Abstract
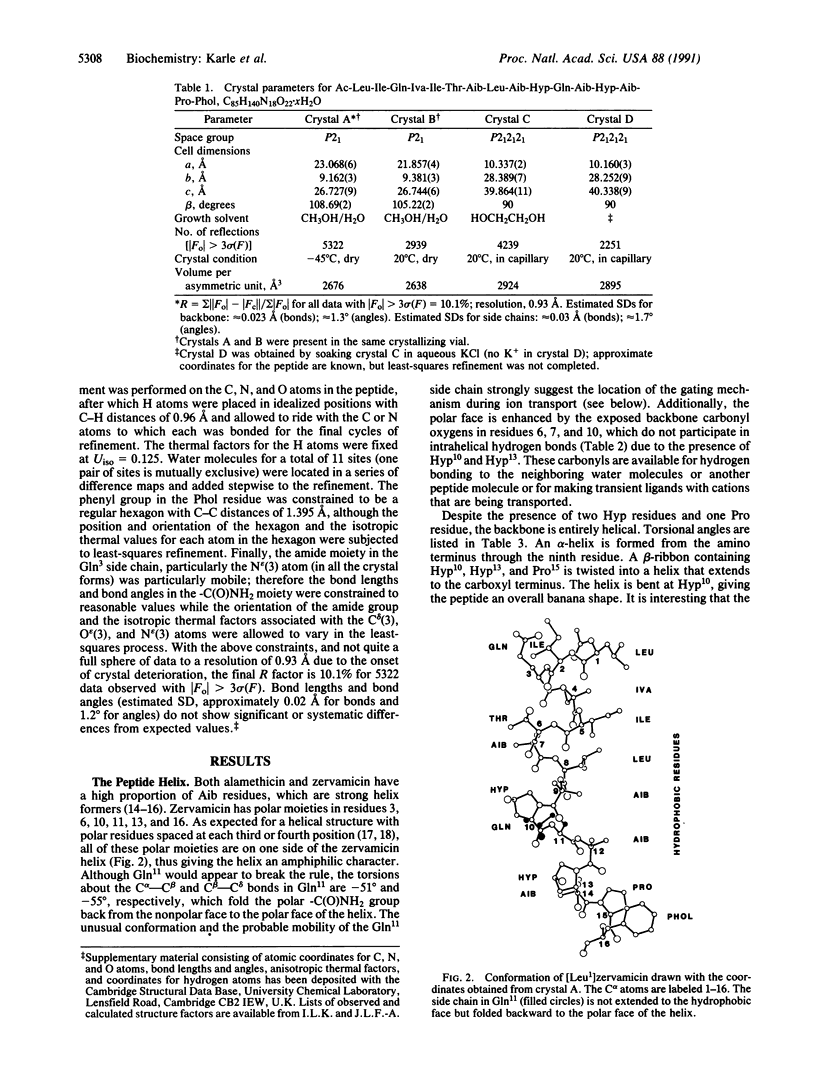
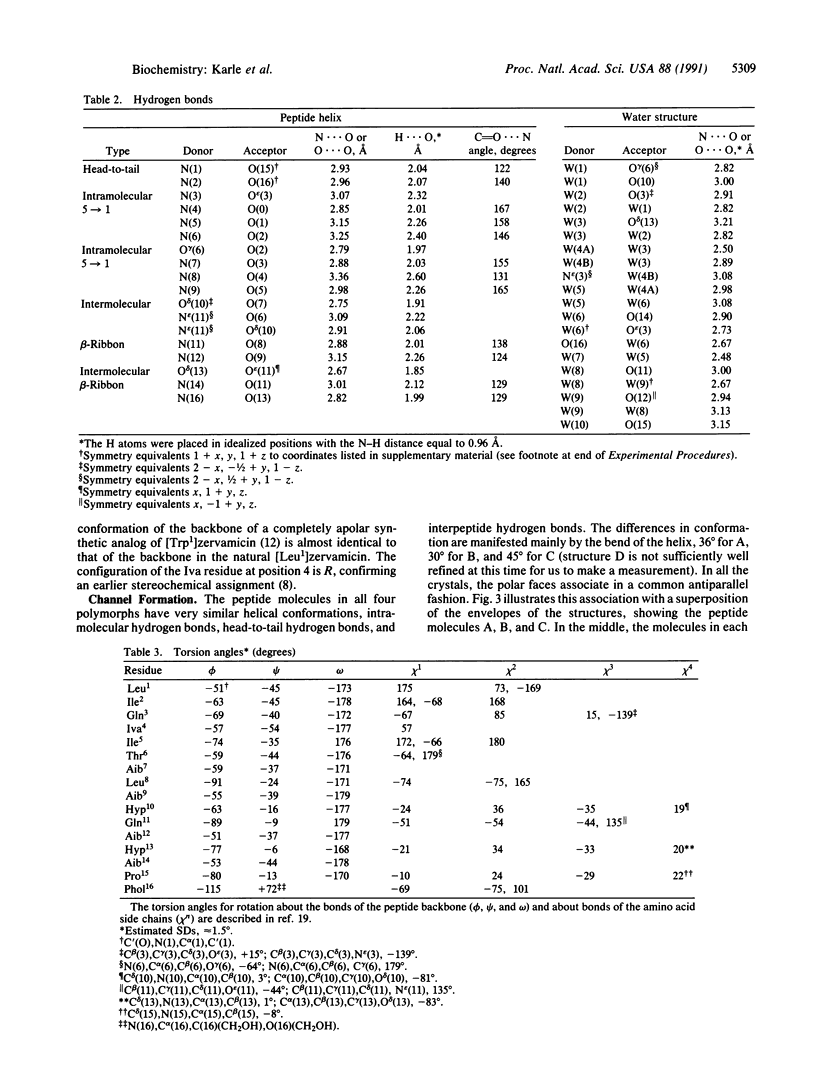
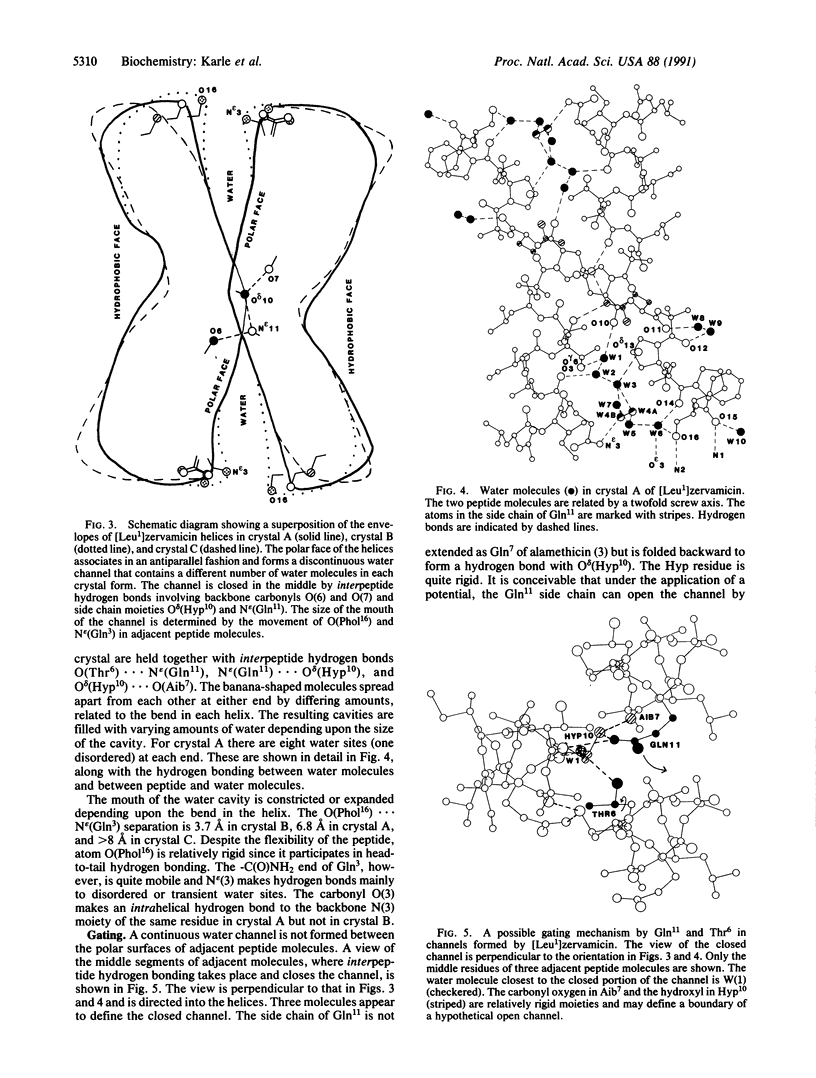
Structures in four different crystal forms of [Leu1]zervamicin (zervamicin Z-L, Ac-Leu-Ile-Gln-Iva-Ile5-Thr-Aib-Leu-Aib-Hyp10-Gln-Aib-Hyp-Aib-P ro15-Phol, where Iva is isovaline, Aib is alpha-amino isobutyric acid, Hyp is 4-hydroxyproline, and Phol is phenylalaninol), a membrane channel-forming polypeptide from Emericellopsis salmosynnemata, have been determined by x-ray diffraction. The helical structure is amphiphilic with all the polar moieties on the convex side of the bent helix. Helices are bent at Hyp10 from approximately 30 degrees to approximately 45 degrees in the different crystal forms. In all crystal forms, the peptide helices aggregate in a similar fashion to form water channels that are interrupted by hydrogen bonds between N epsilon H(Gln11) and O delta (Hyp10) of adjacent helices. The Gln11 side chain is folded in an unusual fashion in order to close the channel. Space is available for an extended conformation for Gln11, in which case the channel would be open, suggesting a gating mechanism for cation transport. Structural details are presented for one crystal form derived from methanol/water solution: C85H140N18O22. 10H2O, space group P21, a = 23.068(6) A, b = 9.162(3) A, c = 26.727(9) A, beta = 108.69(2) degrees (standard deviation of last digit is given in parentheses); overall agreement factor R = 10.1% for 5322 observed relfections [magnitude of Fo greater than 3 sigma (F)]; resolution, 0.93 A.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argoudelis A. D., Johnson L. E. Emerimicins II, 3 and IV, antibiotics produced by Emericellopsis microspora in media supplemented with trans-4-n-propyl-L-proline. J Antibiot (Tokyo) 1974 Apr;27(4):274–282. doi: 10.7164/antibiotics.27.274. [DOI] [PubMed] [Google Scholar]
- Fox R. O., Jr, Richards F. M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-A resolution. Nature. 1982 Nov 25;300(5890):325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- Hall J. E., Vodyanoy I., Balasubramanian T. M., Marshall G. R. Alamethicin. A rich model for channel behavior. Biophys J. 1984 Jan;45(1):233–247. doi: 10.1016/S0006-3495(84)84151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser E. T., Kézdy F. J. Amphiphilic secondary structure: design of peptide hormones. Science. 1984 Jan 20;223(4633):249–255. doi: 10.1126/science.6322295. [DOI] [PubMed] [Google Scholar]
- Karle I. L., Balaram P. Structural characteristics of alpha-helical peptide molecules containing Aib residues. Biochemistry. 1990 Jul 24;29(29):6747–6756. doi: 10.1021/bi00481a001. [DOI] [PubMed] [Google Scholar]
- Karle I. L., Flippen-Anderson J. L., Sukumar M., Balaram P. Parallel and antiparallel aggregation of alpha-helices. Crystal structures of two apolar decapeptides X-Trp-Ile-Ala-Aib-Ile-Val-Aib-Leu-Aib-Pro-OMe (X = Boc, Ac). Int J Pept Protein Res. 1990 Jun;35(6):518–526. [PubMed] [Google Scholar]
- Karle I. L., Flippen-Anderson J. L., Uma K., Balaram P. Helix aggregation in peptide crystals: occurrence of either all parallel or antiparallel packing motifs for alpha-helices in polymorphs of Boc-Aib-Ala-Leu-Ala-Leu-Aib-Leu-Ala-Leu-Aib-OMe. Biopolymers. 1990 Dec;29(14):1835–1845. doi: 10.1002/bip.360291414. [DOI] [PubMed] [Google Scholar]
- Karle I. L., Flippen-Anderson J., Sukumar M., Balaram P. Conformation of a 16-residue zervamicin IIA analog peptide containing three different structural features: 3(10)-helix, alpha-helix, and beta-bend ribbon. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5087–5091. doi: 10.1073/pnas.84.15.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall G. R., Hodgkin E. E., Langs D. A., Smith G. D., Zabrocki J., Leplawy M. T. Factors governing helical preference of peptides containing multiple alpha,alpha-dialkyl amino acids. Proc Natl Acad Sci U S A. 1990 Jan;87(1):487–491. doi: 10.1073/pnas.87.1.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew M. K., Balaram P. Alamethicin and related membrane channel forming polypeptides. Mol Cell Biochem. 1983;50(1):47–64. doi: 10.1007/BF00225279. [DOI] [PubMed] [Google Scholar]
- Menestrina G., Voges K. P., Jung G., Boheim G. Voltage-dependent channel formation by rods of helical polypeptides. J Membr Biol. 1986;93(2):111–132. doi: 10.1007/BF01870804. [DOI] [PubMed] [Google Scholar]
- Miller C. Genetic manipulation of ion channels: a new approach to structure and mechanism. Neuron. 1989 Mar;2(3):1195–1205. doi: 10.1016/0896-6273(89)90304-8. [DOI] [PubMed] [Google Scholar]
- Mueller P., Rudin D. O. Action potentials induced in biomolecular lipid membranes. Nature. 1968 Feb 24;217(5130):713–719. doi: 10.1038/217713a0. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H., Nilsson L., Rigler R., Voges K. P., Jung G. Structural fluctuations of a helical polypeptide traversing a lipid bilayer. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5067–5071. doi: 10.1073/pnas.85.14.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]


