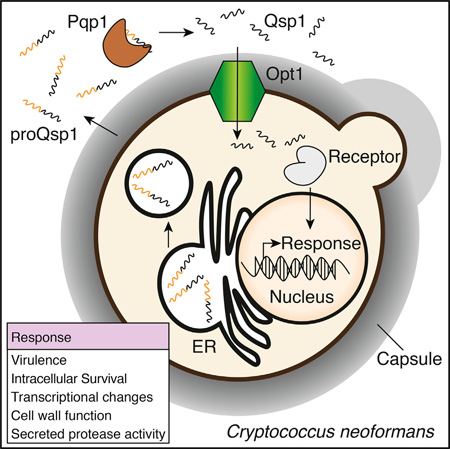
SUMMARY
Quorum sensing (QS) is a bacterial communication mechanism in which secreted signaling molecules impact population function and gene expression. QS-like phenomena have been reported in eukaryotes with largely unknown contributing molecules, functions, and mechanisms. We identify Qsp1, a secreted peptide, as a central signaling molecule that regulates virulence in the fungal pathogen Cryptococcus neoformans. QSP1 is a direct target of three transcription factors required for virulence, and qsp1Δ mutants exhibit attenuated infection, slowed tissue accumulation, and greater control by primary macrophages. Qsp1 mediates autoregulatory signaling that modulates secreted protease activity and promotes cell wall function at high cell densities. Peptide production requires release from a secreted precursor, proQsp1, by a cell-associated protease, Pqp1. Qsp1 sensing requires an oligopeptide transporter, Opt1, and remarkably, cytoplasmic expression of mature Qsp1 complements multiple phenotypes of qsp1Δ. Thus, C. neoformans produces an autoregulatory peptide that matures extracellularly but functions intracellularly to regulate virulence.
Graphical Abstract
INTRODUCTION
Cryptococcus neoformans is the most common cause of fungal meningitis. This species causes 950,000 cases and 625,000 deaths annually, an estimated 40% of all HIV-related mortality (Armstrong-James et al., 2014). C. neoformans is a member of a pathogen species complex that includes C. neoformans var. grubii (serotype A) and C. neoformans var. neoformans (serotype D). We study C. neoformans var grubii, which is responsible for ~90% of infections in HIV/AIDS patients. Investigations of C. neoformans pathogenesis have focused on three virulence traits: melanin production, growth at human body temperature, and production of a polysaccharide capsule.
Many bacterial pathogens require autoregulatory quorum sensing (QS) systems for virulence, which are regarded as attractive therapeutic targets. In classical bacterial QS paradigms, cells secrete a signal and monitor its concentration to sense cell number and/or diffusion. Bacteria use QS to regulate diverse biological functions (Bassler and Miller, 2013). In Gram-negative bacteria, the most common secreted signals are acyl homoserine lactones (AHLs) (Schuster et al., 2013). Gram-positive bacteria generally utilize peptides (Ng and Bassler, 2009).
QS-like phenomena in fungi have been proposed based on the effects of conditioned media (e.g., Albuquerque and Casadevall, 2012; Albuquerque et al., 2014). However, QS systems with specialized, species-specific biosynthetic pathways and specific reception mechanisms akin to those of bacteria have not yet been elucidated in any eukaryote. A candidate for a bona fide QS signal in C. neoformans was identified in studies of a mutant C. neoformans serotype D strain lacking the co-repressor Tup1 (Lee et al., 2007). The authors observed that tup1Δ strains, but not wild-type strains, failed to grow when a low number of cells were plated. Purification of the activity from tup1Δ culture supernatants revealed an 11 amino acid peptide that was able to rescue this defect. They suggested that this peptide could be part of a QS system in C. neoformans and named it Qsp1 (Quorum Sensing-Like Peptide 1). Unfortunately, no phenotype for a qsp1Δ strain was reported, and the reason tup1Δ strains require Qsp1 is not known. A similar low-density plating defect was not observed in a serotype A strain lacking TUP1 (Lee et al., 2009). Thus, the normal function of Qsp1 remained unknown.
RESULTS
QSP1 Is a Direct Target of Three Virulence Regulators
We reasoned that virulence factors would be enriched in the regulatory targets of Gat201, a key transcription factor required for virulence (Chun et al., 2011; Liu et al., 2008). Our previous ChIP-on-chip studies of Gat201 identified important targets (Chun et al., 2011). To develop an extended network, we employed ChIP-seq (Figure 1A). Previous studies suggested genes whose promoters are bound by more than one transcription factor regulating a particular process often share that function (Beyhan et al., 2013; Nobile et al., 2012; Pérez et al., 2013). Thus, we selected Gat204 and Liv3, transcription factors previously identified as Gat201 targets and implicated in virulence (Chun et al., 2011; Liu et al., 2008), for ChIP-seq in tissue culture conditions.
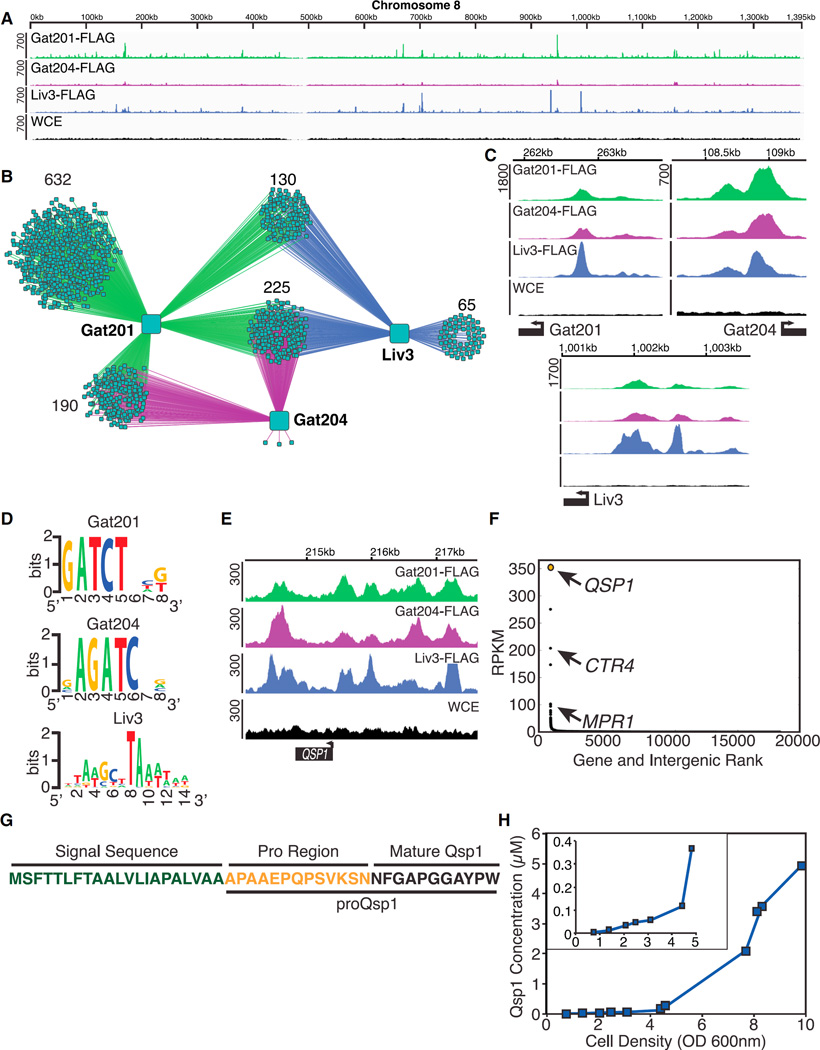
Figure 1. QSP1 Is a Direct Target of the Gat201-Gat204-Liv3 Regulatory Network.
(A) ChIP-seq data from chr8. y axis: number of reads.
(B) Network diagram.
(C) Gat201, Gat204, and Liv3 binding at each other’s promoter.
(D) Binding motifs derived from ChIP-seq datasets.
(E) ChIP-Seq data at the QSP1 promoter.
(F) Rank and RPKM for each gene in a wild-type strain grown in tissue cultures conditions.
(G) Predicted Qsp1 precursor.
(H) Qsp1 accumulation in wild-type culture supernatants. Inset: Qsp1 accumulation at lower cell densities.
Gat201, Gat204, and Liv3 control a transcription factor network, sharing characteristics of published regulatory networks (Hughes and de Boer, 2013; Sorrells and Johnson, 2015). Gat201, Gat204, and Liv3 targets (Table S1) overlap significantly (p < 10−300 by chi-square test), with 225 “central targets” shared by all three regulators (Figure 1B). RNA-seq analysis indicates 42% of binding events in the network cause a change in target transcript levels (Figures S1A and S1B and Table S1). Each transcription factor binds its own promoter and the promoter of the other two regulators (Figure 1C). De novo motif finding using the ChIP-seq data (Figure 1D) shows that Gat201 and Gat204 have typical GATA family transcription factor specificities (Ko and Engel, 1993) (Figure S1). Liv3, a Wor1-ortholog, produced a DNA binding motif reminiscent of the C. albicans Wor1 motif (Lohse et al., 2010) (Figure S1C).
A prominent central target, QSP1 is a gene encoding a small secreted peptide (Lee et al., 2007) (Figures 1E and S1D). RNA-seq analysis revealed that QSP1 produces the most abundant transcript in cells grown in tissue culture conditions (Figure 1F). QSP1 is predicted to encode a peptide precursor containing a signal sequence and a 24 amino acid pro-peptide (proQsp1). The C-terminal 11 amino acids of proQsp1 encode the mature Qsp1 peptide (Lee et al., 2007) (Figure 1G). To determine whether high amounts of the peptide are produced, we raised polyclonal rabbit antisera against Qsp1 (Figure S1E). Qsp1 is detected in wild-type culture supernatants by immunoblotting, producing a signal that comigrates with the synthetic peptide (Figure S1F). We developed a quantitative ELISA, which revealed that the peptide accumulates to micromolar concentrations in culture supernatants as a function of cell density (Figure 1H).
QSP1 Is Required for Virulence
As a highly expressed, central target of the Gat201-Gat204-Liv3 network, QSP1 was a candidate virulence factor. We assessed whether qsp1Δ mutants display a growth defect in vitro and observed no defects in the growth of qsp1Δ cells on media containing oxidative, nitrosative, pH, iron starvation, osmotic, or cell wall stressors (Figures S1G and S1H). Next, we assessed virulence of two independent qsp1Δ mutants using an intranasal infection model in A/J mice (Chun and Madhani, 2010; Lim et al., 1980). We found that qsp1Δ mutants are significantly attenuated (Figure 2A). Since A/J mice have a C5 complement deficiency (Nilsson and Müller-Eberhard, 1967), we tested whether qsp1Δ mutants were also attenuated in C57Bl/6 mice (Huffnagle et al., 1998). We obtained similar results in this mouse background (Figure S2A).
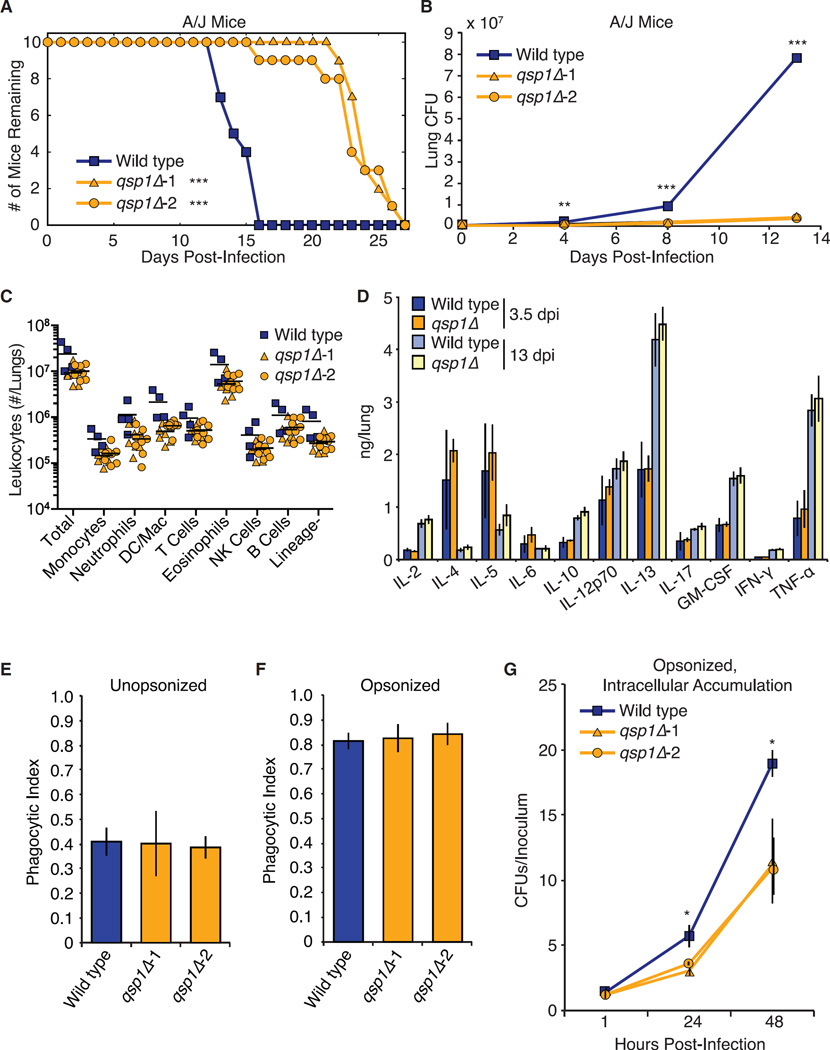
Figure 2. Qsp1 Is Required for Virulence and Accumulation within Macrophages.
(A) Survival analysis. 10 A/J mice infected per genotype. **p < 10−4 (log-rank test).
(B) Lung burden analysis. 9 A/J mice infected per genotype. **p < 10−3, ***p < 10−4 (t test).
(C) Leukocyte recruitment. 4 C57Bl/6 mice infected with wild-type, 8 C57Bl/6 mice infected per qsp1Δ strain.
(D) Cytokine analysis. 10 C57Bl/6 mice infected per genotype. Error bars are SD.
(E and F) Phagocytic index of unopsonized (E) and opsonized (F) C. neoformans. >1,200 BMDMs quantified per genotype. Error bars are SD.
(G) Intracellular accumulation. CFUs from BMDM cell lysates, normalized to starting inoculum. Error bars represent 95% confidence intervals constructed by bootstrapping. *p < 0.05 (bootstrap).
In C. neoformans, virulence-attenuated mutants fall into several classes. One group comprises mutants deficient in early pulmonary accumulation. Another group corresponds to mutants that accumulate normally in the lung but are deficient in dissemination. To differentiate between these two groups, we quantified the cryptococcal viable cell counts (CFUs) in lung homogenates obtained from mice infected with either wild-type or qsp1Δ strains. qsp1Δ mutants accumulated to approximately 3-fold fewer CFUs at day 4 after infection and to 20-fold fewer CFUs at day 13 (Figure 2B). These defects were not due to a deficiency in initial seeding of the tissue (Figure S2B). We examined CNS infection using an intracisternal infection model in immunosuppressed rabbits (Perfect et al., 1980). We detected no difference in cerebrospinal fluid CFUs between wild-type and qsp1Δ (Figure S2C).
Cryptococcal infections that result in a Th2-dominant response tend to favor C. neoformans (Herring et al., 2004; Piehler et al., 2011). Th1-dominant responses more effectively clear the fungus (Zhang et al., 2009). We therefore examined whether there is an altered inflammatory response during qsp1Δ infection. We quantified leukocytes recruited to lungs after infection with wild-type or qsp1Δ strains using flow cytometry-based profiling (Figure 2C). Consistent with the lower pulmonary burden found after infection with qsp1Δ mutants, there were fewer leukocytes recruited. The majority were eosinophils, indicating a Th2-polarized infection (Akuthota et al., 2008). We assessed the level of cytokines in lungs and found no differences between wild-type and mutant infections (Figure 2D). Type 2 cytokines (IL-4, IL-5, and IL-13) (Herring et al., 2004) dominated at 3.5 days after infection. IL-13 was the most abundant cytokine 13 days after infection. These data are consistent with studies of T cell polarization during wild-type C. neoformans infections (Qiu et al., 2013; Zhang et al., 2010).
We examined the interaction between the qsp1Δ mutant and macrophages, the most abundant immune cells in the alveolar space (Gordon and Read, 2002). Hosts susceptible to cryptococcal infection display decreased survival when depleted of macrophages (Osterholzer et al., 2009; Shao et al., 2005). Macrophages also contribute to cryptococcal dissemination (Coelho et al., 2014). We measured phagocytosis rates of wild-type and qsp1Δ strains using bone marrow-derived macrophages (BMDMs) as described previously (Chun and Madhani, 2010; Nicola and Casadevall, 2012) (Figure S2D). We observed no difference between wild-type and qsp1Δ mutants when cryptococcal cells were unopsonized (Figures 2E and S2E) or opsonized with a monoclonal antibody against the capsule (Figures 2F and S2E). We reasoned that qsp1Δ cells may have altered fitness after phagocytosis in the high-density environment of the phagolysosome and measured the ability of the qsp1Δ mutant to accumulate within macrophages after phagocytosis (Figure S2F). Wild-type cells accumulate intracellularly significantly faster than qsp1Δ cells, which displayed approximately 2-fold fewer CFUs 24 and 48 hr after infection (Figure 2G). This observation raised the possibility that phagosome maturation may be altered in macrophages infected with the qsp1Δ mutant. However, colocalization studies revealed no impact of QSP1 on the level of association between cryptococcal cells and acidic macrophage compartments (Figure S2G).
Intercellular Signaling by Qsp1
We serendipitously observed that qsp1Δ mutants form dry, wrinkled colonies at room temperature, in contrast to smooth, mucoid colonies produced by wild-type (Figure 3A). This colony phenotype provided an opportunity to assay Qsp1 function. We developed a confrontation assay in which recipient strains were grown as colonies near a patch of donor strain cells. When qsp1Δ cells are confronted by a wild-type patch, they grow to produce smooth colonies (Figure 3B). We reasoned that this effect was due to Qsp1 diffusing from the donor. Indeed, synthetic Qsp1 was sufficient to cause qsp1Δ cells to produce smooth colonies (Figure S3A). To determine whether this effect was specific, we synthesized and tested the predicted proQsp1 and pro region peptides. We also synthesized a peptide in which the 11 amino acids of Qsp1 are scrambled (Scrambled Qsp1). We found that proQsp1 is sufficient to complement the qsp1Δ colony phenotype, but the pro region peptide and scrambled Qsp1 are not (Figure S3A). To assess structure-function relationships in Qsp1, we synthesized mutant peptides, including all possible single amino acid substitutions to alanine/glycine for large side chains or to tryptophan for small side chains and N- and C-terminal truncations of Qsp1 (Table S8). None of the truncations display colony phenotype-altering activity (data not shown). All but three positions in Qsp1 are sensitive to substitution (Figure S3B).
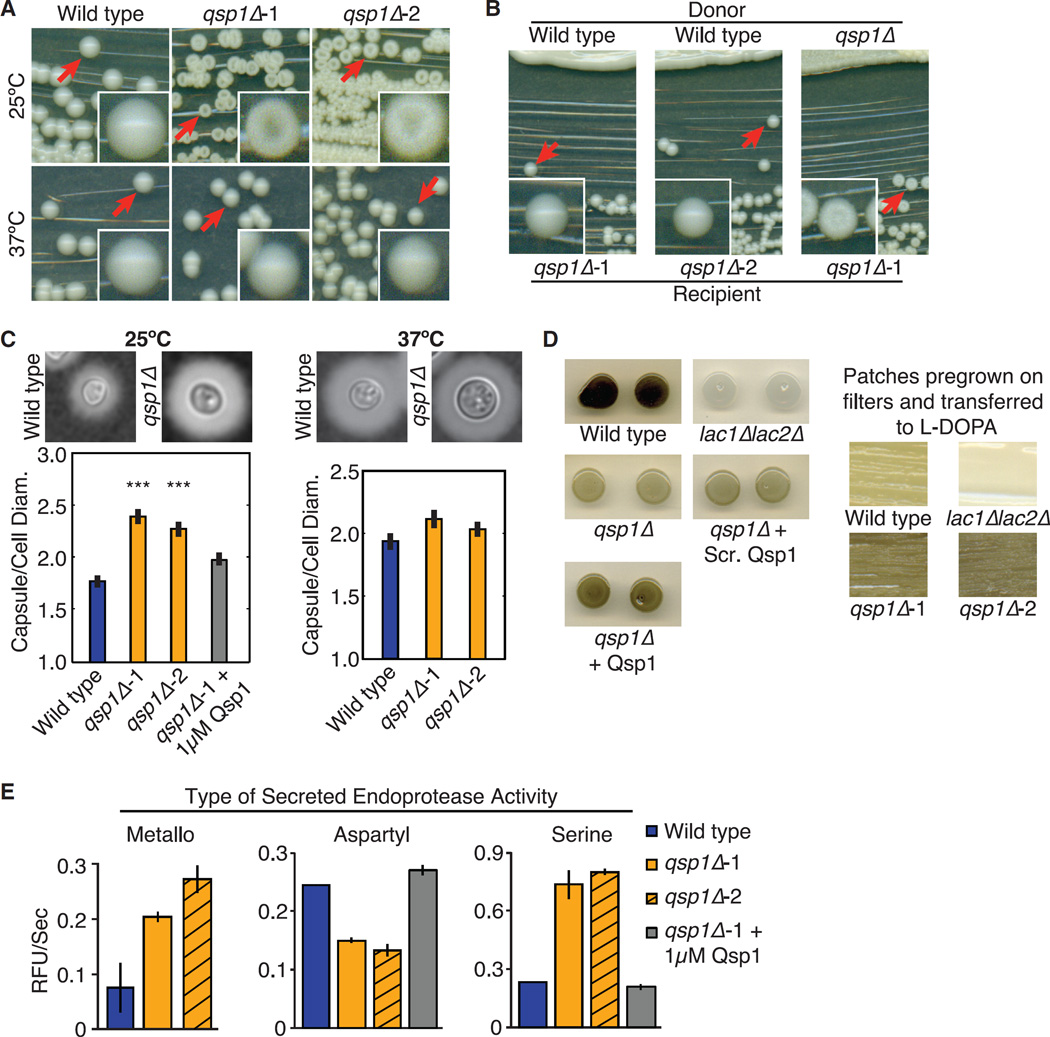
Figure 3. Impact of Qsp1 on Virulence-Related Phenotypes.
(A) Colony morphology.
(B) Complementation assays.
(C) Capsule quantification. Cryptococcal capsule assessed using India ink. ***p < 10−5 (t test). Error bars are SE > 100 cells per genotype quantified.
(D) Melanin assay. Left: cells grown in liquid culture with or without synthetic peptides before spotting. Right: patches grown on filters and transferred to L-DOPA plates.
(E) Secreted protease activities. Error bars are SD.
Modulation of Canonical Virulence Factors by Qsp1
Other cryptococcal mutants have been reported to form dry colonies, mostly mutants lacking capsule (Chang and Kwon-Chung, 1994, 1998) or mutants that are hyphal (Walton et al., 2006). However, qsp1Δ cells grow as yeasts and stain normally with an anti-capsular monoclonal antibody (mAb339) (data not shown). Therefore, we subjected wild-type and qsp1Δ cells to capsule-inducing conditions and quantified capsule size. At 37°C, qsp1Δ mutants formed capsules that were similar to wild-type. At room temperature, qsp1Δ mutant cells had larger capsules than wild-type (Figures 3C and S3C).
Cell wall-associated melanin is another well-studied virulence factor of C. neoformans. The lac1Δlac2Δ mutant lacks both laccase enzymes responsible for producing melanin and therefore remains light-colored on L-DOPA plates. We found that qsp1Δ mutants displayed altered melanization at 37°C, and the phenotype depended on the condition of cultures used to inoculate the assay. Using cells from saturated cultures, we observed that qsp1Δ cells are hypomelanized. This defect could partially be rescued by addition of synthetic Qsp1. Conversely, qsp1Δ cells grown on filters on solid medium and then transferred to L-DOPA plates are hypermelanized (Figure 3D). lac1Δ cells are attenuated for virulence, and mouse lungs infected with this mutant show altered leukocyte recruitment and cytokine levels (Qiu et al., 2012). As the qsp1Δ mutant does not display such changes, it is unlikely that qsp1Δ’s attenuated virulence is due solely to changes in melanization.
Qsp1 Impacts Multiple Secreted Endoprotease Activities
A secreted metalloprotease was shown to be required for cryptococcal invasion of the central nervous system (Vu et al., 2014). We have recently identified a secreted aspartyl protease required for cryptococcal virulence (S.C.C., P.A. Dumesic, C.M.H., A.J. O’Donoghue, L. Pallova, P. Majer, H.D.M., and C.S.C., unpublished data). In work to be described elsewhere, we developed fluorogenic peptide substrates (Figure S3D) to assay secreted serine, aspartyl, and metallo endoprotease activities in C. neoformans. We identified the genes encoding each activity (S.C.C., P.A. Dumesic, C.M.H., A.J. O’Donoghue, L. Pallova, P. Majer, H.D.M., and C.S.C., unpublished data). Then, we examined the impact of Qsp1 on these activities in C. neoformans. In wild-type culture supernatants from tissue culture conditions, secreted metalloprotease and serine endoprotease activities dominate. Conversely, secreted aspartyl endoprotease activity dominates when cultures are grown in minimal media. qsp1Δ mutants have decreased secreted aspartyl endoprotease activity in minimal media and increased secreted serine endoprotease and metalloprotease activities in tissue culture conditions compared to wild-type. Growing cultures with synthetic Qsp1 restored serine and aspartyl endoprotease activities (Figure 3E).
Qsp1 Regulates the Transcriptome at High Cell Density
To determine whether Qsp1 regulates gene expression, we performed a low-coverage RNA-seq experiment comparing wild-type and qsp1Δ cells grown in log-phase and saturated culture conditions. We observed 100 genes differentially expressed in qsp1Δ compared to wild-type saturated cultures but only five differentially expressed genes in log-phase cultures (Figure 4A and Table S2). This indicates a density-dependent impact on the transcriptome. We performed a high-coverage RNA-seq experiment comparing saturated wild-type and qsp1Δ cultures and identified 1,749 differentially expressed genes. This set of Qsp1-regulated genes is enriched for genes that encode proteins of the cell wall/extracellular proteome of C. neoformans (Figure 4B) (Eigenheer et al., 2007). Also, many QSP1-regulated genes are known or predicted to be involved in cell wall biogenesis or regulation (Figure S4A).
Figure 4. Qsp1 Has Density-Dependent Impacts on Gene Expression and Cell Wall Integrity.
(A) Density-dependent RNA-seq. Expression profiling for qsp1Δ and wild-type log-phase and saturated cultures. The number of differentially expressed genes (padj < 0.05 by DESeq) is shown.
(B) Overlap of QSP1-regulated genes with published cell wall and secreted proteome (Eigenheer et al., 2007). QSP1-dependent genes are 2.3-fold enriched (p < 10−3 by hypergeometric distribution).
(C) Transmission electron microscopy.
(D and E) Quantification of cell wall width (D) and percentage of cells with visible cell wall layers (E). **p < 0.001, *p < 0.01 (t test). Error bars are SD.
(F) Stress assays. Cultures were incubated with stressors and plated for viability. The concentration where qsp1Δ viability is most different from wild-type is shown in red.
(G) LIV3-dependent genes. Expression profiling qsp1Δ and qsp1Δliv3Δ saturated cultures grown with or without synthetic Qsp1. Differentially expressed genes shown, with genes differentially expressed in qsp1Δ versus wild-type in yellow.
(H) LIV3 suppression of the qsp1Δ dry colony phenotype.
qsp1Δ Mutants Display Density-Dependent Cell Wall Defects
We assessed cell wall structure of wild-type and qsp1Δ cells by electron microscopy as described previously (O’Meara et al., 2013) (Figure 4C). We observed that qsp1Δ cells display significantly thinner cell walls than wild-type (Figure 4D). Previous studies have shown that wild-type cell walls are organized into distinct layers, typical of fungi (Doering, 2009; Free, 2013). The number and width of layers is dynamic and condition dependent (Sakaguchi et al., 1993; Shepardson and Cramer, 2013). We observed distinct layers in 90% of the wild-type cell walls examined. In contrast, we observed layers in about 60% of qsp1Δ cell walls (Figure 4E).
As described, qsp1Δ mutants do not display plating defects on agar containing the cell wall perturbants Calcofluor White, caffeine, or SDS. However, we reasoned that this assay would not capture phenotypes that are specific to high cell density, since single cells are deposited. Instead, we analyzed cell wall function in dense conditions by incubating saturated cultures with cell wall stressors and examining the viability of cells after incubation by plating. qsp1Δ mutants display a loss of viability relative to wild-type when exposed to high concentrations of SDS and caffeine (Figure 4F). This phenotype could be rescued if cells were grown with synthetic Qsp1 before the assay. In contrast, exponentially growing cultures display no difference in viability between the two genotypes. We did not observe increased killing of saturated qsp1Δ cultures upon acidified nitrite or hydrogen peroxide treatment (Figures S4B and S4C). Since the qsp1Δ capsule and cell wall phenotypes are reminiscent of mutants lacking chitin or chitosan, we quantified chitin and chitosan content of cell walls but found that wild-type and qsp1Δ mutants display similar profiles (Figure S4D).
A Transcription Factor Functions Downstream of Qsp1
Of the 100 most-changed transcripts in qsp1Δ cells, we observed a significant enrichment for regulatory targets of the transcription factor Liv3 (Figure S4E). To test whether Liv3 mediates some or all of the Qsp1 response, we constructed a qsp1Δliv3Δ deletion mutant. We performed RNA-seq of qsp1Δ and qsp1Δliv3Δ both grown to saturation in either the presence or absence of synthetic Qsp1. We identified 851 differentially expressed genes in qsp1Δ mutants grown in the presence of synthetic Qsp1 compared to qsp1Δ mutants grown in the absence of synthetic Qsp1 (Figure 4G and Table S2). These strongly overlap with genes identified when wild-type and qsp1Δ cells are compared. In contrast, we observed only 282 differentially expressed genes in the qsp1Δliv3Δ mutant grown with synthetic Qsp1 compared to without (Figure 4G). Thus, Liv3 mediates a majority (66%) of the transcriptional response to Qsp1.
We assessed whether liv3Δ mutants share the dry colony phenotype of qsp1Δ and found that liv3Δ mutants have a dry colony phenotype at room temperature (data not shown). The phenotype is most pronounced at 30°C (Figure S4F). liv3Δ mutants form smooth colonies when grown at 37°C (Figure S4F). Supporting a downstream role for Liv3, liv3Δ mutants cannot be complemented by a nearby patch of wild-type cells (Figure S4G) or by the synthetic Qsp1 peptides (Figure S4H). In addition, expression of Liv3 using the copper-regulated pCTR4 promoter (Ding et al., 2013) bypasses the dry colony phenotype of qsp1Δ cells (Figure 4H).
Forward Genetic Screen for Mutants in the QSP1 Pathway
We sought additional components of the Qsp1 signaling pathway. We attempted to construct a deletion mutant for each of the 1,010 direct targets of Gat201, Gat204, and/or Liv3. We screened the 705 mutants successfully created for a dry colony phenotype. We profiled melanin, capsule, and cell morphology for the nine mutants identified (Table S3). One of the mutants is liv3Δ. We screened an additional 1,407 mutants produced while constructing a deletion collection for C. neoformans (2,112 strains constructed and screened). Of these, we identified six additional mutants with the dry colony phenotype (Table S3). These include three known acapsular mutants (cap10Δ, cap60Δ, and cap64Δ; Chang and Kwon-Chung, 1998, 1999; Chang et al., 1996) and one mutant defective in capsule attachment (pbx1Δ; Kumar et al., 2014; Liu et al., 2007).
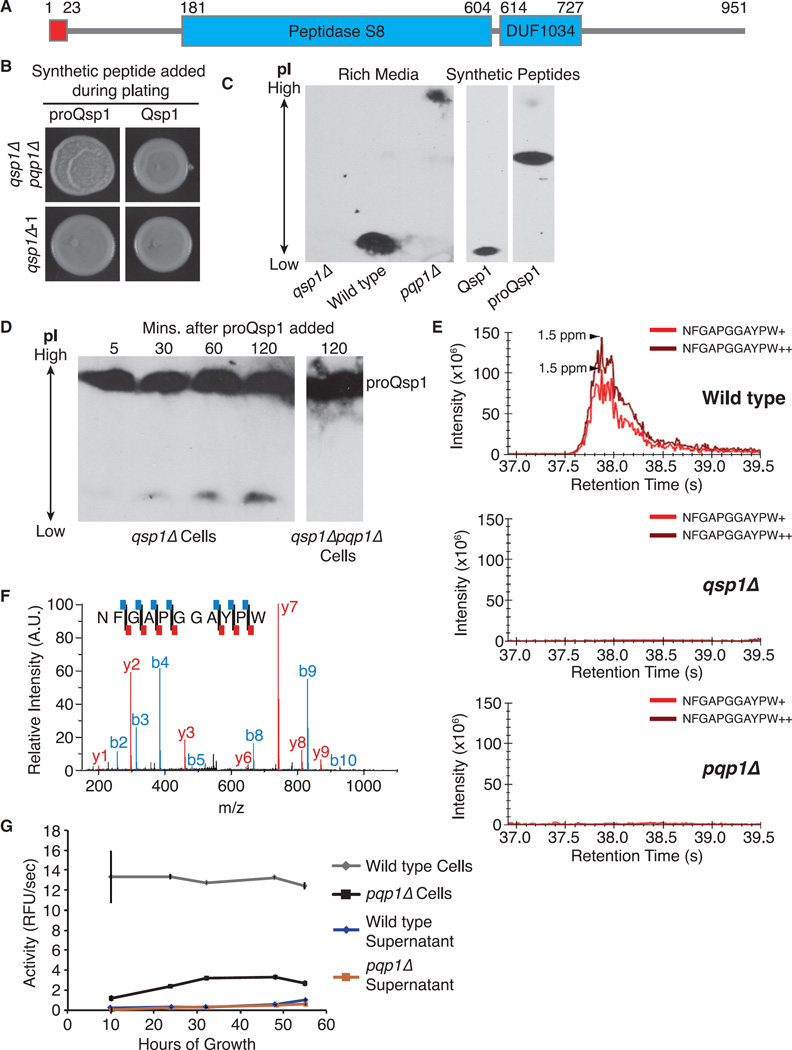
Pqp1 Is a Protease Required for Qsp1 Processing
Among the mutants with a dry colony phenotype was a predicted secreted subtilisin-like serine protease encoded by CNAG_00150 (Figure 5A). Only a peptide corresponding to the C-terminal 11 amino acids of the predicted proQsp1 peptide was detected in culture supernatants in prior studies. Therefore, we hypothesized that CNAG_00150 encodes the protease responsible for cleaving the predicted proQsp1 species. We refer to this protease as proQsp1 Protease 1 (Pqp1). The colony phenotype of a qsp1Δpqp1Δ mutant can be complemented by synthetic Qsp1 but not by synthetic proQsp1 (Figure 5B). Thus, cells lacking PQP1 are defective in processing synthetic proQsp1 into Qsp1 but are able to respond to Qsp1 itself.
Figure 5. Pqp1 Is Required for Qsp1 Precursor Processing.
(A) Pqp1 protein structure.
(B) Spotting assays.
(C) Secretion assays. IEFGE and immunoblotting against Qsp1/proQsp1 in culture supernatants and Qsp1/proQsp1 synthetic peptides. Middle and right panels displaying mobilities of synthetic peptides are from the same blot, with irrelevant lanes removed.
(D) Predicted Qsp1 precursor cleavage assays. Synthetic proQsp1 was incubated with cultures. Supernatants were isolated and then analyzed for cleavage by IEFGE and immunoblotting against Qsp1/proQsp1. Extracted ion chromatogram of Qsp1 from LC/MS of culture supernatants.
(E) Qsp1 MS2 spectra. Each fragment detected is shown with red (y ions) or blue (b ions) lines on the peptide sequence.
(F) Pqp1 activity assay. Error bars are SD.
Using synthetic Qsp1 and proQsp1, we determined that while Qsp1 antisera recognize both peptides, they cannot be distinguished by SDS-PAGE and immunoblotting. Therefore, we used one-dimensional isoelectric focusing gel electrophoresis (IEFGE). In both rich and minimal media, we identified a band with identical mobility as synthetic Qsp1 in wild-type supernatants but not in pqp1Δ or qsp1Δ supernatants. In pqp1Δ supernatants from rich media, we observed a band migrating at a more basic pI, but its mobility differed from synthetic proQsp1 (Figure 5C). The larger band appearing in pqp1Δ supernatants may be a modification to the predicted precursor. Since synthetic proQsp1 complements the colony phenotype of qsp1Δ cells, this modification may not be necessary. To test this explicitly, we added synthetic proQsp1 to qsp1Δ cultures and analyzed the products by IEFGE and immunoblotting (Figure S5A). Consistent with processing, we observed the appearance of a band that comigrated with synthetic Qsp1. This band did not appear when synthetic proQsp1 was added to qsp1Δpqp1Δ cultures (Figure 5D).
We analyzed peptide species in wild-type culture supernatants by LC-MS/MS and identified peaks (Figure 5E) and spectra (Figure 5F) corresponding to the Qsp1 species. However, we did not observe peaks or spectra corresponding to Qsp1 in qsp1Δ or pqp1Δ culture supernatants (Figure 5E), supporting the model that Pqp1 is required for maturation of the endogenous precursor. No peak corresponding to proQsp1 was identified in culture supernatants from any genotype examined, again consistent with a modification of this putative precursor.
Pqp1 was previously identified as an extracellular, but surface-associated, protein (Eigenheer et al., 2007). We quantified proQsp1 processing activity by intact cells and cell-free supernatants using a proQsp1-like substrate (Figure S5B). Approximately 90% of protease activity against this substrate is cell associated (Figure 5G). A small amount of residual cleavage (10%–20% of wild-type) was found in pqp1Δ mutant cells, presumably due to one or more unknown proteases that cleave at some position in the substrate. We analyzed nine additional mutants lacking predicted secreted proteases (Table S4); none display a dry colony phenotype.
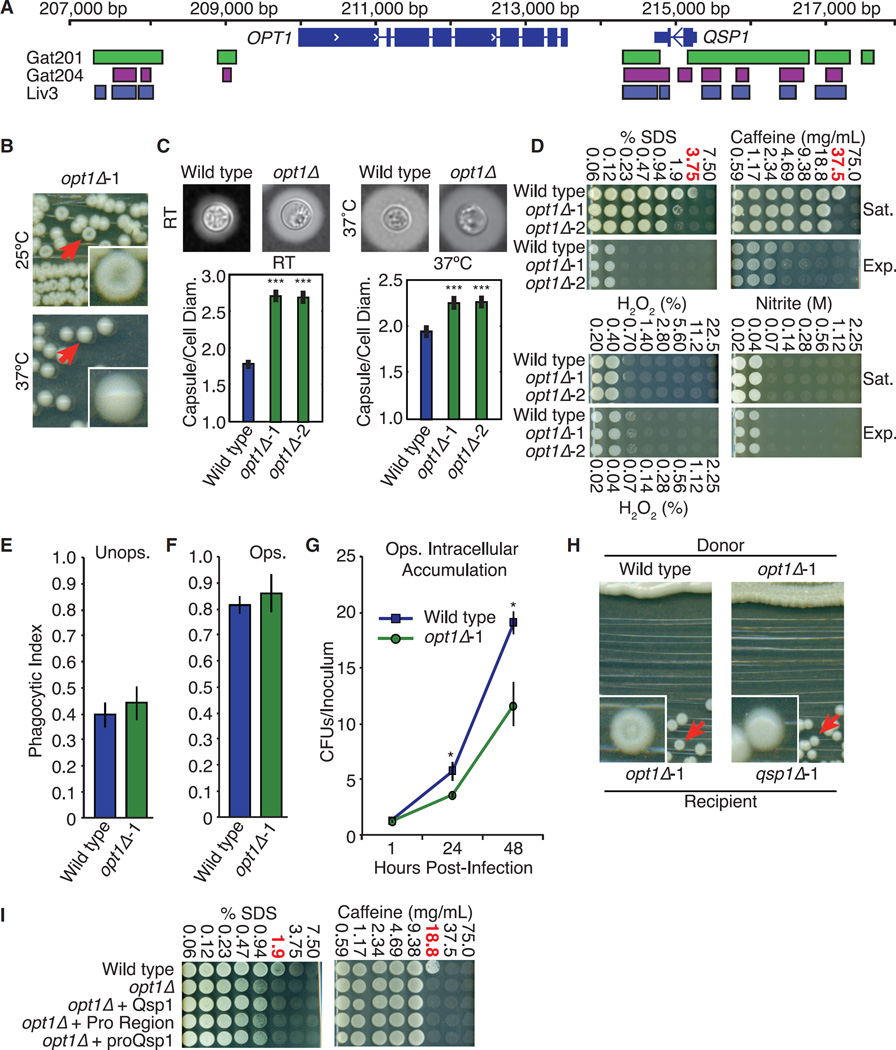
A Predicted Oligopeptide Transporter Required for Peptide Sensing
We searched downstream components in the QSP1 signaling pathway by analyzing published mutants in G protein-coupled receptors and two-component receptors (Table S4). None showed a dry colony phenotype (data not shown). However, our screen identified a gene encoding a predicted oligopeptide transporter, which we named OPT1 (Figure S6A). The QSP1 and OPT1 genes are located next to each other and both display binding of Gat201, Gat204, and Liv3 to their promoters (Figure 6A). Significantly, opt1Δ mutants show nearly all of the phenotypes of qsp1Δ mutants, including dry colonies and increased capsule size at room temperature (Figures 6B, 6C, and S6B). opt1Δ mutants display the same melanization pattern as qsp1Δ mutants (Figure S6C). Additionally, opt1Δ mutants show viability defects in saturated cultures exposed to SDS and caffeine that are indistinguishable from those of qsp1Δ mutants (Figures 6D and S6D). Likewise, opt1Δ mutants have no plating defects on any stress conditions tested (Figures S6E and S6F). Finally, opt1Δ mutants display no differences in unopsonized (Figure 6E) or opsonized (Figure 6F) uptake by BMDMs compared to wild-type. opt1Δ mutants have a 2-fold intracellular proliferation defect after phagocytosis by BMDMs, similar to qsp1Δ (Figure 6G).
Figure 6. A Predicted Oligopeptide Transporter Is Required for Cells to Respond to Qsp1.
(A) Transcription factor binding sites near QSP1 and OPT1.
(B) Colony morphology.
(C) Capsule sizes. Cryptococcal capsule stained with India ink. >100 cells per genotype quantified. ***p < 10−5 (t test). Wild-type data from Figure 3 included for comparison. Error bars are SE.
(D) Stress assays. The concentration where qsp1Δ viability is most different from wild-type is shown in red. Wild-type data from Figure 4 included for comparison.
(E and F) Phagocytic index of unopsonized (E) and opsonized (F) C. neoformans. >1,200 BMDMs quantified per genotype. Wild-type data from Figure 2 included for comparison. Error bars are SD.
(G) Macrophage intracellular accumulation assay. CFUs isolated from BMDMs, normalized to starting inoculum. Wild-type data from Figure 2 included for comparison. Error bars are 95% confidence intervals. *p < 0.05 (bootstrap).
(H) Complementation assays.
(I) Stress assays as described in (D).
Although the phenotypes of opt1Δ mutants closely mirror those of qsp1Δ mutants, opt1Δ mutants differ in two important respects: (1) they can produce Qsp1 and (2) they fail to respond to Qsp1. The dry colony phenotype of opt1Δ mutants cannot be complemented by wild-type patches (Figure 6H). However, opt1Δ patches can complement nearby qsp1Δ colonies, indicating that opt1Δ mutants still produce Qsp1. By ELISA, we found that opt1Δ mutants produce wild-type amounts of Qsp1 in culture supernatants at cell densities up to OD 4.5. At higher densities, we observed less accumulation (Figure S6G). However, the colony phenotype of opt1Δ mutants and the sensitivity of saturated opt1Δ cultures to cell wall stressors are not complemented by addition of synthetic Qsp1 (Figures 6I and S6H). Therefore, these phenotypes are not due to a defect in Qsp1 production.
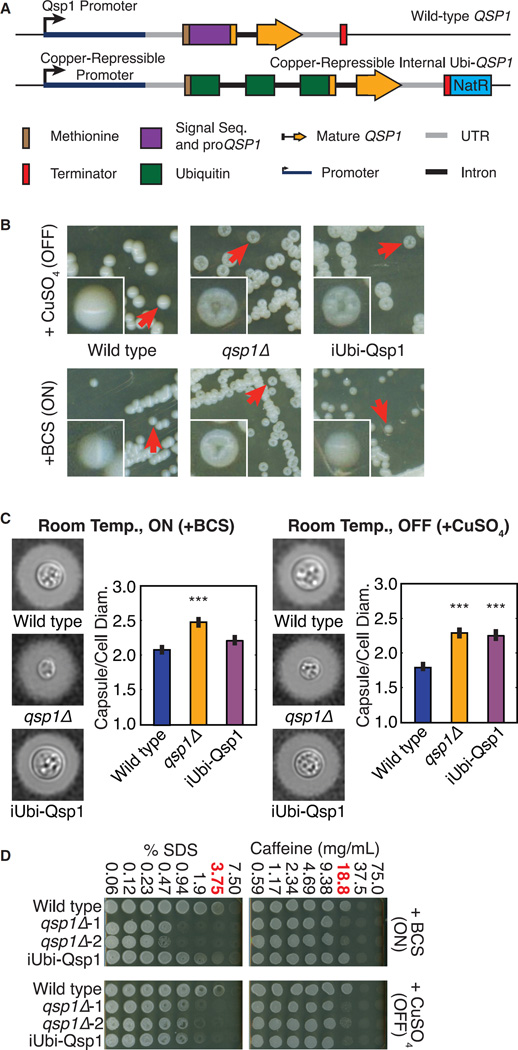
Qsp1 Functions Intracellularly
The requirement of Opt1, a predicted oligopeptide transporter, for the response to Qsp1, but not for its production, suggests that Qsp1 may be imported to act within cells. Thus, we replaced the QSP1 gene with an N-terminal fusion of ubiquitin to the mature Qsp1 peptide. This construct expresses a precursor (iUbi-Qsp1) predicted to be cleaved by endogenous ubiquitin proteases and release the mature Qsp1 peptide cytoplasmically (Varshavsky, 2000) (Figure 7A). We controlled the expression of this precursor using the pCTR4 promoter. These strains did not produce detectable extracellular Qsp1 when assayed using confrontation assays (data not shown). Strikingly, expression of the iUbi-Qsp1 construct by growth on inducing media produces a wild-type colony phenotype. Repression of the gene recapitulates the dry colony phenotype of the qsp1Δ mutant (Figure 7B). The media conditions used to regulate the pCTR4 promoter do not alter the colony phenotypes of wild-type or qsp1Δ cells (Figure 7B). Furthermore, we observed that induction of iUbi-Qsp1 also complements the capsule size changes (Figure 7C) and SDS/caffeine sensitivity (Figure 7D) of the qsp1Δ mutant. The requirement of OPT1 for Qsp1 action and the ability of iUbi-Qsp1 to complement multiple qsp1Δ phenotypes indicates an intracellular site of action for Qsp1.
Figure 7. Cytoplasmic Expression of Mature Qsp1 Complements Multiple qsp1Δ Phenotypes.
(A) Schematic of wild-type QSP1 and iUbi-Qsp1.
(B) Colony morphologies.
(C) Capsule quantification. Cryptococcal capsule stained with India ink. >100 cells per genotype/condition quantified. ***p < 10−5 (t test). Error bars are SE.
(D) Stress assays.
DISCUSSION
We describe a peptide-based cell-cell signaling system required for the virulence of Cryptococcus neoformans, the most common cause of fungal meningitis and a major driver of mortality in HIV/AIDS. This work provides clear evidence that autoregulatory signaling is required for fungal virulence. In addition, our studies reveal that the signaling peptide, Qsp1, is formed by extracellular proteolysis of a precursor but that sensing of Qsp1 occurs intracellularly, a signaling mechanism without precedent in eukaryotes.
A Secreted Peptide Required for Virulence in a Eukaryotic Pathogen
We found that qsp1Δ cells are substantially attenuated in a murine inhalation model. We ruled out a number of possibilities for the attenuated virulence phenotype of qsp1Δ cells, including stress sensitivity and general growth defects. Additionally, our studies did not reveal any detectable change in the dysfunctional Th2 polarization that characterizes cryptococcal disease. As macrophages are key determinants of the outcome of C. neoformans infection, we compared the interactions of wild-type and qsp1Δ cells with primary macrophages and observed a defect in intracellular accumulation of qsp1Δ cells compared to wild-type. The intracellular accumulation defect of qsp1Δ cells likely contributes to its virulence defect as other cryptococcal mutants with decreased accumulation inside macrophages are attenuated in an intranasal model of infection (Evans et al., 2015; Liu and Xue, 2014; Ma et al., 2009). An intriguing possibility is that confinement in the phagolysosome leads to the accumulation of high local concentrations of Qsp1 (Carnes et al., 2010), thereby triggering an adaptive program. We observed no changes in the phagosomal acidification of macrophages infected with qsp1Δ mutants when compared to those infected with wild-type, indicating that phagosome maturation per se is not blocked. Any number of antimicrobial mechanisms that act after phagosome acidification may be interdicted by Qsp1 signaling. Alternatively, the Qsp1 system may simply enable fungal mechanisms that resist killing such as cell wall remodeling.
Qsp1 Mediates Autoregulatory Signaling
Our identification of phenotypes for qsp1Δ cells enabled us to demonstrate that the 11 amino acid mature Qsp1 peptide is a cell-cell signaling molecule produced by C. neoformans that acts on cryptococcal cells. RNA-seq analysis confirms this view that Qsp1 acts on cryptococcal cells themselves and that the impact of QSP1 on the transcriptome is density dependent. These observations are consistent with our finding that only saturated cultures show sensitivity to stress conditions. We observed Qsp1-dependent changes in all three major endoprotease activities. Qsp1 promotes an aspartyl protease activity that we have found in other studies to be required for virulence (S.C.C., P.A. Dumesic, C.M.H., A.J. O’Donoghue, L. Pallova, P. Majer, H.D.M., and C.S.C., unpublished data). In contrast, Qsp1 limits metalloprotease and serine protease activities, which may be deleterious at high cell densities. The outer surface of fungi is the cell wall, a dynamic and highly organized protective organelle that impacts virtually all transactions with the environment. It was therefore notable that genes encoding extracellular/cell wall proteins were significantly enriched among those regulated by QSP1 at high cell density. In addition, PLB1, a gene encoding a phospholipase required for cell wall integrity and virulence, was among genes activated by Qsp1 (Cox et al., 2001; Siafakas et al., 2007). Indeed, our studies of cell wall ultra-structure revealed changes in thickness and organization in qsp1Δ mutants. Furthermore, we observed a concomitant sensitivity of saturated qsp1Δ cultures to killing by cell wall stressors. Our studies provide a clear example in fungi of a mutant that does not synthesize a signaling molecule and exhibits a density-dependent phenotype.
Production of the Signal Requires a Cell-Associated Serine Protease
We used the qsp1Δ dry colony phenotype to perform a forward genetic screen for additional components of the pathway. The analysis identified a predicted subtilase-like serine protease, PQP1. Our data suggest that Pqp1 is the protease that cleaves a Qsp1 precursor to produce the 11 amino acid mature Qsp1 peptide. (1) pqp1Δ cells fail to produce Qsp1. (2) While qsp1Δ’s colony phenotype can be complemented by either Qsp1 or a predicted precursor, proQsp1, only Qsp1 can complement pqp1Δ cells. (3) An exogenously added predicted precursor (proQsp1) can be processed by a cell-associated activity present in wild-type and qsp1Δ strains but not mutants lacking Pqp1. (4) Deletion of genes coding for all other predicted secreted endoproteases did not yield any mutants with a dry colony phenotype.
Reception Requires a Predicted Oligopeptide Transporter: Intracellular Action of Qsp1
Additional mutants isolated based on their dry colony phenotypes were not complemented by synthetic Qsp1, making them candidates for factors involved in the response to Qsp1. Among these, the opt1Δ mutant shares the full constellation of qsp1Δ phenotypes. The notable difference is that, rather than being defective in producing the signal, the opt1Δ mutant is defective in responding to the signal. We reasoned that Opt1 functions in the response to Qsp1. Opt1 is a member of a family of oligopeptide transporters (Gomolplitinant and Saier, 2011), leading us to hypothesize that Opt1 transports Qsp1 into cells.
We tested this hypothesis by constructing a strain to conditionally express the mature Qsp1 peptide intracellularly (iUbi-Qsp1). As expected, the iUbi-Qsp1 strain does not secrete Qsp1. Nonetheless, it displays a wild-type colony phenotype upon induction, indicating that expression of mature Qsp1 intracellularly complements the dry colony phenotype. Significantly, we observed that iUbi-Qsp1 also complements additional phenotypes of the qsp1Δ mutant: capsule size alterations and sensitivity to cell wall stressors. Together with the requirement of a predicted oligopeptide transporter for the cellular response to Qsp1, these data indicate that Qsp1 functions intracellularly after being imported.
Evolution of Peptide-Based Autoregulatory Signaling
QSP1 and OPT1 form a two-gene cluster in C. neoformans, raising the possibility that they were recently acquired by horizontal gene transfer. However, phylogenetic analysis (Figure S6I) indicates that OPT1 was present in the ancestor of present day Tremellaceae, the basidiomycete family that harbors the Cryptococcus species complex. Moreover, genes encoding candidate Qsp1 precursors lie adjacent to OPT1 in Filobasidiella depauparata, Tsuchiyaea wingfieldii, and Cryptococcus amylolentus (Table S5). Thus, the Qsp1 system likely evolved prior to the emergence of the pathogenic Cryptococcus neoformans/Cryptococcus gattii species complex. This conclusion is germane because, while bacterial QS molecules are typically sensed by cell surface receptors, in a handful of Gram-positive species the response to a QS peptide requires a multisubunit oligopeptide permease (Lazazzera and Grossman, 1998; Rutherford and Bassler, 2012). The permease enables import of the peptide and its binding to intracellular receptors. Furthermore, in some of these species, the mature peptide is produced by C-terminal cleavage of a precursor by a cell wall-associated peptidase. As none of the components involved appear to have a proximal bacterial ancestor, this eukaryotic system arose via convergent evolution.
EXPERIMENTAL PROCEDURES
Yeast strains
Yeast strains used in this study are listed in Table S6. All C. neoformans strains were derived from the described parent using published procedures (Chun and Madhani, 2010). Gat201, Gat204, and Liv3 strains are derived from the CM18 parent for consistency with previous publications. The rest of the strains reported are derived from the KN99α wild-type since that isolate is more closely related to the H99 clinical isolate (Janbon et al., 2014).
Chromatin Immunoprecipitation and RNA-Seq Cultures
C. neoformans cultures of 50 ml were grown overnight. 50 ODs were washed 1× with water and resuspended in 15 ml DMEM. This suspension was transferred to 150 mm × 25 mm tissue culture-treated plates already containing 5 ml DMEM. Cultures were incubated sitting at 37°C, 5% CO2 for 8 hr for ChIP-seq and 24 hr for RNA-Seq.
Chromatin Immunoprecipitation
C. neoformans cultures were grown and immunoprecipitation was performed as previously published (Chun et al., 2011). Libraries were prepared for high-throughput sequencing and analyzed using Bowtie1 (Langmead, 2010), SAMtools (Li et al., 2009), and the MACS2 peak caller (Zhang et al., 2008) as described in the Supplemental Experimental Procedures. Overlap between biological replicates was high (Table S7), but for subsequent analysis we only included genes whose promoters had at least one ChIP-seq peak in each of the three biological replicates analyzed. Binding motifs were analyzed using Gimsan (Ng and Keich, 2008).
RNA Expression Profiling
Total RNA was isolated and libraries prepared as described previously (Dumesic et al., 2015). Analysis was performed using Tophat (Trapnell et al., 2012) and the TMM algorithm (Oshlack et al., 2010) or DESeq (Anders and Huber, 2010) as described in the Supplemental Experimental Procedures.
Mass Spectrometry
Peptides were enriched in samples from culture supernatants by precipitation with 90% methanol and 1% acid. LC-MS/MS identification was performed as described in the Supplemental Experimental Procedures.
Colony Phenotype Patch Complementation Assays
All assays were performed at room temperature unless otherwise specified. Peptides for Qsp1 mutational analysis (Table S8) were synthesized by Peptide 2.0.
ELISA and Immunoblotting Assays
Cell-free supernatants were analyzed by ELISA or immunoblotting as described in the Supplemental Experimental Procedures using 1:1,000 diluted anti-Qsp1 serum.
Fluorescent Substrate Protease Activity Assay
Protease assays were performed with the specified substrates as previously described (Jambunathan et al., 2012).
Growth, Viability, Capsule, and Melanin Assays
Growth on various medias, capsule, and melanin were analyzed as previously described (Liu et al., 2008) with minor changes described in the Supplemental Experimental Procedures.
Cell Wall Analysis
24 hr DMEM cultures were analyzed as previously described (Reese et al., 2007).
Intranasal Infection Model
Mouse lung infections were performed as previously described (Chun and Madhani, 2010), under the supervision of the UCSF IACUC Protocol AN091509.
Rabbit Meningitis Model
Rabbit intrathecal infections were performed as previously described (Perfect et al., 1980), according to the Duke University IACUC protocol A003-14-01.
Leukocyte Recruitment and Cytokine Analysis
Lung leukocytes and cytokines were isolated and analyzed as previously described (Wiesner et al., 2015), according to University of Minnesota IACUC protocols 1010A91133 and 1207A17286.
Macrophage Interaction Analysis
Macrophage uptake and intracellular proliferation assays were performed as previously described (Nicola and Casadevall, 2012) with minor changes described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Qsp1 is an autoregulatory signaling peptide required for fungal virulence
Qsp1 controls intracellular growth, cell wall, and protease activities
Opt1 is a peptide transporter required for Qsp1 action in receiving cells
Intracellular expression of mature Qsp1 complements qsp1Δ phenotypes
In Brief.
Bacterial quorum sensing (QS) often controls virulence. Homer et al. reveal a peptide-based QS system in the fungal pathogen C. neoformans. This system requires signaling mechanisms in which the exported peptide is imported and acts intracellularly to control aspects of virulence, including growth, cell surface properties, and secreted protease activities.
Acknowledgments
We thank members of the lab for helpful discussions, N. Nguyen for media preparation, members of the J. Cox lab for kind gifts of BMDMs and reagents, and J. Heitman for strains. Supported by R01AI096869, F30HL120496-01A1, P41RR011823, P41GM103533, R01AG027463, and R01AI73896.
Footnotes
ACCESSION NUMBERS
The accession number for the ChIP-seq and RNA-seq data reported in this paper is GEO: GSE73203.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2016.05.001.
AUTHOR CONTRIBUTIONS
C.M.H. and H.D.M. designed the study. D.K.S. performed experiments in Figures 7 and S3B. A.I.G. performed experiments in Figures 3D, 4H, 5B–5D, S1F, and S6C and Table S3. A.I.G., I.C., and S.P. created knockouts for screening. S.C.C. and C.S.C. performed experiments in Figure 3E and provided protocols for assays in Figure 5G. D.W. and K.N. performed experiments in Figures 2C and 2D. J.K.D., J.J.M., and J.R.Y. performed MS analyses in Figure 5. D.T. and J.P. performed the experiment in Figure S2C. R.U. and J.K.L. performed the experiment in Figure S4D. C.A.C. performed sequence and phylogeny analysis in Figure S6I and Table S5. V.P. provided protocols and guidance for the assay in Figure S2G. C.M.H. performed experiments in Figures 1, 2, 3, 4, 5, and 6 and S1–S6. C.M.H. and H.D.M. wrote the manuscript. All authors contributed to editing the manuscript.
REFERENCES
- Akuthota P, Wang HB, Spencer LA, Weller PF. Immunoregulatory roles of eosinophils: a new look at a familiar cell. Clin. Exp. Allergy. 2008;38:1254–1263. doi: 10.1111/j.1365-2222.2008.03037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque P, Casadevall A. Quorum sensing in fungi–a review. Med. Mycol. 2012;50:337–345. doi: 10.3109/13693786.2011.652201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque P, Nicola AM, Nieves E, Paes HC, Williamson PR, Silva-Pereira I, Casadevall A. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. MBio. 2014;5:e00986–e13. doi: 10.1128/mBio.00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol. 2014;22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Miller MB. The prokaryotes. Berlin: Springer; 2013. Quorum sensing; pp. 495–509. [Google Scholar]
- Beyhan S, Gutierrez M, Voorhies M, Sil A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013;11:e1001614. doi: 10.1371/journal.pbio.1001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes EC, Lopez DM, Donegan NP, Cheung A, Gresham H, Timmins GS, Brinker CJ. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 2010;6:41–45. doi: 10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Penoyer LA, Kwon-Chung KJ. The second capsule gene of cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Madhani HD. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzymol. 2010;470:797–831. doi: 10.1016/S0076-6879(10)70033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun CD, Brown JC, Madhani HD. A major role for capsule-independent phagocytosis-inhibitory mechanisms in mammalian infection by Cryptococcus neoformans. Cell Host Microbe. 2011;9:243–251. doi: 10.1016/j.chom.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C, Bocca AL, Casadevall A. The intracellular life of Cryptococcus neoformans. Annu. Rev. Pathol. 2014;9:219–238. doi: 10.1146/annurev-pathol-012513-104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- Ding C, Festa RA, Chen YL, Espart A, Palacios Ò, Espín J, Capdevila M, Atrian S, Heitman J, Thiele DJ. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe. 2013;13:265–276. doi: 10.1016/j.chom.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 2009;63:223–247. doi: 10.1146/annurev.micro.62.081307.162753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Homer CM, Moresco JJ, Pack LR, Shanle EK, Coyle SM, Strahl BD, Fujimori DG, Yates JR, 3rd, Madhani HD. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell. 2015;160:204–218. doi: 10.1016/j.cell.2014.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 2007;7:499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Li Z, Hughes WS, Djordjevic JT, Nielsen K, May RC. Cryptococcal phospholipase B1 is required for intracellular proliferation and control of titan cell morphology during macrophage infection. Infect. Immun. 2015;83:1296–1304. doi: 10.1128/IAI.03104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free SJ. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- Gomolplitinant KM, Saier MH., Jr Evolution of the oligopeptide transporter family. J. Membr. Biol. 2011;240:89–110. doi: 10.1007/s00232-011-9347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SB, Read RC. Macrophage defences against respiratory tract infections. Br. Med. Bull. 2002;61:45–61. doi: 10.1093/bmb/61.1.45. [DOI] [PubMed] [Google Scholar]
- Herring AC, Hernández Y, Huffnagle GB, Toews GB. Role and development of TH1/TH2 immune responses in the lungs. Semin. Respir. Crit. Care Med. 2004;25:3–10. doi: 10.1055/s-2004-822300. [DOI] [PubMed] [Google Scholar]
- Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6) J. Immunol. 1998;160:2393–2400. [PubMed] [Google Scholar]
- Hughes TR, de Boer CG. Mapping yeast transcriptional networks. Genetics. 2013;195:9–36. doi: 10.1534/genetics.113.153262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan K, Watson DS, Kodukula K, Galande AK. Proteolytic fingerprinting of complex biological samples using combinatorial libraries of fluorogenic probes. Curr. Protoc. Protein Sci. 2012;Chapter 21:22. doi: 10.1002/0471140864.ps2122s70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ, 3rd, Yadav V, Chatterjee G, Mullapudi N, Hon CC, Billmyre RB, Brunel F, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Heiss C, Santiago-Tirado FH, Black I, Azadi P, Doering TL. Pbx proteins in Cryptococcus neoformans cell wall remodeling and capsule assembly. Eukaryot. Cell. 2014;13:560–571. doi: 10.1128/EC.00290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010;Chapter 11:7. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA, Grossman AD. The ins and outs of peptide signaling. Trends Microbiol. 1998;6:288–294. doi: 10.1016/s0966-842x(98)01313-4. [DOI] [PubMed] [Google Scholar]
- Lee H, Chang YC, Nardone G, Kwon-Chung KJ. TUP1 disruption in Cryptococcus neoformans uncovers a peptide-mediated density-dependent growth phenomenon that mimics quorum sensing. Mol. Microbiol. 2007;64:591–601. doi: 10.1111/j.1365-2958.2007.05666.x. [DOI] [PubMed] [Google Scholar]
- Lee H, Chang YC, Varma A, Kwon-Chung KJ. Regulatory diversity of TUP1 in Cryptococcus neoformans. Eukaryot. Cell. 2009;8:1901–1908. doi: 10.1128/EC.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim TS, Murphy JW, Cauley LK. Host-etiological agent interactions in intranasally and intraperitoneally induced Cryptococcosis in mice. Infect. Immun. 1980;29:633–641. doi: 10.1128/iai.29.2.633-641.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TB, Xue C. Fbp1-mediated ubiquitin-proteasome pathway controls Cryptococcus neoformans virulence by regulating fungal intracellular growth in macrophages. Infect. Immun. 2014;82:557–568. doi: 10.1128/IAI.00994-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Kelly MJ, Chow ED, Madhani HD. Parallel beta-helix proteins required for accurate capsule polysaccharide synthesis and virulence in the yeast Cryptococcus neoformans. Eukaryot. Cell. 2007;6:630–640. doi: 10.1128/EC.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Zordan RE, Cain CW, Johnson AD. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA. 2010;107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. USA. 2009;106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P, Keich U. GIMSAN: a Gibbs motif finder with significance analysis. Bioinformatics. 2008;24:2256–2257. doi: 10.1093/bioinformatics/btn408. [DOI] [PubMed] [Google Scholar]
- Nicola AM, Casadevall A. In vitro measurement of phagocytosis and killing of Cryptococcus neoformans by macrophages. Methods Mol. Biol. 2012;844:189–197. doi: 10.1007/978-1-61779-527-5_14. [DOI] [PubMed] [Google Scholar]
- Nilsson UR, Müller-Eberhard HJ. Deficiency of the fifth component of complement in mice with an inherited complement defect. J. Exp. Med. 1967;125:1–16. doi: 10.1084/jem.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio. 2013;4:4. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholzer JJ, Milam JE, Chen GH, Toews GB, Huffnagle GB, Olszewski MA. Role of dendritic cells and alveolar macrophages in regulating early host defense against pulmonary infection with Cryptococcus neoformans. Infect. Immun. 2009;77:3749–3758. doi: 10.1128/IAI.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez JC, Kumamoto CA, Johnson AD. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol. 2013;11:e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- Piehler D, Stenzel W, Grahnert A, Held J, Richter L, Köhler G, Richter T, Eschke M, Alber G, Müller U. Eosinophils contribute to IL-4 production and shape the T-helper cytokine profile and inflammatory response in pulmonary cryptococcosis. Am. J. Pathol. 2011;179:733–744. doi: 10.1016/j.ajpath.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Davis MJ, Dayrit JK, Hadd Z, Meister DL, Osterholzer JJ, Williamson PR, Olszewski MA. Immune modulation mediated by cryptococcal laccase promotes pulmonary growth and brain dissemination of virulent Cryptococcus neoformans in mice. PLoS ONE. 2012;7:e47853. doi: 10.1371/journal.pone.0047853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Dayrit JK, Davis MJ, Carolan JF, Osterholzer JJ, Curtis JL, Olszewski MA. Scavenger receptor A modulates the immune response to pulmonary Cryptococcus neoformans infection. J. Immunol. 2013;191:238–248. doi: 10.4049/jimmunol.1203435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AJ, Yoneda A, Breger JA, Beauvais A, Liu H, Griffith CL, Bose I, Kim MJ, Skau C, Yang S, et al. Loss of cell wall alpha(1–3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 2007;63:1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi N, Baba T, Fukuzawa M, Ohno S. Ultrastructural study of Cryptococcus neoformans by quick-freezing and deep-etching method. Mycopathologia. 1993;121:133–141. doi: 10.1007/BF01104068. [DOI] [PubMed] [Google Scholar]
- Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acylhomoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- Shao X, Mednick A, Alvarez M, van Rooijen N, Casadevall A, Goldman DL. An innate immune system cell is a major determinant of species-related susceptibility differences to fungal pneumonia. J. Immunol. 2005;175:3244–3251. doi: 10.4049/jimmunol.175.5.3244. [DOI] [PubMed] [Google Scholar]
- Shepardson KM, Cramer RA. Fungal cell wall dynamics and infection site microenvironments: signal integration and infection outcome. Curr. Opin. Microbiol. 2013;16:385–390. doi: 10.1016/j.mib.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siafakas AR, Sorrell TC, Wright LC, Wilson C, Larsen M, Boadle R, Williamson PR, Djordjevic JT. Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J. Biol. Chem. 2007;282:37508–37514. doi: 10.1074/jbc.M707913200. [DOI] [PubMed] [Google Scholar]
- Sorrells TR, Johnson AD. Making sense of transcription networks. Cell. 2015;161:714–723. doi: 10.1016/j.cell.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. Ubiquitin fusion technique and its descendants. Methods Enzymol. 2000;327:578–593. doi: 10.1016/s0076-6879(00)27303-5. [DOI] [PubMed] [Google Scholar]
- Vu K, Tham R, Uhrig JP, Thompson GR, 3rd, Na Pombejra S, Jamklang M, Bautos JM, Gelli A. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. MBio. 2014;5:e01101–e01114. doi: 10.1128/mBio.01101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol. Biol. Cell. 2006;17:3768–3780. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner DL, Specht CA, Lee CK, Smith KD, Mukaremera L, Lee ST, Lee CG, Elias JA, Nielsen JN, Boulware DR, et al. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog. 2015;11:e1004701. doi: 10.1371/journal.ppat.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang F, Tompkins KC, McNamara A, Jain AV, Moore BB, Toews GB, Huffnagle GB, Olszewski MA. Robust Th1 and Th17 immunity supports pulmonary clearance but cannot prevent systemic dissemination of highly virulent Cryptococcus neoformans H99. Am. J. Pathol. 2009;175:2489–2500. doi: 10.2353/ajpath.2009.090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang F, Bhan U, Huffnagle GB, Toews GB, Standiford TJ, Olszewski MA. TLR9 signaling is required for generation of the adaptive immune protection in Cryptococcus neoformans-infected lungs. Am. J. Pathol. 2010;177:754–765. doi: 10.2353/ajpath.2010.091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.