Abstract
Background/Aims:
p16 is tumor suppressor gene acting as a cell cycle regulator. The present study was conducted to compare p16 expression in normal, dysplastic, and malignant colonic mucosae, and to explore its relation to clinicopathological variables and follow-up data in colorectal carcinoma (CRC).
Patients and Methods:
Tissue microarrays were performed from 25 normal colonic mucosae, 41 colonic adenomas, and 191 CRC, with corresponding 50 nodal metastases. Immunohistochemistry was performed using anti-p16 antibody, sections were scored, and statistical analysis was performed. K-ras mutation detection was also performed.
Results:
Immunoexpression of p16 was significantly higher in CRC than in adenomas (P = 0.033) and normal colonic mucosa (P = 0.005). There was no statistically significant difference between p16 expression in CRC and nodal metastasis. There was no significant association between p16 immunoexpression in CRC and all clinicopathological data and survival probability. K-ras mutations were detected in 34% of CRC. However, there was no correlation between K-ras status and p16 expression (P = 0.325).
Conclusion:
Absence of p16 expression is correlated to a benign course of CRC adenomas. p16 has a key role in CRC progression and can be used as a marker for colorectal adenoma. On the other hand, it has no role as a predictive and/or prognostic factor in CRC. Further extended studies are required to explore the role of p16 as indicator of premalignant lesions in the colon and to test its relation with CRC histological grade, as well as to test its value as a new therapeutic target.
Keywords: Adenomas, clinicopathological characteristics, colorectal carcinoma, p16, prognosis
It is well established that colorectal carcinoma (CRC) is a genetic disease with complex and diverse pathways. CRC develops from the normal colonic mucosa through variable molecular mechanisms;firstly, is the chromosomal instability pathway associated with cumulative genetic mutations in many oncogenes and tumor suppressor genes as K-ras, p53, c-Myc, and cyclin-D, accounting for approximately 80% of sporadic cases. Second, is the microsatellite instability pathway characterized by genetic alterations in DNA mismatch repair genes (responsible for repairing insertions, deletions, and misincorporation of bases during DNA replication and recombination avoiding frameshift mutation and base substitution) occurring in 10–15% of sporadic cases of CRC. Third, is the lynch pathway accounting for 3%, and lastly, is the familial adenomatous polyposis syndrome accounting for 1% of all CRC cases.[1,2,3,4,5]
p16 is a tumor-suppressor gene now recognized to be the second most common molecular defect in human cancer.[6] The role of p16 tumor suppressor gene is to bind to CD4/6 preventing its interaction with cyclin D; ultimately, this reaction inhibits cell cycle progression from G1 to S phase. Hence, the p16 pathway of action connects the process of oncogenesis and cell aging; downregulation of p16 by hypermethylation, point mutation, or deletion of the gene leads to the progression of cell cycle whereas activation of the gene will stimulate cellular aging or senescence.[7,8,9] Inactivation of p16 is well-documented in several malignancies as squamous cell carcinoma of cervix,[10] oropharynx,[11] and esophagus,[12] non-small cell carcinoma of lung,[13] mesothelioma,[14] and pancreatic carcinoma.[15]
The participation of p16 in carcinogenesis and its role as a diagnostic, prognostic, or predictive factor in CRC is still controversial. The present study was conducted to compare p16 expression in normal, dysplastic, and malignant colonic mucosa, and to explore its relation with clinicopathological variables and follow-up data in CRC.
PATIENTS AND METHODS
Patients
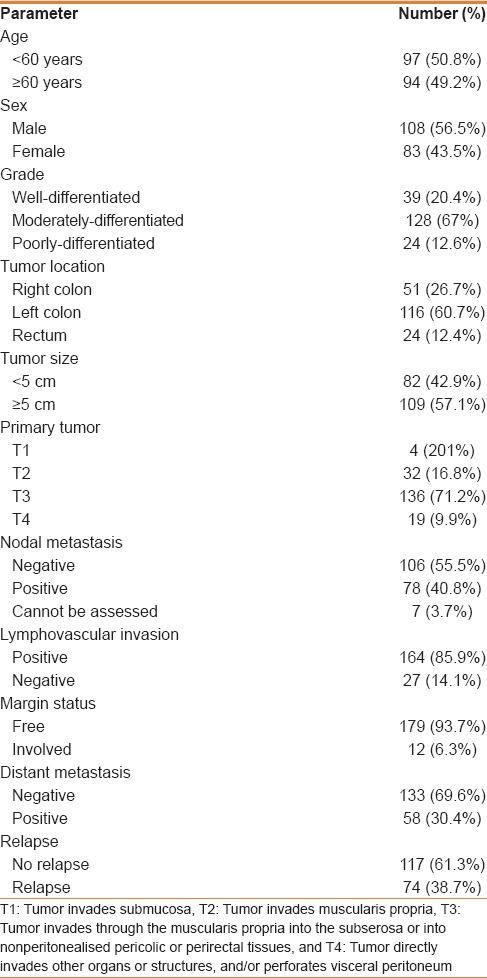
Archival tissues from 25 normal colonic mucosa, 41 colonic adenomas, 191 CRCs, and 50 local nodal metastases were used in the present study. Patients were admitted, diagnosed, and managed in King Abdulaziz University Hospital (KAUH), Jeddah in the period between 2000 and 2012. Clinical history and follow-up data were collected from the hospital medical records after obtaining appropriate approval from the ethics committee. Paraffin blocks of all cases were collected from the archives of Pathology Department, KAUH. Serial sections were cut from paraffin blocks, stained with Hematoxylin and Eosin and revised for routine histological classification, and grading according to World Health Organization (WHO) subtyping criteria.[16] Carcinomas were staged following the American Joint Committee on Cancer (AJCC) staging system.[17] Details of clinicopathological findings are listed in Table 1. The study was approved by the Research Committee of the Biomedical Ethics Unit, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia. All patients included in this study gave an informed written consent for utilization of their material in research.
Table 1.
Clinicopathological parameters of CRC cases
Tissue microarray
A tissue microarray was constructed, as previously described.[18,19,20] Two cylindrical cores of 1.5 mm in diameter were selected from donor paraffin blocks and arrayed in recipient paraffin blocks using the automated tissue arrayer (Master 3D Histech). Normal placenta tissue was used as control tissue to help orientation of samples in each tissue microarray block. Tissue microarray blocks were sectioned into 4-μm thick sections and mounted on positive charged slides to be stained by immunohistochemical staining. Normal colorectal mucosae were taken from unremarkable mucosa in patients with diverticular disease, ulcerative colitis, ischemic colitis, or Hirschsprung disease.
Immunohistochemistry
Immunohistochemistry procedure was carried out using an automatic immunostainer (Ventana Bench Mark XT, Ventana Inc., Tucson, AZ). The ready-to-use anti-p16 antibody (Ventana Medical Systems) was optimally diluted according to the manufacturer's recommendations. In each analysis, positive controls were used consisting of CRC samples previously shown to stain with this antibody. Tris-buffered saline in place of the primary antibody was used as a negative control.
Interpretation of immunohistochemical staining
Positive staining for p16 was interpreted as presence of brown nucleo/cytoplasmic stain of cells. The following semiquantitative scoring system was recorded for percentage of stained cells on a scale from 0 to 3. Immunostaining in >40% of cells (score 3), immunostaining in 10–40% of cells (score 2), immunostaining in <10% of cells (score 1), and score 0 implied negative staining. When dichotomized for statistical assessment, score 0 and 1 were defined as low immunoexpression whereas score 2 and 3 were included in high immunoexpression.
K-ras mutation detection
DNA was extracted from 10-mm thin formalin-fixed paraffin-embedded slices using the Qiagen QIAMP Formalin-fixed Paraffin-embedded Tissue DNA extraction kit, following the manufacturer's guidelines. K-ras mutational status was determined according to the previously published report.[21] However, K-ras mutations were investigated in 50 samples according to the availability of DNA material.
Statistical analysis
Differences between the two groups of patients on one variable were tested by using Mann–Whitney test whereas Kruskal–Wallis test was used for differences between the three groups of patients. Nonparametric Chi-square was used to test the difference along one variable. Binary logistic regression analysis was used to predict lymph node metastasis, distant metastasis, surgical resection margins involvement, lymphovascular invasion, and local disease recurrence in relation to immunoexpression of p16. Estimated odds ratio {exponential (B)}, 95% confidence interval (CI) for exp (B). The Kaplan–Meier procedure was used to calculate the disease-free survival probabilities and the Log rank test was used to compare the difference between survivals. Time was calculated from the date of diagnosis to the appearance of disease relapse (or date of last seen disease-free). Statistical procedures were performed using Statistical Package for the Social Sciences (SPSS® version 16.0, Chicago, IL, USA). Statistical significance was determined at P value of ≤0.05
RESULTS
p16 immunohistochemistry
p16 immunostaining was observed as combined nucleocytoplasmic. p16 nuclear localization was generally associated with strong cytoplasmic staining. It showed a very minimal expression (score + 0) in most normal colonic mucosa [Figure 1a]. In colorectal adenoma, high p16 immunoexpression was observed in 14.6% [Figure 1b]. p16 expression in colorectal adenoma was significantly higher than in normal mucosa (P = 0.046) [Table 2]. In regards to CRC, positivity was observed in 142/193 of patients (73.6%). Positive p16 immunostaining in CRC was distributed as follows; 84/142 (59.1%) showed score + 1 positivity, 40/142 (28.2%) score + 2, and finally, 18/142 (12.7%) score + 3. Negative p16 immunostaining was observed in 51/193 of patients (26.4%). Representative figures are shown in Figure 1c. p16 was overexpressed in 28% of nodal metastases [Figure 1d]. p16 immunostaining was significantly higher in CRC than in adenoma (P = 0.033) and normal colonic mucosa (P = 0.005}. There was no statistically significant difference between p16 expression in CRC and nodal metastasis [Table 3].
Figure 1.
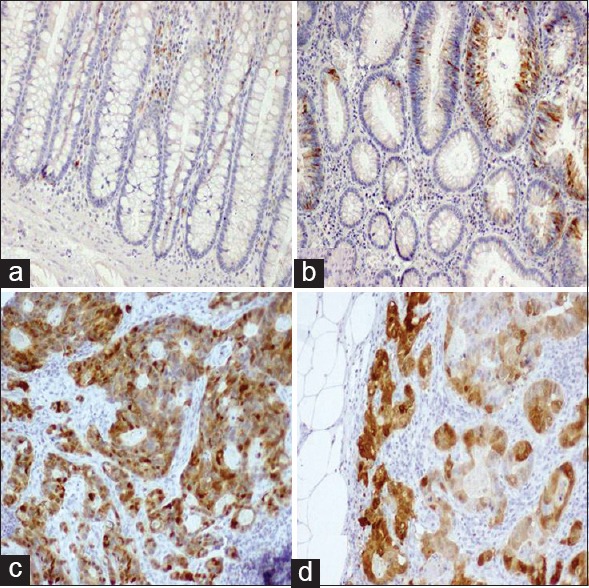
Immunostaining of p16. (a) A normal colonic mucosa showing no p16 immunostaining. (b) Low combined nucleocytoplasmic immunoexpression is shown in an adenoma. (c) A moderately differentiated colorectal carcinoma (CRC) showing high p16 immunostaining. (d) A metastatic CRC in lymph node shows a high p16 immunostaining. Original magnification used is 200×. Immunohistochemistry was done using anti-p16 antibody, diaminobenzidine as the chromogen, and hematoxylin as a counterstain
Table 2.
Categories of p16 immunoexpression in different tissues (One sample Chi-square test)
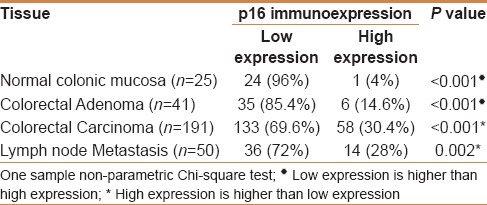
Table 3.
Comparison of p16 immunoexpression tissues examined (Mann–Whitney test)

Relationship between p16 expression and clinicopathological features of colorectal cancer
There was no statistically significant association between p16 immunoexpression in CRC and clinicopathological data, except for a borderline significant relation to tumor grade [Table 4]. In addition, p16 immunoexpression in CRC failed to predict nodal metastasis, distant metastasis, margin status, lymphovascular invasion, or tumor relapse [Table 5]. There was no relation between p16 immunostaining and survival probabilities (disease free survival; log-rank = 0.149, P = 0.700, and overall survival; log-rank = 0.158, P = 0.209) [Figures 2 and 3].
Table 4.
Correlation of p16 immunoexpression in primary colorectal carcinoma with clinicopathological parameters

Table 5.
Regression analysis for p16 immunoexpression
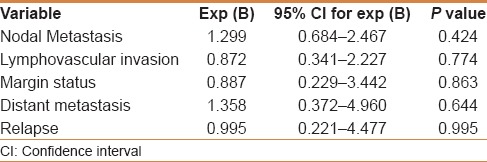
Figure 2.
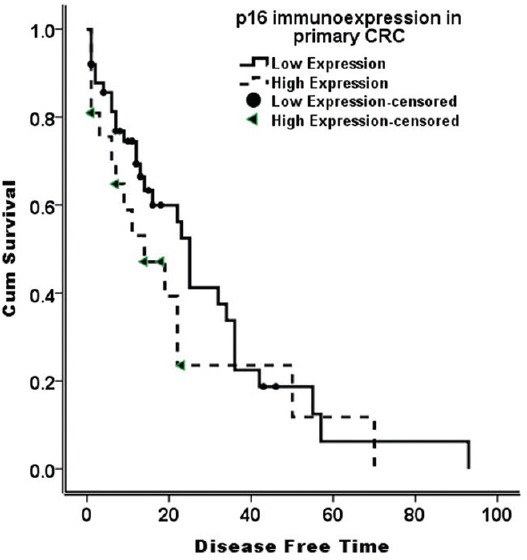
Disease-free survival curve (Kaplan–Meier) according to p16 immunostaining. There is no difference of survival probability between low and high p16; immunoexpression (log-rank = 0.149, P = 0.700)
Figure 3.
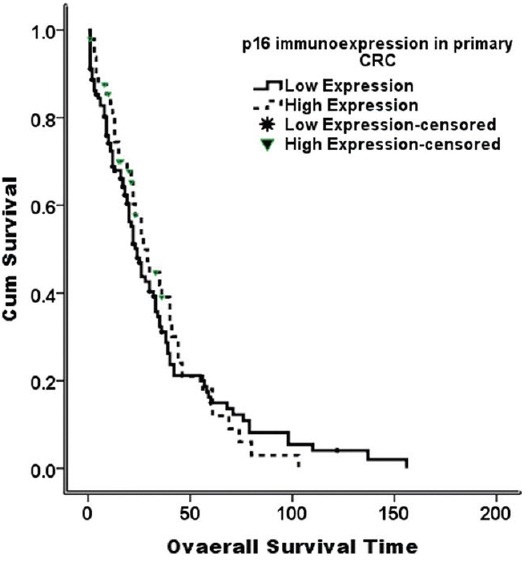
Overall survival curve (Kaplan–Meier) according to p16 immunostaining. There is no difference of survival probability between low and high p16; immunoexpression (log-rank = 0.158, P = 0.209)
p16 immunoexpression and k-ras mutation
Mutations were detected in 18 out of 53 cases (34%). However, the correlation between k-ras status and p16 immunostaining profile was investigated only in 50 cases. There were no statistically significant differences in p16 immunoexpression in k-ras mutant and nonmutant CRC patients (P = 0.325).
DISCUSSION
Various genetic alterations are involved in the carcinogenesis of CRC and the adenoma-carcinoma sequence theory was established to reveal these.[22] Gene silencing by promoter CpG islands methylation as an epigenetic alteration has attracted attention as one of the aberrant gene expressions and the aberrant methylation involved in the carcinogenesis of CRC.[23,24] Several studies have been conducted to investigate molecular alterations underlying the carcinogenesis process in CRC to predict prognosis and develop new targeted therapeutic agents.[2,4,5,9]
Abnormal function of p16 is reported to be one of the earliest events occurring in cancer progression.[1,2,3] Recent clinical practice reported consistent association between p16 and premalignant lesions and used p16 successfully as predictor of progression to high grade dysplasia or cancer in cervical biopsies and in Barrett's oesophagus.[10,12]
The aim of the current study was to compare p16 expression in normal colonic epithelium, colorectal adenoma, and CRC, and to explore its relation to clinicopathological variables and follow-up data in CRC. p16 was found to have a combined nuclear and cytoplasmic localization in all cases, as previously reported.[25,26] In another study, it was expressed in the cytoplasm with absent nuclear expression.[27] The importance of the cytoplasmic translocation of p16 needs to be stressed regarding its prognostic significance.
In our study, the differential expression of p16 showed statistically significant overexpression in CRC, supporting its contributory role in CRC carcinogenesis and progression. Several studies have been conducted to explore the relation between p16 overexpression and clinicopathological characteristics of CRC.[25,28,29,30] However, none of these studies found any relation between p16 and tumor location, size, grade, and stage. In the present study, we did not find any relation between p16 and clinicopathological variables, except for borderline relation to tumor grade. p16 overexpression was reduced as tumor grade was increased, however, it was not statistically significant.
Previous reports documented a positive correlation between p16 expression and tumor progression and poor prognosis in CRC;[6,31,32,33] however, other studies failed to find any significant role of p16 as a useful marker of prognosis or a predictor of CRC.[34,35] The inconsistency of results may be related to several factors including number of patients tested for p16 immunostaining, p16 antibody used, scoring cutoff point,[33,36,37,38] and immunohistochemistry technique.
Ras genes encode for membrane-attached small guanine triphosphate bound proteins that play a key role in signal transduction of extracellular mitogenic signals (such as growth factors) to the nucleus. Ras mutations are frequent in human tumors. K-ras mutations (mainly at codons 12 and 13) that constitutively activate their function are present in up to 40% of colorectal adenomas and carcinomas.[39,40] The impact of ras mutations in colorectal tumorigenesis is high, and several studies have suggested that k-ras mutations might accumulate during tumor progression and associate with poorer survival.[41] In the present study, k-ras mutations were detected in 34% of CRC, which is similar to previous reports.[6,42] The presence of k-ras mutation did not correlate with p16 immunostaining, which is contrary to a previous report, which observed a strong correlation between k-ras and p16 with poor outcome.[6]
CONCLUSION
Absence of p16 expression is correlated to the course of colorectal adenoma. p16 may play a key role in CRC progression and can be used as a marker for dysplasia and colonic adenomatous polyps. On the other hand, it has no role as a predictive or a prognostic factor in CRC. Further studies are required to explore the role of p16 as an indicator of premalignant lesions in the colon and to test its relation to CRC histological grade, as well as to test its value as a new targeted therapeutic target.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH) – King Abdulaziz City for Science and Technology - the Kingdom of Saudi Arabia – award number (11-BIO1524-03). The authors also, acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support.
REFERENCES
- 1.Shi C, Washington K. Molecular testing in colorectal cancer: Diagnosis of Lynch syndrome and personalized cancer medicine. Am J Clin Pathol. 2012;137:847–59. doi: 10.1309/AJCPI83DINULUJNI. [DOI] [PubMed] [Google Scholar]
- 2.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–85. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka T. Colorectal carcinogenesis: Review of human and experimental animal studies. J Carcinog. 2009;8:5. doi: 10.4103/1477-3163.49014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2012;138:2073–87-e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M, Gonzalez S, Risques RA, Marcuello E, Mangues R, Germa JR, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001;19:299–304. doi: 10.1200/JCO.2001.19.2.299. [DOI] [PubMed] [Google Scholar]
- 7.Romagosa C, Simonetti S, Lopez-Vicente L, Mazo A, Lleonart ME, Castellvi J, et al. p16(Ink4a) overexpression in cancer: A tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087–97. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 8.Lee MH, Choi BY, Cho YY, Lee SY, Huang Z, Kundu JK, et al. Tumor suppressor p16(INK4a) inhibits cancer cell growth by downregulating eEF1A2 through a direct interaction. J Cell Sci. 2013;126:1744–52. doi: 10.1242/jcs.113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2010;60:116–29. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesnikova I, Lidang M, Hamilton-Dutoit S, Koch J. p16 as a diagnostic marker of cervical neoplasia: A tissue microarray study of 796 archival specimens. Diagn Pathol. 2009;4:22. doi: 10.1186/1746-1596-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer CA, Zlobec I, Green E, Probst S, Storck C, Lugli A, et al. Is the improved prognosis of p16 positive oropharyngeal squamous cell carcinoma dependent of the treatment modality? Int J Cancer. 2010;126:1256–62. doi: 10.1002/ijc.24842. [DOI] [PubMed] [Google Scholar]
- 12.Wang JS, Guo M, Montgomery EA, Thompson RE, Cosby H, Hicks L, et al. DNA promoter hypermethylation of p16 and APC predicts neoplastic progression in Barrett's esophagus. Am J Gastroenterol. 2009;104:2153–60. doi: 10.1038/ajg.2009.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panani AD, Maliaga K, Babanaraki A, Bellenis I. Numerical abnormalities of chromosome 9 and p16CDKN2A gene deletion detected by FISH in non-small cell lung cancer. Anticancer Res. 2009;29:4483–7. [PubMed] [Google Scholar]
- 14.Lopez-Rios F, Chuai S, Flores R, Shimizu S, Ohno T, Wakahara K, et al. Global gene expression profiling of pleural mesotheliomas: Overexpression of aurora kinases and P16/CDKN2A deletion as prognostic factors and critical evaluation of microarray-based prognostic prediction. Cancer Res. 2006;66:2970–9. doi: 10.1158/0008-5472.CAN-05-3907. [DOI] [PubMed] [Google Scholar]
- 15.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–42. [PubMed] [Google Scholar]
- 16.Hamilton S, Rubio C, Vogelstein B, Sobin L, Kudo S, Fogt F, et al. In: Carcinoma of the colon and rectum In WHO Classification of Tumours: Pathology and Genetics of Tumours of Digestive System. Hamilton SR, Aaltonen LA, editors. Lyon (France): 2000. pp. 105–119. [Google Scholar]
- 17.ECompton C, Byrd D, Garcia-Aguilar J, Kurtzman S, Olawaiye A, Washington M. AJCC Cancer Staging Handbook. 7th ed. New York: Springer; 2012. pp. 185–203. [Google Scholar]
- 18.Al-Maghrabi J, Emam E, Gomaa W, Saggaf M, Buhmeida A, Al-Qahtani M, et al. c-MET immunostaining in colorectal carcinoma is associated with local disease recurrence. BMC Cancer. 2015;15:676. doi: 10.1186/s12885-015-1662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Maghrabi J, Gomaa W, Buhmeida A, Qari Y, Al-Qahtani M, Al-Ahwal M. Prognostic significance of VEGFR1/Flt-1 immunoexpression in colorectal carcinoma. Tumour Biol. 2013;35:9045–51. doi: 10.1007/s13277-014-2124-5. [DOI] [PubMed] [Google Scholar]
- 20.Gomaa W, Ke Y, Fujii H, Helliwell T. Tissue microarray of head and neck squamous carcinoma: Validation of the methodology for the study of cutaneous fatty acid-binding protein, vascular endothelial growth factor, involucrin and Ki-67. Virchows Arch. 2005;447:701–9. doi: 10.1007/s00428-005-0002-7. [DOI] [PubMed] [Google Scholar]
- 21.Schulten HJ, Al-Maghrabi J, Al-Ghamdi K, Salama S, Al-Muhayawi S, Chaudhary A, et al. Mutational screening of RET, HRAS, KRAS, NRAS, BRAF, AKT1, and CTNNB1 in medullary thyroid carcinoma. Anticancer Res. 2011;31:4179–83. [PubMed] [Google Scholar]
- 22.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 24.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 25.Wieser RJ, Faust D, Dietrich C, Oesch F. p16INK4 mediates contact-inhibition of growth. Oncogene. 1999;18:277–81. doi: 10.1038/sj.onc.1202270. [DOI] [PubMed] [Google Scholar]
- 26.Wehmuth C, Santos EMM, Wernek I, Coudry RA, Soares FA, Lopes A, et al. P16 and p27 Immunohistochemical Expression in Colorectal Cancer: Analysis of 128 Patients. App Cancer Res. 2007;27:150–5. [Google Scholar]
- 27.Zhao P, Mao X, Talbot IC. Aberrant cytological localization of p16 and CDK4 in colorectal epithelia in the normal adenoma carcinoma sequence. World J Gastroenterol. 2006;12:6391–6. doi: 10.3748/wjg.v12.i39.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishiguro A, Takahata T, Saito M, Yoshiya G, Tamura Y, Sasaki M, et al. Influence of methylated p15 and p16 genes on clinicopathological features in colorectal cancer. J Gastroenterol Hepatol. 2006;21:1334–9. doi: 10.1111/j.1440-1746.2006.04137.x. [DOI] [PubMed] [Google Scholar]
- 29.Huh JW, Lee JH, Kim HR. Expression of p16, p53, and Ki-67 in colorectal adenocarcinoma: A study of 356 surgically resected cases. Hepatogastroenterology. 2010;57:734–40. [PubMed] [Google Scholar]
- 30.Von Stockmar-Von Wangenheim CA, Monig SP, Schneider PM, Landsberg S, Drebber U, Holscher AH, et al. p16, cyclin D1 and Rb expression in colorectal carcinomas: Correlations with clinico-pathological parameters and prognosis. Mol Med Rep. 2008;1:27–32. [PubMed] [Google Scholar]
- 31.Lam AK, Ong K, Giv MJ, Ho YH. p16 expression in colorectal adenocarcinoma: Marker of aggressiveness and morphological types. Pathology. 2008;40:580–5. doi: 10.1080/00313020802320713. [DOI] [PubMed] [Google Scholar]
- 32.Zhao P, Hu YC, Talbot IC. Expressing patterns of p16 and CDK4 correlated to prognosis in colorectal carcinoma. World J Gastroenterol. 2003;9:2202–6. doi: 10.3748/wjg.v9.i10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui X, Shirai Y, Wakai T, Yokoyama N, Hirano S, Hatakeyama K. Aberrant expression of pRb and p16(INK4), alone or in combination, indicates poor outcome after resection in patients with colorectal carcinoma. Hum Pathol. 2004;35:1189–95. doi: 10.1016/j.humpath.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Prall F, Ostwald C, Weirich V, Nizze H. p16(INK4a) promoter methylation and 9p21 allelic loss in colorectal carcinomas: Relation with immunohistochemical p16(INK4a) expression and with tumor budding. Hum Pathol. 2006;37:578–85. doi: 10.1016/j.humpath.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Yi J, Wang ZW, Cang H, Chen YY, Zhao R, Yu BM, et al. p16 gene methylation in colorectal cancers associated with Duke's staging. World J Gastroenterol. 2001;7:722–5. doi: 10.3748/wjg.v7.i5.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kratzke RA, Greatens TM, Rubins JB, Maddaus MA, Niewoehner DE, Niehans GA, et al. Rb and p16INK4a expression in resected non-small cell lung tumors. Cancer Res. 1996;56:3415–20. [PubMed] [Google Scholar]
- 37.Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Immunohistochemical overexpression of p16 protein associated with intact retinoblastoma protein expression in cervical cancer and cervical intraepithelial neoplasia. Pathol Int. 1998;48:580–5. doi: 10.1111/j.1440-1827.1998.tb03954.x. [DOI] [PubMed] [Google Scholar]
- 38.Palmqvist R, Rutegard JN, Bozoky B, Landberg G, Stenling R. Human colorectal cancers with an intact p16/cyclin D1/pRb pathway have up-regulated p16 expression and decreased proliferation in small invasive tumor clusters. Am J Pathol. 2000;157:1947–53. doi: 10.1016/S0002-9440(10)64833-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata D, Schaeffer J, Li ZH, Capella G, Perucho M. Genetic heterogeneity of the c-K-ras locus in colorectal adenomas but not in adenocarcinomas. J Natl Cancer Inst. 1993;85:1058–63. doi: 10.1093/jnci/85.13.1058. [DOI] [PubMed] [Google Scholar]
- 40.Tortola S, Marcuello E, Gonzalez I, Reyes G, Arribas R, Aiza G, et al. p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. J Clin Oncol. 1999;17:1375–81. doi: 10.1200/JCO.1999.17.5.1375. [DOI] [PubMed] [Google Scholar]
- 41.Ahnen DJ, Feigl P, Quan G, Fenoglio-Preiser C, Lovato LC, Bunn PA Jr, et al. Ki-ras mutation and p53 overexpression predict the clinical behavior of colorectal cancer: A Southwest Oncology Group study. Cancer Res. 1998;58:1149–58. [PubMed] [Google Scholar]
- 42.Kim SH, Park KH, Shin SJ, Lee KY, Kim TI, Kim NK, et al. p16 Hypermethylation and KRAS Mutation Are Independent Predictors of Cetuximab Plus FOLFIRI Chemotherapy in Patients with Metastatic Colorectal Cancer. Cancer Res Treat. 2016;48:208–15. doi: 10.4143/crt.2014.314. [DOI] [PMC free article] [PubMed] [Google Scholar]


