal forward primers, located in relatively conserved regions 38 and 90 bases upstream of Vpu, and a single universal reverse primer downstream of Vpu, which are predicted to cover the vast majority of global HIV-1 group M sequence diversity. We show that inclusion of up to 90 upstream bases of HIV-1 genomic sequence does not significantly influence in vitro Vpu expression or function when a Rev/Rev Response Element (RRE)-dependent expression system is used. We further assess the function of four diverse HIV-1 Vpu sequences, revealing reproducible and significant differences between them. Our approach represents a scalable option to measure the in vitro function of genetically diverse natural Vpu isolates in a CD4+ T-cell line.
Keywords: HIV-1 Vpu, sequence diversity, CD4, BST2/tetherin, downregulation, Rev/RRE
1. Introduction
Human Immunodeficiency Virus type 1 (HIV-1) isolates belonging to the Main (M) group are responsible for the pandemic currently infecting 36.7 million individuals worldwide (Faria et al., 2014; Hemelaar, 2012; UNAIDS, 2016). HIV-1 group M sequences exhibit extensive diversity both within and between hosts. Globally, HIV-1 group M strains are presently classified phylogenetically into nine subtypes and 79 circulating recombinant forms that differ by up to 30% in their envelope amino acid sequence (Hemelaar, 2012). Given this diversity, it is not surprising that natural (patient-derived) HIV-1 sequences also exhibit differences in their in vitro function, which may in turn influence viral pathogenesis. For example, it has been hypothesized that HIV-1 subtype C isolates, which comprise more than >50% of infections worldwide (Hemelaar, 2012), may be inherently more transmissible - possibly because of subtype-specific motifs within Env-gp120 that modulate interactions with host cell entry receptors (Walter et al., 2009) - though this remains controversial (Kahle et al., 2014). Subtype-specific differences in HIV-1 entry efficiency (Marozsan et al., 2005), replication capacity (Aralaguppe et al., 2016; Konings et al., 2006), and the function of viral accessory proteins Vif (Binka et al., 2012; Iwabu et al., 2010) and Nef (Mann et al., 2013) have also been described. Methods to assess the function of other HIV-1 proteins encoded by natural sequences are thus warranted.
The HIV-1 accessory protein Vpu is a multifunctional ~16kDa transmembrane protein that enhances viral replication. Vpu's most well-characterized functions are downregulation of the HIV entry receptor CD4 (Lama, Mangasarian, and Trono, 1999; Levesque, Zhao, and Cohen, 2003; Willey et al., 1992) and the antiviral protein tetherin (also known as BST2 or CD317) (Dube et al., 2010; Neil, Zang, and Bieniasz, 2008; Sauter et al., 2009; Van Damme et al., 2008), which enhance viral egress (Douglas et al., 2010) and promote viral evasion of innate (Galao et al., 2012) and adaptive (Pham et al., 2014; Veillette et al., 2014) immune responses. Vpu-mediated downregulation of HLA-C (Apps et al., 2016), as well as innate immunomodulatory receptors, including NTB-A (Shah et al., 2010), CD1d (Moll et al., 2010) and PVR (CD155) (Matusali et al., 2012), has also been described. The high global sequence diversity of the vpu gene region however has limited the development of scalable methods to clone and express Vpu from natural sequences, and thus the functional range of this protein remains incompletely characterized.
Vpu is expressed late in the HIV-1 life cycle from a partially spliced bicistronic mRNA encoding both vpu and env. In vivo, HIV-1 protein expression from partially- (and un-) spliced viral mRNA transcripts is dependent on the HIV-1 regulatory protein Rev, which facilitates the nuclear export of these transcripts by binding to a ~350 base RNA sequence within env termed the Rev response element (RRE). Though autonomous (i.e. non-Rev/RRE-dependent) in vitro Vpu expression can be achieved by cloning the vpu open reading frame directly at the start codon (Chen et al., 2015; Douglas et al., 2013; Galaski et al., 2016; Mwimanzi et al., 2016; Verma et al., 2013), protein expression is generally not robust. Moreover, the extensive genetic diversity in and upstream of vpu's 5' coding sequence necessitates the design of primers specific for each HIV-1 isolate (Chen et al., 2015; Douglas et al., 2013; Galaski et al., 2016), or panels of primers specific for each HIV-1 subtype (Verma et al., 2013), which limits scalability. Robust autonomous Vpu expression can be achieved by codon optimizing the vpu sequence to maximize expression in mammalian cells (Anson and Dunning, 2005; Nguyen et al., 2004), but this negates the purpose of assessing vpu sequences in their natural form.
For these reasons, in vitro expression of natural Vpu sequences is generally achieved using Rev/RRE-dependent systems. Nevertheless, some studies employing Rev/RRE-dependent systems to assess natural vpu sequences have still cloned the genedirectly at the start site using isolate-specific primers (Jafari, Guatelli, and Lewinski, 2014). Alternatively, the Rev/RRE-dependent expression plasmid pCRV1ΔVpu, which employs a forward primer located in a conserved upstream region, has been used to assess the function of larger numbers of natural vpu sequences, but only in epithelial cell lines that are not the natural targets of HIV-1 infection (Apps et al., 2016; Pickering et al., 2014). Here, we describe a Rev/RRE-dependent method for robust expression and functional assessment of natural HIV-1 group M vpu sequences in a CD4+ T-cell line. The method's main innovation is the use of universal (pan-HIV-1 group M) primers for vpu amplification and cloning, thus removing the need for isolate-specific primer design.
2. Methods
2.1 Plasmids, cell lines and HIV-1 sequences
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: A plasmid encoding the entire genome of the HIV-1 subtype B reference strain NL4.3 (pNL4.3) Cat #114 from Dr. M. Martin (Adachi et al., 1986); a plasmid encoding the entire genome of the HIV-1 subtype C infectious molecular clone MJ4 (pMJ4), Cat#6439 from Drs. T. Ndung'u, B. Renjifo and M. Essex (Ndung'u et al., 2000); an expression plasmid encoding a codon-optimized version of the NL4.3 vpu gene (pcDNA-Vphu), Cat#110079 from Dr. S. Bour and Dr. K. Strebel (Nguyen et al., 2004), and the clonal Human T4-lymphoblastoid cell line CEM-SS, Cat #776 from Dr. P. Nara (Nara and Fischinger, 1988). The plasmids pCRV1ΔVpu and pCRV1ΔVpu-NL4.3 Vpu were provided by Dr. R. Apps with permission from Dr. P. Bieniasz and Dr. S.Neil (Apps et al., 2016; Jouvenet et al., 2009; Pickering et al., 2014). The GFP expressing plasmid pMAX-GFP was purchased from Amaxa.
The pSELECT-GFPzeo dual expression vector (InvivoGen; hereafter referred to as “pSel-GFP” or simply “pSel”), which contains one transcription unit that expresses the gene of interest and another that expresses a gfp:zeocin resistance gene fusion, was modified as follows to create pSelRRE-GFP, which incorporates the HIV-1 RRE at the 3' end of gene-of-interest transcripts. First, pSel-GFP's multiple cloning site was expanded to include additional restriction sites including AscI and SacII, and the RRE from the HIV-1 subtype B reference strain NL4.3 was cloned into the Sac II and existing downstream Nhe I site using the forward primer 5’-AGAGCCGCGGAGTAGCACCCACCAAGGCAAA-3’ and reverse primer 5’-GCCTGCTAGCACTAGCATTCCAAGGCACAGC-3’ (restriction sites underlined). pSel-RevΔGFP, a plasmid expressing the HIV-1 Rev protein without concomitant GFP was engineered by eliminating GFP from pSel-GFP by PCR-mediated deletion then cloning the Rev gene from the HIV-1 subtype B strain SF2 (synthesized as a gBlock; IDT DNA technologies, Coralville USA) into its BamHI and Nhe I sites.
2.2 Universal Vpu primer design and amplification
Two forward primers containing degenerate bases capturing HIV-1 group M sequence diversity were designed 90 and 38 bases upstream of the vpu start site (Figure 1), with sequences 5’ -AGAGCACCGGCGCGCCGGCAGGAAGAAGCGRRGACASMGA - 3’ (“−90 primer”; SgrI and AscI sites underlined; bold bases span nucleotides 5972 to 5995 of the HIV-1 subtype B genomic reference strain HXB2; GenBank Acc# K03455) and 5’-AGAGGGCGCGCCATCAARHTYCTVTAYCAAAGCAGTAAGTA-3’ (“−38 primer”; Asc I site underlined; bold bases span HIV-1 nucleotides 6024 to 6052). A single universal reverse primer (5'-GCCTCCGCGGATCGATGGTACCCCATARTAGACHGTRACCCA-3' (SacII and ClaI sites underlined; bold bases span HIV-1 nucleotides 6352 to 6327) was used to amplify all control and natural vpu sequences. Forward primers containing SgrI and AscI sites were also designed to amplify the control HIV-1 NL4.3 and codon-optimized vpu sequences at their respective start codons.
Figure 1. Location of universal forward primers for Vpu amplification.
Alignment of HIV-1 group M reference sequences (www.hiv.lanl.gov) spanning HIV-1 genomic nucleotides 5970-6095 (HIV-1 HXB2 genomic reference numbering convention) illustrates the extensive genetic diversity within and upstream of Vpu's initial 5' transmembrane domain. NL4.3 is used as the alignment reference; others are identified by their subtype and Genbank Accession number. At each position in the alignment, “.” indicates an identical base to the reference, letters identify bases that differ from it, and “-” indicates a deleted base. The sequences of universal forward primers located 90 and 38 bases upstream of Vpu, and a forward primer matching the first 25 bases of NL43 Vpu (“0”) are indicated above the alignment. Universal primer sequences contain degenerate bases designated by IUPAC ambiguity codes. Locations of the HIV-1 Rev and Vpu start codons are also shown.
Using these primers, HIV-1 NL4.3 vpu sequences containing 0, 38 and 90 upstream bases were PCR amplified from pNL4.3 for cloning into pSel-GFP and pSelRRE-GFP. The codon-optimized vpu sequence was amplified from pcDNA-Vphu and sub-cloned into pSel-GFP only. The “−38” forward and universal reverse primers were also used to amplify one HIV-1 sequence each from subtypes A, B, C and a subtype A/D recombinant for cloning into pSelRRE-GFP. The subtype B sequence was isolated from a participant of the Research in Access to Care Among the Homeless (REACH) Study (Robertson et al., 2004); the subtype A and A/D recombinant were isolated from participants of the Uganda AIDS Rural Treatment Outcomes (UARTO) study (Hunt et al., 2011). Participants provided written informed consent and approval was obtained from the relevant institutional review boards. Briefly, total nucleic acids were extracted from blood plasma and subjected to nested RT-PCR using HIV-1 subtype-optimized primers in the first round and the above primers in the second round. The subtype C vpu sequence was PCR amplified from the infectious molecular clone MJ4 (Ndung'u et al., 2000) using the above primers.
2.3 Cloning and Vpu sequence verification
Vpu amplicons were verified by gel electrophoresis, gel extracted (GeneJET; ThermoFisher Scientific), digested with AscI and SacII (New England Biolabs) and ligated at a 3:1 molar ratio with linearized pSel-GFP and pSelRRE-GFP vectors. Ligations were transformed into E. cloni competent cells (Lucigen), plated on zeocin-containing LB agar and grown overnight at 37°C. Single colonies were grown in liquid LB containing zeocin for ~16 hours at 37°C, plasmid DNA was purified using the E.Z.N.A. Plasmid Mini Kit (Omega Bio-tek) and the presence of inserts was verified by AscI/SacII digestion. Plasmids were sequenced on an ABI 3130xl Genetic Analyzer using the BigDye Terminator3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed using Sequencher 5.0.1 (Genecodes). DNA from sequence-validated colonies was maxiprepped (Qiagen). Sequence alignments were performed using MUSCLE (Edgar, 2004); maximum-likelihood phylogenies were inferred using phyML (Guindon et al., 2009) and trees were visualized using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
2.4 Assessment of Vpu-mediated CD4 and Tetherin downregulation
Assessment of Vpu-mediated CD4 and tetherin downregulation in CEM T-cells were performed using a range of DNA and cell concentrations; for conciseness we describe one protocol then briefly discuss modifications. A total of 5 μg Vpu DNA (in pSel-GFP or pSelRRE-GFP) with or without 7 μg of pSel-Rev-ΔGFP were transfected into 2.5 million CEM T-cells in a final volume of 150 μl using the GenePulser MXCell™ (Bio-Rad) in 96-well plate format (square wave protocol, 250 V, 2000 μF, infinite Ω, 25 millisecond single pulse). Transfected cells were resuspended in 350μl of R10+ media (RPMI-1640 supplemented with 2mM L-glutamine, 1000 U/ml Penicillin and 1 mg/ml Streptomycin, all from Sigma-Aldrich, Co.) plus 10% fetal bovine serum (Life Technologies) and incubated for ~20 hours at 37°C at 5% CO2. Following this, 500,000 cells were stained with Allophycocyanin (APC)-labeled anti-human CD4 and Phycoerythrin (PE)-labeled anti-human CD317 (i.e. tetherin) antibodies (BD Biosciences) for 30 minutes at 4°C. Cell surface CD4 and tetherin expression, along with intracellular GFP were measured by flow cytometry (EasyCyte 8HT Flow Cytometer; Millipore) and analyzed using the software FlowJo. We further optimized this protocol to require co-transfection of only 1.5 μg of Vpu (in pSelRRE-GFP) and 2 μg pSel-Rev-ΔGFP into 500,000 CEM cells, with the same electroporation, resuspension and flow cytometric conditions as above. The ability of each Vpu clone to downregulate CD4 and Tetherin on CEM cells was expressed as the Median Fluorescence Intensity (MFI) of receptor expression in GFPhigh versus GFPneg gates. This value was then normalized to that obtained for the first experimental replicate of a designated control Vpu such that values of <100, 100 and >100% indicate inferior, equal and superior receptor downregulation function compared to the control, respectively.
2.5 Western Blot
A total of 5 μg Vpu-containing plasmid DNA (pSel-GFP or pSelRRE-GFP) with or without 7 μg of pSel-Rev-ΔGFP was transfected into 2.5 million CEM T-cells in a total volume of 150 μL and recovered for 20 hours at 37°C with 5% CO2. Cells were pelleted then lysed with 150 μL Radioimmunoprecipitation assay (RIPA) buffer containing 10% protease inhibitor cocktail P8340 (both from Sigma-Aldrich) on ice for 1 hour, vortexing every 15 minutes. A total of 50 μL of lysate was mixed with 50 μl 2X Laemmli buffer containing 5% beta-mercaptoethanol, incubated at 99°C for 5 minutes and subjected to SDS-PAGE using Mini-Protean TGX 4%-20% polyacrylamide gels (Bio-Rad); proteins were then transferred onto a Polyvinylidene Diflouride (PVDF) membrane. The membrane was blocked in Tris buffered saline solution containing Tween (TBST) containing 5% bovine serum albumin (BSA) for 30 minutes, stained with the primary antibody (rabbit anti-VPU, Fab-Gennix VPU-101AP; 1:500 dilution) in 5% BSA-TBST overnight at 4°C, washed 3 times with TBST and stained for 30 minutes with secondary donkey anti-rabbit HRP (GE Healthcare) (1:30,000) in 5% BSA-TBST at room temperature. Proteins were detected using Clarity western ECL substrate (Bio-Rad) and visualized on an ImageQuant LAS 4000 luminescent imager (GE healthcare).
3. Results
We sought to develop a universal method for amplification of diverse natural HIV-1 group M vpu sequences, and functional assessment of their encoded Vpu proteins, in a CD4+ T-cell line. Substantial genetic variation (in terms of insertions/deletions and single nucleotide polymorphisms) immediately upstream of vpu made it impossible to design a universal forward primer here; however, we identified relatively conserved regions at 38 and 90 bases upstream of vpu that were amenable to primer design (Figure 1). The latter location represents the most distal upstream position to locate a forward primer due to the presence of the rev start codon 2 bases further upstream. A single universal reverse primer was located in the first relatively conserved downstream position (3' end of primer is 17 bases downstream of vpu). The primers incorporated 3-5 degenerate bases to enhance coverage of HIV-1 group M sequence diversity and restriction sites were introduced at their 5' ends to allow directional cloning into pSel-GFP and its derivatives. Analysis of 2843 HIV-1 full genomes (www.hiv.lanl.gov; accessed Sep 14, 2016) indicated that >78% of HIV-1 isolates contained one or fewer base mismatches to these primers (each region analyzed separately).
Both universal forward primers and the single reverse primer were used to amplify vpu from the HIV-1 subtype B reference strain NL4.3 for cloning into pSel-GFP, yielding pSel-NL4.3-38Vpu and pSel-NL4.3-90Vpu. NL4.3 vpu was also cloned directly at its start codon, yielding pSel-NL4.3Vpu. An NL4.3 vpu sequence that was codon optimized for expression in mammalian cells (pSel-COVpu) and empty vector (pSel-GFP) served as positive and negative controls respectively.
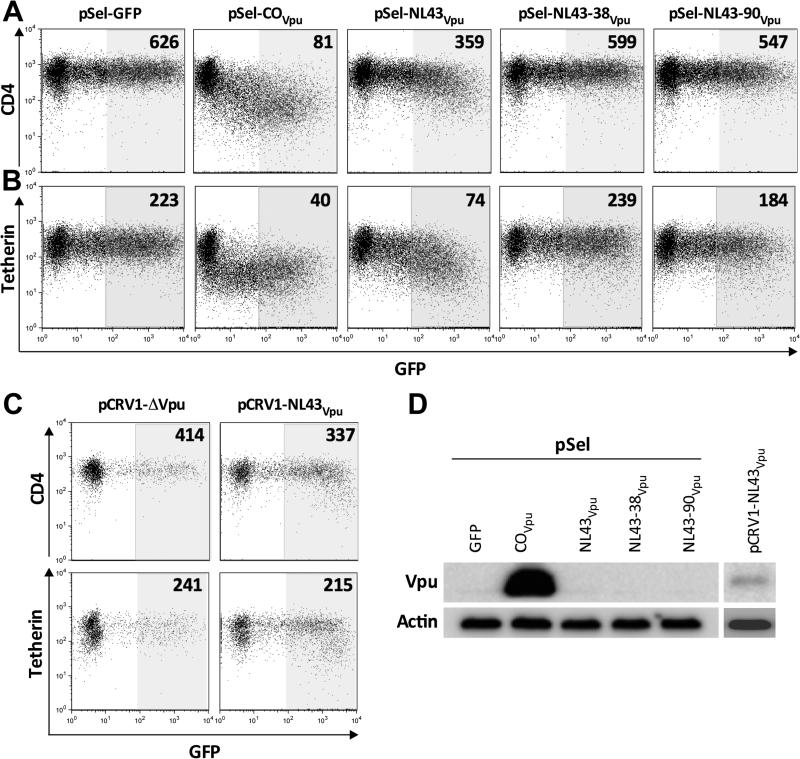
As expected, transfection of up to 5 μg of plasmid DNA into 2.5 million CEM CD4+ T-cells consistently yielded robust Vpu-mediated CD4 downregulation by pSel-COVpu (e.g. nearly 8-fold reduction in CD4 median fluorescence intensity [MFI] between GFPhigh and GFPneg gates in the representative experiment shown in Figure 2A), and no receptor downregulation by empty vector. In line with studies that have assessed Vpu function by cloning the gene directly at the start codon (Chen et al., 2015; Galaski et al., 2016; Mwimanzi et al., 2016), pSel-NL4.3Vpu displayed modest (e.g. less than two-fold) CD4 downregulation function (Figure 2A). In contrast, vpu sequences cloned with 38 or 90 upstream bases consistently displayed no or limited CD4 downregulation ability, respectively (Figure 2A). These observations remained consistent when Vpu-mediated tetherin downregulation was assessed (Figure 2B). Moreover, while codon-optimized Vpu was detected by western blot, Vpu was not detected for any of the natural (unmodified) NL4.3 constructs (Figure 2D). These results indicate that natural Vpu was poorly expressed and that the presence of upstream bases further inhibited expression.
Figure 2. The presence of upstream bases severely impairs autonomous Vpu function/expression; cytotoxicity of pCRV1 in our T-cell line.
Panel A: Representative flow cytometry plots of Vpu-mediated CD4 downregulation by empty pSel-GFP and Vpu sequences cloned into this plasmid (COVpu = codon optimized Vpu; NL43Vpu, NL43-38Vpu and NL43-90Vpu = NL43 Vpu cloned with 0, 38 and 90 upstream bases, respectively). Numbers represent the median fluorescence intensity [MFI] of CD4 staining in the shaded gate. Experiments were performed by transfecting 5 μg plasmid DNA into 2.5 million CEM T-cells. Panel B: same as A, except for Tetherin downregulation. Panel C: Representative flow cytometry plots of Vpu-mediated CD4 (top) and tetherin (bottom) downregulation by empty pCRV1ΔVpu (left) and pCRV1-NL43Vpu, each cotransfected with 1μg of a GFP-expressing plasmid. Numbers represent the median fluorescence intensity [MFI] of CD4 staining in the shaded gate. Panel D: Detection of Vpu levels from these constructs by western blot, with actin as the housekeeping control.
The pCRV1ΔVpu plasmid is a Vpu expression system that is amenable to universal primer design (Apps et al., 2016; Jouvenet et al., 2009; Pickering et al., 2014). This plasmid features a partial HIV-1 5' LTR that preserves the universal 5' donor splice site, followed by a multiple cloning site, intact HIV-1 tat and rev coding regions and intervening RRE (but where Vpu and other viral proteins have been inactivated, truncated or deleted). Vpu sequences are cloned into this plasmid using a forward primer in the same conserved region as our “−90” primer, in order to capture cis-acting elements required for plasmid-encoded tat and rev expression. However, while this plasmid has been used to express Vpu in epithelial (HeLa) cell lines (Apps et al., 2016; Pickering et al., 2014), it caused substantial cytotoxicity in CEM T-cells, where fewer than 5% viable cells (as assessed by forward and side scatter profiles) would routinely remain following electroporation with as little as 1 μg of plasmid (not shown). Moreover, among remaining live cells, Vpu-mediated CD4 and tetherin downregulation was consistently weak, even when up to 6 μg plasmid was delivered (e.g. 20% and 10% reductions in receptor MFI respectively; Figure 2C). Though Vpu was weakly detectable by western blot when expressed in pCRV1ΔVpu (Figure 2D), high toxicity precluded further investigation of this plasmid in our T cell line.
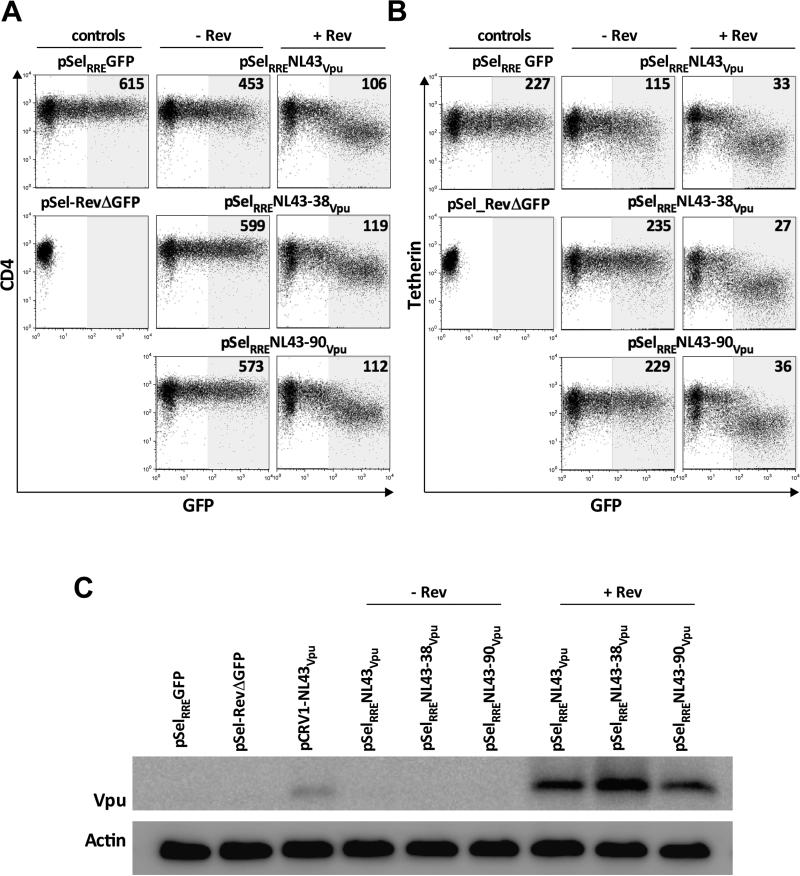
Robust Vpu expression of from natural sequences has been achieved using Rev/RRE-dependent systems (Jafari et al., 2014), but to our knowledge no studies have evaluated whether expression is affected by the presence or quantity of upstream bases. We thus inserted the RRE sequence from HIV-1 NL4.3 downstream of the vpu cloning site in pSel-GFP, so that resulting transcript would contain the RRE at its 3' end. We then cloned vpu with 0, 38 and 90 upstream bases into this modified plasmid, yielding pSelRRE-NL4.3Vpu, pSelRRE-NL4.3-38Vpu and pSelRRE-NL4.3-90Vpu. Consistent with previous observations, pSelRRE-NL4.3Vpu displayed modest ability to downregulate CD4 and tetherin when expressed alone in CEM T-cells (~25% and ~50% reductions in receptor MFI, respectively, when 5 μg plasmid was transfected into 2.5 million cells) whereas pSelRRE-NL4.3-38Vpu and pSelRRE-NL4.3-90Vpu displayed limited or no function (Figure 3A, 3B).
Figure 3. Robust Vpu function/expression, regardless of the presence of upstream bases, using a Rev/RRE-dependent system.
Panel A: Representative flow cytometry plots of Vpu-mediated CD4 downregulation by control (left column) and NL43 Vpu sequences cloned into pSelRRE-GFP, cotransfected without (middle column) and with (right column) a plasmid encoding HIV-1 Rev. Controls pSELRRE-GFP and pSel_RevΔGFP are shown separately to demonstrate GFP expression from the former but not the latter. Numbers on flow plots represent the MFI of CD4 staining in shaded gate. Experiments were performed by transfecting 5 μg Vpu with 7 μg Rev DNA into 2.5 million cells. Panel B: same as A, except for tetherin downregulation. Panel C: Detection of Vpu levels from pSELRRE-GFP constructs, with or without Rev, by western blot, with actin as the housekeeping control.
In contrast, when 5 μg of pSelRRE-NL4.3Vpu, pSelRRE-NL4.3-38Vpu or pSelRRE-NL4.3-90Vpu were co-transfected with 7 μg of a Rev expression vector (pSel-RevΔGFP), robust CD4 and tetherin downregulation (~6 and ~7-fold, respectively) was observed for all three constructs (Figures 3A and 3B). Similarly, Vpu protein was readily detectable in the presence of Rev, but not when expressed alone (Figure 3C). Together, these observations indicate that vpu sequences containing up to 90 upstream bases can be robustly expressed using a Rev/RRE-dependent system.
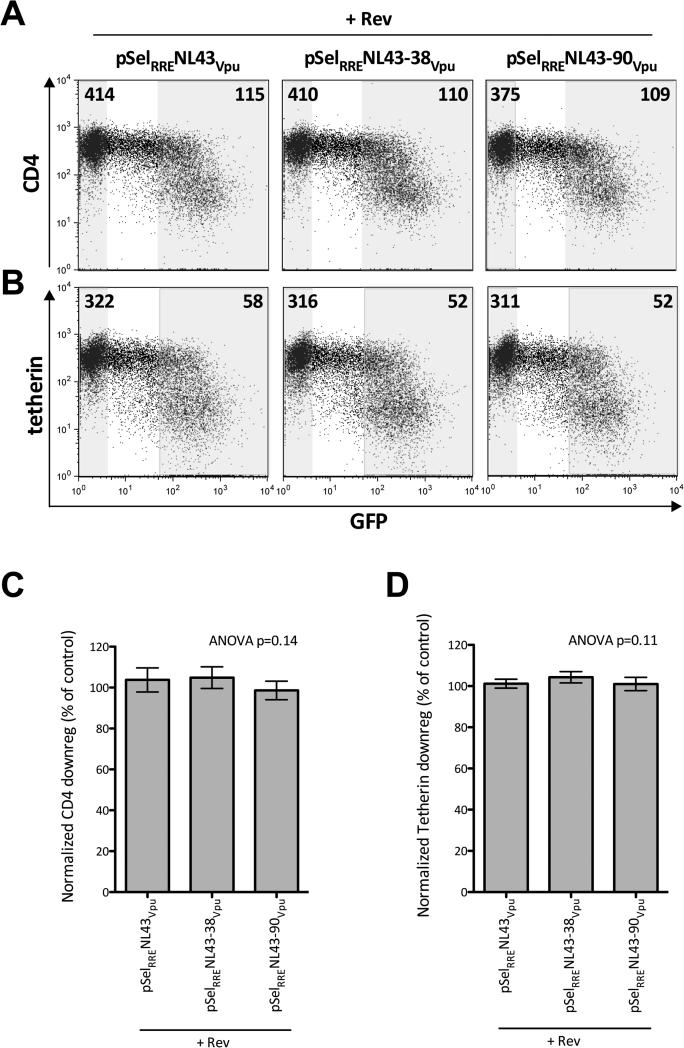
The experiments shown in Figure 3 were undertaken with up to 12 μg total DNA and 2.5 million cells, which is quite resource intensive. We thus optimized the assay to require less DNA and only 500,000 cells. To do this, we tested a range of Vpu_RRE and Rev input plasmid ratios and concentrations up to a maximum of 6 μg total DNA. Broadly, higher Vpu function was observed with increasing amounts of total DNA, but this was associated with higher cytotoxicity (not shown). Balancing these two considerations, we found that 1.5 μg Vpu_RRE and 2 μg Rev yielded consistent Vpu protein expression and function without major toxicity. Using this optimized protocol, we consistently observed robust Vpu-mediated CD4 and tetherin downregulation by all three vpu constructs (e.g. ~4-fold and ~6 fold respectively in the representative experiment shown in Figures 4A and 4B).
Figure 4. Robust and reproducible Vpu function in an optimized assay.
Panel A: Representative flow cytometry plots showing CD4 downregulation when 1.5 μg of pSELRRE-GFP plasmid containing Vpu with 0 (pSelRRENL43Vpu) 38 (pSelRRE-38-NL43Vpu) or 90 (pSelRRE-90-NL43Vpu) upstream bases is cotransfected with 2 μg of pSel_RevΔGFP into 500,000 CEM T-cells. Numbers on flow plots represent the MFI of CD4 staining in shaded GFPneg and GFPhi gates. Panel B: same as A, except for tetherin. Panel C: Normalized CD4 downregulation function of NL43 Vpu constructs containing 0, 38 or 90 upstream bases averaged over four experiments. Histograms and error bars denote means and 95% confidence intervals, respectively. Applying one-way ANOVA yields p=0.14, indicating that the presence and quantity of upstream bases does not significantly alter Vpu function. Panel D: same as C, except for tetherin downregulation.
To further examine the impact of upstream bases on Vpu function, we performed 4 independent experiments each containing a minimum of 2 replicates using the optimized protocol. To combine results across experiments, the downregulation ability of each vpu construct was normalized to that of the first experimental replicate of pSelRRE_NL4.3Vpu such that values of <100, 100% and >100% indicate inferior, equal and superior receptor downregulation function to the control, respectively. Application of one-way ANOVA to the resulting data confirmed that the function of pSelRRE_NL4.3Vpu, pSelRRE_NL4.3-38Vpu or pSelRRE_NL4.3-90Vpu did not differ significantly when co-delivered with Rev (p=0.14 for CD4 downregulation, p=0.11 for tetherin downregulation; Figures 4C, 4D). This indicates that the presence and quantity of upstream bases included in a given vpu sequence does not significantly alter its protein function in this assay.
Finally, we applied our optimized assay to assess the receptor downregulation capacities of Vpu proteins encoded by four natural vpu sequences, representing subtypes A, B and C and a recombinant A/D isolate (Figure 5A). Their functions varied quite widely. In one representative experiment, CD4 downregulation ranged from 42% (i.e. less than two-fold) for the recombinant A/D Vpu to 72% (i.e. more than three-fold) for the subtype C Vpu (Figure 5B), while tetherin downregulation ranged from 62% for the subtype B Vpu to 78% for the subtype C Vpu (Figure 5C). Importantly, their relative functional hierarchy was reproducible across three independent experiments, each featuring a minimum of two replicates. For these experiments, we normalized all results to the first experimental replicate of the subtype B isolate. One-way ANOVA confirmed that the observed functional differences between Vpu proteins were statistically significant (p<0.0001 for both CD4 and tetherin downregulation; Figures 5D, 5E). Specifically, for CD4 downregulation, the subtype B and recombinant A/D Vpus displayed lower function while the subtype A and C Vpus displayed higher function. For tetherin downregulation, the functional hierarchy from lowest to highest was subtype A = B < A/D recombinant < C. These data indicate that our assay is capable of discerning functional differences between Vpu proteins encoded by diverse natural sequences.
Figure 5. Optimized assay functionally differentiates Vpu proteins from diverse HIV-1 subtypes.
Panel A: Unrooted maximum-likelihood phylogeny inferred from an alignment of our subtype A, B, C and A/D recombinant Vpu sequences (denoted “A_isolate”, “B_isolate”, “C_isolate” and “AD.Recomb_isolate”) with published HIV-1 subtype A, B C and D reference sequences (identified by the subtype followed by the Genbank Accession number). Panel B: Representative flow cytometry plots showing CD4 downregulation when 1.5 μg of pSELRRE-GFP plasmid containing the subtype-specific Vpu sequence cloned with 38 upstream bases is cotransfected with 2 μg of pSel_RevΔGFP into 500,000 cells. Numbers represent the MFI of CD4 staining in shaded GFPneg and GFPhi gates. Panel C: same as A, except for tetherin downregulation. Panel D: Normalized CD4 downregulation function of each Vpu clone averaged over three independent experiments. Histograms and error bars denote means and 95% confidence intervals respectively. One-way ANOVA p-value was <0.0001, indicating significant differences in Vpu function between isolates. Tukey's test was used to compare all pairs of isolates; asterisks indicate significance levels (** = p<0.01; *** = p<0.001). Panel E: Same as D, except for tetherin downregulation.
4. Discussion
While inclusion of upstream sequence information severely affected autonomous in vitro Vpu expression, we show that vpu sequences containing up to 90 upstream bases can be robustly expressed using a Rev/RRE-dependent system with no significant alteration to Vpu's CD4 or tetherin downregulation functions. Our method, which features a choice of two universal (pan-HIV-1 group M) forward primers, thus represents a more rapid and scalable approach since it alleviates the need to design isolate-specific primers within the genetically diverse region immediately upstream of vpu. As such, our method should facilitate larger-scale genotype/phenotype assessments of diverse vpu sequences.
Application of our assay to a small yet diverse panel of natural vpu sequences revealed functional differences between isolates, supporting the notion that the dynamic range of Vpu function may be substantial. It is intriguing that the subtype C Vpu (MJ4 strain) exhibited the highest CD4 and tetherin downregulation function overall. Whether this observation is specific to this isolate or whether it is a general feature of the subtype will require assessment of large numbers of natural vpu sequences, which is now feasible using this assay.
To our knowledge, the only other in vitro Vpu expression system amenable to universal priming is the Rev/RRE-dependent plasmid pCRV1ΔVpu (Apps et al., 2016; Jouvenet et al., 2009; Pickering et al., 2014). In this system, the vpu sequence of interest is cloned with 90 upstream bases, and the plasmid produces a long mRNA transcript (encoding vpu, followed by tat and rev exon 1, the RRE, and tat and rev exon 2) which is subsequently spliced to generate these three viral proteins. Due to high cytotoxicity however, we were not able to employ this expression system in our CEM T-cell line. Another potential limitation of pCRV1ΔVpu for the assessment of diverse HIV-1 sequences is that, since cis-acting elements in the upstream bases regulates splicing of the resulting mRNA transcript, it is possible that cloning non-subtype B sequences into this plasmid could introduce genetic incompatibilities between these elements and the subtype B plasmid backbone. Granted, this concern is theoretical, and pCRV1ΔVpu has been used successfully to evaluate subtype C vpu sequences (Apps et al., 2016). Nevertheless, the protocol presented here should not yield subtype incompatibilities despite employing HIV-1 subtype B Rev and RRE, because the sole function of these elements is to facilitate nuclear export of the mRNA transcript. Indeed, our system could theoretically be used to produce other difficult-to-express proteins in CEM T-cells.
A limitation of the present study is that only Vpu's two best characterized functions, CD4 and tetherin downregulation, were assessed; further work would be required to adapt the protocol to assess other Vpu functions. Primer coverage of HIV-1 group M diversity was estimated in silico only; nevertheless our observations suggest that these primers should amplify a diverse range of HIV-1 isolates, and that the resulting Vpu constructs be comparable in function. Our observations also suggest that, if necessary, minor optimizations to primers (in terms of position or degenerate base composition) should not significantly affect Vpu function. It also worth noting that, in contrast to the Rev/RRE-dependent assay developed by Jafari et al (Jafari et al., 2014) where GFP is expressed from the Rev expression vector, GFP in our system is expressed from the Vpu expression vector. Though this should not affect assay performance, it does allow the user to gate on Vpu-expressing cells.
5. Conclusion
Our method represents a universal and thus scalable option for in vitro Vpu expression from genetically diverse HIV-1 group M sequences - whether these be individual unique variants isolated from within a single infected host, or variants isolated from different hosts - in a CD4+ T-cell line. This system should enhance our ability to measure the breadth and range of Vpu-mediated functions in natural isolates both within and between hosts and to assess the potential impact of vpu diversity on HIV-1 pathogenesis.
Highlights.
- Robust, scalable method for in vitro expression of natural HIV-1 vpu sequences
- Utilizes universal primers covering the majority of global HIV-1 group M diversity
- Allows more rapid assessment of Vpu-mediated CD4 and tetherin downregulation
Acknowledgements
We thank Kyle Cobarrubias and Jason Wang for technical assistance, and Denis Chopera, Ravesh Singh and Thumbi Ndung'u for helpful discussions.
Funding statement
This work was supported by an operating grant (HOP-115700) and a project grant (PJT - 148621) from the Canadian Institutes for Health Research to ZLB/MAB, and the Canada-Sub Saharan Africa (CANSSA) HIV/AIDS Network, through funding provided by the Global Health Research Initiative, itself a collaborative partnership of IDRC, CIHR, and Global Affairs Canada to MAB/ZLB. The UARTO cohort is supported by grants from the National Institutes of Health P30 AI027763, R01 MH054907 and UM1 CA181255 to DRB and JNM. MAB is supported by a Canada Research Chair in Viral Pathogenesis and Immunity. ZLB holds a Michael Smith Foundation for Health Research (MSFHR) Scholar award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Los Alamos HIV database. http://www.hiv.lanl.gov/
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson DS, Dunning KR. Codon-optimized reading frames facilitate high-level expression of the HIV-1 minor proteins. Mol Biotechnol. 2005;31:85–8. doi: 10.1385/MB:31:1:085. [DOI] [PubMed] [Google Scholar]
- Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA, Neil S, Pickering S, Schneider DK, Piechocka-Trocha A, Walker BD, Thomas R, Shaw GM, Hahn BH, Keele BF, Lifson JD, Carrington M. HIV-1 Vpu Mediates HLA-C Downregulation. Cell Host Microbe. 2016;19:686–95. doi: 10.1016/j.chom.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aralaguppe SG, Winner D, Singh K, Sarafianos SG, Quinones-Mateu ME, Sonnerborg A, Neogi U. Increased replication capacity following evolution of PYxE insertion in Gag-p6 is associated with enhanced virulence in HIV-1 subtype C from East Africa. J Med Virol. 2016 doi: 10.1002/jmv.24610. [DOI] [PubMed] [Google Scholar]
- Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tibroni N, Sauter D, Galaski J, Miura T, Alter G, Mueller B, Haller C, Walker BD, Kirchhoff F, Brumme ZL, Ueno T, Fackler OT. Modest attenuation of HIV-1 Vpu alleles derived from elite controller plasma. PLoS One. 2015;10:e0120434. doi: 10.1371/journal.pone.0120434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Bai Y, Gustin JK, Moses AV. A comparative mutational analysis of HIV-1 Vpu subtypes B and C for the identification of determinants required to counteract BST-2/Tetherin and enhance viral egress. Virology. 2013;441:182–96. doi: 10.1016/j.virol.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Fruh K. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Binette J, Mercier J, Chiasson A, Cohen EA. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 2010;6:e1000856. doi: 10.1371/journal.ppat.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe. 2012;12:633–44. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaski J, Ahmad F, Tibroni N, Pujol FM, Muller B, Schmidt RE, Fackler OT. Cell Surface Downregulation of NK Cell Ligands by Patient-Derived HIV-1 Vpu and Nef Alleles. J Acquir Immune Defic Syndr. 2016;72:1–10. doi: 10.1097/QAI.0000000000000917. [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating Maximum Likelihood Phylogenies with PhyML. Methods Mol Biol. 2009;537:113–37. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182–92. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, Kembabazi A, Neilands TB, Bangsberg DR, Deeks SG, Martin JN. Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS. 2011;25:2123–31. doi: 10.1097/QAD.0b013e32834c4ac1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabu Y, Kinomoto M, Tatsumi M, Fujita H, Shimura M, Tanaka Y, Ishizaka Y, Nolan D, Mallal S, Sata T, Tokunaga K. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J Biol Chem. 2010;285:35350–8. doi: 10.1074/jbc.M110.173286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari M, Guatelli J, Lewinski MK. Activities of transmitted/founder and chronic clade B HIV-1 Vpu and a C-terminal polymorphism specifically affecting virion release. J Virol. 2014;88:5062–78. doi: 10.1128/JVI.03472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–44. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle E, Campbell M, Lingappa J, Donnell D, Celum C, Ondondo R, Mujugira A, Fife K, Mugo N, Kapiga S, Mullins JI, Baeten JM, Partners in Prevention HTST. HIV-1 subtype C is not associated with higher risk of heterosexual HIV-1 transmission: a multinational study among HIV-1 serodiscordant couples. AIDS. 2014;28:235–43. doi: 10.1097/QAD.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings FA, Burda ST, Urbanski MM, Zhong P, Nadas A, Nyambi PN. Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes A and G. J Med Virol. 2006;78:523–34. doi: 10.1002/jmv.20572. [DOI] [PubMed] [Google Scholar]
- Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu inhibitable manner. Curr Biol. 1999;9:622–31. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- Levesque K, Zhao YS, Cohen EA. Vpu exerts a positive effect on HIV-1 infectivity by down-modulating CD4 receptor molecules at the surface of HIV-1-producing cells. J Biol Chem. 2003;278:28346–53. doi: 10.1074/jbc.M300327200. [DOI] [PubMed] [Google Scholar]
- Mann JK, Byakwaga H, Kuang XT, Le AQ, Brumme CJ, Mwimanzi P, Omarjee S, Martin E, Lee GQ, Baraki B, Danroth R, McCloskey R, Muzoora C, Bangsberg DR, Hunt PW, Goulder PJ, Walker BD, Harrigan PR, Martin JN, Ndung UT, Brockman MA, Brumme ZL. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology. 2013;10:100. doi: 10.1186/1742-4690-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan AJ, Moore DM, Lobritz MA, Fraundorf E, Abraha A, Reeves JD, Arts EJ. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J Virol. 2005;79:7121–34. doi: 10.1128/JVI.79.11.7121-7134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol. 2012;86:4496–504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll M, Andersson SK, Smed-Sorensen A, Sandberg JK. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood. 2010;116:1876–84. doi: 10.1182/blood-2009-09-243667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Tietjen I, Miller SC, Shahid A, Cobarrubias K, Kinloch NN, Baraki B, Richard J, Finzi A, Fedida D, Brumme ZL, Brockman MA. Novel acylguanidine-based inhibitor of HIV-1. J Virol. 2016;90:9495–508. doi: 10.1128/JVI.01107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara PL, Fischinger PJ. Quantitative infectivity assay for HIV-1 and-2. Nature. 1988;332:469–70. doi: 10.1038/332469a0. [DOI] [PubMed] [Google Scholar]
- Ndung'u T, Renjifo B, Novitsky VA, McLane MF, Gaolekwe S, Essex M. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology. 2000;278:390–9. doi: 10.1006/viro.2000.0583. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nguyen KL, llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, Bour S. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology. 2004;319:163–75. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Pham TN, Lukhele S, Hajjar F, Routy JP, Cohen EA. HIV Nef and Vpu protect HIV-infected CD4+ T cells from antibody-mediated cell lysis through down-modulation of CD4 and BST2. Retrovirology. 2014;11:15. doi: 10.1186/1742-4690-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering S, Hue S, Kim EY, Reddy S, Wolinsky SM, Neil SJ. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog. 2014;10:e1003895. doi: 10.1371/journal.ppat.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MJ, Clark RA, Charlebois ED, Tulsky J, Long HL, Bangsberg DR, Moss AR. HIV seroprevalence among homeless and marginally housed adults in San Francisco. Am J Public Health. 2004;94:1207–17. doi: 10.2105/ajph.94.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–21. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8:397–409. doi: 10.1016/j.chom.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS UNAIDS fact sheet 2016. 2016.
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–52. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. Interaction with Cellular CD4 Exposes HIV-1 Envelope Epitopes Targeted by Antibody-Dependent Cell-Mediated Cytotoxicity. J Virol. 2014;88:2633–44. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Ronsard L, Kapoor R, Banerjea AC. Genetic characterization of natural variants of Vpu from HIV-1 infected individuals from Northern India and their impact on virus release and cell death. PLoS One. 2013;8:e59283. doi: 10.1371/journal.pone.0059283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Armitage AE, Graham SC, de Oliveira T, Skinhoj P, Jones EY, Stuart DI, McMichael AJ, Chesebro B, Iversen AK. Functional characteristics of HIV-1 subtype C compatible with increased heterosexual transmissibility. AIDS. 2009;23:1047–57. doi: 10.1097/QAD.0b013e32832a1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]