Abstract
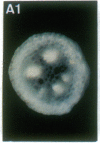
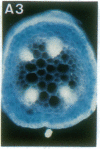
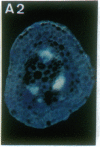
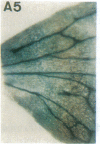
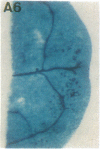
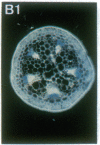
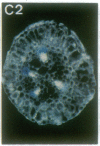
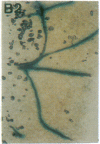
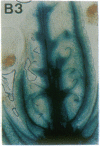
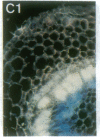
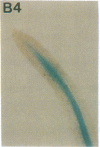
Vectors were constructed for the isolation of random transcriptional and translational beta-glucuronidase gene fusions in plants. This system is based on the random integration of the transferred DNA (T-DNA) into the plant nuclear genome. The Escherichia coli beta-glucuronidase coding sequence without promoter, and also devoid of its ATG initiation site in the translational gene fusion vector, was inserted in the T-DNA with its 5' end at a distance of 4 base pairs from the right T-DNA border sequence. Transgenic plants can be selected by using a chimeric (P35S-nptII-3' ocs) kanamycin-resistance gene present in the same T-DNA. Subsequent screening of these for beta-glucuronidase expression allows the identification of clones harboring a fusion of the beta-glucuronidase coding sequence with plant 5' regulatory sequences. After transformation of Arabidopsis thaliana C24 root explants, beta-glucuronidase expression was detected in 54% and 1.6% of the plants transformed with the transcriptional and translational fusion vectors, respectively. Several different patterns of tissue-specific beta-glucuronidase expression were identified. The plant upstream sequence of a beta-glucuronidase fusion that is specifically expressed in the phloem of all organs was cloned and sequenced. After introduction in A. thaliana C24 and Nicotiana tabacum SR1, this sequence mediates the same highly phloem-specific beta-glucuronidase expression pattern as in the original transgenic plant from which it was isolated. These data demonstrate that this system facilitates the isolation and analysis of plant DNA sequences mediating regulated gene expression.
Full text
PDF
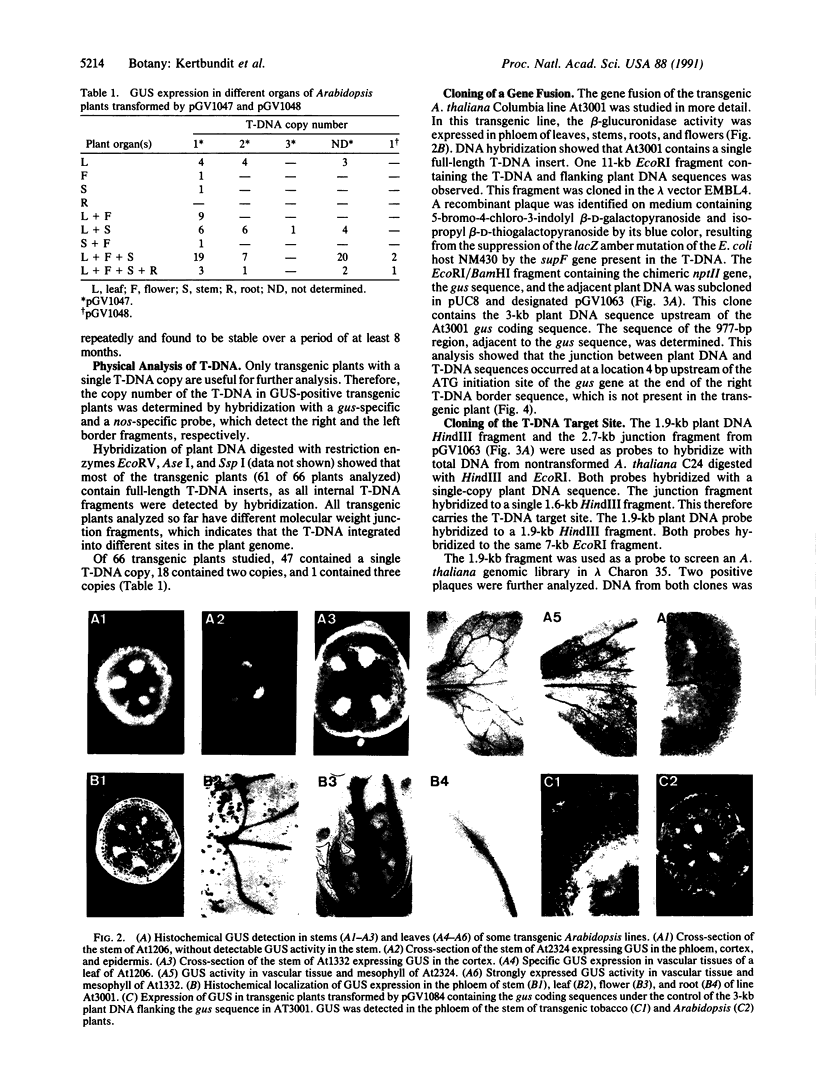
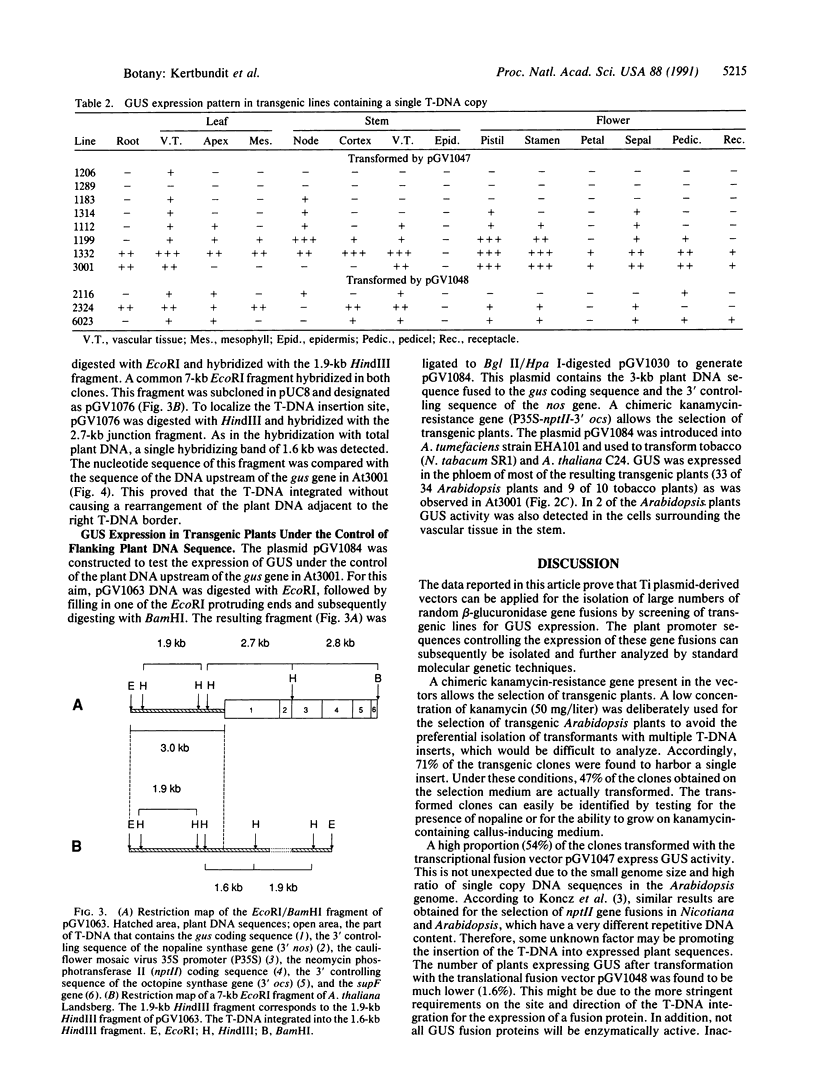

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Benfey P. N., Ren L., Chua N. H. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989 Aug;8(8):2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Colson C., Glover S. W., Symonds N., Stacey K. A. The location of the genes for host-controlled modification and restriction in Escherichia coli K-12. Genetics. 1965 Nov;52(5):1043–1050. doi: 10.1093/genetics/52.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greve H., Dhaese P., Seurinck J., Lemmers M., Van Montagu M., Schell J. Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J Mol Appl Genet. 1982;1(6):499–511. [PubMed] [Google Scholar]
- Depicker A., Stachel S., Dhaese P., Zambryski P., Goodman H. M. Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet. 1982;1(6):561–573. [PubMed] [Google Scholar]
- Fang R. X., Nagy F., Sivasubramaniam S., Chua N. H. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989 Jan;1(1):141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley R. T., Rogers S. G., Horsch R. B., Sanders P. R., Flick J. S., Adams S. P., Bittner M. L., Brand L. A., Fink C. L., Fry J. S. Expression of bacterial genes in plant cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4803–4807. doi: 10.1073/pnas.80.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella L., Block M. D., Messens E., Hernalsteens J. P., Montagu M. V., Schell J. Chimeric genes as dominant selectable markers in plant cells. EMBO J. 1983;2(6):987–995. doi: 10.1002/j.1460-2075.1983.tb01532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood E. E., Helmer G. L., Fraley R. T., Chilton M. D. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol. 1986 Dec;168(3):1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga G., Jefferson R. A., Bevan M. W. Endoplasmic reticulum targeting and glycosylation of hybrid proteins in transgenic tobacco. Plant Cell. 1989 Mar;1(3):381–390. doi: 10.1105/tpc.1.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Martini N., Mayerhofer R., Koncz-Kalman Z., Körber H., Redei G. P., Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers M., De Beuckeleer M., Holsters M., Zambryski P., Depicker A., Hernalsteens J. P., Van Montagu M., Schell J. Internal organization, boundaries and integration of Ti-plasmid DNA in nopaline grown gall tumours. J Mol Biol. 1980 Dec 15;144(3):353–376. doi: 10.1016/0022-2836(80)90095-9. [DOI] [PubMed] [Google Scholar]
- Lloyd A. M., Barnason A. R., Rogers S. G., Byrne M. C., Fraley R. T., Horsch R. B. Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens. Science. 1986 Oct 24;234(4775):464–466. doi: 10.1126/science.234.4775.464. [DOI] [PubMed] [Google Scholar]
- Lojda Z. Indigogenic methods for glycosidases. II. An improved method for beta-D-galactosidase and its application to localization studies of the enzymes in the intestine and in other tissues. Histochemie. 1970;23(3):266–288. doi: 10.1007/BF00306428. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M. Arabidopsis, a useful weed. Cell. 1989 Jan 27;56(2):263–269. doi: 10.1016/0092-8674(89)90900-8. [DOI] [PubMed] [Google Scholar]
- Nagy F., Boutry M., Hsu M. Y., Wong M., Chua N. H. The 5'-proximal region of the wheat Cab-1 gene contains a 268-bp enhancer-like sequence for phytochrome response. EMBO J. 1987 Sep;6(9):2537–2542. doi: 10.1002/j.1460-2075.1987.tb02541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell J. T., Nagy F., Chua N. H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. 1985 Feb 28-Mar 6Nature. 313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeri T. H., Herrera-Estrella L., Depicker A., Van Montagu M., Palva E. T. Identification of plant promoters in situ by T-DNA-mediated transcriptional fusions to the npt-II gene. EMBO J. 1986 Aug;5(8):1755–1760. doi: 10.1002/j.1460-2075.1986.tb04423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu M., Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute E., Joos H., Maes M., Warren G., Van Montagu M., Schell J. Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for the reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J. 1983;2(3):411–417. doi: 10.1002/j.1460-2075.1983.tb01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]