Abstract
Small heat shock proteins (sHsps) are a family of ATP-independent molecular chaperones that are important for binding and stabilizing unfolded proteins. In this task, the sHsps have been proposed to coordinate with ATP-dependent chaperones, including heat shock protein 70 (Hsp70). However, it isn’t yet clear how these two important components of the chaperone network are linked. We report that the Hsp70 co-chaperone, BAG3, is a modular, scaffolding factor to bring together sHsps and Hsp70s. Using domain deletions and point mutations, we confirmed that BAG3 uses both of its IPV motifs to interact with sHsps, including Hsp27 (HspB1), αB-crystallin (HspB5), Hsp22 (HspB8) and Hsp20 (HspB6). BAG3 does not appear to be a passive scaffolding factor; rather, its binding promoted de-oligomerization of Hsp27, likely by competing for the self-interactions that normally stabilize large oligomers. BAG3 bound to Hsp70 at the same time as either Hsp22, Hsp27 or αB-crystallin, suggesting that it might physically bring the chaperone families together into a complex. Indeed, addition of BAG3 coordinated the ability of Hsp22 and Hsp70 to refold denatured luciferase in vitro. Together, these results suggest that BAG3 physically and functionally links Hsp70 and sHsps.
Graphical Abstract
Introduction
The mammalian genome contains multiple classes of molecular chaperones that are named according to their approximate size, including heat shock protein 90 (Hsp90), heat shock protein 70 (Hsp70), heat shock protein 40 (Hsp40) and the small heat shock proteins (sHsps) [1]. Each of the categories of chaperones appears to be partly dedicated to individual aspects of protein quality control [2]. For example, Hsp70 and Hsp90 are ATPases that use nucleotide hydrolysis to regulate reversible binding to unfolded or partially folded proteins, stabilizing many “clients” and favoring their folding and/or trafficking [3; 4]. Other chaperones, such as the Hsp40s, appear to recruit specific clients to the Hsp70 and Hsp90 systems [5]. The importance of having multiple, non-overlapping categories of chaperones is highlighted by their incredible conservation across all kingdoms of life [6–8].
Together, the chaperones constitute a network that maintains global protein folding and function [1]. Within the chaperone network, many of the major chaperones make physical interactions with each other. For example, the prokaryotic orthologs of Hsp70 and Hsp90 directly bind to each other [9; 10], as do the orthologs of Hsp70 and Hsp110 [11]. In mammals, Hsp70s are physically linked to the Hsp90s, but through the scaffolding protein, HOP (Hsc70-Hsp90 organizing factor). HOP is required for the Hsp70/Hsp90-mediated activation of steroid hormone receptors [12–15], perhaps by facilitating client transfer between the chaperones [16; 17]. Thus, the mammalian chaperone network appears to be held together, in part, by protein-protein interactions between chaperones and scaffolding proteins. This feature might allow unfolded clients to be protected by a closely-knit series of physically interacting factors, possibly limiting their exposure to bystander proteins.
Within the chaperone network, small heat shock proteins (sHsps) are a large and enigmatic class [18]. In humans, there are ten members of the sHsp family, denoted HSPB1 through HSPB10 [19]. Unlike other molecular chaperone families, such as Hsp70 or Hsp90, sHsps do not possess enzymatic activity; instead, sHsps function as “holdases”, a term that refers to their ability to bind and stabilize denatured or non-native proteins against aggregation [20; 21]. Individual sHsps range in size from 12 to 43 kDa and they are defined by the presence of a conserved α-crystallin domain. In each sHsp, the α-crystallin domain that is flanked by variable, disordered N- and C-terminal domains that contain phosphorylation sites and may bind to clients [22]. The structure of the α-crystallin domain of Hsp27 has been solved by NMR and it features an antiparallel beta-sheet that mediates stable dimerization [23–25]. In addition, the α-crystallin domain contains two highly conserved beta sheets, termed β4–β8, that form a hydrophobic groove. In some sHsps, such as Hsp27 and α-crystallin, an IXI motif in the C-terminus binds to this groove and stabilizes higher order oligomers [26–29] Specifically, the IXI motif from one unit of the sHsp seems to reach back onto the β4–β8 groove of another. This groove can even support hetero-oligomers between different sHsp family members [30; 31]. The oligomers are typically polydisperse and range in size from ~12 to 40 dimer subunits [30; 32; 33].
The method by which sHsps bind and stabilize clients is still being explored. One model suggests that the sHsp oligomers dissociate in the presence of clients, and then re-form into a new oligomeric form containing bound client [34; 35]. This idea is supported by electron microscopy [20; 36] and mass spectrometry studies [37–39]. It has also been shown that smaller oligomers of sHsp are more potent holdases in vitro [39; 40] and that even fragments of sHsps can still bind clients [41]. These observations suggest that smaller oligomers might be better able to bind clients. In cells, phosphorylation of some sHsps forces larger oligomers into smaller ones, perhaps providing a way for cell stress signaling to increase chaperone function [39; 42; 43]. Similarly, some sHsps can also sense pH [44; 45], adjusting their structure to protect clients.
One major gap in our understanding of the chaperone network is how the sHsps link to the other major components. This question is important because sHsps lack the enzymatic activity that appears to be required for active remodeling or refolding by the other categories of chaperones. Rather, sHsps have been proposed to work in conjunction with other chaperone systems, including Hsp70, in cells [46; 47]. In this model, sHsp oligomers might interact with clients and keep them in a state that is competent for re-folding by Hsp70.
Bcl-2 Associated Anthanogene-3 (BAG3) is a stress-inducible, 61 kDa protein that is characterized by the presence of multiple protein-protein interaction motifs (Fig 1a), separated by long regions of predicted disorder. BAG3 is a member of the BAG family of co-chaperones (BAG1–5), which are characterized by a conserved BAG domain. The BAG domain is well known to bind the nucleotide-binding domain (NBD) of members of the Hsp70 family [48; 49]. This protein-protein interaction helps release ADP from the chaperone to facilitate nucleotide cycling. BAG3 binds to Hsp70 tighter than BAG1 or BAG2 [50], and it is the most potent nucleotide exchange factor (NEF), suggesting that it is a functionally important partner for Hsp70s. Outside of the BAG domain, the members of the BAG family have a variety of other motifs that are not shared. For example, BAG3 contains a WW domain that has been shown to be important for binding PPxY motif proteins, such as RAPGEF6 [51] and SYNPO2 [52]. The PXXP region of BAG3 has been implicated in interactions with SH3 domain-containing proteins, including Src [53] and PLC-γ [54], an interaction that might link BAG3 and Hsp70 to signaling pathways. Finally, and most importantly for this study, BAG3 contains two IPV motifs separated by ~100 amino acids in its N-terminus. BAG3 has been proposed to interact with sHsps through its IPV motifs [55–57], possibly mimicking the intra-molecular interactions that normally occur between the IXI motif and the β4–β8 grooves.
Figure 1.
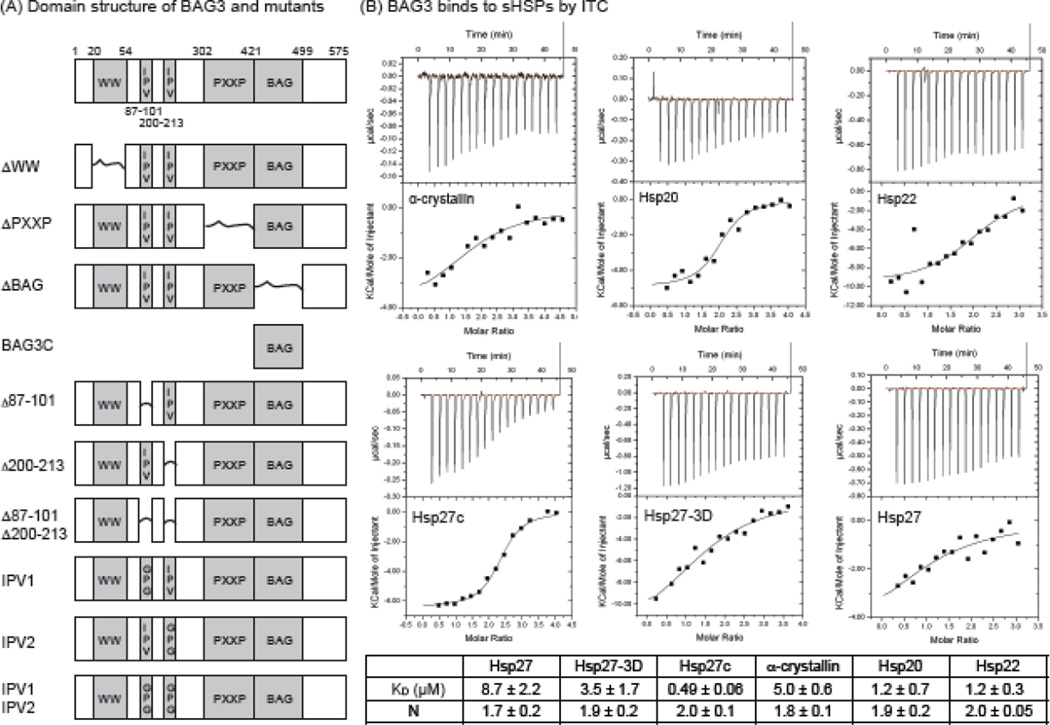
BAG3 binds to small heat shock proteins. (A) Schematic of the domain architecture of BAG3, plus the deletions and point mutations used in this study. Deleted domains are indicated by a connecting line. (B) ITC results confirm the relative affinities of BAG3 for sHsps. Results are representative of experiments performed in triplicate. Error is SEM.
Based on these interactions, it seemed plausible that BAG3 could be a scaffolding protein that physically links sHsps to the Hsp70 system. To test this model in vitro, we generated a suite of BAG3 constructs with individual domains mutated or deleted. We found that BAG3 interacts with multiple members of the sHsp family using both of its IPV motifs. Interestingly, we found that BAG3 binding was not passive; rather it disrupts the Hsp27 oligomers, as judged by chromatography and electron microscopy. This finding suggests that BAG3 might actively remodel sHsps, potentially activating them or changing their interactions with clients. Indeed, we found that BAG3 could coordinate the ability of Hsp22 and Hsp70 to refold denatured luciferase. Finally, we were able to show that BAG3 can bridge the two chaperone families at the same time and that this physical interaction is dependent on the IPV and BAG domains of BAG3. Based on these data, we propose that BAG3 is the link between these two important components of the chaperone network.
Results
BAG3 binds multiple sHsps
To study the interactions with BAG3, we selected four members of the sHsp family that are ubiquitously expressed in all human tissues [19]: Hsp27 (HSPB1), α-crystallin (HSPB5), Hsp20 (HSPB6), and Hsp22 (HSPB8). Of these proteins, Hsp20, Hsp22 and α-crystallin have been reported to interact with BAG3 by co-immunoprecipitation and pulldown studies [55; 57]; however, the affinities of these interactions were not known. Therefore we employed isothermal titration calorimetry (ITC) to better understand them (Fig 1b). Interestingly, BAG3 bound tightest to Hsp22 (1.2 ± 0.3 µM) and Hsp20 (1.2 ± 0.7 µM), In contrast, BAG3 had a relatively weaker affinity for Hsp27 (8.7 ± 2.2 µM) and α-crystallin (5.0 ± 0.6 µM). Because of the possibility of competing interactions and the structural heterogeneity of the sHsps, the reported KD values are best used as relative values. Regardless, these results support the idea that BAG3 uses IPV motifs to interact with sHsps because Hsp22 and Hsp20 lack their own IXI motifs and primarily exists as dimers or tetramers in solution [56; 58], while Hsp27 and αB-crystallin have IXI motifs and form larger oligomers.
To better understand whether BAG3 might compete with the IXI motifs, we studied the interaction of BAG3 with two Hsp27 variants. Hsp27-3D is a triple phospho-mimetic mutant that forms smaller oligomers in solution [39], while Hsp27c is a truncated form that contains only the core α-crystallin domain and is exclusively a dimer in solution [25]. We found that Hsp27c bound with the tightest affinity (KD ~ 0.49 ± 0.06 µM), followed by Hsp27-3D (3.5 ± 1.7 µM), and then Hsp27 (8.7 ± 2.2 µM). These results suggest that larger Hsp27 oligomers may have the weakest affinity for BAG3, while dimers bind the tightest. We hypothesize that this difference is because of competition between IXI motifs in the sHSPs and IPV motifs in BAG3 (see below). Further, the ITC results suggested that the stoichiometry of each of the sHsp:BAG3 interactions is approximately 2:1 (Fig 1b). This was an intriguing result because BAG3 contains two IPV motifs, so it could potentially interact with both of the conserved β4–β8 grooves on either side of a sHsp dimer.
BAG3 uses IPV motifs to interact with the β4–β8 region
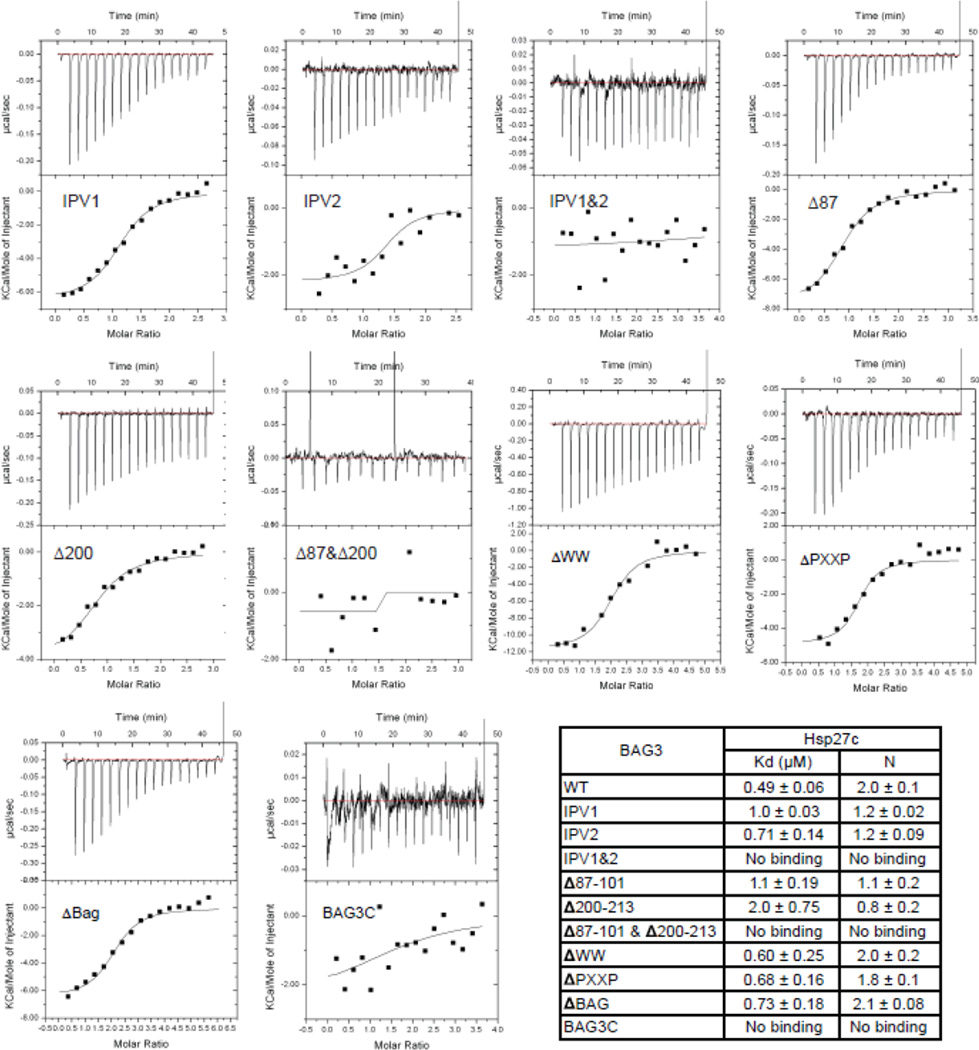
To address this question more definitively, we prepared labeled Hsp27c and examined binding to BAG3 by 15N-1N HSQC NMR. We observed selective chemical shift perturbations (CSPs) in the β4–β8 region (Supplemental Fig S1A), consistent with the idea that BAG3 competes with the native IXI motifs. Further, mutation of valine to alanine in a short IPV-containing peptide weakened its apparent affinity (Supplemental Fig S1B), showing that the IPV was important. Next, we generated BAG3 variants in which individual domains were systematically deleted. These deletions included the BAG3 variants: ΔWW, Δ87–101, Δ200–213, Δ87–101 & Δ200–213, ΔPXXP, ΔBAG and BAG3C (see Fig. 1a). The truncations Δ87–101, Δ200–213, Δ87–101 & Δ200–213 removed the first IPV motif, the second IPV motif or both, respectively, while the other truncations removed known domains involved in other protein-protein interactions. Finally, BAG3C was composed of only the BAG domain, so we could use it to understand the role of this region. We assayed the truncated proteins for binding to Hsp27c using ITC (Fig 2) and found that the ΔWW, ΔPXXP, or ΔBAG proteins bound with approximately the same affinity as full length (~ 0.5 to 0.7 µM). BAG3C construct did not bind to Hsp27c, as expected. These results are consistent with the role of IPV motifs in binding to sHsps and they suggest that other domains are not involved. More interestingly, we found that deletion of individual IPV motifs (Δ87–101, Δ200–213) or mutation of the first IPV motif to GPG (named as IPV1) weakened affinity by at least 2-fold (~1 to 2 µM). The mutation or deletion of individual IPV motifs also reduced the stoichiometry (N) of the interaction from 2:1 to approximately 1:1 (Fig 2b). When both IPV motifs were deleted (Δ87–101&Δ200–213) or mutated (IPV1 and IPV2 to GPG), binding was abolished. The simplest explanation for these results is that BAG3 uses both of its IPV motifs to engage the Hsp27c dimer.
Figure 2.
BAG3 uses both IPV motifs to bind sHsps. Deletions and point mutations in BAG3 reveal that the IPV motifs are important for binding to Hsp27c, using ITC. Experiments were performed in duplicate and the error bars are SEM. See Supplemental Fig S2 for binding to the other sHsps.
To explore whether this bi-dentate binding mode might be conserved in the interaction with other sHsps, we measured binding of each of the BAG3 mutants to Hsp27, αB-crystallin, Hsp22 and Hsp20 using a flow cytometry protein interaction assay (FCPIA). Consistent with the results obtained using Hsp27c, deletion or mutation of one IPV motif weakened the apparent binding affinity (typically by about 2-fold) for each of the full-length sHsps (Supplemental Fig S2). Removing both IPV motifs prevented measurable binding to Hsp27, HSP27-3D, Hsp20 and αB-crystallin (KD > 50 µM) and weakened the affinity for Hsp27c and Hsp22 by more than 25-fold (KD ~10 to 20 µM). Replacing both IPV motifs with GPG had a similar effect; binding to Hsp27, HSP27-3D and αB-crystallin was unmeasurable (KD > 50 µM) and the affinity for Hsp27c, Hsp20 and Hsp22 was weakened (KD ~ 3 µM). Together, these results suggest that BAG3 uses both of its IPV motifs to interact with sHsps. These interactions do not necessarily have to involve two IPV motifs binding to a single dimer, especially in the context of the more complex, polydisperse sHsp oligomers.
BAG3 reduces the size of Hsp27 oligomers
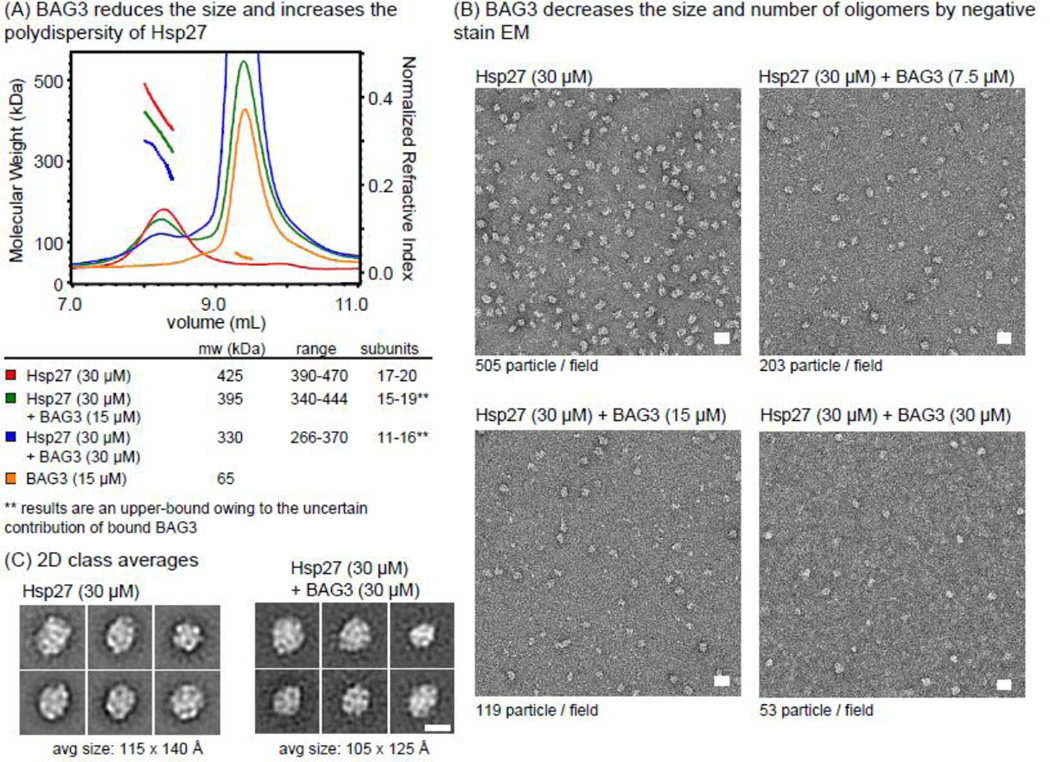
Knowing that BAG3 uses its IPV motifs to interact with sHsps and that sHsps also use their IXI motifs to regulate their oligomer size, we wondered if BAG3 could disrupt sHsp oligomers. To test this hypothesis, we first used size exclusion chromatography with multi-angle light scattering (SEC-MALS). This technique allows molecular weight determination of a sample based on the intensity of light scattering as a function of angle. In these experiments, we focused on Hsp27 because it is known to form large oligomers that can be visualized by electron microscopy (see below) [44]. Injection of Hsp27 (30 µM) alone yielded an SEC-MALS trace with an average mass of 425 kDa (Fig. 3a). This mass corresponds to ~18 monomeric subunits, which is consistent with literature values [59]. In addition, the Hsp27 peak was broad (mass range 390–470 kDa), suggesting a polydisperse ensemble of oligomers that has been observed previously [60]. Adding BAG3 to this sample effectively reduced the apparent oligomer size from 425 kDa to 330 kDa (Fig. 3a). This change represents a predicted drop in average subunit size from approximately 18 monomers to ~14 monomers, supporting the hypothesis that BAG3 can disassemble oligomers of Hsp27. Importantly, this estimate is a conservative upper bound because BAG3 is also likely to contribute to the apparent molecular mass. If, based on the ITC results, we assume a 2:1 stoichiometry then the oligomers would be composed of ~6 monomers. Although a definitive conclusion is difficult, owing to the heterogeneity of the samples, the qualitative conclusion is that BAG3 disrupts the Hsp27 oligomers.
Figure 3.
BAG3 binding reduces the size of Hsp27 oligomers. (A) Hsp27 (30 µM) was incubated with increasing concentrations of BAG3 and the mixtures were analyzed by SEC-MALS. BAG3 reduced the average molecular weight of the Hsp27 peak and increased the polydispersity of the samples. Experiments were repeated in duplicate and average MW is reported with SEM. Importantly, the estimate of the number of Hsp27 subunits within the complex does not include the contributed mass of bound BAG3 because the stoichiometry is not yet clear, so it should be considered a conservative upper bound. (B) 2D class averages of oligomer particles show a modest decrease in the average particle size. Scale bar is 10 nm. (C) BAG3 decreases the number of Hsp27 oligomers, as measured by negative stain electron microscopy. Scale bar is 20 nm.
To test this idea in a different way, we used electron microscopy (EM). We found that oligomers of Hsp27 are visible using negative stain EM, with an average size of 115 × 140 angstroms (Fig. 3b). Importantly, we expect that smaller oligomers, such as dimers, are “invisible” by this method because they don’t readily accumulate stain. In fact, this feature allowed us to ask whether BAG3 could convert samples of Hsp27 into a smaller oligomer size by two approaches. In the first, we examined all of the visible particles and measured their dimensions to see if BAG3 could favor smaller structures. Indeed, we found that the Hsp27 particle size decreased to 105 × 125 angstroms when treated with 30 µM BAG3 (Fig. 3b). Next, we counted the total number of visible Hsp27 particles in the grids to estimate whether BAG3 was altering the overall oligomer population. Small oligomers and dimers of Hsp27 would not be expected to retain stain and, therefore, would not be visible by this method. Strikingly, we found that the total number of visible particles was strongly reduced by BAG3 in a dose-dependent manner (Fig. 3c). Specifically, the average number of oligomers observed in micrographs of Hsp27 alone was 505 particles/field. With increasing amounts of BAG3 at a constant concentration of Hsp27 (30 µM), the number of large particles was drastically d: 203 in the presence of 7.5 µM BAG3, 119 in the presence of 15 µM BAG3, and only 53 oligomer particles in the presence of 30 µM BAG3. At this stage, the exact stoichiometry of the Hsp27-BAG3 complexes and the size distribution of treated samples are not clear. However, these results point to a model in which competition for IXI motifs during binding to BAG3 is not static – it results in a decrease in average Hsp27 oligomer size.
BAG domain is essential for Hsp70 NEF function
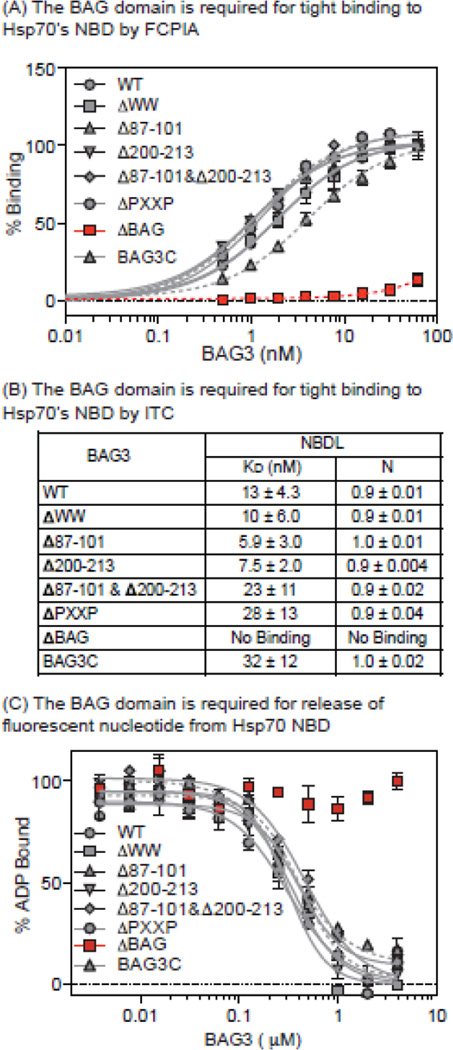
BAG3 forms a tight interaction with the NBD of Hsp70 (KD ~10 nM) to stimulate ADP release [50]. While it is known that this process requires the BAG domain, it wasn’t yet clear if other regions of BAG3, especially the IPV motifs, might contribute to Hsp70 binding. To explore this idea, we measured binding of the BAG3 deletion mutants to Hsp70. Using FCPIA, we first confirmed that ΔBAG can no longer bind to Hsp70 (Fig 4a), consistent with the literature. This result was also confirmed by ITC (Fig 4b). The other deletions or mutations had a less dramatic effect on the BAG3-Hsp70 affinity (typically less than 2-fold reduction), suggesting that the majority of the binding energy originates from the BAG domain.
Figure 4.
BAG3 binds Hsp70 through its BAG domain and stimulates client release. Binding of labeled BAG3 to immobilized Hsp70 by (A) FCPIA and (B) ITC. Results are the average of experiments performed in triplicate. Error bars represent EM. (C) BAG3 releases fluorescent nucleotide from Hsp70 NBD. Deletion of the BAG domain blocks this activity. Results are the average of experiments performed in triplicate. Error is SEM.
To verify this finding in a different platform, we turned to a nucleotide release assay. In this assay, Hsp70 is loaded with a fluorescent ATP analog and then the ability of BAG3 to release the tracer is measured by fluorescence polarization (FP) [50]. We found that all of the domain deletion constructs were approximately equivalent in their ability to promote nucleotide release from Hsp70 (Fig. 4c), with the exception of the ΔBAG construct. Together, these results point to a model in which the BAG domain interaction with Hsp70’s NBD is largely “insulated” from other parts of BAG3, including the IPV motifs.
Hsp70-BAG3-sHsp form a ternary complex
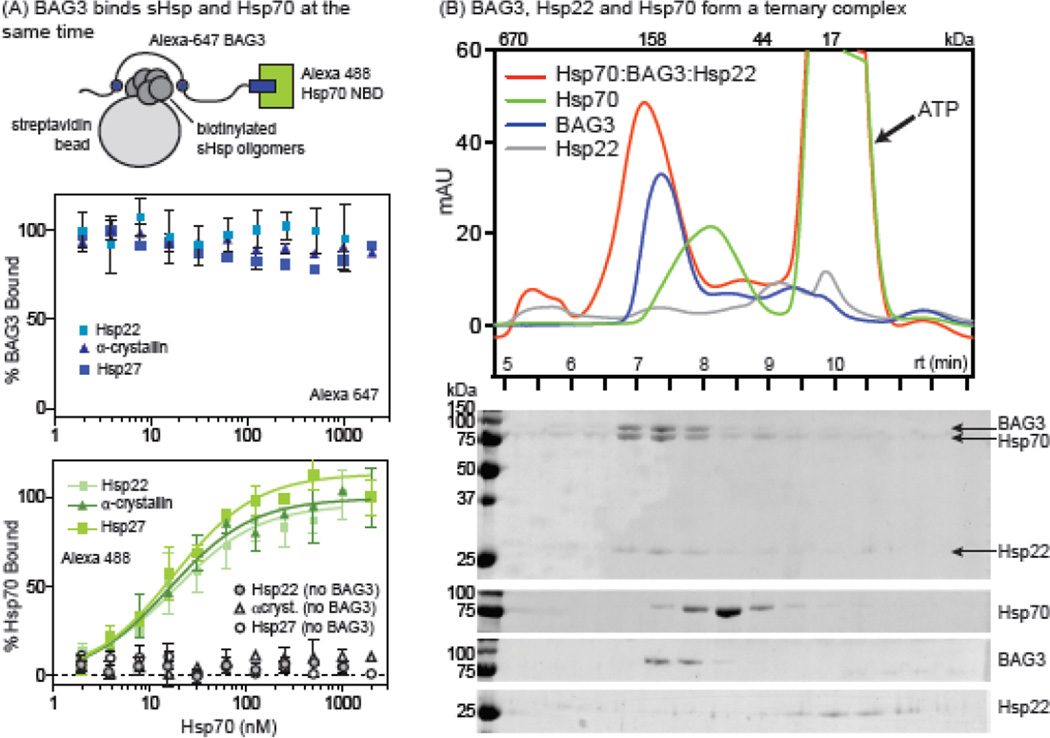
After characterizing the individual, binary interactions between BAG3-sHsp and BAG3-Hsp70, we set out to determine if a ternary Hsp70-BAG3-sHsp complex could be formed. The results of the binding studies thus far suggested that BAG3 could be a modular scaffolding protein, leading to the prediction that binding to Hsp70 would not impact binding to sHsps and vice versa. To ask this question, we immobilized Hsp22, α-crystallin or Hsp27 on streptavidin beads, incubated them with a constant concentration of Alexa 647 labeled BAG3 (50 nM), and then added increasing amounts of Alexa 488 labeled Hsp70NBD (Fig. 5a). Unfortunately, we were not able to immobilize sufficient levels of Hsp20 for this study. If Hsp70NBD could compete with sHsp for binding to BAG3, we would expect to see a decrease in Alexa 647 signal upon titration. However, we observed no decrease in fluorescence in the presence of Hsp70NBD (Fig. 5a; top). Moreover, since we labeled BAG3 and Hsp70NBD with fluorophores that have distinct spectral properties, we were also able to confirm that both proteins were bound at the same time by monitoring the increase in Alexa 488 signal (Fig 5a, bottom). Lending further support to the idea of a modular interaction, the apparent affinity of the Hsp70NBD-BAG3 interaction (~15 nM) was unchanged in the presence of Hsp22, α-crystallin or Hsp27, when compared to the binary interaction. These findings suggest that sHsps and Hsp70 do not interfere with (or promote) binding to the other partner. In important control studies, we found that Hsp70NBD did not bind Hsp22, α-crystallin or Hsp27 in the absence of BAG3 (Fig. 5a). Thus, Bag3 appears to be a modular scaffolding protein for the two chaperone families.
Figure 5.
BAG3 stabilizes a ternary complex with Hsp70. (a) Alexa Fluor 488-labeled Hsp70 NBD was titrated against a solution of Alexa Fluor 647-labeled BAG3 in the presence of sHsp (a-crystallin, Hsp27 or Hsp22) coated beads. Binding was detected using a flow cytometer. Experiments were performed in triplicate and error is SEM. A schematic of binding experiment is shown. Note that the sHsp oligomer size is heterogeneous. (B) SEC and SDS-PAGE analysis of a solution of Hsp22 (6 µM), BAG3 (12 µM) and Hsp70 (6 µM) or solutions of individual components. In all experiments, ATP was added (1 mM) to favor tight binding of Hsp70 to BAG3.
To confirm this idea, we analyzed solutions of Hsp70, BAG3, Hsp22 or a mixture of the three by size exclusion chromatography (SEC). Hsp22 was chosen for this experiment because of its tight affinity (see Fig 1b) and its relatively homogeneous oligomer size distribution on SEC (Fig 5b). Also, ATP (1 mM) was added to favor tight binding of Hsc70NBD to BAG3. Other sHsps were too heterogeneous in this platform to allow conclusions. However, we found that Hsp70, BAG3 and Hsp22 were clearly distinguished from each other on the SEC traces when injected individually (Fig 5b). Moreover, pre-incubation of these components showed a conversion to a higher order complex (Fig 5b). All three proteins were part of this complex, as judged by collecting fractions and subjecting them to SDS-PAGE. Thus, BAG3 appears to be an adapter protein that links Hsp22, and likely other sHSPs, to Hsp70.
Hsp22 promotes the co-chaperone activity of BAG3 in vitro
To explore possible functional implications of the ternary complex, we measured two chaperone activities of Hsp70: ATP turnover and firefly luciferase refolding. ATPase activity is typically measured using a colorimetric, malachite green assay, while refolding activity is measured by treating denatured firefly luciferase with chaperones and monitoring the recovery of luminescence. It is known that substoichiometric concentrations of BAG3 promote Hsp70’s nucleotide turnover because it overcomes the rate-limiting step in cycling. Likewise, low levels of BAG3 are thought to balance the binding-and-release of luciferase in the refolding assay, increasing the yield of re-folded enzyme. However, in both assays, higher concentrations of BAG3 seem to stall Hsp70, such that it converts from being a stimulator into an apparent inhibitor [50].
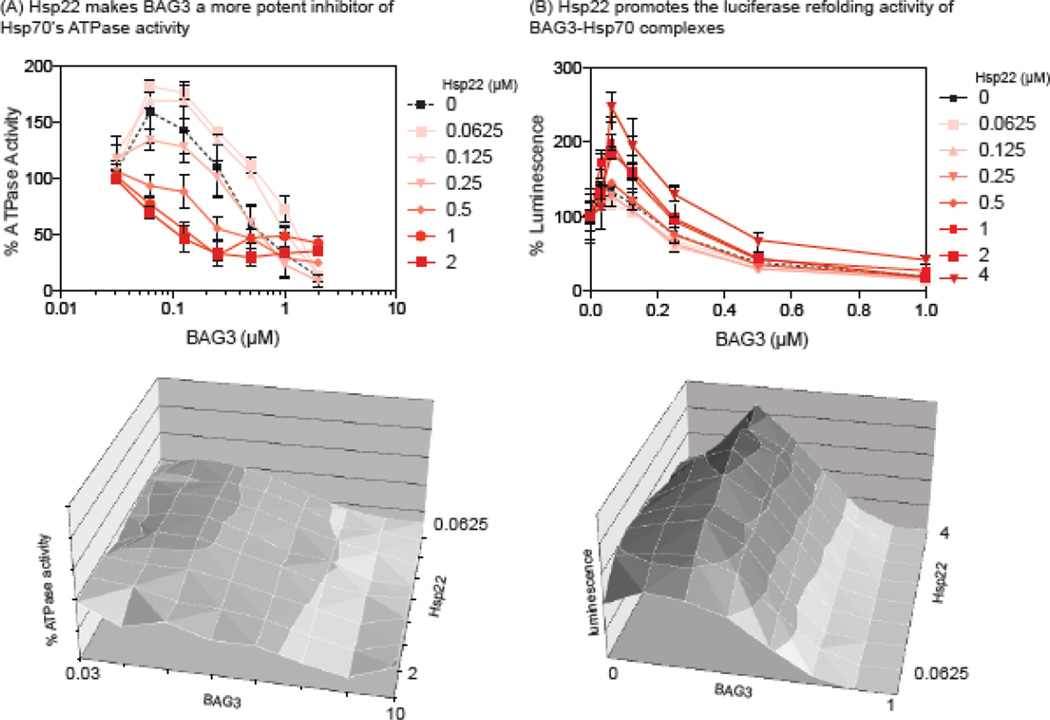
Using these assays, we varied the concentration of both BAG3 and Hsp22, holding the levels of Hsc70 (1 µM) constant. Importantly, we found that Hsp22 had no significant effect on either ATPase activity or luciferase refolding in the absence of BAG3 (Fig 6A and 6B), so this sHsp was ideal for these experiments. In the ATPase assay, we observed a striking ability of Hsp22 to promote the activity of BAG3 (Fig 6A). For example, at 0.5 µM of Hsp22, BAG3 became a potent suppressor of cycling. At 2 µM, this effect was even more pronounced, allowing BAG3 to be a strong inhibitor at concentrations at which it was normally inactive (e.g. 0.125 µM; Fig 6A). We noted a trend of Hsp22 to modestly promote the ability of low levels of BAG3 to activate turnover when the Hsp22 was also present at low concentrations (0.125 and 0.0625 µM of Hsp22; Fig 6A); however, this trend was not statistically significant (p value > 0.01).
Figure 6.
Hsp22 affects the chaperone functions of Hsp70 through BAG3. (A) The ATPase activity of Hsp70 was measured by malachite green assays. Addition of BAG3 suppresses ATPase activity, consistent with previous reports. Hsp22 promotes this activity and makes BAG3 more effective at lower concentrations. Results are the average of at least three independent experiments performed in triplicate. Error bars represent SD. Importantly, Hsp22 has no effect on Hsp70’s ATPase activity in the absence of BAG3. (B) Hsp22 coordinates with Hsp70-BAG3 to refold denatured luciferase. Titration of BAG3 into Hsp70 initially promotes luciferase refolding, but it then becomes inhibitory at higher concentrations. Addition of Hsp22 promotes luciferase refolding. Results are the average of at least three independent experiments performed in triplicate. The error bars represent SD. In both A and B, the results are shown as a two- and three-dimensional display of the same data.
The possible effects of BAG3 and Hsp22 on luciferase refolding were measured using a similar approach. Chaperone-mediated refolding of firefly luciferase requires a careful balance of client binding and release, which eventually favors the folded, enzymatically active state. We observed that titration of Hsp22 into the Hsc70-BAG3 system strongly promoted the refolding of luciferase, with luminescence increased by 2- to 3-fold when Hsp22 was above 1 µM (Fig 6B). Again, this activation was highly dependent on coordination with BAG3, because Hsp22 was unable to promote refolding in the absence of BAG3 or at high concentrations of BAG3. These results suggest that BAG3 regulates the chaperone functions of Hsc70 and Hsp22 in the context of refolding damaged luciferase.
Discussion
BAG3 binds sHsps using both of its IPV motifs
Using pulldowns, BAG3 had been reported to interact with some sHsps through its IPV motifs [56; 57]. Using NMR, we first confirmed that BAG3 binds the conserved β4–β8 region of the core crystallin domain, where the IXI motifs normally reside (see Supplemental Fig S1). Further, deleting or mutating either of the IPV motifs of BAG3 weakened binding to Hsp27, Hsp20, Hsp22 and αB-crystallin and deleting or mutating them both largely abolished binding (see Fig 1). These results suggest that BAG3 uses both of its IPV motifs to engage the two β4–β8 grooves. In the simplest case (such as Hsp27c), such an interaction could occur through a “clamp-like” mechanism, with both of the IPV motifs making contact within a single dimer. There is a sufficient distance between the IPV motifs (~100 amino acids; see Fig 1a) to allow for such an interaction and the 2:1 stoichiometry, calculated from ITC, is supportive. However, the sHsps that form larger oligomers, such as those formed by full length Hsp27 or αB-crystallin, may interact with BAG3 in a more complex way. Indeed, it seems likely that a single BAG3 might use both IPV motifs to bind β4–β8 regions in nearby protomers within the oligomer, which aren’t necessarily part of a single homodimer.
One important implication of this model is that BAG3 might be expected to inhibit oligomerization of some sHsps. Specifically, it is known that the IXI motifs of some sHsps are important in linking dimers together into higher order oligomers. This idea is supported by the fact that Hsp20 and Hsp22 both lack their own IXI motifs and have reported to form relatively smaller oligomers in solution [56; 58]. Indeed, we found that incubation of full length Hsp27 with BAG3 lead to an overall decrease in both oligomer size and frequency, as judged by SEC-MALS and EM. Thus, BAG3 does not appear to be a “passive” scaffolding protein. Rather, its interactions with sHsps compete for IXI binding and partially disrupt quaternary structure. As we observed in the luciferase refolding experiments (see below), this structural change may have functional implications. It is important to note that interactions outside the IXI motifs also contribute to the formation of higher order oligomers, so it seems unlikely that BAG3 could convert oligomers to an entirely dimeric state.
BAG3 is a modular scaffolding protein for two components of the chaperone network
Hsp70 and the sHsps constitute an ancient system for protecting proteins under conditions of proteotoxic stress. sHsps are capable of binding and stabilizing unfolded/denatured proteins and keeping them in a refolding-competent form [21]. Due to their lack of intrinsic refolding ability, sHsps must then collaborate with ATP-powered refolding systems, such as the Hsp70 system [61]. Indeed, it has been shown both in vitro and in vivo that sHsp substrates can be refolded by the Hsp70 chaperone system [62–65]. However, a mechanistic understanding of how sHsps might communicate with the Hsp70 system has been elusive, especially for the mammalian systems.
Using FCPIA and SEC, we found that BAG3 could bind to Hsp70 at the same time as Hsp27, Hsp22 and αB-crystallin. Multiple pieces of evidence suggest that the strength of the two protein-protein interactions within this ternary complex (e.g. BAG3-Hsp70, BAG3-sHsp) are not influenced by the other. For example, the apparent affinity of the BAG3 interaction with Hsp27, Hsp22 and αB-crystallin was not significantly different in the presence of Hsp70 (see Fig 5). Moreover, deletion of the BAG domain or the IPV motifs from BAG3 did not impact binding to the other partner (see Fig 2 and Fig 4). Thus, it appears that BAG3 is a modular scaffolding protein that physically links sHsps to Hsp70. In this way, BAG3 may act like the scaffolding protein, HOP, which coordinates the Hsp70-Hsp90 axis [66]. In that system, HOP is thought to organizing “hand-off” of partially folded clients, such as nuclear hormone receptors, between the chaperones. We speculate that the mammalian chaperone network may contain additional examples of such scaffolding proteins.
BAG3 coordinates the functions of Hsp22 and Hsp70 during the refolding of denatured luciferase
Because smaller oligomers of Hsp27 and α-crystallin had been shown to be more potent chaperones [39; 40] and we found that BAG3 could compete for the IXI motifs that help hold together oligomers (see Fig 3), we wondered whether BAG3 might promote sHsp chaperone activity. Further, BAG3 is well known to regulate Hsp70’s chaperone function through its BAG domain interactions, so it might be expected to coordinate the activities of both sHsps and Hsp70s. Indeed, we found that the combination of BAG3, Hsp22 and Hsp70 (at the proper ratio) was a potent refolding machine (See Fig 6). The mechanistic details of this process are not yet known and it remains to be seen whether this ternary complex could be functionally important in cells and animals. Yet, these results suggest that BAG3 is both a physical and functional link between the sHsps and Hsp70s. In this way, BAG3 is not a “passive” scaffolding protein, but one that remodels the activity of its chaperone partners.
BAG3 binds to Hsp70 with an affinity of ~13 nM (see Fig 4) in the ATP state, while its affinity for sHsps is between ~8700 to 350 nM (see Fig 1). Although there is much to learn about how “client” proteins, expression level and post-translational modifications might impact these apparent affinity constants in the cell, it is interesting that BAG3 has a tighter affinity for Hsp70 than sHsps. This observation suggests that BAG3 and Hsp70 form a relatively more stable pair, which might only transiently coordinate with sHsps.
BAG3 may be a stress-inducible regulator of the chaperone network
BAG3 is the only stress-inducible member of the BAG family, so we propose that its unique scaffolding function may be especially important during conditions that favor protein unfolding and aggregation. In this scenario, the stress-inducible expression of BAG3 might bring together sHsps with Hsp70s, while simultaneously “activating” both systems. It is clear that other stress-inducible mechanisms, such as phosphorylation of sHsps [42; 43], also contribute to adaptive proteostasis. However, the expression of BAG3 may also play a role in this process through its ability to coordinate two major components of the chaperone network.
Methods
Cloning and Recombinant Protein Production
All domain deletion constructs were subcloned from the BAG3 pMCSG7 parent vector and confirmed with DNA sequencing. Mutations were constructed using standard mutagenesis protocols. Constructs for Hsp27, Hsp27c, Hsp27-3D, and α-crystallin were all received from the Klevit laboratory. Hsp22 was a kind gift from Jean-Marc Fontaine, and Hsp20 was received from the Conklin laboratory and subsequently cloned into the pMCSG7 vector. All constructs were transformed into BL21(DE3) cells and single colonies were used to inoculate TB medium containing ampicillin (50 µg/mL). Cultures were grown at 37 °C for 5 hours, cooled to 20 °C and induced overnight with 200 µM IPTG. BAG3 full length and IPV mutants were purified as previously described [50]. BAG3 deletion constructs were pelleted and re-suspended in His-binding buffer (50 mM Tris, 300 mM NaCl, 10 mM imidazole, pH 8.0) + 3M Urea. Samples were sonicated and then applied to the Ni-NTA resin. After Ni-NTA columns, all proteins were subjected to TEV cleavage overnight, concentrated and applied to a Superdex S200 (GE Healthcare) size exclusion column in BAG buffer (25 mM HEPES, 5 mM MgCl2, 150 mM KCl pH 7.5). Hsp72 and Hsp72NBD were purified as described elsewhere [67]. sHsp in plasmids containing an N-terminal His tag (Hsp27-3D, α-crystallin, Hsp22, and Hsp20) were all purified using a His column and subsequent SEC on a Superdex S200 in BAG buffer or PBS, as previously reported [33]. Hsp27 and Hsp27c were in tagless vectors, so they were purified using a two step ammonium sulfate precipitation followed by MonoQ and SEC. Briefly, ammonium sulfate was added to a final concentration of 16.9% (w/v), centrifuged, pellet discarded, and then an additional 16.9% (w/v) ammonium sulfate was added to the supernatant to precipitate the protein from solution. Precipitated protein was brought up and dialyzed into MonoQ Buffer A (20 mM Tris, pH 8.0) overnight, followed by a MonoQ column (0–1 M NaCl gradient), and finally an SEC on a Superdex S75 (Hsp27c) or Superdex S200 (GE Healthcare) equilibrated in 50 mM sodium phosphate, 100 mM NaCl, pH 7.5 buffer. Throughout the manuscript, the concentration of sHSPs is reported based on effective monomer concentration and not oligomer concentration.
Proteins were labeled with Alexa Fluor® 488 5-SDP ester or Alexa Fluor® 647 NHS ester (Life Technologies) according to the suppliers instructions. Hsp70 and sHsp were biotinylated using EZ-link NHS-Biotin (Thermo Scientific) according to the supplier instructions. After labeling, the proteins were subjected to gel filtration to remove unreacted label.
Isothermal Titration Calorimetry (ITC)
BAG3 constructs, Hsp72NBD and sHsps were dialyzed overnight against ITC buffer (25 mM HEPES, 5 mM MgCl2, 100 mM KCl pH 7.5). Concentrations were determined using a BCA Assay (Thermo Scientific), and the experiment was performed with a MicroCal VP-ITC (GE Healthcare) at 25 °C. Hsp72NBD (100 µM) or indicated sHsp (200 µM) in the syringe was titrated into a 10 µM cell solution of BAG3 protein. Calorimetric parameters were calculated using Origin® 7.0 software and fit with a one-site binding model.
Flow Cytometry Protein Interaction Assay (FCPIA)
The assay procedure was adopted from previous reports[50]. Briefly, biotinylated Hsp70 was immobilized (1h at room temperature) on streptavidin coated polystyrene beads (Spherotech). After immobilization, beads were washed to remove any unbound protein and then incubated with labeled BAG3 proteins at the indicated concentrations. Binding was detected using an Accuri™ C6 flow cytometer to measure median bead-associated fluorescence. Beads capped with biocytin were used as a negative control, and non-specific binding to beads was subtracted from signal.
For ternary complex formation experiments, Hsp27 or α-crystallin was immobilized on beads with constant concentration (50 nM) of Alexa 647-labeled BAG3 present. Increasing concentrations of Alexa 488-labeled Hsp72 NBD were titrated against the sHsp-BAG3 solution and fluorescence was measured using an Accuri™ C6 flow cytometer. Again, beads capped with biocytin were used as a negative control, and non-specific binding to beads was subtracted from signal.
Nucleotide Release Assay
A fluorescent ATP analogue, N6-(6-Amino)hexyl-ATP-5-FAM (ATP-FAM) (Jena Bioscience) was used to measure BAG3 induced nucleotide dissociation from Hsp72, as previously described [50]. In black, round-bottom, low-volume 384-well plates (Corning), 1 µM Hsp72 and 20 nM ATP-FAM were incubated with varying concentrations of BAG3 protein for 10 minutes at room temperature in assay buffer (100 mM Tris, 20 mM KCl, 6 mM MgCl2 pH 7.4). After incubation fluorescence polarization was measured (excitation: 485 nm emission: 535 nm) using a SpectraMax M5 plate reader.
SEC-MALS
Solutions of Hsp27 (15 µM) and BAG3 (either 15 or 30 µM) were pre-mixed and resolved by analytical size exclusion chromatography on a Shodex 804 column on an Ettan LC (GE Healthcare). Molecular weights were determined by multiangle laser light scattering using an in-line DAWN HELEOS detector and an Optilab rEX differential refractive index detector (Wyatt Technology Corporation). The column was equilibrated overnight in BAG buffer prior to analysis. Samples were run at the indicated concentrations. Calculation of molecular weights was performed using the ASTRA software package (Wyatt Technology Corporation).
Size Exclusion Chromotography
Solutions of BAG3 (6 µM), Hsp72 (6 µM), Hsp22 (12 µM), or BAG3-Hsp72-Hsp22 (6 µM : 6 µM : 12 µM) were examined using a Superdex S200 (GE Healthcare) size exclusion column. Indicated fractions were collected and analyzed using SDS-PAGE analysis. Samples were separated using 4–15% Tris-Tricine gels (Bio-Rad) and stained with Coomassie Blue. Image color was changed to grayscale for clarity.
Electron Microscopy
Solutions of Hsp27 (30 µM) and BAG3 (either 7.5, 15 or 30 µM) were pre-incubated, diluted and applied to a thin carbon coated copper grid for negative staining using uranyl formate at pH ~6.0 as previously described70. Electron micrographs were collected on a Tecnai T12 Microscope (FEI) equipped with a LaB6 filament operated at 120 kV. Images were collected at 50,000× magnification with a 2.2 Å /pixel spacing and 1.0–1.3 A defocus range on a 4k × 4k CCD camera (Gatan). Particle images were selected and extracted from micrographs, subsequently phase-corrected from the estimated CTF parameters using EMAN2 software71. 2D reference free alignment and classification was then performed using the SPIDER software package72. A set of 15 class averages representing ~1500 particles for each BAG3 concentration were used to measure the longest dimension observed as well as the distance perpendicular to obtain an average size of the particles.
Chaperone Assays
The steady state ATPase activity of Hsc70 was measured by malachite green and the refolding of chemically denatured firefly luciferase was measured by recovered luminescence, as previously reported [50]. Hsc70 was used at 1 µM. DnaJA2 was used at 0.5 µM and ATP at 1 mM. In the ATPase experiments, results were normalized to the signal from Hsc70 and DnaJA2 with 30 nM BAG3 (100%). In the luciferase refolding experiments, results were normalized to the luminescence signal produced by addition of Hsc70 and DnaJA2 alone (100%). At these concentrations and incubation times, this value represents only ~40% of the total luminescence signal that can be recovered. We selected these conditions to allow for BAG3 and Hsp22 to have the possibility of further improving the refolding activity.
Supplementary Material
Highlights.
BAG3 binds small heat shock proteins and Hsp70 at the same time
BAG3 reduces the size of small heat shock protein oligomers
BAG3 coordinates the ability of sHsp and Hsp70 refold denatured luciferase
Acknowledgments
This work was supported by the NIH NS059690 (to J.E.G.) and NIH EY017370 (to D.R.S.). We thank David Agard, Rachel Klevit, Bruce Conklin and Michael Welch for plasmids and access to equipment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009;19:52–61. doi: 10.1016/j.tcb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 chaperone proteins. Topics in current chemistry. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Quarterly reviews of biophysics. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell JC, Craig EA. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci U S A. 1984;81:848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 9.Genest O, Hoskins JR, Kravats AN, Doyle SM, Wickner S. Hsp70 and Hsp90 of E. coli Directly Interact for Collaboration in Protein Remodeling. J Mol Biol. 2015;427:3877–3889. doi: 10.1016/j.jmb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamoto H, Fujita K, Ohtaki A, Watanabe S, Narumi S, Maruyama T, et al. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. J Biol Chem. 2014;289:6110–6119. doi: 10.1074/jbc.M113.524801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, Zahn R, et al. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol. 2012;19:1347–1355. doi: 10.1038/nsmb.2442. [DOI] [PubMed] [Google Scholar]
- 12.Schmid AB, Lagleder S, Grawert MA, Rohl A, Hagn F, Wandinger SK, et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 2012;31:1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 15.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 16.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Alvira S, Cuellar J, Rohl A, Yamamoto S, Itoh H, Alfonso C, et al. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nature communications. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 18.Morrow G, Hightower LE, Tanguay RM. Small heat shock proteins: big folding machines. Cell Stress Chaperones. 2015;20:207–212. doi: 10.1007/s12192-014-0561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappe G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1–10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, et al. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem. 2004;279:7566–7575. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- 21.Haslbeck M, Vierling E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol. 2015;427:1537–1548. doi: 10.1016/j.jmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mainz A, Peschek J, Stavropoulou M, Back KC, Bardiaux B, Asami S, et al. The chaperone alphaB-crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat Struct Mol Biol. 2015;22:898–905. doi: 10.1038/nsmb.3108. [DOI] [PubMed] [Google Scholar]
- 23.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 24.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 25.Hochberg GK, Ecroyd H, Liu C, Cox D, Cascio D, Sawaya MR, et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci U S A. 2014;111:E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun N, Zacharias M, Peschek J, Kastenmuller A, Zou J, Hanzlik M, et al. Multiple molecular architectures of the eye lens chaperone alphaB-crystallin elucidated by a triple hybrid approach. Proc Natl Acad Sci U S A. 2011;108:20491–20496. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasta SY, Raman B, Ramakrishna T, Rao Ch M. The IXI/V motif in the C-terminal extension of alpha-crystallins: alternative interactions and oligomeric assemblies. Molecular vision. 2004;10:655–662. [PubMed] [Google Scholar]
- 28.Jehle S, Vollmar BS, Bardiaux B, Dove KK, Rajagopal P, Gonen T, et al. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc Natl Acad Sci U S A. 2011;108:6409–6414. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Studer S, Obrist M, Lentze N, Narberhaus F. A critical motif for oligomerization and chaperone activity of bacterial alpha-heat shock proteins. Eur J Biochem. 2002;269:3578–3586. doi: 10.1046/j.1432-1033.2002.03049.x. [DOI] [PubMed] [Google Scholar]
- 30.Arrigo AP. Human small heat shock proteins: protein interactomes of homo- and hetero-oligomeric complexes: an update. FEBS Lett. 2013;587:1959–1969. doi: 10.1016/j.febslet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Bova MP, McHaourab HS, Han Y, Fung BK. Subunit exchange of small heat shock proteins. Analysis of oligomer formation of alphaA-crystallin and Hsp27 by fluorescence resonance energy transfer and site-directed truncations. J Biol Chem. 2000;275:1035–1042. doi: 10.1074/jbc.275.2.1035. [DOI] [PubMed] [Google Scholar]
- 32.Aquilina JA, Benesch JL, Bateman OA, Slingsby C, Robinson CV. Polydispersity of a mammalian chaperone: mass spectrometry reveals the population of oligomers in alphaB-crystallin. Proc Natl Acad Sci U S A. 2003;100:10611–10616. doi: 10.1073/pnas.1932958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makley LN, McMenimen KA, DeVree BT, Goldman JW, McGlasson BN, Rajagopal P, et al. Pharmacological chaperone for alpha-crystallin partially restores transparency in cataract models. Science. 2015;350:674–677. doi: 10.1126/science.aac9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shashidharamurthy R, Koteiche HA, Dong J, McHaourab HS. Mechanism of chaperone function in small heat shock proteins: dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. J Biol Chem. 2005;280:5281–5289. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- 35.Giese KC, Vierling E. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J Biol Chem. 2002;277:46310–46318. doi: 10.1074/jbc.M208926200. [DOI] [PubMed] [Google Scholar]
- 36.Stromer T, Ehrnsperger M, Gaestel M, Buchner J. Analysis of the interaction of small heat shock proteins with unfolding proteins. J Biol Chem. 2003;278:18015–18021. doi: 10.1074/jbc.M301640200. [DOI] [PubMed] [Google Scholar]
- 37.Sobott F, Benesch JL, Vierling E, Robinson CV. Subunit exchange of multimeric protein complexes. Real-time monitoring of subunit exchange between small heat shock proteins by using electrospray mass spectrometry. J Biol Chem. 2002;277:38921–38929. doi: 10.1074/jbc.M206060200. [DOI] [PubMed] [Google Scholar]
- 38.Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, et al. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci U S A. 2010;107:2007–2012. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jovcevski B, Kelly MA, Rote AP, Berg T, Gastall HY, Benesch JL, et al. Phosphomimics destabilize Hsp27 oligomeric assemblies and enhance chaperone activity. Chem Biol. 2015;22:186–195. doi: 10.1016/j.chembiol.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Peschek J, Braun N, Rohrberg J, Back KC, Kriehuber T, Kastenmuller A, et al. Regulated structural transitions unleash the chaperone activity of alphaB-crystallin. Proc Natl Acad Sci U S A. 2013;110:E3780–E3789. doi: 10.1073/pnas.1308898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nahomi RB, DiMauro MA, Wang B, Nagaraj RH. Identification of peptides in human Hsp20 and Hsp27 that possess molecular chaperone and anti-apoptotic activities. Biochem J. 2015;465:115–125. doi: 10.1042/BJ20140837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arrigo AP, Gibert B. HspB1 dynamic phospho-oligomeric structure dependent interactome as cancer therapeutic target. Current molecular medicine. 2012;12:1151–1163. doi: 10.2174/156652412803306693. [DOI] [PubMed] [Google Scholar]
- 43.Welsh MJ, Gaestel M. Small heat-shock protein family: function in health and disease. Ann N Y Acad Sci. 1998;851:28–35. doi: 10.1111/j.1749-6632.1998.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 44.Rajagopal P, Tse E, Borst AJ, Delbecq SP, Shi L, Southworth DR, et al. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. eLife. 2015;4 doi: 10.7554/eLife.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleckenstein T, Kastenmuller A, Stein ML, Peters C, Daake M, Krause M, et al. The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes. Mol Cell. 2015;58:1067–1078. doi: 10.1016/j.molcel.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 46.Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. Journal of structural biology. 2012;179:152–160. doi: 10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Calderwood SK, Gong J. Heat Shock Proteins Promote Cancer: It's a Protection Racket. Trends Biochem Sci. 2016 doi: 10.1016/j.tibs.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, et al. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- 49.Rampelt H, Mayer MP, Bukau B. Nucleotide exchange factors for Hsp70 chaperones. Methods Mol Biol. 2011;787:83–91. doi: 10.1007/978-1-61779-295-3_7. [DOI] [PubMed] [Google Scholar]
- 50.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J Biol Chem. 2014;289:1402–1414. doi: 10.1074/jbc.M113.521997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwasaki M, Tanaka R, Hishiya A, Homma S, Reed JC, Takayama S. BAG3 directly associates with guanine nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell adhesion. Biochem Biophys Res Commun. 2010;400:413–418. doi: 10.1016/j.bbrc.2010.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013;23:430–435. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- 53.Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, et al. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731–4740. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doong H, Price J, Kim YS, Gasbarre C, Probst J, Liotta LA, et al. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70. Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 55.Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, Charette SJ, et al. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2010;425:245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- 56.Carra S, Seguin SJ, Landry J. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008;4:237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 57.Hishiya A, Salman MN, Carra S, Kampinga HH, Takayama S. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PLoS One. 2011;6:e16828. doi: 10.1371/journal.pone.0016828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van de Klundert FA, Smulders RH, Gijsen ML, Lindner RA, Jaenicke R, Carver JA, et al. The mammalian small heat-shock protein Hsp20 forms dimers and is a poor chaperone. Eur J Biochem. 1998;258:1014–1021. doi: 10.1046/j.1432-1327.1998.2581014.x. [DOI] [PubMed] [Google Scholar]
- 59.Theriault JR, Lambert H, Chavez-Zobel AT, Charest G, Lavigne P, Landry J. Essential role of the NH2-terminal WD/EPF motif in the phosphorylation-activated protective function of mammalian Hsp27. J Biol Chem. 2004;279:23463–23471. doi: 10.1074/jbc.M402325200. [DOI] [PubMed] [Google Scholar]
- 60.Aquilina JA, Benesch JL, Ding LL, Yaron O, Horwitz J, Robinson CV. Subunit exchange of polydisperse proteins: mass spectrometry reveals consequences of alphaA-crystallin truncation. J Biol Chem. 2005;280:14485–14491. doi: 10.1074/jbc.M500135200. [DOI] [PubMed] [Google Scholar]
- 61.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 62.Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem. 2003;278:31033–31042. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- 63.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2000;122:189–198. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 67.Thompson AD, Scaglione KM, Prensner J, Gillies AT, Chinnaiyan A, Paulson HL, et al. Analysis of the tau-associated proteome reveals that exchange of Hsp70 for Hsp90 is involved in tau degradation. ACS Chem Biol. 2012;7:1677–1686. doi: 10.1021/cb3002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.