Abstract
The importance of the condensin complex for mitotic chromosome organization has been appreciated for over 20 years, although exactly what the chromosomal architecture assembled by condensin is, and how that structure contributes to chromosomal segregation remain largely unclear. Here, we discuss our recent findings on how fission yeast condensin mediates interactions among genes and how condensin-dependent interactions play dual roles in the chromosome territory arrangement during interphase and in mitotic chromosome organization, which supports the fidelity of chromosome segregation.
Keywords: Condensin, Fission yeast, 3D genome organization, Mitotic chromosome assembly, Chromosome segregation, Chromosome territory
Introduction
It has been widely accepted that the three-dimensional (3D) chromosome/genome organization is linked to important nuclear processes, such as gene regulation, DNA replication and repair, and mitotic chromosome segregation (Misteli 2007; Dekker and Mirny 2016). The condensin and cohesin complexes consisting of Structural Maintenance of Chromosomes (SMC) proteins are known to function in mitotic chromosome assembly and in sister-chromatid cohesion, respectively (Hirano 2006). Accumulating evidence demonstrates that these SMC complexes also function in interphase genome organizations (Wood et al. 2010; Hirano 2012). We have recently shown that a direct interaction between the TATA box-binding protein (TBP) and the Cnd2 kleisin subunit of condensin mediates the specific chromosome organization, which is required for the faithful segregation of mitotic chromosomes in fission yeast, Schizosaccharomyces pombe (Iwasaki et al. 2015a). During interphase, condensin-mediated gene interactions contribute to the formation of chromosome territories (Iwasaki et al. 2015b). Based on our recent findings, here, we discuss how the general transcription factor, TBP, and condensin coordinate global transcriptional activity with higher-order chromosome organizations, i.e., gene interactions, chromosome territory arrangement, and mitotic chromosome assembly.
Physical interaction between condensin and TBP
It has previously been shown that a kleisin subunit of condensin physically interacts with all other condensin subunits, thereby serving as a central scaffold of this protein complex (Onn et al. 2007). Fission yeast condensin comprises 5 subunits: two SMC subunits (Cut3 and Cut14) and three non-SMC subunits (Cnd1, Cnd2, and Cnd3); Cnd2 is the kleisin subunit. The C-terminal domains of the kleisin subunits in the condensin and cohesin complexes fold into a specific protein structure referred to as the winged-helix domain, which forms an interaction surface for the SMC subunit (Haering et al. 2004; Fennell-Fezzie et al. 2005). We have shown that Cnd2 interacts with TBP, and that the point mutations in the Cnd2 winged-helix domain inhibit this condensin-TBP interaction (Iwasaki et al. 2015a). Biochemical analyses with a series of Cnd2 mutant proteins further indicate that one side of the helix within the winged-helix domain mediates the Cnd2 interaction with Cut3 (Smc4 condensin subunit), while the other side of the same helix is involved in the interaction with TBP. The cnd2-C703R point mutation does not affect the Cnd2 interactions with any of the other condensin subunits, but specifically diminishes the interaction with TBP.
Condensin localization and regulation
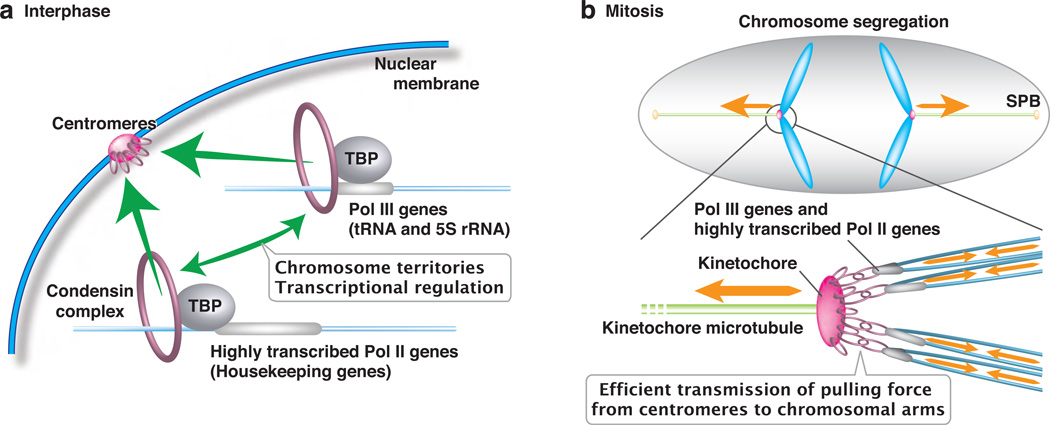
Condensin localizes to RNA polymerase III-transcribed genes (Pol III genes), such as tRNA and 5S rRNA, and highly transcribed Pol II genes (Iwasaki et al. 2015a). We have shown that condensin molecules are recruited by TBP onto these highly active genes through the Cnd2-TBP interaction (Fig. 1a; Iwasaki et al. 2015a). Condensin is loaded to centromeres through a mechanism involving kinetochore proteins (Nakazawa et al. 2008; Tada et al. 2011); CENP-B and Ku cooperatively recruit condensin to retrotransposons (Tanaka et al. 2012). These results demonstrate that distinct genetic elements rely on different factors for condensin loading.
Fig. 1.
Condensin-mediated gene interactions and their functions in fission yeast. a Schematic of the TBP-dependent condensin loading. Highly transcribed genes including Pol III genes and Pol II-transcribed housekeeping genes are bound by TBP, which recruits condensin to gene loci. Condensin in turn mediates interactions (green arrows) among highly transcribed genes present on the same chromosomes and centromeres. This genome organization is coupled to the chromosome territory arrangement and gene regulation. b Model explaining how condensinmediated gene interactions with centromeres bolsters the fidelity of chromosome segregation during mitosis.
We have shown that condensin binding to chromatin during S phase is inhibited by histone H3 lysine 56 acetylation (Tanaka et al. 2012). It also has been shown that cdc2-dependent phosphorylation of the Cut3 condensin subunit is required for the nuclear accumulation of condensin during mitosis (Sutani et al. 1999). And, aurora B-dependent phosphorylation of the Cnd2 subunit promotes its binding to the histone H2A during mitosis (Tada et al. 2011). These studies collectively demonstrate that condensin phosphorylation and epigenetic histone modifications regulate the efficiency of condensin loading and the stability of condensin binding to chromatin in a cell cycle-dependent manner.
Condensin and chromosome territory arrangement
The fission yeast condensin complex is known to have both interphase and mitotic functions (Saka et al. 1994; Aono et al. 2002). We have recently shown that condensin mediates the interactions among Pol III genes during interphase, and that Pol III genes present on the same chromosomes tend to associate via condensin-condensin interactions (Iwasaki et al. 2015b). Since condensin is recruited to many highly transcribed genes—Pol III genes and Pol II-transcribed housekeeping genes—through the condensin-TBP interaction, we predict that the intra-chromosomal interactions occur preferentially among not only Pol III genes, but also highly transcribed Pol II genes (Fig. 1a). When these intra-chromosomal gene interactions are disrupted by condensin mutations, the territorial arrangement of chromosomes is compromised (Iwasaki et al. 2015b). Therefore, we propose that condensin connects highly transcribed genes on the same chromosomes, and that these intra-chromosomal interactions contribute to the formation of chromosome territories (Fig. 1a).
In terms of cell cycle-dependent regulation, chromosome territories become most indistinct during S phase, when condensin-mediated intra-chromosomal gene interactions occur to the least degree, whereas the territories are most clearly separated during mitosis, when the interactions are promoted. Note that territory formation is coordinated with the intrachromosomal interactions throughout the cell cycle (Iwasaki et al. 2015b). These results suggest that chromosome territories are regulated during the cell cycle through the modulation of condensin-mediated intra-chromosomal gene interactions.
Condensin and transcriptional regulation
Since condensin connects highly transcribed genes, it is possible that condensin-mediated genome organization affects gene expression. In support of this premise, when interactions among Pol III genes and centromeres are disrupted by condensin mutations, and transcription of Pol III genes are enhanced (Iwasaki et al. 2010). The temperature-sensitive mutation of the sfc3 TFIIIC (Pol III transcription factor) gene, which we generated and termed sfc3-1, reduces Pol III transcription and concomitantly facilitates interactions of Pol III genes with centromeres. Similarly, inhibition of Pol III transcription by chemical treatment facilitates interactions of Pol III genes with centromeres. These results suggest that condensin-mediated interactions and gene transcription are reciprocally regulated, although detailed mechanisms remain to be explored.
Our current hypothesis is that condensin-mediated interactions force highly transcribed genes to cluster near centromeres, and that the subnuclear environment around centromeres, densely occupied by heterochromatin components, is unfavorable for optimal transcription. On the other hand, we posit that while transcription contributes to the dissociation of condensin molecules from highly transcribed genes, transcription factors such as TFIIIC and TBP remain bound to their target genes after transcriptional inhibition. Therefore, transcriptional attenuation can stabilize the binding of TBP-recruited condensin to chromatin and promote condensin-mediated gene interactions.
Mitotic chromosome assembly
Interactions among highly transcribed genes and centromeres are promoted during mitosis (Iwasaki et al. 2010; Iwasaki et al. 2015a). Centromeres are the chromosomal regions where kinetochore microtubules are attached and where the pulling force is generated. The tethering of chromosomal arm regions to centromeres consequently facilitates the transmission of physical force at centromeres to chromosomal arms (Fig. 1b). This mechanism allows chromosomes to behave as units, thereby improving the fidelity of chromosome segregation. Since highly transcribed genes, including Pol III genes and Pol II-transcribed housekeeping genes, tend to associate with centromeres, we propose that this chromosome-organizing mechanism contributes to the effective transfer of important genetic materials to daughter nuclei during mitosis.
Condensin-TBP mechanism links transcription and chromosome organization
As delineated above, condensin plays multifaceted roles in the 3D chromosome organization (interphase and mitosis), chromosome territory arrangement, gene regulation, and chromosome segregation. Since TBP is the general transcription factor (Vannini and Cramer 2012) and recruits condensin to different types of highly transcribed genes, i.e., Pol II and Pol III genes, global transcriptional activities can influence condensin-mediated genome organization. For instance, when Pol III transcription is impaired by the sfc3-1 gene mutation or chemical treatment, the intra-chromosomal gene interactions mediated by condensin and chromosome territory arrangement are concomitantly facilitated, indicating that global transcriptional activities are negatively correlated with territory formation (Iwasaki et al. 2010; Iwasaki et al. 2015b).
Transcription and mitotic chromosome assembly are also reciprocally regulated by condensin-mediated interactions. Mitotic defects caused by the condensin mutation are suppressed by inhibiting Pol III and Pol II transcription (Iwasaki et al. 2010; Sutani et al. 2015). Note that this condensin mutation is temperature-sensitive and partially impairs condensin activity. The condensin mutation compromises the interactions among highly transcribed genes and centromeres, but the transcriptional reduction restores the interactions. Since the interactions among highly transcribed gene loci and centromeres are facilitated when transcription is attenuated, global transcriptional activities play an inhibitory role in the mitotic chromosome assembly required for faithful chromosome segregation.
Concluding remarks and future perspectives
Condensin is dispensable for global chromosome compaction during mitosis, but is indispensable for mitotic chromosome organization in higher eukaryotes (Hudson et al. 2003; Belmont 2006). It remains enigmatic how condensin mediates the mitotic chromosome assembly that is required for proper chromosome segregation in mammalian cells. Our study suggests that the kleisin subunit of the human condensin I complex, hCAP-H (Cnd2 cognate), interacts with human TBP (Iwasaki et al. 2015a). As in fission yeast cells, condensin is enriched at highly transcribed genes in higher eukaryotes (Kim et al. 2013; Sutani et al. 2015). It has recently been shown that condensin mediates gene looping and activation in human cells, and that condensin mutations are significantly accumulated in various cancer types (Li et al. 2015; Leiserson et al. 2015). Further work in the fission yeast model, addressing how the condensin-TBP interaction contributes to higher-order genome organization, gene regulation, and chromosome segregation, will help elucidate the evolutionarily conserved mechanisms of how condensin organizes eukaryotic genomes and serves as a functional ligature among important cellular processes.
Acknowledgments
We thank Drs. Louise Showe and Rachel Locke for critically reading the manuscript, and Sylvie Shaffer for editorial assistance. This work was supported by the G. Harold & Leila Y. Mathers Charitable Foundation and the NIH Director’s New Innovator Award Program of the National Institutes of Health under award number [DP2-OD004348 to K.N.]. Support for Shared Resources utilized in this study was provided by Cancer Center Support Grant (CCSG) P30CA010815 to The Wistar Institute.
Reference
- Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- Belmont AS. Mitotic chromosome structure and condensation. Current opinion in cell biology. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J. 2005;24:1921–1930. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nature reviews. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Tanizawa H, Kim KD, Yokoyama Y, Corcoran CJ, Tanaka A, Skordalakes E, Showe LC, Noma K. Interaction between TBP and condensin drives the organization and faithful segregation of mitotic chromosomes. Mol Cell. 2015a;59:755–767. doi: 10.1016/j.molcel.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Corcoran CJ, Noma KI. Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle. Nucleic Acids Res. 2015b doi: 10.1093/nar/gkv1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P, Hudson DF. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun. 2013;4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, Lawrence MS, Gonzalez-Perez A, Tamborero D, Cheng Y, Ryslik GA, Lopez-Bigas N, Getz G, Ding L, Raphael BJ. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hu Y, Oh S, Ma Q, Merkurjev D, Song X, Zhou X, Liu Z, Tanasa B, He X, Chen AY, Ohgi K, Zhang J, Liu W, Rosenfeld MG. Condensin I and II Complexes License Full Estrogen Receptor alpha-Dependent Enhancer Activation. Mol Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Aono N, Hirano M, Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Sakata T, Nakato R, Masuda K, Ishibashi M, Yamashita D, Suzuki Y, Hirano T, Bando M, Shirahige K. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun. 2015;6:7815. doi: 10.1038/ncomms8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E, Noma K. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell. 2012;48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]