Abstract
Cross talk between light signaling and cold signaling has been elucidated in the model plant Arabidopsis and tomato, but little is known about their relationship in rice. Here, we report that phytochrome B (phyB) mutants exhibit improved cold tolerance compared with wild type (WT) rice (Oryza sativa L. cv. Nipponbare). The phyB mutants had a lower electrolyte leakage index and malondialdehyde concentration than the WT, suggesting that they had greater cell membrane integrity and less lipid peroxidation. Real-time PCR analysis revealed that the expression levels of dehydration-responsive element binding protein 1 (OsDREB1) family genes, which functions in the cold stress response in rice, were increased in the phyB mutant under normal and cold stress conditions. PIFs are central players in phytochrome-mediated light signaling networks. To explore the relationship between rice PIFs and OsDREB1 gene expression, we produced overexpression lines of rice PIF genes. OsDREB1 family genes were up-regulated in OsPIL16-overexpression lines, which had improved cold tolerance relative to the WT. Chromatin immunoprecipitation (ChIP)-qPCR assay revealed that OsPIL16 can bind to the N-box region of OsDREB1B promoter. Expression pattern analyses revealed that OsPIL16 transcripts were induced by cold stress and was significantly higher in the phyB mutant than in the WT. Moreover, yeast two-hybrid assay showed that OsPIL16 can bind to rice PHYB. Based on these results, we propose that phyB deficiency positively regulates OsDREB1 expression through OsPIL16 to enhance cell membrane integrity and to reduce the malondialdehyde concentration, resulting in the improved cold tolerance of the phyB mutants.
Keywords: cold stress, rice, OsDREB1, phytochrome B, phytochrome interacting factor-like protein
Introduction
Light is a major environmental signal influencing a multitude of steps in plant development such as seed germination, carbon assimilation, stem elongation, leaf morphology, and flowering (Carvalho et al., 2011). Plants use phytochromes to sense red light-depleted (shade) and red light-enriched (full sun) conditions (Rockwell and Lagarias, 2006). Phytochromes are light-absorbing photoreceptors that exist in two fundamental forms: the red (R)-light-absorbing form, designated Pr, and the FR-light-absorbing form, designated Pfr. The inactive Pr form is converted to the active Pfr form by exposure to R-light, and is converted back to the inactive Pr form by exposure to FR-light or through dark reversion. Phytochromes interact with members of the basic bHLH family of PIFs to regulate the expression of a large number of light-responsive genes and thus influence many photomorphogenic events (Franklin and Quail, 2010; Nagatani, 2010; Gu et al., 2011). There are five phytochrome genes (PHYA to PHYE) and eight PIF genes (PIF1, PIF3, PIF4, PIF5, PIF6, PIF7, PIF8, and PIL1) in Arabidopsis thaliana, and three phytochrome genes (PHYA to PHYC) and six PIL genes (PIL11 to PIL16) in Oryza sativa (Nakamura et al., 2007; Leivar and Quail, 2011; Jeong and Choi, 2013; Luo et al., 2014).
In addition to their roles in plant photomorphogenesis, cross-talk between phytochrome-mediated light signals and cold signaling pathways has been identified in the model plant Arabidopsis and tomato (Williams et al., 1972; Kim et al., 2002; Wang et al., 2016). As early as 1972, Williams et al. noticed that phytochrome could mediate short day enhancement of cold acclimation, but the molecular basis for phytochrome regulation of cold tolerance remained unknown until recent years (Williams et al., 1972). CBF/DREB1 genes, which are rapidly and transiently induced by low temperatures, play an important role in cold stress responses (Gilmour et al., 2004; Heidarvand and Amiri, 2010; Mao and Chen, 2012). CBF/DREB1s can up-regulate COR genes that contain a C-repeat/drought-responsive promoter element, and thus cause metabolic changes to enhance cold tolerance (Thomashow, 2001). A low R/FR light signal increases CBF gene expression in Arabidopsis in a circadian clock-dependent manner. The transcript abundance of COR15a in a phytochrome-deficient mutant analysis revealed that phyB and phyD repress the CBF regulon in high R/FR light in a non-redundant manner, and the phyD mutant showed enhanced cold tolerance (Franklin and Whitelam, 2007). Kidokoro et al. (2009) found that PIF7 specifically binds to the G-box (CACGTG) of the DREB1C and DREB1B promoters in Arabidopsis. Transactivation analysis using mesophyll protoplasts revealed that PIF7 functions as a transcriptional repressor of DREB1B and DREB1C expression under circadian control, and its activity is regulated by the PIF7-interacting factors TOC1 and phyB (Kidokoro et al., 2009). Genetic analysis indicated that the CBF pathway is repressed by phyB, PIF4, and PIF7 under a warm long-day (LD) growing season, and this repression is relieved by short-day conditions. As a result, the freezing tolerance of short-day plants was increased relative to that of LD plants in Arabidopsis (Lee and Thomashow, 2012). Wang et al. (2016) found that FR and R light perceived by phyA and phyB positively and negatively regulated cold tolerance, respectively, in tomato (Solanum lycopersicum). FR light-induced activation of phyA triggers ABA signaling and, subsequently, JA signaling, leading to activation of the CBF pathway and a cold response in tomato plants (Wang et al., 2016).
Cold stress is an important factor limiting rice yield in many areas of high latitude and altitude. There are 10 putative DREB1 homologs (DREB1A to DREB1I) in rice, six of which (OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G) were similarly expressed in response to chilling acclimation and cold stress. Moreover, these genes were co-expressed with genes involved in cold signaling, suggesting that they might be involved in the cold response in rice (Mao and Chen, 2012). Functional analysis revealed that over-expression of OsDREB1A, OsDREB1B, or OsDREB1F in rice and over-expression of OsDREB1D or OsDREB1F in Arabidopsis conferred enhanced cold tolerance in transgenic plants (Dubouzet et al., 2003; Wang et al., 2008; Zhang et al., 2009). PhyB deficiency alleviated chilling-induced photoinhibition in rice, probably through a more stabilized chloroplast structure and higher unsaturated fatty acid content in membrane lipids in the phyB mutant (Yang et al., 2013). Cordeiro et al. (2016) observed that OsPIF14 could bind to the OsDREB1B promoter through two N-boxes (CACG(A/C)G) and that the flanking regions of the hexameric core were essential for protein-DNA interaction and stability. Transactivation assays using Arabidopsis protoplasts and rice protoplasts showed that OsPIF14 downregulates OsDREB1B gene expression (Cordeiro et al., 2016). Furthermore, yeast two-hybrid and Co- immunoprecipitation analyses revealed that OsPIF14 preferentially binds to the active Pfr form of rice phyB (Cordeiro et al., 2016).
In this work, we observed that phyB mutants exhibited enhanced cold tolerance compared with the WT, suggesting that phyB may be involved in the regulation of tolerance to cold stress in rice. To investigate the mechanism by which phyB regulates cold tolerance, we analyzed the ELI and MDA content in WT and phyB mutant plants. Rice plants deficient in phyB exhibited reduced ELI and MDA values, presumably as a result of increased membrane integrity. We also analyzed the expression of DREB1 genes related to cold responses in the WT and phyB mutant. Genes acting upstream of the OsDREB1 gene family were further dissected.
Materials and Methods
Plant Materials and Stress Treatment
OsPIL16 overexpression, phyB1 mutant, phyB2 mutant and WT (O. sativa L., cv. Nipponbare) rice plants were used in this study. The genetic background of the phyB1 and phyB2 mutants was O. sativa cv. Nipponbare, which was described previously (Takano et al., 2001, 2005). Seeds were surface sterilized in 75% (v/v) ethanol for 30 s and then in 5% NaClO (v/v) for 20 min. The seeds were then rinsed six times in sterile double-distilled water. The sterilized seeds were soaked in water at room temperature for 2 days and then germinated for 1 day at 28°C. Seedlings were grown hydroponically in Yoshida’s culture solution as described previously (Zhou et al., 2013). Plants were cultured in a growth chamber at 28°C/28°C (day/night) under a 15-h-light/9-h-dark photoperiod. For cold stress, three-leaf stage seedlings were placed in a climatic chamber at 4°C. All treatments were repeated three times. For R light treatment, seeds from WT and phyB were de-husked, surface-sterilized, and grown in 0.4% agar. After incubating at 4°C overnight, the seeds were transferred to darkness for 7 days at 28°C, and then transferred to R light for another 24 h. The above-ground parts were harvested for RNA isolation.
Cell Membrane Permeability
The cell membrane permeability of plantlets was assessed by the ELI of their tissues according to Sutinen et al. (1992) with the following modifications. The second leaves were collected from three individual plants at the three-leaf stage grown under normal conditions or treated at 4°C for different time periods. After rinsing them three times in deionized water, the samples were cut into 1 cm long sections and placed in deionized water. After the leaf sections were placed in a vacuum for 10 min and kept at room temperature for 20 min, the electrical conductivity value (named C1) was determined. Then, the samples were boiled for 30 min, and the total conductivity value (named C2) was determined after the solution was cooled to room temperature. The ELI was expressed as ELI (%) = (C1/C2) × 100.
MDA Concentration Analysis
The MDA concentration was measured as described previously with the following modifications (Kim and Tai, 2011). The leaves were weighed and homogenized in 5 ml of 10% trichloroacetic acid solution. The homogenate was centrifuged, and 2 ml of the supernatant was added to 2 ml 0.67% thiobarbituric acid. The mixture was incubated in boiling water for 30 min, and the reaction was stopped in an ice bath. The MDA concentration was expressed as mol g-1 fresh mass (FM).
Quantitative PCR
Surface-sterilized seeds of the WT and phyB1 mutant were incubated under controlled photoperiodic conditions (15 h light, 28°C/9 h dark, 28°C) until the three-leaf stage. Samples from unstressed plants were collected as controls (0 h). Samples were taken throughout the cold stress treatment at 1, 4, 12, and 24 h after exposing the seedlings to cold stress. The collected samples were immediately flash frozen in liquid nitrogen and stored at -80°C for further analysis. Total RNA was isolated from the third leaves using the RNAiso reagent (TaKaRa, Dalian, China). First-strand cDNAs were synthesized from total RNA using a PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time), DNase treatment was applied before cDNAs synthesis to reduce DNA contamination according to the manufacturer’s instructions. Quantitative PCR was performed on the Thermal Cycler DiceTM Real Time System (TaKaRa) using SYBR Premix Ex TaqTM (TaKaRa). Each reaction contained 10 μl of SYBR Premix Ex TaqTM (TaKaRa), 2 μl of cDNA sample, and 0.2 μl 20 μM gene-specific primer pairs in a final volume of 20 μl. The PCR thermal cycle used was as follows: denaturation at 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 30 s. As an internal control, the rice elongation factor gene (OsEF1α, AK061464) was used to quantify the relative transcript level of each target gene. Three biological replicates were performed. The specific primers used in this study are listed in Supplementary Table 1.
Construction of the Plant Expression Vector and Rice Transformation
To construct the OsPIL16-overexpression (OsPIL16-OX) vector, the ORF region of OsPIL16 was amplified by PCR from cDNA using the primers OsPIL16-F and OsPIL16-R, and cloned into the p1390-Ubi vector between the maize ubiquitin promoter and the nos terminator (Li et al., 2012). To construct the OsPIL16-HA overexpression vector, the ORF region of OsPIL16 was amplified by PCR from cDNA using the primers OsPIL16-F and OsPIL16-HA-R, and cloned in frame at its 3′ end with HA tag in the modified p1390-Ubi vector between the maize ubiquitin promoter and the nos terminator. The plasmid was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Rice (O. sativa cv. Nipponbare) was transformed via the agroinfection method as described previously (Hood et al., 1993; Hiei et al., 1994).
DNA Extraction and Southern Blot Analysis
Genomic DNA was isolated and purified from the OsPIL16-OX lines and WT plants at the six-leaf stage following Murray and Thompson (Murray and Thompson, 1980). Approximately 100 μg of DNA was digested with HindIII and subjected to electrophoresis on a 0.8% agarose gel. The DNA was then transferred to a nylon membrane (Hybond-N+; Amersham, Buckinghamshire, UK). PCR primers for amplification of the HptII gene (the selectable marker gene) are listed in Supplementary Table 1. The amplification conditions were one cycle of 3 min at 94°C, 35 cycles of 30 s at 94°C, 1 min at 56°C and 1 min at 72°C, and finally one cycle of 5 min at 72°C. The amplified fragment was purified using a PCR clean-up system. DIG-labeled probe preparation and southern blotting were performed according to the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany) instructions.
Yeast Two-Hybrid Assay
The full coding sequence of OsPIF16 was amplified by PCR from cDNA using the primers OsPIL16-AD-F and OsPIL16-AD-R, and cloned into the vector pGADT7. The C-terminal non-photoactive region of PHYB (1891–3513 bp) was amplified by PCR from cDNA using the primers C-phyB-F and C-phyB-R, and cloned into pGBKT7. The bait and prey were cotransformed into yeast strain AH109 according to the Yeast Protocols Handbook. Then, positive yeast colonies were plated on SD -Leu -Trp -His media containing 20 mM 3-AT. pGADT7 was used as a prey for the negative control.
Cis-Acting Element Analysis
The promoter sequences of OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G were downloaded from the Rice Genome Annotation Project1 and used for cis-acting element analysis.
Western Blot
Seedlings of WT and OsPIL16-HA overexpression lines were ground in liquid nitrogen and homogenized in a denaturing buffer (100 mM NaP2PO4, 10 mM Tris-Cl, and 8 M urea) by vigorous vortexing. Cell debris was removed by centrifugation at 14, 000 g for 10 min at 4°C. For western blot analysis, the supernatants were separated on 7.5% SDS-polyacrylamide gel, and the separated proteins were transferred to Immobilon-P-Transfer Membrane (Millipore, Bedford, USA). For detection of OsPIL16-HA, the membrane was incubated with rabbit polyclonal anti-HA antibody (Abcam, Cambridge, UK) in PBS buffer containing 0.05% Tween-20. Bands were visualized with BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, Nanjing, China), according to the manufacturer’s instructions.
ChIP -qPCR Assay
ChIP assay was performed using an EpiQuik Plant Chip Kit (Epigentek, San Jose, California, USA). 14-day-old seedlings were incubated in the dark for 12 h after light irradiation and then crosslinked in 1% formaldehyde by vacuum filtration in the dark. ChIP analysis was performed by using an affinity purified anti-HA polyclonal antibody (ab9110, Abcam, Cambridge, UK) and normal IgG was used as negative control. Primers OsDREB1B-G-F and OsDREB1B-G-R, OsDREB1B-N-F and OsDREB1B-N-R that anneal to the G-box and N-box motifs in the OsDREB1B promoters, respectively, were used for qPCR. qPCR was performed using SYBR Green reagent (Takara, Dalian, Japan). Results were presented as percent of input.
All primers used in this study are listed in Supplementary Table 1.
Results
phyB Mutants Exhibit Enhanced Cold Tolerance
To test their cold tolerance, phyB mutant and WT rice plants were grown to the three-leaf stage in the same tray under well-watered conditions; the phyB1 and phyB2 mutants grew and developed normally (Figure 1A, Control). After 4°C treatment for 4 days, both the WT and phyB mutant plants showed significant wilting symptoms (Figure 1A, Cold). The cold-stressed seedlings were then removed to normal growth conditions for 7 days. The phyB1 and phyB2 plants recovered almost completely, whereas the WT plants did not (Figure 1A, Recovered); 95 and 94% of the phyB1 and phyB2 plants, respectively, survived and grew new leaves, while 0% of the WT plants grew new leaves 7 days after being removed to normal conditions (Figure 1B). These results unambiguously demonstrated that the phyB-deficient mutants had improved cold tolerance.
FIGURE 1.
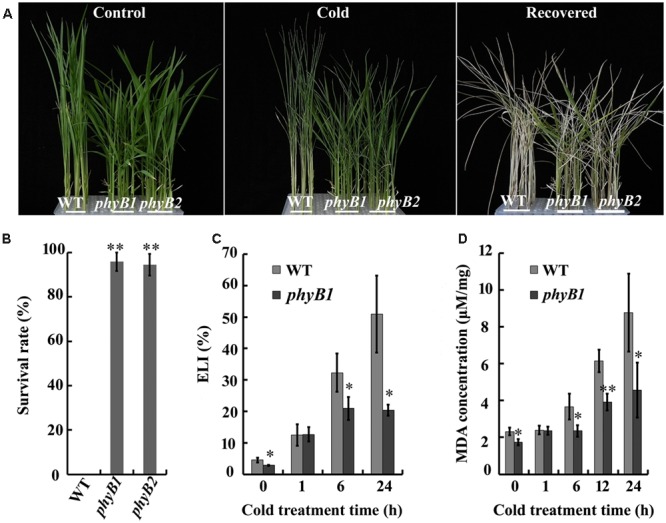
phyB mutants have enhanced cold tolerance. (A) Representative photographs of wild-type (WT) and phyB mutants (phyB1 and phyB2) under cold stress. Seedlings at the three-leaf stage were treated with cold (4°C) for 4 days and then allowed to recover at 28°C for an additional 7 days. (B) Survival rate of seedlings. (C) Electrolyte leakage index. (D) MDA concentration. Asterisk (∗) indicates that the mean value is significantly different from that of the control: ∗P < 0.05; ∗∗P < 0.01.
phyB Deficiency Results in Lower ELI and MDA Concentration
To investigate the mechanism of cold tolerance, we monitored changes in the ELI, an indicator of membrane integrity. We first monitored the changes in the ELI over 24 h of cold stress in the WT and phyB1 mutant. Under normal growth conditions, the ELI values of the WT and phyB1 mutant were 4.5 and 2.8%, respectively (Figure 1C). After cold stress treatment, ELI values increased in both the phyB1 mutant and the WT (Figure 1C). However, the phyB1 mutant exhibited an obviously lower ELI at 6 and 24 h after cold treatment compared with the WT (Figure 1C). We speculated that phyB deficiency probably increased the ability to maintain membrane integrity under cold stress, which is likely to be one of the factors enabling the phyB mutants to cope with cold stress.
Malondialdehyde is the end product of lipid peroxidation that is generated from the oxidation of polyunsaturated fatty acids by ROS and other free radicals in membranes, and acts as an indicator of lipid peroxidation (Liu et al., 2013). We measured the MDA concentration in the WT and the phyB1 mutant after cold stress treatment. The MDA concentration increased to about twofold in the phyB1 mutant and to about fourfold in the WT after 24 h of cold stress (Figure 1D), consistent with the changes in ELI. The lower MDA concentration in the phyB mutant indicated reduced membrane lipid peroxidation relative to the WT, which probably contributed to the enhanced membrane integrity under cold stress.
phyB Altered OsDREB1 Expression Patterns
Dehydration-responsive element binding protein 1 play an important role in cold responses, as indicated by the observation that overexpression of OsDREB1s can enhance cold tolerance in rice (Mao and Chen, 2012). To explore whether OsDREB1s were involved in the enhanced cold tolerance of the phyB mutant, we analyzed the expression patterns of OsDEREB1s in WT and phyB mutant plants grown under normal conditions and cold stress. Six of the nine OsDREB1 family members, including OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G, were rapidly induced by cold stress in both the WT and phyB mutant, but their expression levels were relatively higher in the phyB mutant compared with the WT under normal conditions and cold stress (Figure 2). Although OsDREB1D was induced by cold treatment in the WT but not the phyB mutant, its expression was about twofold higher in the phyB1 mutant relative to the WT under normal conditions (Figure 2). OsDREB1J and OsDREB1I expression was not obviously induced by cold stress, but their transcription levels were clearly higher in the phyB mutant than in the WT. These results demonstrated that phyB deficiency promoted OsDREB1 family gene expression. Considering that overexpression of OsDREB1A, OsDREB1B, and OsDREB1F enhanced the cold tolerance of rice and that overexpression of OsDREB1F or OsDREB1D in Arabidopsis conferred enhanced cold tolerance (Ito et al., 2006; Wang et al., 2008), we speculated that increased expression of OsDREB1 family genes probably contributed to the enhanced cold tolerance of the phyB mutants.
FIGURE 2.
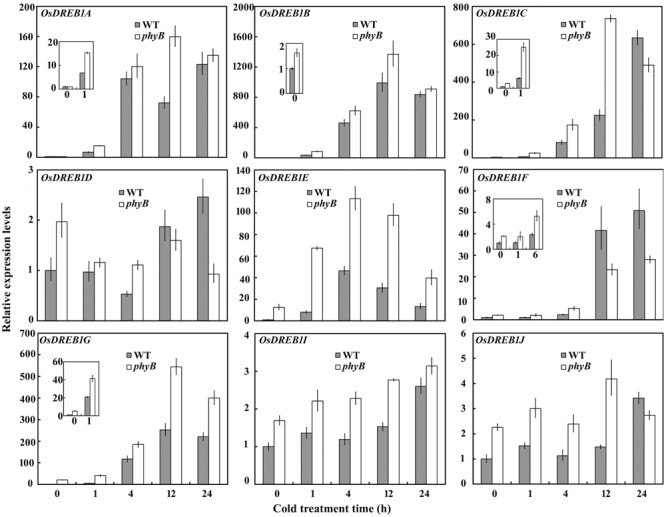
OsDREB1 expression patterns in the WT and phyB mutant. Seedlings at the three-leaf stage were treated with cold (4°C) for 0, 1, 4, 12, or 24 h. Real-time PCR was performed using cDNA derived from leaves. Error bars indicate SE (n = 3).
phyB Altered the Expression Pattern of OsPILs
How does phyB regulate OsDREB1 expression? Because PIFs play a pivotal role in phytochrome-mediated light signaling networks (Castillon et al., 2007), we wondered whether OsPIFs were involved in the regulation of the cold response. Nakamura et al. (2007) identified six PIL homologs in rice, designated OsPIL11 through OsPIL16, by extensively evaluating all of the rice databases (Nakamura et al., 2007). To dissect the role of OsPILs in cold responses, we compared their expression patterns in WT and phyB mutant plants grown under normal conditions and cold stress. OsPIL12 expression levels were not affected by cold stress in the WT or phyB mutant, while OsPIL11, OsPIL13, and OsPIL14 were induced by cold stress in the phyB mutant but not the WT (Figure 3). OsPIL15 was induced by cold stress in both the WT and phyB mutant, whereas cold stress induced OsPIL16 expression only in the WT, not in the phyB mutant (Figure 3). The expression levels of OsPIF15 and OsPIF16 were much higher in the phyB mutant than in the WT under normal conditions and cold stress (Figure 3). In particular, OsPIL16 transcript levels were about 400-fold higher in the phyB mutant than in the WT. To understand how phyB regulates OsPIL16 gene, we compared the transcript levels between WT and phyB mutant seedlings that were grown in the dark, or grown in the dark but irradiated with 24 h of continuous R light before harvest. OsPIL16 transcript level was decreased by R light in WT but not in phyB (Supplementary Figure 1), suggesting that phyB perceive R light to inhibit OsPIL16 gene expression at the transcriptional level. Based on these results, we deduce that the expression of multiple OsPILs was regulated by cold stress, and that phyB was involved in modulating the OsPIL gene expression.
FIGURE 3.
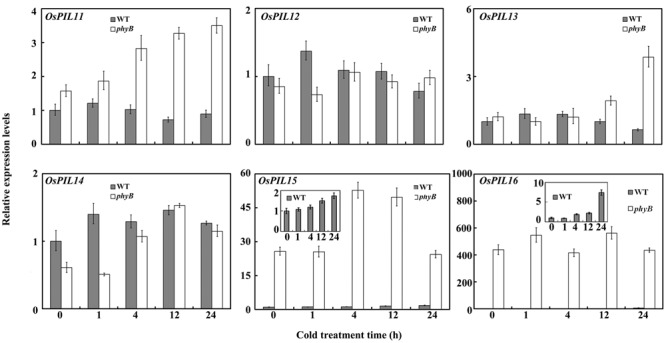
OsPIL expression patterns in the WT and phyB mutant. Seedlings at the three-leaf stage were treated with cold (4°C) for 0, 1, 4, 12, or 24 h. Real-time PCR was performed using cDNA derived from leaves. Error bars indicate SE (n = 3).
OsPIL16-Overexpression Lines Have Enhanced Cold Tolerance and Increased Expression Levels of OsDREB1s
To investigate the role of OsPILs in regulating cold tolerance in rice, we produced the overexpression lines of OsPIL15 and OsPIL16 because of the significant difference in their transcript levels between the WT and phyB (Figure 3). The OsPIL15-overexpression (OsPIL15-OX) lines were reported in our previous study (Zhou et al., 2014). In this study, we produced transgenic rice lines overexpressing OsPIL16 (Figure 4A). Two independent homozygous lines of the T4 progeny (#1 and #13) were used to analyze the role of OsPIL16 in rice based on their high expression levels (Figures 4B,C). Seedlings of the OsPIL16-OX lines and WT at the three-leaf stage were hydroponically grown under cold conditions for 4 days and then allowed to recover for 7 days. The OsPIL16-OX lines exhibited enhanced cold tolerance, as more seedlings survived compared with the WT (Figure 4D). However, OsPIL15-OX lines exhibited the same sensitivity as the WT (data not shown). These results suggested that OsPIL16 was involved in the cold response.
FIGURE 4.
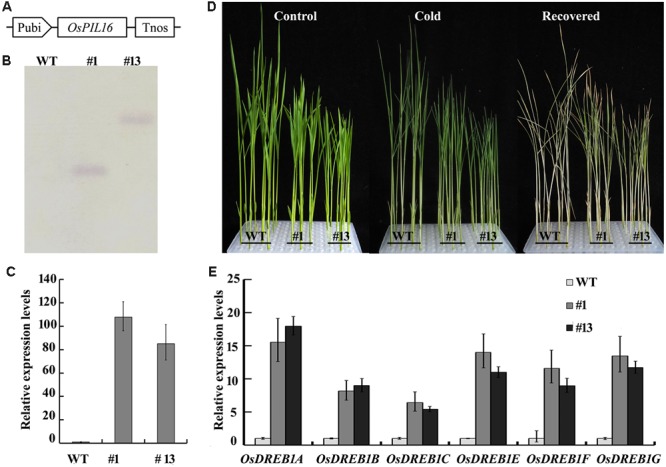
Function of OsPIL16 in regulating cold stress. (A) Schematic diagram of the construct used to obtain OsPIL16-OX lines. The OsPIL16 gene was driven by the maize ubiquitin promoter (Pubi). (B) Southern blot analysis of OsPIL16-OX lines. (C) The expression levels of OsPIL16 in overexpression lines. Real-time PCR was performed using cDNA derived from leaves. Error bars indicate SE (n = 3). (D) Representative photographs of the WT and OsPIL16-OX lines under cold stress. Three-leaf stage seedlings were treated with cold (4°C) for 4 days and then allowed to recover at 28°C for an additional 7 days. (E) OsDREB1 expression levels in the OsPIL16-OX lines and WT. Real-time PCR was performed using cDNA derived from the leaves of three-leaf stage seedlings. Error bars indicate SE (n = 3).
In Arabidopsis, PIF4 and PIF7 down regulate the CBF pathway and freezing tolerance under LD conditions (Lee and Thomashow, 2012). To clarify the relationship between the OsPILs and OsDREB1s, we compared the transcript levels of the OsDREB1s between the WT and OsPIL16-OX or OsPIL15-OX lines. As shown in Figure 4E, the expression levels of OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1E, OsDREB1F, and OsDREB1G were increased in the OsPIL16-OX lines compared with the WT. These results suggested that OsPIL16 enhanced cold tolerance by up regulating OsDREB1 expression. However, the transcript levels of the OsDREB1 genes were similar in the WT and OsPIL15-OX lines (Supplementary Figure 2). The expression patterns of the OsDREB1s were consistent with the cold tolerance of the OsPIL16-OX and OsPIL15-OX lines.
A previous study revealed that PIFs can bind to the G-box, PBE (CACATG) or N-box (CACGCG or CACGAG) (Cordeiro et al., 2016; Kim et al., 2016). Promoter analysis of OsDREB1A, OsDREB1B, OsDREB1C, OsDREB1D, OsDREB1E, OsDREB1F, and OsDREB1G revealed that they all contained a G-box, PBE, or N-box (Table 1). This result suggested that OsPIL16 probably up-regulates the expression of OsDREB1 genes by directly binding to their promoter regions to enhance cold tolerance. To test this hypothesis, we produced the OsPIL16-HA overexpression (OsPIL16-HA OX) lines and detected OsPIL16-HA protein using western blotting (Supplementary Figure 3). ChIP-qPCR assays using primers that anneal to the G-box and N-box motifs in the OsDREB1B promoters were performed. The results show that N-box regions were enriched in the chromatin fractions by anti-HA antibody compared to that by control IgG (Figure 5).
Table 1.
Promoter analysis of OsDREB1s.
| Gene | Cis-acting element | Position | Sequence |
|---|---|---|---|
| OsDREB1A | G-box | –563 to –558 | catgctccCACGTGccatagat |
| N-box | –1551 to –1546 | tcgcggagCACGCGgtgttgtg | |
| OsDREB1B | G-box | –2896 to –2891 | tatcaatCACGTGgcaatttct |
| N-box | –1526 to –1521 | ggggagatCACGAGatgaatct | |
| N-box | –180 to –175 | tgagctgcCACGCGggcccacc | |
| OsDREB1C | G-box | –2095 to –2090 | tggatggtCACGTGgcaggaaa |
| G-box | –1439 to –1434 | tactccctCACGTGcggctgga | |
| G-box | –473 to –303 | gccatctcCACGTGgccacccc | |
| G-box | –181 to –176 | tctcccgcCACGTGcgcgccgc | |
| OsDREB1E | PBE | –674 to –668 | ggagtgtgCACATGaagctcgt |
| N-box | –552 to –547 | caacgaatCACGCGctctccaa | |
| N-box | –534 to –529 | ccaactcaCACGCGtccgcatc | |
| N-box | –397 to –392 | cagagaagCACGAGcccaagcc | |
| OsDREB1F | G-box | –291 to –296 | cacgggcgCACGTGtttcatcc |
| G-box | –260 to –254 | tgaatcccCACGTGacgatcga | |
| N-box | –463 to –457 | ttattttaCACGAGaaatttaa | |
| OsDREB1G | PBE | –2769 to –2764 | caaaaataCACATGtagaggtg |
| N-box | –382 to –373 | cgccgtccCACGAGacaacgag | |
| N-box | –91 to –86 | tccctctcCACGCGctaaacta |
The N-box, PBE (PIF binding E-box), and G-box are marked by capital letters.
FIGURE 5.
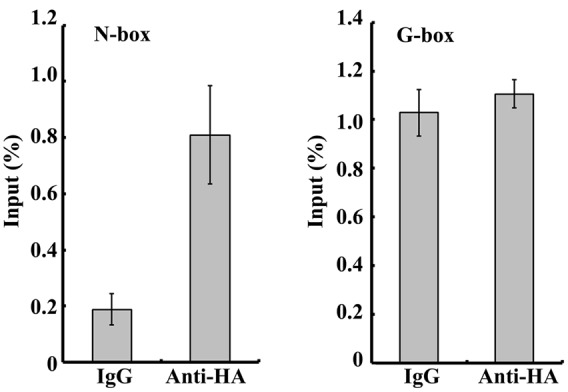
Chromatin immunoprecipitation assays of 14-day-old seedlings grown under long-day conditions and shifted to dark for 12 h. Relative enrichment in the G-box region and N-box region of OsDREB1B promoter was quantified by qPCR.
phyB Interacts with OsPIL16 In vivo
Recent evidence suggests that phytochrome signaling is initiated by the direct interaction of the biologically active forms of phytochromes with PIF members in Arabidopsis (Castillon et al., 2007). To examine the physical interaction between phyB and OsPIL16, we performed a yeast two-hybrid assay, using the coding region of OsPIF16 as prey and the C-terminal non-photoactive coding region of phyB as bait. Three independent clones were used for each sample. As shown in Figure 6 and Supplementary Figure 4, OsPIL16 interacted with the OsPHYB C-terminal domain. Thus, we speculated that phyB affects cold responses through an OsPIL16-mediated pathway.
FIGURE 6.
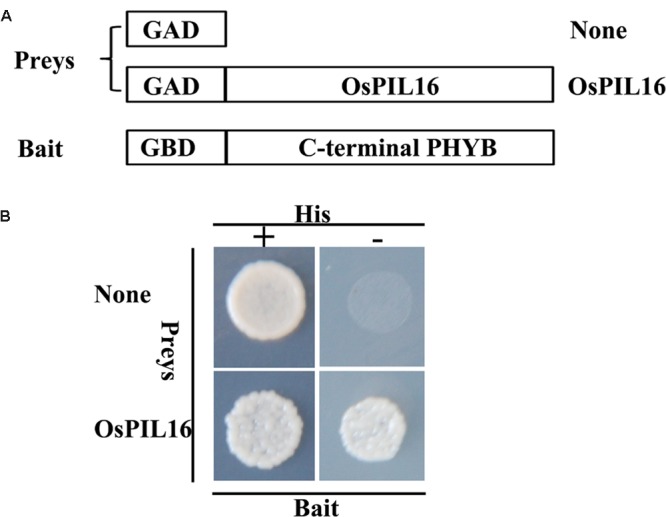
Analysis of the interaction between OsPIF16 and OsPHYB using a yeast two-hybrid assay. (A) Protein constructs used in the yeast two-hybrid assay. OsPIF16 was used as a prey in a translational fusion with the GAL4 AD. The C-terminal non-photoactive region of rice PHYB was fused with the GAL4 BD to use as a bait. (B) Analysis of protein–protein interactions in yeast growing in medium with (left) or without (right) histidine. The negative control was the interaction between OsPIF16 and GBD.
Discussion
phyB plays an important role in rice photomorphogenesis (Takano et al., 2005; Jumtee et al., 2009). In this study, we demonstrated that phyB deficiency resulted in improved cold tolerance compared with the WT though the regulation of OsDREB1 gene expression via an OsPIL16-mediated pathway (Figure 7).
FIGURE 7.
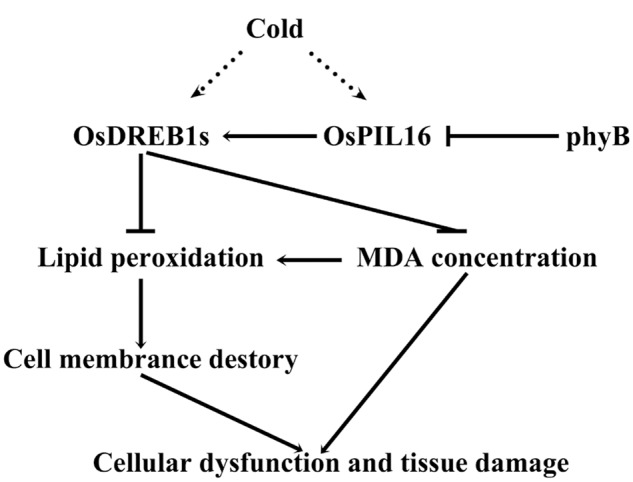
Proposed model of the relationship between phytochrome B and cold stress. Cold stress induces the expression of OsDREB1s. Phytochrome B negatively regulates OsDREB1 genes by inhibiting the expression of OsPIL16. OsDREB1s inhibit lipid peroxidation to alleviate cold-induced membrane damage under cold stress. OsDREB1s also inhibit MDA accumulation to reduce cold damage and enhance cold tolerance. Arrows denote positive effects, bars indicate negative roles, the dotted line indicates indirect regulation, and solid lines indicate direct regulation.
The Enhanced Cold Tolerance of phyB Mutants Is Mainly Attributable to Increased Membrane Integrity and Reduced MDA Content
Cold stress in rice seedlings can negatively affect growth and development or even cause death (Zhang et al., 2014). In this study, phyB deficiency enhanced cold tolerance compared with the WT. One of the major forms of cold damage is severe membrane injury. ELI is an indicator of membrane integrity (Heidarvand and Maali-Amiri, 2013). The ELI was clearly decreased in the phyB mutants compared with the WT (Figure 1C), indicating that phyB deficiency increased membrane integrity under cold stress. This result is consistent with our previous report that a phyB mutant had a more stable chloroplast structure under cold stress (Yang et al., 2013). MDA is the degradation product of polyunsaturated lipids, and the increased membrane integrity of the phyB mutant can be partly attributed to reduced lipid peroxidation as shown by the reduced MDA content (Figure 1D). However, Yang et al. (2013) revealed that phyB mutants had higher unsaturated fatty acid contents in their membrane lipids, which also partly contributes to the increased membrane integrity of the phyB mutant (Yang et al., 2013). Therefore, we speculate that phyB deficiency improves membrane integrity in rice, which is an important factor for enhanced cold tolerance in phyB mutants. Additionally, MDA is a reactive aldehyde that initiates toxic stress in cells and subsequently causes cellular dysfunction and tissue damage (Zhang et al., 2014). Thus, we speculate that the reduced MDA content under cold stress caused by phyB deficiency is likely one of the factors contributing to cold tolerance in the phyB mutants.
phyB Regulates OsDREB1 Expression through OsPIFs to Affect Rice Cold Tolerance
C repeat binding factor/DREB1s play an important role in cold signal transduction (Cook et al., 2004; Zhao et al., 2016). Overexpression of rice or Arabidopsis DREB1s can enhance cold tolerance (Jagloottosen et al., 1998; Dubouzet et al., 2003; Zhang et al., 2009). Interestingly, we found that the phyB mutant had higher expression levels of OsDREB1s (Figure 2), which is probably related to the enhanced cold tolerance in the phyB mutant. Phytochromes have been reported to regulate DREB1 gene expression in Arabidopsis. One study identified phyB as a positive regulator of cold-inducible DRE-controlled expression (Kim et al., 2002). However, the results of a subsequent study indicated a repressive role for phyB and phyD in the control of the CBF-responsive regulon (Franklin and Whitelam, 2007). Kidokoro et al. (2009) also showed that phyB-mediated light signaling had a repressive effect on DREB1 gene expression in Arabidopsis, consistent with our results in this study (Kidokoro et al., 2009). Xu et al. (2011) found that overexpression of ZmCBF3 in rice reduced the MDA concentration (Xu et al., 2011). In this context, we speculate that the reduced MDA concentration in the phyB mutant probably contributed to the upregulation of OsDREB1 genes in our study.
How does phytochrome-mediated pathway regulate DREB1 family gene expression? PIFs, as central players in phytochrome-mediated light signaling networks, have been reported to regulate DREB1 gene expression in Arabidopsis. PIF7 can bind to the promoter of DREB1C, and the pif7 null mutant was shown to have altered transcript levels of DREB1B and DREB1C (Kidokoro et al., 2009). PIF4 can downregulate the CBF pathway and freezing tolerance under LD conditions (Lee and Thomashow, 2012). There are six OsPIFs (OsPIL11–OsPIL16) in rice (Nakamura et al., 2007). The functions of OsPIL13 and OsPIL15 in rice growth and development have been reported. Overexpression of OsPIL13 in transgenic rice plants promoted internode elongation (Todaka et al., 2012). OsPIL15 represses seedling growth in the dark, and when exposed to R or FR light, OsPIL15-OX lines can relieve growth retardation and promote seedling elongation (Zhou et al., 2014). However, to our knowledge, the roles of OsPIL13 and OsPIL15 in regulating OsDREB1 gene expression have not been reported. In this study, our results showed that OsPIL16 upregulated the expression of multiple OsDREB1 genes (Figure 4E), which is consistent with the enhanced cold tolerance of OsPIL16-OX transgenic lines (Figure 4D). OsPIL14 was reported to downregulate OsDREB1B gene expression in rice protoplasts by binding to the N-box motif of the OsDREB1B promoter (Cordeiro et al., 2016). How does OsPIL16, as a bHLH transcription factor, regulate the expression of DREB1s? bHLH proteins are able to bind to hexameric E-box (CANNTG) or N-box motifs, depending on their transcriptional activity (Cordeiro et al., 2016). A previous study revealed that PIFs not only bind to the G-box in vitro and in vivo, and also bind weakly in vitro to PBE (Kim et al., 2008, 2016; Zhang et al., 2013; Pfeiffer et al., 2014). We found that there were N-boxes, PBE or G-boxes in the promoter regions of the OsDREB1 genes (Table 1). ChIP-qPCR assay confirmed that OsPIL16 can bind to the N-box of OsDREB1B promoter (Supplementary Figure 3), thus to regulate the expression of OsDREB1B.
To dissect the relationship between phyB and OsPILs, we compared the transcript levels of OsPILs in the WT and phyB mutant. The transcript levels of both OsPIL15 and OsPIL16 were significantly higher in the phyB mutant than in the WT (Figure 2). However, OsPIL15-OX did not show higher OsDREB1 transcript levels (Supplementary Figure 2), which is probably consistent with the similar cold response of the OsPIL15-OX line to the WT. Unlike OsPIL15-OX, OsPIL16-OX had higher OsDREB1 transcript levels (Figure 4D), which contributed to the enhanced cold tolerance in the OsPIL16-OX lines (Figure 4C). To explain how phyB affects OsDREB1s family gene expression through OsPIL16, it is necessary to clarify the relationship between phyB and OsPIL16. Therefore, we examined the regulation of phyB on OsPIL16 transcript levels and physical interaction between phyB and OsPIL16 proteins. As shown in the results (Figure 3; Supplementary Figure 1), phyB perceives the R light to inhibit OsPIL16 expression at the transcriptional level. Physical interaction between OsPIL16 and OsPHYB were also confirmed by Y2H (Figure 6; Supplementary Figure 4). There is a well-known model of PIF function in phytochrome signaling pathways in Arabidopsis (Castillon et al., 2007; Leivar and Quail, 2011). Light signals induce photoconversion of phytochromes to the active Pfr forms before nuclear migration. In the nucleus, phytochromes physically interact with PIFs, which results in phosphorylation of PIFs. The phosphorylated forms of PIFs are subsequently degraded by the 26S proteasome. The light-induced proteolytic removal of PIFs results in relieving the negative regulation of photomorphogenesis. However, Park et al. (2012) reported that phyB inhibits the regulatory activity of PIF1 and PIF3 by two different modes of action: by releasing them from their target promoters and by mediating their degradation. In this study, OsPIL16 induced OsDREB1s gene expression probably through binding to the N-box of their promoters. Thus, OsPIL16 is the positive regulator of OsDREB1 gene expression and rice cold tolerance. However, the physical interaction between phyB and OsPIL16 proteins inhibits the OsDREB1 gene expression by either degradation of OsPIL16 protein or by inhibiting the bind of OsPIL16 to N-box of OsDREB1 gene promoter. This hypothesis can explain the higher transcript levels of OsDREB1s in the phyB mutants than in the WT.
In summary, we determined the function of phyB in the cold tolerance of rice using genetic and physiological approaches. Although our findings suggest that phyB affect rice cold tolerance through OsPIL16 pathway in this study, some other pathways, for example ABA pathway and ICE pathway, are probably also involved in the phyB-regulated cold responses in rice. ABA has been shown to increase levels of CBF1 transcript and protein (Knight et al., 2004). Our previous result revealed that light signals mediated by phytochrome B affect ABA pathway in rice (Gu et al., 2012). To test this possibility, we must produce the null mutants of pil16 and phyBpil16 double mutant in the future experiments. In addition, further work is needed to decipher the mechanisms by which OsPIL16 regulates OsDREB1s.
Author Contributions
YH and XX conceived the idea, led the study design, data analysis, and manuscript writing. YL performed southern blot, assisted in quantitative PCR and yeast two-hybrid. CZ and LX performed vector construction and rice transformation. LC and GZ assisted in the measurement of ELI and MDA. LC and YH performed ChIP-qPCR. JZ assisted in rice growing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- bHLH
helix-loop-helix
- CBF
C repeat binding factor
- ChIP
chromatin immunoprecipitation
- COR
COLD REGULATED
- d
day
- DREB
dehydration-responsive element binding protein
- ELI
electrolyte leakage index
- FR
far-red
- IgG
immunoglobulin G
- MDA
malondialdehyde
- OX
overexpression
- PBE
PIF binding E-box
- phyB
phytochrome B
- PIF
phytochrome-interacting factor
- PIL
phytochrome-interacting factor-like
- R
red
- WT
wild type
Funding. This work was partly supported by grants from the Chinese National Natural Science Foundation (31300232 and 31270232), the Chinese Ministry of Agriculture (2014ZX08001-005B), Training Program of the Major Scientific and Technological Achievements of Shandong Academy of Agricultural Sciences (2014CGPY08) and Yang Talents Training Program of Shandong Academy of Agricultural Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01963/full#supplementary-material
References
- Carvalho R. F., Campos M. L., Azevedo R. A. (2011). The role of phytochrome in stress tolerance. J. Integr. Plant Biol. 53 920–929. 10.1111/j.1744-7909.2011.01081.x [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. 10.1016/j.tplants.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Cook D., Fowler S., Fiehn O., Thomashow M. F. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 15243–15248. 10.1073/pnas.0406069101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro A. M., Figueiredo D. D., Tepperman J., Borba A. R., Lourenco T., Abreu I. A., et al. (2016). Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim. Biophys. Acta 1859 393–404. 10.1016/j.bbagrm.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E. G., Miura S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33 751–763. 10.1046/j.1365-313X.2003.01661.x [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Quail P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61 11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Whitelam G. C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39 1410–1413. 10.1038/ng.2007.3 [DOI] [PubMed] [Google Scholar]
- Gilmour S. J., Fowler S. G., Thomashow M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54 767–781. 10.1023/B:PLAN.0000040902.06881.d4 [DOI] [PubMed] [Google Scholar]
- Gu J. W., Liu J., Xue Y. J., Zang X., Xie X. Z. (2011). Functions of phytochrome in rice growth and development. Rice Sci. 18 231–237. 10.1016/S1672-6308(11)60032-2 [DOI] [Google Scholar]
- Gu J. W., Zhang F., Zhao J., Zhou J. J., Qian F. Q., Yan L. H., et al. (2012). Light signals mediated by phytochrome B affect abscisic acid pathway in rice (in Chinese). Chin. Sci. Bull. 57 2371–2379. 10.1360/972011-2561 [DOI] [Google Scholar]
- Heidarvand L., Amiri R. M. (2010). What happens in plant molecular responses to cold stress? Acta Physiol. Plant. 32 419–431. 10.1007/s11738-009-0451-8 [DOI] [Google Scholar]
- Heidarvand L., Maali-Amiri R. (2013). Physio-biochemical and proteome analysis of chickpea in early phases of cold stress. J. Plant Physiol. 170 459–469. 10.1016/j.jplph.2012.11.021 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. 10.1046/j.1365-313X.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Hood E. E., Gelvin S. B., Melchers L. S., Hoekema A. (1993). New Agrobacterium helper plasmid for gene transfer to plants (EHA105). Transgenic Res. 2 208–218. 10.1007/BF01977351 [DOI] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47 141–153. 10.1093/pcp/pci230 [DOI] [PubMed] [Google Scholar]
- Jagloottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Jeong J., Choi G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35 371–380. 10.1007/s10059-013-0135-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumtee K., Okazawa A., Harada K., Fukusaki E., Takano M., Kobayashi A. (2009). Comprehensive metabolite profiling of phyA phyB phyC triple mutants to reveal their associated metabolic phenotype in rice leaves. J. Biosci. Bioeng. 108 151–159. 10.1016/j.jbiosc.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Kidokoro S., Maruyama K., Nakashima K., Imura Y., Narusaka Y., Shinwari Z. K., et al. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151 2046–2057. 10.1104/pp.109.147033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20 1260–1277. 10.1105/tpc.108.058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim Y. K., Park J. Y., Kim J. (2002). Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 29 693–704. 10.1046/j.1365-313X.2002.01249.x [DOI] [PubMed] [Google Scholar]
- Kim J., Kang H., Park J., Kim W., Yoo J., Lee N., et al. (2016). PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28 1388–1405. 10.1105/tpc.16.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Tai T. H. (2011). Evaluation of seedling cold tolerance in rice cultivars: a comparison of visual ratings and quantitative indicators of physiological changes. Euphytica 178 437–447. 10.1007/s10681-010-0343-4 [DOI] [Google Scholar]
- Knight H., Zarka D. G., Okamoto H., Thomashow M. F., Knight M. R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135 1710–1717. 10.1104/pp.104.043562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Thomashow M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109 15054–15059. 10.1073/pnas.1211295109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16 19–28. 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ljung K., Breton G., Schmitz R. J., Pruneda-Paz J., Cowing-Zitron C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26 785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Liu Y., Rao J., Wang G., Li H., Ge F., et al. (2013). Overexpression of the glutathione S-transferase gene from Pyrus pyrifolia fruit improves tolerance to abiotic stress in transgenic tobacco plants. Mol. Biol. 47 515–523. 10.1134/S0026893313040109 [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H. L., He S. B., Li L., Jia K. P., Yang H. Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26 2441–2456. 10.1105/tpc.113.121657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D., Chen C. (2012). Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 7:e47275 10.1371/journal.pone.0047275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2010). Phytochrome: structural basis for its functions. Curr. Opin. Plant Biol. 13 565–570. 10.1016/j.pbi.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kato T., Yamashino T., Murakami M., Mizuno T. (2007). Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 71 1183–1191. 10.1271/bbb.60643 [DOI] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J. C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72 537–546. 10.1111/j.1365-313X.2012.05114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J. M., Zhang Y., Quail P. H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7 1598–1618. 10.1093/mp/ssu087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N. C., Lagarias J. C. (2006). The structure of phytochrome: a picture is worth a thousand spectra. Plant Cell 18 4–14. 10.1105/tpc.105.038513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutinen M.-L., Palta J. P., Reich P. B. (1992). Seasonal differences in freezing stress resistance of needles of Pinus nigra and Pinus resinosa: evaluation of the electrolyte leakage method. Tree Physiol. 11 241–254. 10.1093/treephys/11.3.241 [DOI] [PubMed] [Google Scholar]
- Takano M., Inagaki N., Xie X., Yuzurihara N., Hihara F., Ishizuka T., et al. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17 3311–3325. 10.1105/tpc.105.035899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M., Kanegae H., Shinomura T., Miyao A., Hirochika H., Furuya M. (2001). Isolation and characterization of rice phytochrome A mutants. Plant Cell 13 521–534. 10.1105/tpc.13.3.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F. (2001). So what’s new in the field of plant cold acclimation? Lots! Plant Physiol. 125 89–93. 10.1104/pp.125.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D., Nakashima K., Maruyama K., Kidokoro S., Osakabe Y., Ito Y., et al. (2012). Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. U.S.A. 109 15947–15952. 10.1073/pnas.1207324109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Guo Z., Li H., Wang M., Onac E., Zhou J., et al. (2016). Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 170 459–471. 10.1104/pp.15.01171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67 589–602. 10.1007/s11103-008-9340-6 [DOI] [PubMed] [Google Scholar]
- Williams B. J., Pellett N. E., Klein R. M. (1972). Phytochrome control of growth cessation and initiation of cold acclimation in selected woody plants. Plant Physiol. 50 262–265. 10.1104/pp.50.2.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li L., Fan Y., Wan J., Wang L. (2011). ZmCBF3 overexpression improves tolerance to abiotic stress in transgenic rice (Oryza sativa) without yield penalty. Plant Cell Rep. 30 1949–1957. 10.1007/s00299-011-1103-1 [DOI] [PubMed] [Google Scholar]
- Yang J. C., Li M., Xie X. Z., Han G. L., Sui N., Wang B. S. (2013). Deficiency of phytochrome B alleviates chilling-induced photoinhibition in rice. Am. J. Bot. 100 1860–1870. 10.3732/ajb.1200574 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen Q., Wang S., Hong Y., Wang Z. (2014). Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice (N. Y.) 7:24 10.1186/s12284-014-0024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Jin X. F., Xiong A. S., Peng R. H., Hong Y. H., et al. (2009). Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 42 486–492. 10.5483/BMBRep.2009.42.8.486 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J. M., Speed T. P., et al. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9:e1003244 10.1371/journal.pgen.1003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J. K. (2016). Mutational evidence for the critical role of CBF genes in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu Q., Zhang F., Wang Y., Zhang S., Cheng H., et al. (2014). Overexpression of OsPIL15, a phytochrome-interacting factor-like protein gene, represses etiolated seedling growth in rice. J. Integr. Plant Biol. 56 373–387. 10.1111/jipb.12137 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang F., Deng P., Jing W., Zhang W. (2013). Characterization and mapping of a salt-sensitive mutant in rice (Oryza sativa L.). J. Integr. Plant Biol. 55 504–513. 10.1111/jipb.12048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


