Abstract
The human APOBEC3G (A3G) enzyme restricts HIV-1 in the absence of the viral accessory protein viral infectivity factor (Vif) by deaminating viral cDNA cytosines to uracils. These uracil lesions base-pair with adenines during the completion of reverse transcription and result in A3G signature G-to-A mutations in the viral genome. Vif protects HIV-1 from A3G-mediated restriction by forming an E3-ubiquitin ligase complex to polyubiquitinate A3G and trigger its degradation. Prior studies indicated that Vif may also directly block the enzymatic activity of A3G and, provocatively, that Vif derived peptides, Vif 25–39 and Vif 105–119, are similarly inhibitory. Here, we show that Vif 25–39 does not inhibit A3G enzymatic activity and that the inhibitory effect of Vif 105–119 and that of a shorter derivative Vif 107–115, although recapitulated, is non-specific. We also elaborate a simple method for assaying DNA cytosine deaminase activity that eliminates potential PCR-induced biases. Our results show that these Vif-derived peptides are unlikely to be useful as tools to study A3G function or as leads for the development of future therapeutics.
Keywords: DNA cytosine deamination, Evolvability, HIV-1, Mutation, Vif-derived peptides
Graphical Abstract
Introduction
APOBEC3G (A3G) is a human innate immune effector enzyme with demonstrated potent activity against a wide variety of exogenous and endogenous parasitic elements (reviewed by [1–4]). Most studies have focused on the mechanism of A3G-mediated restriction of HIV-1. In the absence of viral countermeasures, A3G packages into nascent viral particles and then, during the early stages reverse transcription in susceptible target cells, A3G deaminates viral cDNA cytosines to uracils. These uracil lesions template the insertion of adenine bases during subsequent viral DNA synthesis and result in viral genomic strand G-to-A mutations. If unhindered, A3G and related enzymes (A3D, A3F, and A3H) can combine to convert up to 10% of viral cDNA cytosines to uracils in a single round of virus replication, effectively inactivating HIV-1 by hypermutation (lethal mutagenesis) [5, 6].
The viral infectivity factor (Vif) of HIV-1 preserves viral genomic integrity by neutralizing A3G and related APOBEC3 enzymes (A3D, A3F, and A3H). Vif heterodimerizes with the transcription co-factor CBF-β, recruits a CULLIN-RING ubiquitin ligase complex, directly binds these APOBEC3 enzymes, and triggers their polyubiquitination and degradation (reviewed by [1–4]). However, many studies have indicated that A3G-mediated hypermutation and Vif-mediated counteraction are at opposing ends of a wide spectrum of outcomes and that in reality, in infected patients in vivo, is likely to lie somewhere in between. First, the viral genomic strand is depleted of guanines (enriched for adenines) resulting in nucleobase proportions that differ from human genomic DNA, suggesting continuous evolutionary pressure from APOBEC3 enzymes. Second, many common drug-resistance mutations are G-to-A changes at APOBEC3-preferred sites [7, 8]. Particularly noteworthy are deep-sequencing studies showing that preexisting drug resistance mutations could be attributed to A3G even in the presence of a fully functional Vif counter-defense [9]. Additional deep-sequencing studies compared APOBEC3-preferred sites in cell culture experiments with the viral mutation spectra in patient-derived viral sequences and found evidence for APOBEC3-mediated immune escape [10].
The aforementioned observations support the idea that HIV-1 may be “addicted” to the pro-mutagenic activity of the APOBEC3 enzymes in order to maintain high mutation rates and evade potent adaptive immune responses [11]. Thus, in addition to the obvious goal of inhibiting Vif and promoting lethal mutagenesis (therapy by hypermutation), an alternative therapeutic strategy may be blocking APOBEC3 mutagenesis and thereby starving HIV-1 of necessary genetic variation (therapy by hypomutation) [11]. One approach to APOBEC3 inhibition is in developing peptides or peptidomimetics of regions of Vif that directly bind to A3G and related APOBEC3 enzymes. In support of this idea, HIV-1 Vif expression in E. coli was reported to inhibit the activity of the AID, which is an APOBEC3-related DNA deaminase essential for antibody gene diversification [12]. HIV-1 Vif has also been shown to block the single-stranded (ss)DNA deaminase activity of A3G in cells and in viral particles [13–15]. These observations motivated screening of Vif-derived peptides for A3G inhibitory activity [14, 16]. Two peptides spanning Vif regions 25–39 and 105–119 showed low micromolar inhibitory activities in biochemical assays and even evidence for A3G antagonism in living cells [14–18].
Given these promising activities, we aimed to use Vif-derived peptides as lead molecules for therapeutic development. However, we did not find significant A3G inhibitory activity for Vif peptide 25–39 in our assays, in contrast to the small molecule A3G inhibitors that have previously identified [19, 20]. Moreover, although Vif peptide 105–119 and a shorter derivative spanning Vif residues 107–115 showed reproducible A3G inhibitory activity, this proved to be non-specific as an amino acid scrambled derivative of Vif peptide 107–115 still showed similar activity suggesting a general non-specific inhibitory mechanism. In light of the A3G-interacting surfaces of Vif, now appreciated to span several discontinuous secondary structures [21–24], we conclude that these Vif-derived peptides are unlikely to be useful for further development as A3G inhibitors.
Results
Vif peptide screen
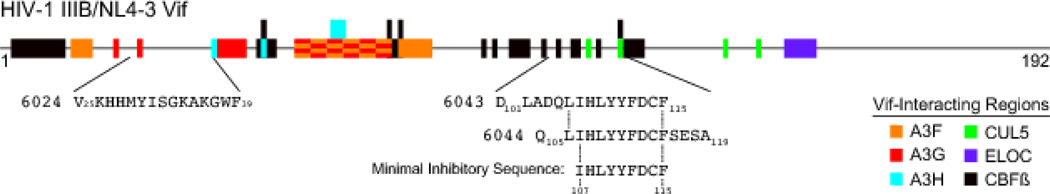
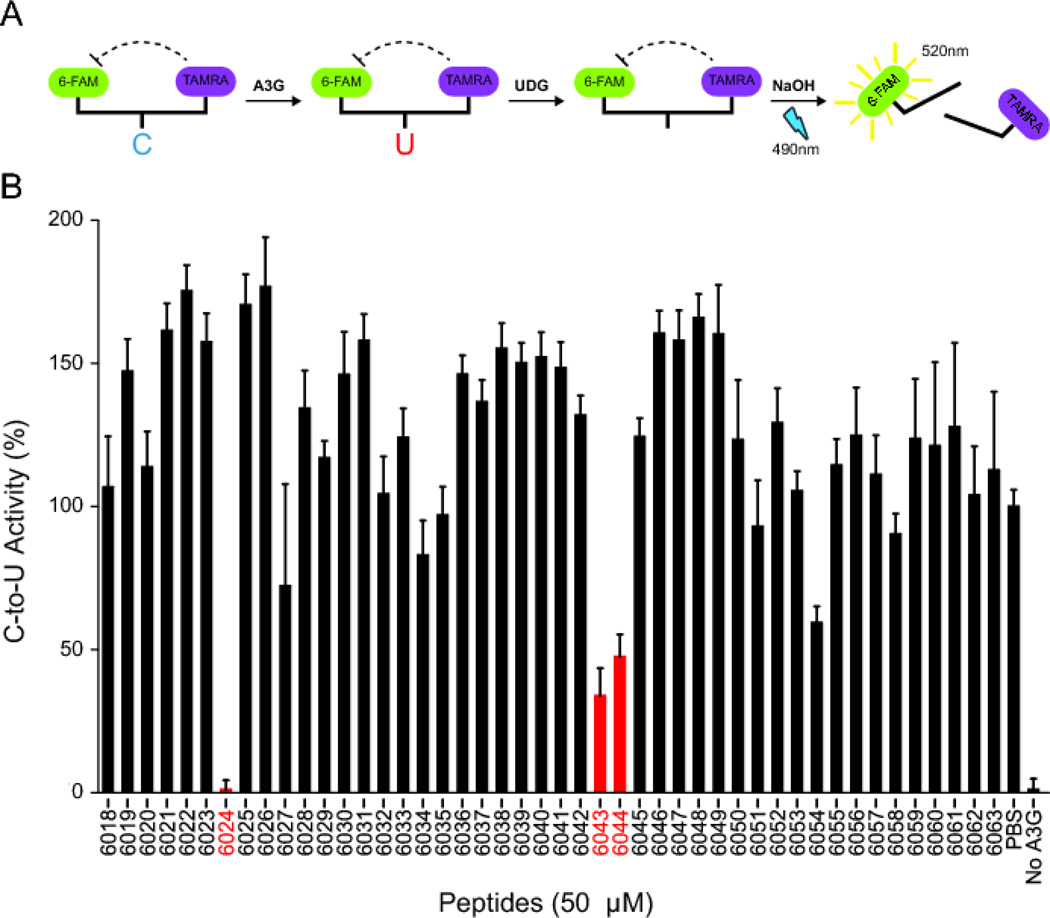
HIV-1 Vif has been subjected to extensive mutation experiments that have defined regions required for interaction with host cellular proteins including the APOBEC3s (A3F, A3G, A3H), CBF-β, and ubiquitin ligase components (CUL5, and ELOC) (Figure 1) (reviewed by [1–4]). A previous screen of Vif peptides identified several with A3G inhibitory activity [14, 16]. We started our studies by attempting to reproduce this screen using the same library (HIV-1 lab strain NL4-3 Vif peptide library Cat#6446), a fluorescence-based DNA deaminase activity assay [19], and A3G expressed in the context of a soluble extract from HEK-293 cells (assay schematic in Figure 2A). The majority of Vif peptides failed to significantly alter the activity of A3G in this assay; however, three elicited strong and reproducible inhibition (Figure 2B). Vif peptide 6024 (H-VKHHMYISGKAKGWF-OH), which spans residues 25–39, showed the strongest inhibition (>10-fold), and overlapping Vif peptides 6043 (H-DLADQLIHLYYFDCF-OH) and 6044 (H-QLIHLYYFDCFSESA-OH), which span residues 101–115 and 105–119, respectively, showed weaker inhibition. These data indicate that two, non-overlapping regions of Vif have A3G inhibition activity, consistent with prior studies [14, 16, 18].
Figure 1. HIV-1 IIIB Vif schematic depicting relevant interacting regions and peptides.
Please see the text for details.
Figure 2. Effects of Vif-derived peptides on the single-stranded DNA deaminase activity of cellular A3G.
(A) Schematic of the fluorescence-based ssDNA deaminase activity assay. 6-FAM fluorescence is quenched by TAMRA in intact substrate ssDNA. The concerted action of ssDNA cytosine deamination by A3G, uracil excision by uracil DNA glycosylase (UDG), and abasic site cleavage by a hydroxide ion releases the 6-FAM from TAMRA quench and yields a strong fluorescence signal.
(B) A3G activity in the presence of 50 µM of the indicated 15-mer Vif peptides. Reactions with 293T cell extract lacking A3G (no A3G) or containing A3G with solvent only (PBS) are shown as controls. Each histogram bar reports the mean A3G activity ± SEM for 2 independent experiments.
Vif25–39 does not inhibit A3G ssDNA cytosine deaminase activity
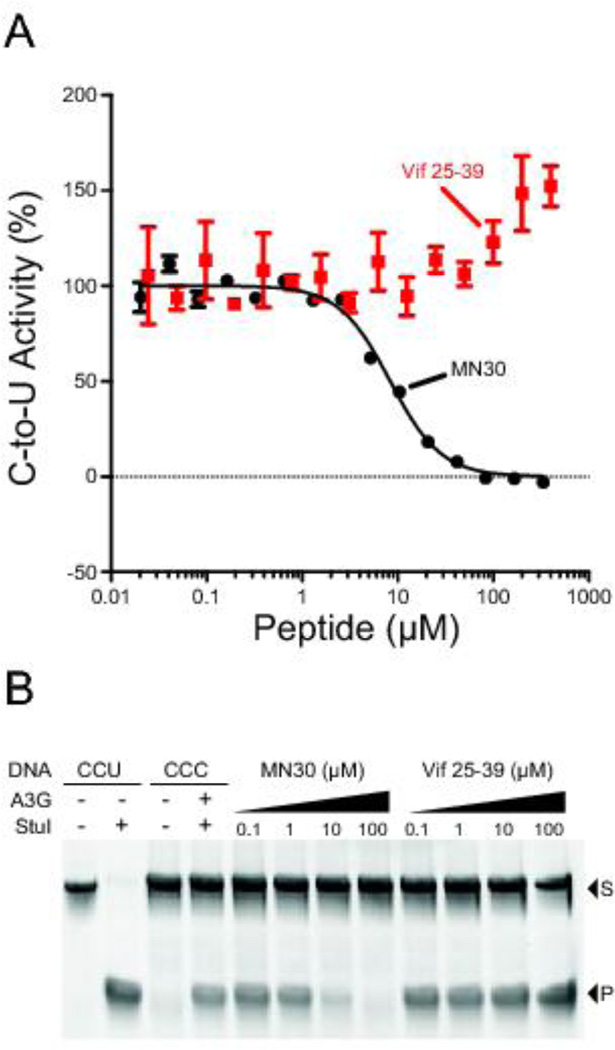
Vif peptide 25–39 overlaps with previously defined A3G-interacting regions (Figure 1) [16, 22, 25–27]. To further investigate the inhibitory potential of this peptide, it was synthesized on solid-phase, purified by reverse-phase HPLC, and characterized by analytical HPLC for purity (99% pure) and the structure validated by mass spectrometry (Table 1). The resulting peptide was then tested in vitro against A3G-myc-His purified from 293T cells using a fluorescently labeled ssDNA substrate with a single A3G-preferred 5'CCC target site. In comparison to the small molecule MN30 used as a positive control [19], previously published data indicating a 0.6 µM IC50 for Vif 25–39 [14], and our own screening data above, the pure Vif peptide 25–39 showed no inhibitory activity and may even stimulate A3G activity at higher concentrations (Figure 3A).
Table 1.
Sequence, purity, and mass spectrometry data for peptides used in this study.
| Peptide Name |
Peptide Sequence* | Mass Calc’d (m/z) |
Mass Found (m/z) |
Purity (215nm / 254nm) |
|---|---|---|---|---|
| Vif25–39 | H-VKHHMYISGKAKGWF-OH | 895.5 (M+2H) |
895.8 (M+2H) |
99% / 99% |
| Vif107–115 | H-IHLYYFDCF-OH | 1220.5 (M+H) |
1220.8 (M+H) |
99% / 97% |
| Vif107-115r | H-FCDFYYLHI-OH | 1220.5 (M+H) |
1220.8 (M+H) |
96% / 97% |
| Vif107-115s | H-ICFLHYDFY-OH | 1220.5 (M+H) |
1220.9 (M+H) |
99% / 90% |
| Purity (220nm)* | ||||
| 6024 | H-VKHHMYISGKAKGWF-OH | 895.5 (M+2H) |
894.56 (M+2H) |
88.8% |
| 6043 | H-DLADQLIHLYYFDCF-OH | 1875.7 (M+H) |
1875.14 (M+H) |
94.2% |
| 6044 | H-QLIHLYYFDCFSESA-OH | 1875.7 (M+H) |
1836.07 (M+H) |
82.3% |
Sequences shown are written left to right: N-terminus → C-terminus. “H-“ designates a free N-terminal amine on the peptide. “-OH” designates the peptide has a C-terminal carboxylic acid.
Purity and mass spectrometry analyses for peptides 6024, 6043, and 6044 were done by Fisher BioServices Inc as part of contracted quality control by the AIDS Reagent Program.
Figure 3. Vif peptide 25–39 does not inhibit A3G ssDNA deaminase activity.
(A) Representative dose response experiment for A3G activity in the presence of the indicated concentration of Vif peptide 25–39. Each data point represents the mean level of A3G activity ± SEM in the fluorescence-based ssDNA deaminase assay (n = 3). Data for the small molecule MN30 are shown as a positive control for inhibition.
(B) A3G activity in the presence of the indicated concentration of Vif25–39 using the PCR/restriction-based assay to quantify ssDNA deaminase activity. MN30 is again used as a positive control for inhibition (S, substrate; P, product). This gel image is representative of >3 independent experiments.
To eliminate the possibility of an assay-specific effect, we repeated the experiment using a PCR and restriction-based ssDNA deaminase assay used previously [28] and still found no inhibitory effect (Figure 3B). Finally, the peptide was again synthesized and purified in-house, as well as ordered from a commercial source, and in both cases again no A3G inhibition was observed. We conclude that Vif peptide 25–39 does not inhibit the ssDNA cytosine deaminase activity of A3G. We note that published results [14] as well our own screening results (Figure 2B) may be due to an impurity because Vif peptides from the AIDS Reagent Program vary in purity from 82.3% to 94.2% according to specifications documentation (Table 1).
Vif-derived peptide 107–115 non-specifically inhibits A3G ssDNA deaminase activity
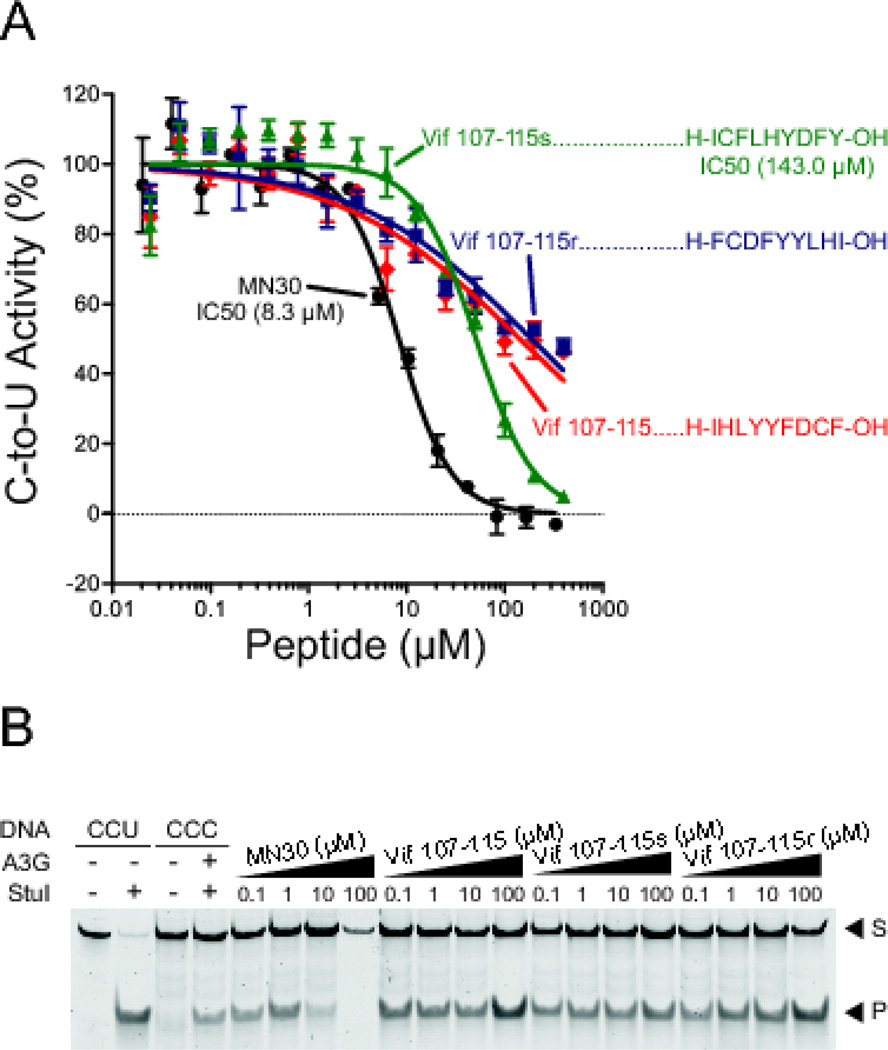
We then turned our attention to Vif peptide 107–115, which is defined by the overlapping region between AIDS Reagent Program peptides 6043 and 6044 and previously shown to similarly inhibit A3G ssDNA deaminase activity [18] (Figure 1). Utilizing the procedure described above, this peptide was synthesized on solid-phase and characterized as >97% pure (Table 1). It was then tested against recombinant A3G-myc-His using both the fluorescence-based ssDNA deamination assay [19] and the PCR and restriction-based assay [28] (Figure 4). With the fluorescence-based assay, Vif peptide 107–115 showed weak inhibitory activity (IC50 > 100 µM), which was considerably less than the positive control MN30 and a recently reported value for the same peptide (IC50 = 0.1 µM [18]). With the PCR and restriction-based assay, Vif peptide 107–115 showed no reproducible inhibitory effect. This differential outcome may be due to the fact that the latter assay is semi-quantitative and potentially susceptible to amplification biases from the PCR procedure.
Figure 4. Vif peptide 107–115 inhibits A3G but this activity is non-specific.
(A) Dose response studies of A3G activity in the fluorescence-based assay with the indicated concentrations of Vif derived peptides or derivatives as described in the main text (red, Vif 107–115; green, Vif 107–115 scrambled; dark blue, Vif 107–115 reverse). Data for the small molecule MN30 are shown as a positive control for inhibition.
(B) A3G activity in the presence of the indicated peptides using the PCR/restriction-based assay to quantify ssDNA deaminase activity. MN30 is again used as a positive control for inhibition (S, substrate; P, product). This gel image is representative of >3 independent experiments.
We next asked whether Vif peptide 107–115 inhibited A3G in a sequence dependent manner. This was done by synthesizing scrambled and reverse versions of Vif 107–115 peptide and testing those peptides in both the fluorescence- and PCR/restriction-based DNA cytosine deaminase assays. With both peptides in both assays, similar weak inhibitory effects were observed (Figure 4A & B). To corroborate these data, the peptides were again synthesized, characterized for purity and correct mass, then retested, ultimately yielding similar results. We conclude that Vif peptide 107–115 is a non-specific inhibitor of A3G ssDNA deaminase activity, most likely by making general hydrophobic interactions with the enzyme.
Order of addition ssDNA binding studies
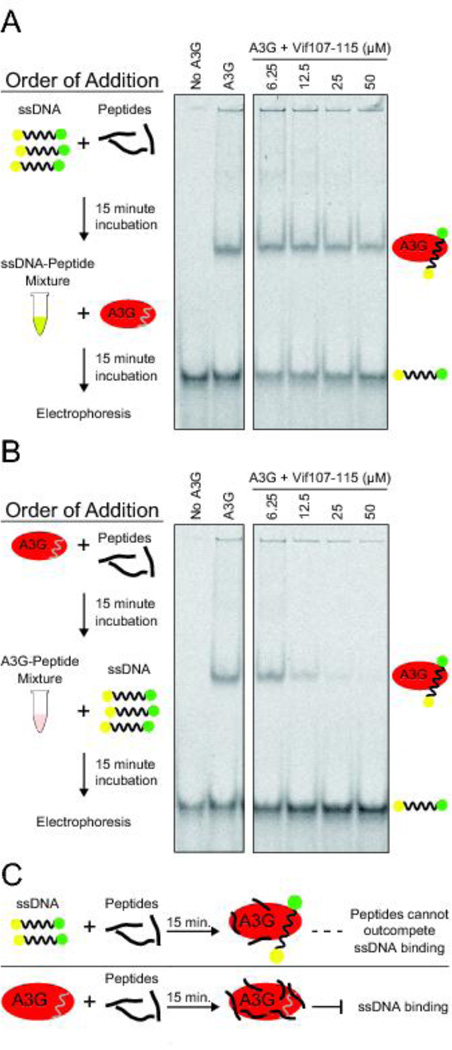
We next performed a series of electrophoretic mobility shift assays (EMSA) to ask whether Vif peptide 107–115 blocks A3G activity by preventing the enzyme from binding to ssDNA substrates. A3G has strong ssDNA binding activity, which is mostly governed by the N-terminal pseudo-catalytic domain with a relatively modest contribution from the C-terminal catalytic domain [19, 28–33]. The initial round of experiments was done by first mixing ssDNA with peptide, incubating 15 minutes, adding enzyme, and then fractionating the complexes on native polyacrylamide gels (EMSA; Figure 5A). This approach yielded uniformly negative data. However, we next wondered if order of addition makes a difference, and first mixed A3G with peptide, incubated 15 minutes to optimize opportunities for inhibitor binding, added ssDNA, and then fractionated the complexes on native polyacrylamide gels (Figure 5B). In this scenario, Vif peptide 107–115 prevented A3G from binding ssDNA in a dose dependent manner. Given the non-specific inhibition mechanism indicated above, one possible explanation for this phenomenon is that hydrophobic peptides like Vif 107–115, given sufficient time, will coat an enzyme non-specifically and result in steric occlusion of potential ssDNA substrates (Figure 5C). In contrast, if stable A3G-ssDNA complexes are first allowed to form then the peptide will not have a chance to bind and will be too weak to outcompete natural ssDNA substrates (Figure 5C).
Figure 5. Reagent addition order influences the ssDNA binding activity of A3G.
(A) Representative EMSA showing the ssDNA binding activity of A3G in reactions in which Vif-derived peptides were first incubated 15 minutes with ssDNA and then A3G was added for an additional 15 minute incubation period, followed by native gel fractionation and imaging to detect free ssDNA (lower band) and ssDNA/A3G complexes (upper band).
(B) Representative EMSA showing the ssDNA binding activity of A3G in reactions in which Vif-derived peptides were first incubated 15 minutes with A3G and then ssDNA was added for an additional 15 minute incubation period, followed by native gel fractionation and imaging to detect free ssDNA (lower band) and ssDNA/A3G complexes (upper band).
Discussion
In this study we pursued the idea that the ssDNA cytosine deaminase activity of A3G may be inhibited by Vif-derived peptides. Prior studies reported sub-micromolar inhibitory activities for Vif peptide 25–39 and Vif peptide 105–119, as well as inhibitory activity for derivative Vif peptide 107–115 [14–18]. Although high concentration screening with crude peptide mixtures initially confirmed these results, subsequent experiments with freshly synthesized and rigorously characterized material were either completely negative (Vif 25–39) or weakly positive through a non-specific mechanism (Vif 107–115). Thus, further development of these Vif-derived peptides for therapeutic inhibition of A3G is unlikely to be fruitful.
However, although the present studies are discouraging, they will hopefully caution others from going down similar paths. In addition, our studies may help to advance the overall concept of inhibiting A3G by exploiting the evolutionarily optimized interaction with HIV-1 Vif. Rapid progress in structural studies of the APOBEC3-Vif interaction is already yielding clues for the rational design of non-linear Vif peptides (e.g., artificially joining polypeptide sequences from different structural regions) [21–24]. However, the success of such structure-guided approaches will ultimately require high-resolution knowledge of the different APOBEC3-Vif interfaces and, although individual structures of APOBEC3 enzymes and of the Vif/CBF-β ligase complex now exist, structures of the larger macromolecular complex with both interaction partners have proven elusive.
Finally, it is noteworthy that APOBEC3 inhibition could be both an effective anti-viral strategy as well as an effective anti-cancer strategy. Recent studies have demonstrated that APOBEC signature mutations are the second largest source of somatic mutation in cancer (2nd only to the “ageing signature” caused by spontaneous, water-mediated deamination of methyl-cytosine nucleobases in CpG motifs) [34–37]. Moreover, some SIV Vif variants are potent inhibitors of human APOBEC3B [38], which is likely the major enzyme responsible for APOBEC signature mutations in cancer. Thus, the overall strategy suggested here of splicing together non-linear Vif peptide epitopes and developing peptidomimetics may still prove useful in the long-term to block APOBEC3B activity, slow rates of tumor evolution, and improve the durability of existing cancer therapies.
Materials & Methods
A3G expression and purification
A3G-mycHis was purified from stably or transiently transfected HEK293T cells using the C-terminal hexahistidine tags, as described [19, 39]. Cells were maintained in DMEM (Invitrogen) with 10% FBS (Hyclone), 50 units/ml penicillin and 50 µg/ml streptomycin (Invitrogen) in 37 °C and 5% CO2. Transfections were done with TransIT-LTI (Mirus Bio). 48 hrs post-transfection, cells were harvested and lysed in 25 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1 mM ZnCl2, 1 mM EDTA, 0.5% Triton X-100, and 10% glycerol. Insoluble materials were removed from crude cell lysates by centrifugation (14,000 rpm, 10 min) and A3G was purified from supernatants.
DNA deaminase activity assays and peptide screens
The fluorescence-based DNA cytosine deamination assay was used to monitor A3G activity in 293T cell lysates, as described [19], and was adapted as follows for work with purified proteins. Recombinant human A3G-mycHis was diluted with 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 10% glycerol, 0.5% triton X-100, 1 mM PMSF (MP Biomedicals), 1 µg/ml Aprotinin (Sigma), 1 µg/ml Leupeptin (MP Biomedicals), and 1 µg/ml Pepstatin A (Fisher Scientific) to working concentrations. 15 µl of enzyme was incubated with 10 pmol ssDNA substrate 5′-6-FAM-AAA-CCC-AAA-GAG-AGA-ATG-TGA-TAMRA-3′ (Biosearch Technologies, Inc.) for the initial peptide screen and 5′-6-FAM-AAATAT-CCC-AAA-GAG-AGA-TAMRA-3′ (Biosearch Technologies, Inc.) was used for follow-up dose response experiments. In both reaction series, ssDNA deamination products were then treated with 0.02 units of UDG (NEB) diluted in 15 µl of 50 mM Tris-Cl, pH 7.4, 10 mM EDTA for 2 hrs at 37 °C in Nunc 384-well black plates. 3 µl of 4 N NaOH was added, followed by mixing and incubating at 37 °C for another 30 min. 3 µl of 4 N HCl and 37 µl of 2 M Tris-Cl (pH 7.9) was then added for neutralization, and the relative deaminase activity was quantified by reading fluorescence with excitation at 490 nm and emission at 520 nm on Synergy Mx Monochromator-Based Multi-Mode (BioTek Instruments, Inc.) or LJL Analyst AD (LJL BioSystems, Inc.) microplate readers. UDG assays omitted the deaminase and used ssDNA substrate with a single uracil in place of the A3G-preferred cytosine (5′-6-FAM-AAACCUAAA-GAG-AGA-ATG-TGA-TAMRA-3′). All compound stocks were suspended in 10 mM DMSO and diluted as indicated. HTS used 0.04 µM A3G, 0.33 µM ssDNA substrate, 10 µM compound, and 0.02 units UDG (NEB). IC50 values were determined using GraphPad Prism 6.0 software.
Electrophoretic Mobility Shift Assays (EMSA)
A3G-mycHis was incubated with ssDNA substrate 5′-biotin-ATT-ATT-ATT-ATT-CCA-ATG-GAT-TTA-TTT-ATT-TATTTA-TTTATT-T-fluoroescein-3′ in 50 mM Tris-Cl, pH 7.8, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.5 mM ZnCl2, and 10% glycerol at 37 °C for 30 min with varying concentrations of compounds in DMSO. The reaction products were subjected to native TBE PAGE. A3G-ssDNA complexes were visualized at 463 nm using a Fuji FLA-5000 fluorescent image analyzer.
PCR-based ssDNA deamination assay
A3G deamination reactions were performed as described [28]. A3G-catalyzed DNA deamination of 5′-AGGCCC-3′ results in 5′-AGGCCU-3′, which after PCR amplification is converted to 5′-AGGCCT-3′ which is the recognition site for Eco147I/StuI restriction digestion. 10 nM A3G was incubated with 1 nM ssDNA (5′-GGATTGGTTGGTTATTTGTTTAAGGAAGGTGGATTAAAGGCCCAATAAGGTGATGGAAGTTATGTTTGGTAGATTGATGG-3′, Integrated DNA Technologies) in a total volume of 10 µl in 25 mM Tris-Cl, pH 7.0 and 0.1 mg/ml BSA at 37° C for 30 min. The reaction was terminated by heating to 95°C for 5 min. 1 µl of the reaction mixture was used for PCR amplification in a 20 µl reaction volume with 1X PCR buffer containing 1.5 mM MgCl2, 1 µM forward 5′-GGATTGGTTGGTTATTTGTTTAAGGA-3′ and reverse 5′-CCATCAATCTACCAAACATAACTTCCA-3′ primers, 0.2 mM dNTP, and 1 unit of Choice Taq DNA polymerase (Denville Scientific Inc.). The PCR program was 1 cycle at 95° C for 3 min followed by 15 cycles of annealing at 61°C for 30s and denaturing at 94°C for 30s. 10 µl of each PCR product was incubated with Eco147I/StuI restriction enzyme (NEB) for 1 hr at 37 °C. Oligo containing CCU instead of CCC was used as positive control for deamination. Restriction-reaction products were loaded onto 14% polyacrylamide gels and separated by electrophoresis in 1X TBE. Gels were stained with SYBR gold nucleic acid stain (Invitrogen) diluted 1:10,000 in 0.5X TBE and imaged in Typhoon FLA7000 laser scanner (GE).
Solid-Phase Peptide Synthesis (SPPS)
Automated SPPS was performed using a Protein Technologies PS3 peptide synthesizer as reported [40]. Pre-loaded resins (H-Phe-2-ClTrt-Resin [200–400 mesh] 0.69 meq g−1; H-Ile-2-ClTrt-Resin [100–200 mesh] 0.67 meq g−1; H-Tyr(tBu)-2-ClTrt-Resin [100–200 mesh] 0.67 meq g−1) purchased from Peptides International were used for SPPS. Protected resin-bound peptides were cleaved from resin using a solution (10 ml) of trifluoroacetic acid : distilled water : triisopropyl silane : ethane dithiol (85 : 5 : 5 : 5) and precipitated with cold diethyl ether (30 ml). The white precipitate was isolated by centrifugation (4000 rpm, 5 min, 25 °C). The crude peptide was purified via semi-preparative HPLC chromatography using distilled and deionized H2O (with 0.1% TFA) and an increasing gradient of MeCN (with 0.1% TFA) on a Zorbax SB-C18 column (21.2 × 250 mm, 7 µm, Agilent Technologies). After purification, the peptide was analyzed by analytical HPLC for purity (215 and 254 nm) and characterized by electrospray ionization (ESI) mass spectroscopy (MS) in positive ion mode on an Agilent 1100 Series LC/MSD Ion Trap instrument. See Table 1 for the purities of individual peptides and their calculated and observed masses.
Synthetic oligonucleotides and purchased peptides
The sequence of the 80-mer ss-deoxyoligonucleotide substrate used in the deamination assays is: 5'-GGATTGGTTGGTTATTTGTTTAAGGAAGGTGGATTAAAGGCCCAATAAGGTGATGGAAGTTATGTTTGGTAGATTGATGG-3' (the A3G-preferred target cytosine is underlined). The positive control ssDNA has the same sequence except the target C is replaced by a U. The following primers were used for PCR amplification of the substrate and positive control nucleotides:
| Forward | 5'-GGATTGGTTGGTTATTTGTTTAAGGA-3'; |
| Reverse | 5'-CCATCAATCTACCAAACATAACTTCCA-3' |
Vif-derived peptides were obtained through the NIH AIDS Research and Reference Reagent Program as lyophilized powderd and were subsequently dissolved in water or a DMSO solution (Vif peptide set Cat#6446; http://www.aidsreagent.org/support_docs/6446_lot_140319_and_12539_lot_140222_Con_B_Vif_Analytical_Data.pdf). Peptides were also purchased from GL Biochem (Shanghai, China) (data from these peptides are not shown, but available upon request).
HIV-1 IIIB/NL4-3 Vif Sequence (GenBank EU541617)
MENRWQVMIVWQVDRMRIRTWKSLVKHHMYISGKAKGWFYRHHYESTHPRISSEVHIPLGDARLVITTYWGLHTGERDWHLGQGVSIEWRKKRYSTQVDPDLADQLIHLYYFDCFSESAIRNAILGHIVSPRCEYQAGHNKVGSLQYLALAALITPKKIKPPLPSVTKLTEDRWNKPQKTKGHRGSHTMNG
Highlights.
HIV-1 Vif is a natural APOBEC3G antagonist.
Published Vif-derived peptides lack specific APOBEC3G inhibitory activity.
Vif peptide 25–39 is simply non-inhibitory.
Vif peptide 107–115 inhibits A3G in vitro but is non-specific.
Acknowledgments
We thank the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, for the HIV-1 Consensus B Vif Peptide Set. This research was supported by grants from the National Institute of General Medical Sciences (NIGMS P01-GM091743 to RSH and R01-GM110129 to DAH) and the National Institute for Allergy and Infectious Disease (NIAID R37-AI064046 to RSH). Partial salary support for CR was provided by the Pharmacology Graduate Program and subsequently by a NIAID training grant (T32-AI83196; IMV Training Program). RSH is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- A3G
APOBEC3G
- AID
activation-induced deaminase
- CBF-β
core-binding factor beta
- G-to-A
guanine-to-adenine
- HPLC
high-performance liquid chromatography
- HTS
high-throughput screening
- IC50
half maximal inhibitory concentration
- MS
mass spectrometry
- PCR
polymerase chain reaction
- ssDNA
single-stranded DNA
- SPPS
Solid-phase peptide synthesis
- UDG
uracil DNA glycosylase
- Vif
virion infectivity factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
DAH and RSH are co-founders, shareholders, and consultants of ApoGen Biotechnologies, Inc.
References
- 1.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480C:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 6.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haché G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- 8.Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris RS. Enhancing immunity to HIV through APOBEC. Nat Biotechnol. 2008;26:1089–1090. doi: 10.1038/nbt1008-1089. [DOI] [PubMed] [Google Scholar]
- 12.Santa-Marta M, Aires da Silva F, Fonseca AM, Rato S, Goncalves J. HIV-1 Vif protein blocks the cytidine deaminase activity of B-cell specific AID in E. coli by a similar mechanism of action. Mol Immunol. 2007;44:583–590. doi: 10.1016/j.molimm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, et al. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britan-Rosich E, Nowarski R, Kotler M. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. J Mol Biol. 2011;410:1065–1076. doi: 10.1016/j.jmb.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowarski R, Wilner OI, Cheshin O, Shahar OD, Kenig E, Baraz L, et al. APOBEC3G enhances lymphoma cell radioresistance by promoting cytidine deaminase-dependent DNA repair. Blood. 2012;120:366–375. doi: 10.1182/blood-2012-01-402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reingewertz TH, Britan-Rosich E, Rotem-Bamberger S, Viard M, Jacobs A, Miller A, et al. Mapping the Vif-A3G interaction using peptide arrays: a basis for anti-HIV lead peptides. Bioorganic & medicinal chemistry. 2013;21:3523–3532. doi: 10.1016/j.bmc.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowarski R, Prabhu P, Kenig E, Smith Y, Britan-Rosich E, Kotler M. APOBEC3G inhibits HIV-1 RNA elongation by inactivating the viral trans-activation response element. J Mol Biol. 2014;426:2840–2853. doi: 10.1016/j.jmb.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prabhu P, Shandilya SM, Britan-Rosich E, Nagler A, Schiffer CA, Kotler M. Inhibition of APOBEC3G activity impedes double-stranded DNA repair. The FEBS journal. 2016;283:112–129. doi: 10.1111/febs.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, et al. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson ME, Li M, Harris RS, Harki DA. Small-molecule APOBEC3G DNA cytosine deaminase inhibitors based on a 4-amino-1,2,4-triazole-3-thiol scaffold. ChemMedChem. 2013;8:112–117. doi: 10.1002/cmdc.201200411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 22.Letko M, Booiman T, Kootstra N, Simon V, Ooms M. Identification of the HIV-1 Vif and human APOBEC3G protein interface. Cell Rep. 2015;13:1789–1799. doi: 10.1016/j.celrep.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards C, Albin JS, Demir O, Shaban NM, Luengas EM, Land AM, et al. The binding interface between human APOBEC3F and HIV-1 Vif elucidated by genetic and computational approaches. Cell Rep. 2015;13:1781–1788. doi: 10.1016/j.celrep.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, et al. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol. 2016;90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albin JS, Hache G, Hultquist JF, Brown WL, Harris RS. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol. 2010;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans SL, Schon A, Gao Q, Han X, Zhou X, Freire E, et al. HIV-1 Vif N-terminal motif is required for recruitment of Cul5 to suppress APOBEC3. Retrovirology. 2014;11:4. doi: 10.1186/1742-4690-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 29.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3' -->5' on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 30.Shindo K, Li M, Gross PJ, Brown WL, Harjes E, Lu Y, et al. A Comparison of two single-stranded DNA binding models by mutational analysis of APOBEC3G. Biology. 2012;1:260–276. doi: 10.3390/biology1020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, et al. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nature chemistry. 2014;6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwatani Y, Chan DSB, Wang F, Maynard KS, Sugiura W, Gronenborn AM, et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Research. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Nanoscale structure and dynamics of ABOBEC3G complexes with single-stranded DNA. Biochemistry. 2012;51:6432–6440. doi: 10.1021/bi300733d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Land AM, Wang J, Law EK, Aberle R, Kirmaier A, Krupp A, et al. Degradation of the cancer genomic DNA deaminase APOBEC3B by SIV Vif. Oncotarget. 2015;6:39969–39979. doi: 10.18632/oncotarget.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins AL, Peterson KL, Beito TG, Flatten KS, Kaufmann SH, Harki DA. Synthesis of a peptide-universal nucleotide antigen: towards next-generation antibodies to detect topoisomerase I-DNA covalent complexes. Org Biomol Chem. 2016;14:4103–4109. doi: 10.1039/c5ob02049b. [DOI] [PMC free article] [PubMed] [Google Scholar]