Abstract
Culture based methods are commonly employed to detect pathogens in food and environmental samples. These methods are time consuming and complex, requiring multiple non-selective and selective enrichment broths, and usually take at least 1 week to recover and identify pathogens. Improving pathogen detection in foods is a primary goal for regulatory agencies and industry. Salmonella detection in food relies on a series of culture steps in broth formulations optimized to resuscitate Salmonella and reduce the abundance of competitive bacteria. Examples of non-selective pre-enrichment broths used to isolate Salmonella from food include Lactose, Universal Pre-enrichment, BPW, and Trypticase Soy broths. Tetrathionate (TT) and Rappaport–Vassiliadis (RV) broths are employed after a 24-h non-selective enrichment to select for Salmonella and hamper the growth of competitive bacteria. In this study, we tested a new formulation of TT broth that lacks brilliant green dye and has lower levels of TT . We employed this TT broth formulation in conjunction with a 6-h non-selective pre-enrichment period and determined that Salmonella recovery was possible one day earlier than standard food culture methods. We tested the shortened culture method in different non-selective enrichment broths, enumerated Salmonella in the non-selective enrichments, and used 16S rRNA gene sequencing to determine the proportional abundances of Salmonella in the TT and RV selective enrichments. Together these data revealed that a 6-h non-selective pre-enrichment reduces the levels of competitive bacteria inoculated into the selective TT and RV broths, enabling the recovery of Salmonella 1 day earlier than standard culture enrichment methods.
Keywords: Salmonella, FDA BAM, metagenomics, 16S rRNA, selective enrichment, tetrathionate broth
Introduction
The impact of Salmonella food contamination on public health has resulted in the development of several comprehensive culture-based methods to detect Salmonella in food. These are found namely in the Food and Drug Administration (FDA) Bacteriological Analytical Manual (BAM), the United States Department of Agriculture (USDA) Microbiological Laboratory Guidebook (MLG), and the ISO 6579:2002 Microbiology of food and animal feeding stuffs- Horizontal method for the detection of Salmonella species (ISO) (ISO, 2002; USDA, 2014; FDA, 2015). The current method used by the FDA, described in the BAM, requires a 24-h resuscitation in non-selective pre-enrichment broth, followed by parallel 24-h selective enrichments in Tetrathionate (TT) and Rappaport–Vassiliadis (RV) broths to reduce the growth of competitive bacteria (FDA, 2015). Aliquots of TT and RV enrichments are then plated on selective agars, which are incubated for 24 to 48-h. Presumptive Salmonella colonies are isolated for confirmatory testing which can take an additional 2 to 3 days.
Important considerations for successful recovery of Salmonella from food include the type of food, the level of stress or injury imparted on the Salmonella by the food matrix or processing environment, the presence of competitive bacteria, and the level of Salmonella contamination (D’Aoust, 1981; Busse, 1995). Previous studies noted the importance of resuscitating stressed and injured Salmonella, so current methods utilize specific non-selective pre-enrichment broths to recover Salmonella from food (Rappaport et al., 1956; D’Aoust and Maishment, 1979; D’Aoust, 1981; Ray, 1989; Chen et al., 2013). Supplements such as bile salts, brilliant green dye, MgCl2, and malachite green dye are added to broths used for selective enrichment to reduce competitive bacteria in foods tested for Salmonella contamination (Teague and Clurman, 1916; Rappaport et al., 1956; Vassiliadis et al., 1978; Peterz et al., 1989). For example, the low levels of Salmonella, enumerated from pine nuts (0.028–0.093 MPN/g) and paprika (0.04–0.05 CFU/g) implicated in recent outbreaks, highlight the need for non-selective pre-enrichment prior to selective enrichment to increase the probability of detection, especially for injured or stressed organisms that would not survive in selective enrichment broths (Lehmacher et al., 1995; Wang et al., 2015).
Common to all of the detection methods employed for the recovery of Salmonella from food, regardless of the type of food or non-selective pre-enrichment broth, is the use of TT and RV selective broths. The selective action of TT includes: bile salts and brilliant green dye to inhibit gram-positive bacteria; an iodine–iodide (I2–KI) solution added to the base broth to induce TT production, providing a metabolic advantage to organisms that have TT reductase such as Salmonella; and calcium carbonate to neutralize the sulfuric acid produced when TT is reduced, thus maintaining the pH (Teague and Clurman, 1916; Knox, 1945; Palumbo and Alford, 1970; Moats et al., 1974; D’Aoust, 1981; Moats, 1981). Salmonella growth in TT broth relies on the ability of Salmonella to reduce TT and withstand the bactericidal activity of brilliant green dye. It is notable that during infection in the human intestine Salmonella induces inflammation of gut tissues to induce TT production which it can use as an energy source through anaerobic respiration, giving this organism a competitive advantage over competing bacteria that use fermentation in this anaerobic environment (Winter et al., 2010).
Rappaport–Vassiliadis broth has different selective properties to improve Salmonella recovery, namely MgCl2 at a concentration that creates hypertonic conditions that inhibit Proteus and Escherichia coli, and malachite green to inhibit other competing bacteria (Rappaport et al., 1956; Vassiliadis et al., 1978; Peterz et al., 1989). It is important to use MgCl2 instead of other Mg salts, such as MgS203, MgBr2, Mg(N03)2, BaCl2, CaS203.Ca(N03)2, CaBr2, Cal2, KNO3, or BeCI2 in RV, as these other salts are not as selective (Rappaport et al., 1956). The low pH of RV, well tolerated by Salmonella, provides additional selection against coliforms (Rappaport et al., 1956; Vassiliadis et al., 1981; Vassiliadis, 1983). Together the attributes of RV broth, MgCl2, low pH, and malachite green, provide a high osmotic pressure, a low pH that declines over time with bacterial metabolism, and inhibition of gram-positive organisms, respectively, that provide favorable conditions for Salmonella growth (van Schothorst and Renaud, 1985; Peterz et al., 1989). An additional selective feature of both RV and TT broths are incubation temperatures above 37°C to reduce competitive bacteria (D’Aoust, 1981; Peterz et al., 1989). The FDA BAM recommends incubation of RV medium for 24 ± 2 h at 42 ± 0.2°C and incubation of TT broth for 24 ± 2 h at 43 ± 0.2°C (FDA, 2015).
Despite the efficacy and proven success of current culture-based methods for Salmonella detection, reducing detection time is a priority for food safety. Approaches to reduced detection time include reducing the length of non-selective and selective enrichments, changing the broth formulations, and altering incubation temperatures. Attempts to alter enrichment times and temperatures have met with mixed results (Mohr et al., 1974; van Schothorst and van Leusden, 1975; D’Aoust and Maishment, 1979). One investigation determined Salmonella was able to resuscitate sufficiently after 5 to 6-h in a non-selective pre-enrichment broth and could overcome the toxic effects of selective enrichment (Chen et al., 1993). However, most early studies with shortened non-selective pre-enrichments were unsuccessful (Mohr et al., 1974; van Schothorst and van Leusden, 1975; D’Aoust and Maishment, 1979). Additional studies to shorten the time for selective enrichment also failed due to false negative results, especially in low moisture foods (D’Aoust et al., 1990).
The choice of non-selective and selective pre-enrichment broths depends on the food or environmental samples being tested. The FDA BAM, USDA MLG, and ISO manuals specify enrichment media formulations that are suitable for Salmonella detection in foods based on characteristics such as pH, high versus low microbial loads in the food, and the moisture content of the food (ISO, 2002; USDA, 2014; FDA, 2015). Foods that are regulated by the FDA are divided into 35 categories that require 15 different pre-enrichment broths (FDA, 2015).
Molecular methods, such as qPCR, and automated platforms, such as the VIDAS Easy, have successfully detected Salmonella in 24-h non-selective pre-enrichments of pine nuts, chili powder, soft cheese, raw fish, and tomatoes, thus reducing the detection time (Cheng et al., 2009; Wang et al., 2015). However, culture based methods are considered the gold standard for regulatory agencies highlighting the importance of reducing the culture-dependent detection time. Methods such as Pulse Field Gel Electrophoresis and whole genome sequencing currently employed for traceback analysis during foodborne outbreak investigations require pure cultures of bacteria (Swaminathan et al., 2001; Allard et al., 2016).
Recent advances in high-throughput DNA sequencing provide opportunities to profile commodity-associated microbiomes either through amplicon sequencing of the 16S rRNA gene or whole-genome shotgun metagenomic sequencing. Metagenomic sequencing provides accurate data about the entire microbial community including organisms that cannot be cultured using traditional methods (Caporaso et al., 2012). Studies aimed at improving foodborne pathogen detection have utilized microbiome profiling to characterize food microbiomes and follow microbial community shifts throughout the BAM culture process (Pettengill et al., 2012; Ottesen et al., 2013; Jarvis et al., 2015; Leonard et al., 2015). These studies also demonstrate the value of microbiome profiling for characterizing the growth of Salmonella and Escherichia coli pathogens amidst the complex bacterial communities that naturally inhabit leafy green produce and tomatoes (Pettengill et al., 2012; Ottesen et al., 2013; Jarvis et al., 2015; Leonard et al., 2015). The approach is particularly suitable for characterizing competitive members of the microbial community that will hamper detection of pathogens in contaminated foods.
In this study, we employed a combination of microbiology and metagenomic methods to characterize and test potential changes to the FDA BAM method for Salmonella detection in leafy greens. First, four non-selective pre-enrichment broths including Lactose broth, Modified Buffered Peptone Water (mBPW), Trypticase Soy Broth (TSB), and Universal Pre-enrichment broth (UP), were compared in the FDA BAM culture method using cilantro as a model for leafy green produce. Second, we tested changes in the FDA BAM TT broth formulation that reduced selective pressure by eliminating brilliant green and reducing the concentration of iodine as follows: (1) TT, formulated according to the FDA BAM with brilliant green and 2% I2-KI solution; (2) TTA, no brilliant green, 2% I2-KI solution; and (3) TTB, no brilliant green, 1% I2–KI solution. Third, we tested the efficacy of 5 and 6-h non-selective pre-enrichment times in conjunction with the different TT broth formulations, for Salmonella recovery from food.
We evaluated the efficacy of these changes to the BAM method by enumerating Salmonella contamination levels in the non-selective pre-enrichments and employed 16S rRNA gene amplicon sequencing to define the microbial communities in selective TT and RV broth enrichments. Finally, we tested the combination of a reduced non-selective pre-enrichment time and reduced TT selectivity for Salmonella recovery from raw chicken thighs, liquid whole eggs, and peanut butter.
This study demonstrates that a less selective formulation of the BAM TT broth, inoculated with a 6-h non-selective pre-enrichment, consistently recovered Salmonella one day earlier than the current FDA BAM culture method. Additionally, we show that this method can be adapted for use with other food commodities, which is advantageous for Public Health Laboratories that test a variety of foods.
Materials and Methods
Foods Matrices Tested in This Study
Four foods were tested in this study including cilantro, raw chicken thighs, liquid whole eggs, and peanut butter. Cilantro samples, provided by the Department of Agriculture and Rural Development in Lansing, Michigan or purchased from a local grocery store, were stored at 4°C prior to testing. Raw chicken thighs and liquid whole eggs, purchased from a local supermarket, were stored at 4°C prior to testing. Peanut butter samples, purchased from a local grocery store, were stored at room temperature.
Bacterial Strains for Food Inoculations
Nine Salmonella enterica strains representing four serovars, Newport, Tennessee, Thompson, and Enteritidis, were used for food inoculation (Table 1). Two of the strains used to inoculate cilantro, S. Newport SALC14 and S. Tennessee SALC 76, were cultured from cilantro in our laboratory and the third strain, S. Thompson SALC 818, was chosen because S. Thompson was implicated in a cilantro outbreak in 1999 (Campbell et al., 2001). The three S. Tennessee strains used to inoculated peanut butter were isolated from peanut butter during a 2007 peanut butter outbreak (Wilson et al., 2016) and the S. Enteritidis strains used for the chicken and egg inoculations were isolated from the respective foods (Table 1). Culture stocks, stored at –80°C, were streaked onto Trypticase Soy Agar plates (Difco, Sparks, MD, USA) and incubated overnight at 35 ± 2°C. Bacterial cell suspensions, equal to 0.48–0.52 McFarland turbidity units (corresponding to approximately 1 × 108 CFU/ml), were prepared in 0.85% sterile saline and serially diluted to approximately 28 CFU/mL for food inoculations.
Table 1.
Salmonella strains used to inoculate foods.
| DVA name | Nickname | Serotype | Origin | State/Country | Food inoculated | Non-selective pre-enrichment broths tested |
|---|---|---|---|---|---|---|
| SALC 14 | N/A | Newport | Cilantro | New York, USA | Cilantro | Lactose, mBPW |
| SALC 76 | N/A | Tennessee | Cilantro | New York, USA | Cilantro | mBPW, TSB, UP |
| SAL 818 | TXAML1201424 | Thompson | N/A | Texas, USA | Cilantro | mBPW, TSB, UP |
| SAL 622 | SL487 | Tennessee | Peanut Butter Outbreak | Minnesota, USA | Peanut Butter | mBPW |
| SAL 623 | SL488 | Tennessee | Peanut Butter Outbreak | Minnesota, USA | Peanut Butter | mBPW |
| SAL 624 | SL489 | Tennessee | Peanut Butter Outbreak | Tennessee, USA | Peanut Butter | mBPW |
| SAL 274 | B7849 | Enteritidis | Chicken | Spain | Liquid whole eggs | TSB with FeSO4 |
| SAL 274 | B7849 | Enteritidis | Chicken | Spain | Raw Chicken thighs | BPW |
| SAL 311 | SL915 | Enteritidis | Egg Yolk | N/A | Liquid whole eggs | TSB with FeSO4 |
| SAL 311 | SL915 | Enteritidis | Egg Yolk | N/A | Raw Chicken thighs | BPW |
| SAL 336 | SL940 | Enteritidis | Egg | N/A | Liquid whole eggs | TSB with FeSO4 |
| SAL 336 | SL940 | Enteritidis | Egg | N/A | Raw Chicken thighs | BPW |
Prior to inoculation, liquid whole eggs were homogenized in sterile beakers, and the raw chicken thighs were cubed into approximately 1-inch squares using sterile knives. All foods were aseptically portioned (25 g) into sterile Whirlpak bags (Nasco; Fort Atkinson, WI, USA), inoculated with approximately 28 CFU Salmonella per 25 g of food, and aged to simulate natural contamination at 4°C for 48–72 h (cilantro, liquid whole eggs, and chicken thighs) or at room temperature for 14 days (peanut butter). Un-inoculated controls samples were also prepared and aged in parallel for each food.
Non-selective Pre-enrichment of Foods
Aged cilantro samples were enriched in Lactose broth, UP or TSB prepared according to the FDA BAM. Peanut butter and additional cilantro samples were enriched in Modified Buffer Peptone Water [mBPW; Buffered Peptone Water (BPW, DifcoTM, Sparks, MD, USA), with 3.5 g of disodium phosphate and 1.5 g of monopotassium phosphate per liter]. Aged liquid whole eggs were enriched in TSB with ferrous sulfate prepared according to the FDA BAM. The aged chicken thighs were enriched in BPW, the non-selective pre-enrichment broth recommended in the MLG method for the isolation and identification of Salmonella from poultry.
Selective Enrichment Broths
Two modifications that reduce the selective strength of TT broth were compared to the standard TT broth formulation used in the FDA BAM. Standard BAM TT broth consists of a base broth to which Iodine-Potassium Iodide (I2-KI) and brilliant green dye solutions are added on the day of use. Both TT modifications used in this study consist of the TT broth base without the addition of brilliant green dye. TT modification A (TTA) consists of the TT broth base with 2.0% of the I2–KI solution as per the BAM, and TT modification B (TTB) contains only 1.0% of the I2–KI solution. The FDA BAM, USDA MLG, and ISO methods all use a second selective enrichment broth called RV for Salmonella detection. The RV used in this study was prepared in accordance with the FDA BAM method.
A panel of 101 Salmonella isolates, derived from the Defense Science Office (DSO) of the Defense Advanced Research Projects Agency (DARPA), Systems and Assays for Food Examination (SAFE) Program, were cultured in selective RV, TT, TTA, and TTB broths to compare efficacy for Salmonella recovery. The SAFE collection is used at the FDA for validating new methods for Salmonella detection in food. The collection is divided into three groups representing a Salmonella subspecies set (55 strains), an outbreak cluster set (26 strains), and a food set (20 strains, Supplementary Table S1). Parallel sets of RV, TT, TTA, and TTB selective broths were inoculated with 103 or 105 CFU of each Salmonella isolate and incubated at 42 ± 2°C (RV) or 43 ± 0.2°C (TT, TTA, and TTB) for 24 ± 2-h. The following day 10 μl of each culture was plated onto Xylose-Lysine-Desoxycholate Agar (XLD, Becton, Dickinson and Company, Sparks, MD, USA) agar, and after a 24 h incubation at 35 ± 2°C, the plates were recorded as positive (black colonies) or negative (no growth) for Salmonella.
24-hour Non-selective Pre-enrichment with TT Modifications
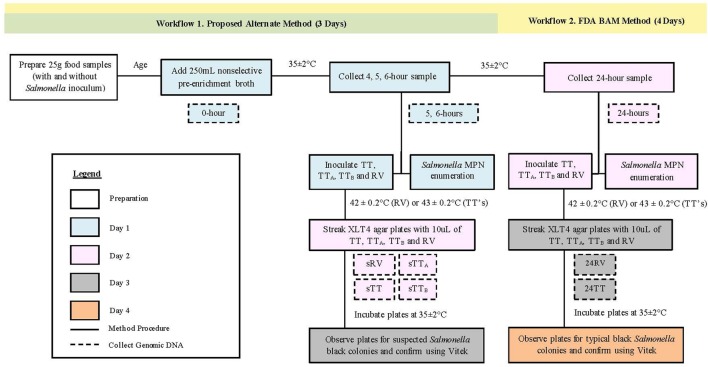
All food samples were processed following the FDA BAM workflow for the detection of Salmonella with a 24-h non-selective pre-enrichment and the standard and modified TT broths, as follows. On day one after aging, food samples were aseptically combined with 250mL of sterile non-selective pre-enrichment broth, massaged for 2 min, and incubated overnight at 35 ± 2°C (Figure 1). On day two, aliquots of the 24-h non-selective pre-enrichments were aseptically transferred to RV (100 μL), TT (1.0 mL), TTA (1.0 mL), and TTB (1.0 mL) selective enrichment broths, and incubated overnight at 42 ± 0.2°C (RV) or 43 ± 0.2°C (TT) (Figure 1). On day three, 10 μL of each selective enrichment was plated on Xylose Lysine TergitolTM 4 (XLT4, Becton, Dickinson and Company, Sparks, MD, USA) agar and incubated overnight at 35 ± 2°C. The remaining volumes of RV, TT, TTA, and TTB selective enrichments from cilantro samples were centrifuged at 7,100 rcf for 30 min to pellet the bacteria for DNA extraction, to represent 24RV, 24TT, 24TTA, and 24TTB time points used for 16S rRNA gene sequencing (Figure 1). Bacterial pellets were stored at –20°C. On day four, suspect black colonies from XLT4 plates were confirmed as Salmonella on the VITEK 2 system (BioMérieux, France).
FIGURE 1.
Workflow for food processing and sample collection.All experiments used Salmonella positive and negative food samples that were aged and processed for Salmonella detection using two workflows. Non-selective enrichment samples were collected at 5, and 6-h (workflow 1, green) and 24-h (workflow 2, yellow) for Salmonella enumeration, inoculation of selective TT and RV enrichments and 16S rRNA gene sequencing. Daily sample processing steps were performed as outline on Day 1 (Blue), Day 2 (Pink), Day 3 (Gray), and Day 4 (Orange).
Reduced Incubation Time for Non-selective Pre-enrichments
In addition to the samples processed according the FDA BAM timeline, replicate samples of each food were incubated for a reduced time in their respective pre-enrichment broths (Figure 1). Specifically, on day one, the non-selective pre-enrichments were incubated at 35 ± 2°C for 5 or 6-h, and aliquots from each time period were transferred to RV (100 μL), TT (1.0 mL), TTA (1.0 mL), and TTB (1.0 mL) selective broths and incubated overnight at 42 ± 0.2°C (RV) or 43 ± 0.2°C (TT). On day two, each selective broth was streaked onto XLT4 agar and incubated overnight. Remaining volumes of RV, TT, TTA, and TTB selective broths from each time point were centrifuged at 7,100 rcf for 30 min to pellet the bacteria for DNA extraction to represent 5 and 6-h RV, TT, TTA, TTB samples (hereafter referred to as sRV, sTT, sTTA, and sTTB) for 16S rRNA gene sequencing (Figure 1). All bacterial pellets were stored at –20°C. On day three, one day earlier than the FDA BAM method, suspect black colonies from XLT4 agar plates were confirmed as Salmonella on the VITEK 2 system.
Enumeration of Salmonella in Cilantro
Cilantro non-selective pre-enrichments incubated for 5, 6, and 24-h were enumerated for Salmonella using the Most Probable Number (MPN) method described in the FDA BAM. Non-selective pre-enrichment samples were serially diluted (10-fold) in 0.85% NaCl. Selective TTB was inoculated, in triplicate, with 1.0 mL of each dilution, incubated at 43 ± 0.2°C for 48-h, and then streaked onto XLT4. The XLT4 agar plates were examined for the presence of typical black Salmonella colonies, which were confirmed in the VITEK 2 system. The FDA BAM MPN calculator was used to compute the results (www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm109656.htm).
Preparation of Genomic DNA
Bacterial pellets from the selective RV, TT, TTA, and TTB enrichment samples were lysed for 20-min at room temperature with 100 ng/uL lysozyme (Sigma–Aldrich; Saint Louis, MO, USA), followed by DNA extraction on the QIACube using the QIAamp DNA mini protocol (QIAGEN, Germany).
16S rRNA Gene Amplification of Selective Enrichments
A two-step PCR amplification strategy was used to generate barcoded 16S rRNA gene amplicons with four sets of PCR primers targeting the Variable 1 to Variable 3 (V1 to V3) regions of the gene. These primers have added base pairs upstream of the 16S rRNA gene aimed at adding nucleotide base diversity to the initial sequencing cycles on the Illumina MiSeq (Supplementary Table S2). Four master mixes were prepared, one for each primer set, consisting of 3.0 ng template DNA, 5.0 μl 5× Omni Klentaq Master Mix (DNA Polymerase Technology, Inc., St. Louis, MO, USA), 10.0 μl PCR Enhancer Cocktail (DNA Polymerase Technology, Inc., St. Louis, MO, USA), and 1.5 μl each forward and reverse primers (600nM final concentration). PCR cycling conditions consisted of a 2-minute denaturation at 94°C, followed by 25 cycles of 40 s at 94°C, 15 s at 56°C, 40 s at 68°C, and a final extension at 68°C for 5 min. The 16S rRNA gene PCR amplicons were visualized on 1% agarose gels and, purified using the QIAquick PCR purification kit (QIAGEN, Germantown, MD, USA) if multiple bands were present.
The second set of PCR reactions used the 16S rRNA gene amplicon PCR products as template with the Illumina Nextera XT indexing primers to generate unique PCR amplicons that were compatible with the Illumina MiSeq sequencing chemistry. Briefly, the 16S rRNA gene amplicons were quantified using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), then amplified in 50 μl reactions with ∼150 ng amplicon products, 5× Omni Klentaq Master Mix, and 5 μl each of the Illumina i7 and i5 indexing primers from the Nextera XT Indexing Kit (Illumina Inc., San Diego, CA, USA). Indexing PCR cycling conditions included initial 3 min and 30 s holds at 72 and 98°C, respectively, followed by 5 cycles of 10 s at 98°C, 30 s at 63°C, and 3 min at 72°C, and a final hold at 10°C.
Library Preparation and 16S rRNA Gene Sequencing
Libraries were size selected using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) to an average library size of 600 bp, and then quantified using the Qubit 2.0 Fluorometer. Library quality was verified on the Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) using the DNA 1000 chip kit. Size selected libraries were normalized to 2 nM using the SequelPrep Normalization Plate Kit, pooled, and then measured using High Sensitivity Qubit Reagents (Life Technologies, Carlsbad, CA, USA). The library pools were denatured with 0.1N NaOH, serially diluted to 8pM with HT1, spiked with 10% 12.5pM PhiX174 bacteriophage DNA, and then sequenced using a 600-cycle MiSeq Reagent Kit v3 (Illumina Inc., San Diego, CA, USA) in four sequencing runs, two with 96 samples, one with 86 samples, and one with 87 samples.
16S rRNA Gene Sequence Data Processing and Analysis
Raw paired-end reads output by the MiSeq platform were merged into consensus fragments by FLASH (Magoc and Salzberg, 2011) and subsequently filtered for quality (max error rate 1%) and length (minimum 200bp) using Trimmomatic and QIIME (Caporaso et al., 2010; Kuczynski et al., 2011; Bolger et al., 2014). Spurious hits to the PhiX control genome were identified using BLASTn and removed. Passing sequences were trimmed of primers and evaluated for chimeras with UCLUST (de novo mode) (Edgar et al., 2011). Chloroplast contaminants were detected and filtered using the RDP classifier with a confidence threshold of 80% (Wang et al., 2007). Sequences were further screened for unknown contaminant using a sensitive BLASTn search against the GreenGenes database (McDonald et al., 2012). To normalize across samples, the 16S rRNA gene sequence profiles were subsampled to an even level of coverage prior to downstream comparisons. High-quality 16S rRNA gene sequences were assigned to a taxonomic lineage using Resphera Insight (Baltimore, MD, USA; www.respherabio.com). Mock community analyses compared Resphera Insight assignments to default UCLUST-Ref and RDP pipelines implemented within QIIME (Caporaso et al., 2010; Kuczynski et al., 2011).
Two Illumina MiSeq runs multiplexed with 96 samples produced a total of 9,631,743 and 6,949,843 quality filtered 16S rRNA gene sequences with average (average ± SD) read lengths of 412±19 and 378±27, respectively. Eighty-seven multiplexed samples in a third Illumina MiSeq run generated 8,875,924 quality-filtered sequences with an average read length of 409±24. A fourth MiSeq run of 86 samples yielded 7,631,971 quality filtered reads with aver age read lengths of 399±24. Raw 16S amplicon sequences reported in this paper have been deposited in the NCBI SRA repository (BioProject accession no. PRJNA343808).
Resphera Insight Validation
To perform an external validation of the species level accuracy of Resphera Insight, we utilized 110 whole-genome shotgun datasets from the GenomeTrakr Project (NCBI Project ID PRJNA183844) designated as novel S. enterica isolates. Raw paired-end sequences were filtered for quality and length, followed by merging of overlapping sequences using FLASH (Magoc and Salzberg, 2011). Merged reads were then screened for 16S rRNA fragments using Bowtie2 (Langmead and Salzberg, 2012) against a broad database of 16S rRNA gene sequences, with additional BLAST-based filtering to confirm location specific query matches to a reference S. enterica 16S rRNA gene (Salmonella enterica ssp. enterica serovar Typhimurium strain LT2; NCBI accession NR_074910.1).
Passing sequences were submitted to Resphera Insight (Baltimore, MD, USA; www.respherabio.com) for high-resolution taxonomic identification. We also compared performance of Insight to the RDP classifier and UCLUST reference algorithms implemented within QIIME (Kuczynski et al., 2012; Navas-Molina et al., 2013). Our primary measure of performance was the Diagnostic True Positive rate (DTP), defined as the percentage of reads with an unambiguous assignment to S. enterica, and we evaluated differences in accuracy associated with changes in read length and gene position.
Statistical Analyses
Z tests were performed to compare the recovery rates of Salmonella SAFE isolates inoculated into TT, TTA, and TTB. Z tests were also used to compare the efficacy of Lactose, mBPW, TSB, and UP non-selective enrichments at 5, 6, and 24 h for recovery of Salmonella from selective RV, TTA, TTB, and TT and to compare the efficacy of the selective enrichment broths. For the ladder comparisons the p values were adjusted for multiple comparisons, using the Benjamini and Hochberg method (R software v3.2) after continuity corrections were implemented (SAS 9.4).
Results
Salmonella Recovery from TT, TTA, TTB, and RV Selective Enrichment Broths
Since a reduced non-selective pre-enrichment will likely have fewer metabolically active cells of Salmonella, as compared to a 24 h pre-enrichment, we first evaluated the sensitivity of standard and modified RV and TT selective enrichment broths at a low inoculum level to mimic these conditions. To do this we directly inoculated RV and TT broths with 103 CFU/mL of each of the 101 SAFE isolates. We also directly inoculated TT and RV broths with a high inoculum of 105 CFU/mL as control samples. Among the selective broths inoculated with 103 CFU/mL Salmonella, the TTB and TTA selective broths had significantly higher recovery rates of 92 and 88% (P ≤ 0.001) compared to the FDA BAM TT formulation (17%) (Table 2; Supplementary Table S1). The Outbreak cluster set had the lowest recovery from the BAM TT broth of 4% (Table 2; Supplementary Table S1). Furthermore, the Outbreak cluster and Food sets of Salmonella SAFE strains were recovered at 100 and 90% in TTB, respectively, both significantly higher than the FDA BAM TT formulations (Table 2; Supplementary Table S1). Interestingly, the recovery of Salmonella from BAM RV (58%) was also lower than TTA (88%) and TTB (92%) for the entire SAFE collection (Table 2; Supplementary Table S1).
Table 2.
Percent recovery of Salmonella from selective enrichment broths.
| Inoculum levels | 103 CFU/mL |
105 CFU/mL |
||||
|---|---|---|---|---|---|---|
| RV (%) | TT (%) | TTBa (%) | TTAa (%) | RV (%) | TT (%) | |
| Total SAFE collection (n = 101) | 58 | 17 | 92∗∗ | 88∗∗ | 97 | 75 |
| Subspecies Set (n = 55) | 60 | 18 | 89∗∗ | 85∗∗ | 95 | 75 |
| Outbreak Cluster Set (n = 26) | 50 | 4 | 100∗∗ | 100∗∗ | 100 | 73 |
| Food Set (n = 20) | 65 | 30 | 90∗∗ | 80∗ | 100 | 80 |
aZ test of significant difference (p < 0.001) between TTA or TTB and TT
∗∗p < 0.001
∗p = 0.004
We inoculated a second set of BAM RV and TT broths with the SAFE Salmonella collection at 105 CFU/mL and were able to recover 97 and 75% of the isolates, respectively, indicating that for some of the SAFE panel strains, the BAM selective broths require more than105 CFU/mL in the food non-selective pre-enrichment for Salmonella recovery (Table 2; Supplementary Table S1). The Subspecies set of Salmonella isolates had the lowest recovery levels in the 105 CFU/mL BAM RV (95%) and TT (75%, Table 2). The Outbreak cluster and Food sets of SAFE strains were recovered in 100% of the BAM RV, and in 73 and 80% in BAM TT broths, respectively, at this inoculum level (Table 2).
In summary, we determined that TTA and TTB broths significantly improved the recovery of Salmonella (inoculated at 103 CFU/mL) and yielded positive results in 88 and 92%, respectively, of the SAFE isolates tested, as compared to the BAM TT and RV recovery rates of 17 and 58%, respectively (Table 2).
Comparison of Lactose Broth to mBPW with TT, TTA, TTB, and RV Selective Broths
Before testing our reduced non-selective pre-enrichment protocol, we first tested two non-selective pre-enrichment broths (Lactose and mBPW), with three TT selective broth formulations (TT, TTA, TTB) and RV, for the recovery of Salmonella from cilantro, to determine the efficacy of these different media formulations on food samples. For these experiments, cilantro samples were inoculated with Salmonella at ∼28 CFU/25 g, and aged at 4°C prior to the 24-h non-selective pre-enrichment. The 24-h non-selective pre-enrichments were inoculated into the four selective RV and TT broths, and then plated on XLT4 for Salmonella isolation and confirmation (Figure 1). The 24-h Lactose broth pre-enrichments (n = 13) had recovery rates of 77% (10/13), 69% (9/13), 92% (12/13), and 92% (12/13) from the RV, TT, TTA, and TTB selective enrichments, respectively (Table 3). In comparison, selective enrichment broths inoculated with 24-h mBPW non-selective pre-enrichments (n = 33) resulted in 100% recovery rates in all four of the selective enrichment broths, suggesting that mBPW improved resuscitation of Salmonella in cilantro (Table 3). Enumeration of three Lactose and two mBPW 24-h non-selective pre-enrichments using the MPN method revealed Salmonella levels of 0.00 in Lactose broth and 7.44 ± 1.15 log MPN g-1 in mBPW, corroborating the lower recovery rates from Lactose broth in 24-h non-selective pre-enrichments (Table 4).
Table 3.
Percent recovery of Salmonella from cilantro non-selective pre-enrichments.
| RV (%) |
TT (%) |
TTA (%) |
TTB (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 6a | 24a | 5a | 6a | 24a | 5a | 6a | 24a | 5a | 6a | 24a | |
| Lactose (n = 13) | 15 | 38 | 77 | 0 | 15 | 69 | 8 | 46 | 92 | 46 | 69 | 92 |
| mBPW (n = 33) | 3 | 9 | 100 | 0 | 3 | 100 | 45∗∗ | 85∗∗ | 100 | 52∗∗ | 97∗∗ | 100 |
| UP (n = 5) | 20 | 40 | 100 | NDb | ND | ND | ND | ND | ND | 100 | 100 | 100 |
| TSB (n = 5) | 40 | 40 | 100 | ND | ND | ND | ND | ND | ND | 100 | 100 | 100 |
aHours
∗∗Z tests of significant difference (p < 0.001) between TTA or TTB and corresponding (same time) RV and TT
using mBPW as non-selective broth
bNot Determined
Table 4.
Average log MPN g-1 Salmonella recovered from cilantro non-selective pre-enrichments.
| 5-h (mean ± SD) | 6-h (mean ± SD) | 24-h (mean ± SD) | |
|---|---|---|---|
| mBPWa | 1.47 ± 0.16 | 2.00 ± 0.42 | 7.44 ± 1.15 |
| Lactoseb | 0.37 ± 0.32 | 0.37 ± 0.32 | 0.00 |
| TSBc | 1.36 ± 1.14 | 2.16 ± 0.28 | 8.10 ± 1.58 |
| UPc | 1.79 ± 0.24 | 2.10 ± 0.74 | 8.74 ± 1.54 |
aFour experiments for 6 and 24-h, two experiments for 5-h
bThree experiments
cTwo experiments
Recovery of Salmonella 1 day Earlier in TTB
To optimize a shortened enrichment time for Salmonella, the four selective broths (RV, TT, TTA, and TTB) were tested in parallel with mBPW and Lactose non-selective pre-enrichments incubated for 5 and 6-h, instead of the 24-h time period recommended in the BAM (Figure 1). Salmonella was recovered one day earlier on XLT4 from 97% (32/33) of cilantro samples when TTB broths were inoculated with 6-h mBPW pre-enrichments, which were found to harbor 2.00 ± 0.42 MPN g-1 Salmonella (Tables 3 and 4). In contrast, Salmonella recovery was only 69% (9/15) when TTB was inoculated with a 6-h Lactose pre-enrichment, which had a correspondingly lower level of Salmonella contamination, 0.37 ± 0.32 MPN g-1 (Tables 3 and 4). In the other selective broths inoculated with 6-h mBPW and Lactose pre-enrichments, recovery, respectively, decreased to 85% (28/33) and 46% (6/13) in TTA, 3% (1/33) and 15% (2/15) in TT, and 9% (3/33) and 38% (5/13) in RV (Table 3). Recovery of typical-Salmonella colonies on XLT4 was possible but not consistent using 5-h resuscitated mBPW and Lactose broth samples, ranging from 0 to 52% depending on the selective broth used (Table 3). It is notable that the XLT4 plates originating from TTB broth inoculated with 5 and 6-h non-selective pre-enrichments had little to no background flora, whereas those from 24-h non-selective pre-enrichments were consistently mixed cultures.
Two additional non-selective pre-enrichment broths, TSB and UP, were tested for their ability of to recover Salmonella one day earlier from selective enrichments. For these studies RV and TTB were inoculated since the data support improved recovery of Salmonella. Similar to the mBPW results, TTB resulted in higher Salmonella recovery compared to RV when plated on XLT4. Both UP and TSB broths resulted in recoveries of 100% (5/5) in TTB and 40% (2/5) in RV using 6-h pre-enrichments that had 2.10 ± 0.52 log MPN g-1 (UP) and 2.16 ± 0.20 log MPN g-1 (TSB) Salmonella, respectively (Tables 3 and 4). Recovery of Salmonella was also 100% when either 5-h UP (n = 5) or TSB (n = 5) non-selective pre-enrichments containing 1.79 ± 0.17 and 1.36 ± 0.81 log MPN g-1 Salmonella, respectively, were inoculated into TTB (Tables 3 and 4). However, recovery decreased to 20% (1/5, UP) and 40% (2/5, TSB) when the same non-selective pre-enrichments were inoculated into RV broth (Table 3). Salmonella was recovered from RV and TTB control samples inoculated with 24-h UP and TSB non-selective pre-enrichments 100% of the time, and similar to the mBPW 24-h non-selective pre-enrichments, the log MPN g-1 Salmonella reached 8.74 ± 1.09 in UP samples and 8.10 ± 1.12 in TSB (Tables 3 and 4).
A final set of experiments with raw chicken thighs (n = 4), liquid whole eggs (n = 3), and peanut butter (n = 3) confirmed early detection of Salmonella in these foods. TTB broths inoculated with 6-h non-selective pre-enrichments of chicken, eggs, or peanut butter were positive for Salmonella in all three foods. Additionally, using 5-h pre-enrichments, 100% of chicken and peanut butter, and 67% (2/3) of egg samples were Salmonella-positive. The recovery of Salmonella from RV broth inoculated with 6-h non-selective pre-enrichments was 100% in chicken thighs, 33% (1/3) in eggs, and 67% (2/3) in peanut butter. All 24-h chicken, egg, and peanut butter non-selective pre-enrichments resulted in positive RV selective enrichments.
In summary we significantly improved Salmonella recovery from cilantro by reducing the length of non-selective pre-enrichment in mBPW from 24 to 6-h, and lowering the selective strength of the BAM TT broth by removing brilliant green dye and reducing the concentration of I2–KI. These changes to the FDA BAM method resulted in the recovery of Salmonella one day early. Additionally, although a larger sample population is required for confirmation, our preliminary results with raw chicken thighs, liquid whole eggs, and peanut butter support that Salmonella recovery can be improved in these commodities. Taken together, our data from 6-h non-selective pre-enrichments of cilantro (excluding lactose replicates), chicken thighs, liquid whole eggs, and peanut butter enabled the recovery of Salmonella 1 day earlier than the BAM method in 98% (52/53) of the samples using TTB and 26% (14/53) using RV supporting an overall improvement. In contrast, the FDA BAM TT broth resulted in very low recoveries of Salmonella from 6-h cilantro non-selective mBPW (3%) and Lactose (15%) pre-enrichments (Table 3).
We also tested cilantro samples in TSB and UP non-selective broths incubated for 5 and 6-h and found that Salmonella recovery was 100% in TTB broth which is higher than the recovery from RV. However, with the small sample sizes (five samples for each broth) tested we did not have the statistical power to show a significant improvement with TTB broth (Table 3).
Resphera Insight Diagnostic True Positive Rates for Salmonella
Among species within the family Enterobacteriaceae, there can be high levels of similarity in 16S rRNA gene sequences, and as a result, many bioinformatics tools maintain poor sensitivity to detect S. enterica at the species level. Therefore, we employed the Resphera Insight algorithm for high-resolution taxonomic assignment, which we validated for accuracy on 110 novel isolates of S. enterica from the GenomeTrakr Project. Overall, between 16S rRNA gene positions 27 and 534, the V1 to V3 region sequenced in this study, Resphera Insight achieves diagnostic true positive rates up to 99.8% for S. enterica with improved accuracy associated with increased read length. In contrast, RDP and UCLUST were unable to achieve a DTP above 0.1% in the V1 to V3 regions (Supplementary Figure S1). It is notable that RDP and UCLUST had higher DTP at the 3′ end of the 16S rRNA gene at positions 800 to 900, and the highest DTP percentages with these algorithms were observed with smaller 16S rRNA gene fragments (Supplementary Figure S1).
16S rRNA Gene Amplicon Sequencing
We employed 16S rRNA gene sequencing to characterize the microbial communities in selective RV, TT, and TTB broth samples originating from 5, 6, and 24-h cilantro non-selective pre-enrichments (Figures 1 and 2). The samples sequenced included RV inoculated with 5, and 6-h non-selective pre-enrichments (sRV, n = 6), or 24-h non-selective pre-enrichments (24RV, n = 9); TTB inoculated with 5, and 6-h non-selective pre-enrichments (sTTB, n = 8) or 24-h non-selective pre-enrichments (24TTB, n = 9); and TT inoculated with 6-h non-selective pre-enrichments (sTT, n = 2), or 24-h non-selective pre-enrichments (24TT, n = 2) (Figures 1 and 2).
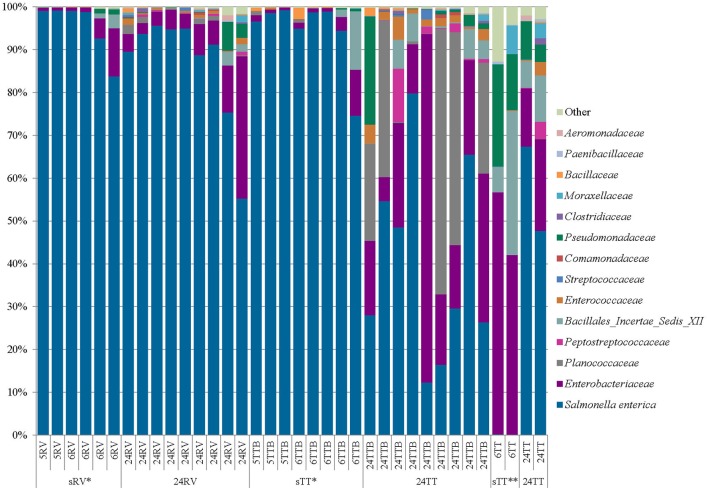
FIGURE 2.
Proportional abundances of bacteria in Tetrathionate (TT) and Rappaport–Vassiliadis (RV) selective enrichments. Proportional abundances of Salmonella and other family members from TT and RV selective enrichments were estimated using 16S rRNA gene sequencing. sRV and sTT (5 and 6-h RV and TT), 24 RV and 24TT (24-h RV and TT), ∗ significant increase in sRV and sTTB verses 24RV or 24TTB, ∗∗ significant decrease in sTT compared to 24 TT, and ∗∗∗ significant increase in 24RV versus 24TTB.
Average S. enterica proportional abundances were 92% for sRV, 87% for 24RV, 92% for sTTB, 40% for 24TTB, 0.07% for sTT, and 58% for 24TT (Figure 2). Although all RV enrichments were similar in appearance when streaked on XLT4 plates, a multivariate linear regression analysis of Salmonella proportional abundances showed that sRV significantly increased Salmonella proportional abundances relative to 24RV (P = 1.58e-04) (Figure 2). Salmonella proportional abundances were also significantly higher in sTTB compared to 24TTB (P = 2.40e-05), supporting the observed reduction in background flora on XLT4 plates derived from the sTTB enrichments (Figure 2, data not shown). Not surprisingly, the Salmonella abundances were significantly lower in sTT than in the 24TT selective enrichments (P = 2.0e-03) (Figure 2). The 24TTB and 24TT selective enrichments had statistically similar proportional abundances of Salmonella while the abundances in 24RV were significantly higher than 24TTB (P = 2.59e-05) (Figure 2). The proportional abundances of Salmonella in the sRV and sTTB enrichments were statistically similar (P = 8.39e-01) as were those in 24RV and 24TT BAM enrichments (P = 1.51e-01). Finally, the proportional abundances of Salmonella in the sTTB enrichments were significantly higher than sTT since TT broth is too harsh for a shortened non-selective pre-enrichment (Figure 2).
Taken together, these 16S rRNA gene sequencing data suggest that a 24-h non-selective cilantro pre-enrichment allows resident bacterial species to resuscitate, thrive, and compete with Salmonella during selective enrichment, whereas the shortened non-selective pre-enrichment is insufficient for resuscitation of resident bacteria and gives Salmonella a competitive edge. In either case, those bacteria that can metabolize the selective agents in RV and TTB will grow and thrive with Salmonella during the selective enrichments (Figure 2). The 16S rRNA gene profiles of TTB samples identified these competitive families as non-Salmonella Enterobacteriaceae, Planococcaceae, Peptostreptococcaceae, and Bacillales Incertae Sedis XII. Although bacteria in these families compete with Salmonella in TTB, most are Gram-positive and were not observed on XLT4 plates. However, members of the non-Salmonella Enterobacteriaceae family (i.e., Citrobacter sp. and Enterobacter sp.) were commonly isolated from XLT4 after enrichment in RV and TTB.
Discussion
Culture based methods for pathogen detection in food are the most sensitive methods available and reducing detection times would be advantageous to food testing laboratories. Here, we were able to detect Salmonella one day earlier than the current FDA BAM method by reducing the non-selective pre-enrichment time in conjunction with a reformulation of TT broth. We tested this new strategy with different non-selective pre-enrichment broths and our results, comparing the efficacy of Lactose broth to mBPW, TSB, and UP, for Salmonella recovery from cilantro match previous studies with cantaloupes, mangos, and tomatoes which suggest that Lactose broth is not always optimal for Salmonella pre-enrichment (Hammack et al., 2006). In our study, even after a 24-h non-selective pre-enrichment in Lactose broth, some cilantro samples were negative for Salmonella but significant improvements were observed using mBPW, and although the sample sizes were too small to support significant improvements with TSA, and UP broths, all of the samples tested in these broths were positive for Salmonella.
An important outcome in this study was defining the enrichment dynamics of Salmonella amidst the complex background microbiome in cilantro. The 16S rRNA gene sequencing data revealed a significant reduction in relative abundances of competitive bacteria in sTTB (8%) and sRV (8%) inoculated with 5 and 6-h non-selective pre-enrichments. The 24RV and 24TTB selective enrichments had 13% and 60% proportional abundances of competitive bacteria, respectively. We also observed that TT broth did not support the recovery of Salmonella from 5 or 6-h non-selective pre-enrichment broths. Our results from the SAFE strain collection suggest that TT broth requires more than 103 CFU/mL Salmonella for successful recovery, which correlates with the 5 and 6-h cilantro non-selective pre-enrichment TT results that were negative since they only contained 1.47 to 2.16 log MPN g-1. Additionally, the negative TT culture results were clearly supported in the 16S rRNA sequencing data where the proportional abundances of Salmonella in the TT broth cultures inoculated with 6-h non-selective pre-enrichments were only 0.07%. Finally, the presence of other Enterobacteriaceae, Planococcaceae, and Pseudomonadaceae in the TT, TTB, and RV enrichments suggests that bacteria in these families are resistant to the inhibitory effects in the selective broths. Further analyses are required to understand the details of the ability of these bacteria to thrive in selective enrichments.
The log MPN g-1 Salmonella levels in 6-h non-selective cilantro pre-enrichments used to inoculated selective TTA, TTB and RV revealed that a log MPN g-1 of 2 to 2.16, obtained after 6-h of non-selective pre-enrichment in mBPW, UP or TSB, was sufficient to recover Salmonella from 97 -100% of cilantro samples. In contrast, the levels of Salmonella at 4-h (0.00 to 1.49 log MPN g-1, data not shown) and 5-h (1.36 to 1.79 log MPN g-1) were insufficient for consistent recovery of Salmonella from TTB broth. There was a well-defined increase in Salmonella recovery from 5 to 6-h of pre-enrichment, indicating that this 6th hour of enrichment sufficiently resuscitated Salmonella. It is evident from the log MPN level increases from 4 to 6-h that Salmonella is proliferating, but for the first time, we are able to see what else is proliferating and how these other bacteria impact proportional abundances of Salmonella during subsequent selective enrichments. Our experiments with raw chicken thighs, peanut butter, and liquid whole eggs resulted in 100% recovery of Salmonella with 6-h non-selective pre-enrichments and TTB suggesting that this method is effective in diverse food matrices.
The increasing use of rapid molecular methods such as qPCR and PCR for Salmonella detection have been highlighted in recent reviews (Park et al., 2014; Bell et al., 2016). qPCR assays require 102 Salmonella/reaction for a positive result and as little as 30 CFU of Salmonella are need for consistent detection with endpoint PCR (Park et al., 2014; Bell et al., 2016). There is a consensus that a 6-h to 24-h pre-enrichment step is required for both of these methods to decrease the negative impacts on PCR and qPCR chemistry inherent to some types of food. Additional problems due to variations in competitive bacteria and inhibition due to the pre-enrichment broth itself were also noted (Park et al., 2014; Bell et al., 2016). Incorporating a 6-h selective TTB enrichment simultaneous to sample preparation for qPCR and PCR would be seamless for laboratories performing these assays, and would significantly increase the overall recovery rate of Salmonella. Additionally, this would reduce detection time in instances where qPCR and PCR fail due to inhibition or low levels of Salmonella.
In conclusion, we propose that an inoculation of TTB with a 6-h non-selective pre-enrichment be incubated overnight in parallel with the standard 24-h non-selective pre-enrichment in the FDA BAM method to enable the detection of Salmonella one day early. Our data also indicates that this will improve recovery rates of Salmonella.
Author Contributions
KJ and ND designed and conducted the experiments and wrote and revised the manuscript. JW performed the Bioinformatic and statistical analyses, and assisted with manuscript preparation and revision. CG contributed to the study design, experimental analysis and manuscript preparation. DH provided funding support and overall supervision. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
ND, CG, and JW are Oak Ridge Institute for Science and Education fellows, and we thank the Department of Energy for their support. We also thank Dr. Lanlan Yin for assistance with statistical analyses.
Funding
The work was funded by the United States Food and Drug Administration and the Oak Ridge Institute for Science and Education.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02103/full#supplementary-material
Performance of Resphera Insight and other tools on novel isolates of S. enterica. Y-axis shows the Diagnostic True Positive Rate (DTP), i.e., the percentage of sequences assigned unambiguously to S. enterica. Lines show the average DTP for all reads covering each gene position for a given read length range. DTP rates over 99.5% are achieved for Resphera Insight with 300bp sequences that span the first 100bp of the 16S rRNA gene.
References
- Allard M. W., Strain E., Melka D., Bunning K., Musser S. M., Brown E. W., et al. (2016). Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 54 1975–1983. 10.1128/JCM.00081-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. L., Jarvis K. G., Ottesen A. R., McFarland M. A., Brown E. W. (2016). Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microb. Biotechnol. 9 279–292. 10.1111/1751-7915.12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse M. (1995). Reviews from the sixth international symposium of the working party for culture media, part IIMedia for Salmonella. Int. J. Food Microbiol. 26 117–131. 10.1016/0168-1605(93)E0030-U [DOI] [PubMed] [Google Scholar]
- Campbell J. V., Mohle-Boetani J., Reporter R., Abbott S., Farrar J., Brandl M., et al. (2001). An outbreak of Salmonella serotype thompson associated with fresh cilantro. Infect. Dis. 183 984–987. 10.1086/319254 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Fraser A. D., Yamazaki H. (1993). Evaluation of the toxicity of Salmonella selective media for shortening the enrichment period. Int. J. Food Microbiol. 18 151–159. 10.1016/0168-1605(93)90219-7 [DOI] [PubMed] [Google Scholar]
- Chen Z., Diao J., Dharmasena M., Ionita C., Jiang X., Rieck J. (2013). Thermal inactivation of desiccation-adapted Salmonella spp. in aged chicken litter. Appl. Environ. Microbiol. 79 7013–7020. 10.1128/AEM.01969-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. M., Van Khanh T., Lin W., Ruby R. M. (2009). Interlaboratory validation of a real-time PCR 24-hour rapid method for detection of Salmonella in foods. J. Food Prot. 72 945–951. 10.4315/0362-028X-72.5.945 [DOI] [PubMed] [Google Scholar]
- D’Aoust J.-Y. (1981). Update on preenrichment and selective enrichment conditions for detection of Salmonella in foods. J. Food Prot. 44 369–374. 10.4315/0362-028X-44.5.369 [DOI] [PubMed] [Google Scholar]
- D’Aoust J. Y., Maishment C. (1979). Preenrichment conditions for effective recovery of Salmonella in foods and feed ingredients. J. Food Prot. 42 153–157. 10.4315/0362-028X-42.2.153 [DOI] [PubMed] [Google Scholar]
- D’Aoust J. Y., Sewell A., Jean A. (1990). Limited sensitivity of short (6 h) selective enrichment for detection of foodborne Salmonella. J. Food Prot. 53 562–625. 10.4315/0362-028X-53.7.562 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2015). Bacteriological Analytical Manual, Salmonella. Available at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm [Google Scholar]
- Hammack T. S., Johnson M. L., Jacobson A. P., Andrews W. H. (2006). Effect of sample preparation and preenrichment media on the recovery of Salmonella from cantaloupes, mangoes, and tomatoes. J. AOAC Int. 89 180–184. [PubMed] [Google Scholar]
- ISO (2002). Microbiology of Food and Animal Feeding stuffs–Horizontal Method for the Detection of Salmonella spp. Available at: http://www.iso.org/iso/catalogue_detail.htm?csnumber = 29315 ISO 6579:2002 [Google Scholar]
- Jarvis K. G., White J. R., Grim C. J., Ewing L., Ottesen A. R., Beaubrun J. J., et al. (2015). Cilantro microbiome before and after nonselective pre-enrichment for Salmonella using 16S rRNA and metagenomic sequencing. BMC Microbiol. 15:160 10.1186/s12866-015-0497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox R. (1945). The effect of tetrathionate on bacterial growth. Br. J. Exp. Pathol. 26 146–150. [Google Scholar]
- Kuczynski J., Stombaugh J., Walters W. A., Gonzalez A., Caporaso J. G., Knight R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics Chap. 10 Unit10.7. 10.1002/0471250953.bi1007s36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Stombaugh J., Walters W. A., Gonzalez A., Caporaso J. G., Knight R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. Chap. 1 Unit1E. 10.1002/9780471729259.mc01e05s27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmacher A., Bockemuhl J., Aleksic S. (1995). Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 115 501–511. 10.1017/S0950268800058660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard S. R., Mammel M. K., Lacher D. W., Elkins C. A. (2015). Application of metagenomic sequencing to food safety: detection of Shiga Toxin-producing Escherichia coli on fresh bagged spinach. Appl. Environ. Microbiol. 81 8183–8191. 10.1128/AEM.02601-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6 610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moats W. A. (1981). Update on Salmonella in foods: selective plating media and other diagnostic media. J. Food Prot. 44 375–380. 10.4315/0362-028X-44.5.375 [DOI] [PubMed] [Google Scholar]
- Moats W. A., Kinner J. A., Maddox S. E., Jr. (1974). Effect of heat on the antimicrobial activity of brillant green dye. Appl. Microbiol. 27 844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr H. K., Trenk H. L., Yeterian M. (1974). Comparison of fluorescent-antibody methods and enrichment serology for the detection of Salmonella. Appl. Microbiol. 27 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Molina J. A., Peralta-Sanchez J. M., Gonzalez A., McMurdie P. J., Vazquez-Baeza Y., Xu Z. (2013). Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 531 371–444. 10.1016/B978-0-12-407863-5.00019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen A. R., Gonzalez A., Bell R., Arce C., Rideout S., Allard M., et al. (2013). Co-enriching microflora associated with culture based methods to detect Salmonella from tomato phyllosphere. PLoS ONE 8:e73079 10.1371/journal.pone.0073079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo S. A., Alford J. A. (1970). Inhibitory action of tetrathionate enrichment broth. Appl. Microbiol. 20 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H., Aydin M., Khatiwara A., Dolan M. C., Gilmore D. F., Bouldin J. L., et al. (2014). Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products. Food Microbiol. 38 250–262. 10.1016/j.fm.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Peterz M., Wiberg C., Norberg P. (1989). The effect of incubation temperature and magnesium chloride concentration on growth of Salmonella in home-made and in commercially available dehydrated Rappaport-Vassiliadis broths. J. Appl. Bacteriol. 66 523–528. 10.1111/j.1365-2672.1989.tb04573.x [DOI] [PubMed] [Google Scholar]
- Pettengill J. B., McAvoy E., White J. R., Allard M., Brown E., Ottesen A. (2012). Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Res. Notes 5:378 10.1186/1756-0500-5-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport F., Konforti N., Navon B. (1956). A new enrichment medium for certain Salmonellae. J. Clin. Pathol. 9 261–266. 10.1136/jcp.9.3.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. (1989). Injured Index and Pathogenic Bacteria: Occurence and Detection in Foods, Water and Feeds. Boca Raton, FL: CRC Press. [Google Scholar]
- Swaminathan B., Barrett T. J., Hunter S. B., Tauxe R. V. (2001). PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7 382–389. 10.3201/eid0703.017303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague O., Clurman A. W. (1916). An improved brilliant-green culture medium for the isolation of typhoid bacilli from stools. J. Infect. Dis. 18 647–652. 10.1093/infdis/18.6.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (2014). FSIS Microbiology Laboratory Guidebook. Available at: http://www.fsis.usda.gov/wps/portal/fsis/topics/science/laboratories-andprocedures/guidebooks-and-methods/microbiology-laboratory-guidebook/microbiology-laboratory-guidebook [Google Scholar]
- van Schothorst M., Renaud A. M. (1985). Malachite green pre-enrichment medium for improved Salmonella isolation from heavily contaminated samples. J. Appl. Bacteriol. 59 223–230. 10.1111/j.1365-2672.1985.tb01783.x [DOI] [PubMed] [Google Scholar]
- van Schothorst M., van Leusden F. M. (1975). Further studies on the isolation of injured Salmonellae from foods. Zentralbl. Bakteriol. Orig A 230 186–191. [PubMed] [Google Scholar]
- Vassiliadis P. (1983). The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of Salmonellas: an overview. J. Appl. Bacteriol. 54 69–76. 10.1111/j.1365-2672.1983.tb01302.x [DOI] [PubMed] [Google Scholar]
- Vassiliadis P., Kalapothaki V., Trichopoulos D., Mavrommatti C., Serie C. (1981). Improved isolation of Salmonellae from naturally contaminated meat products by using rappaport-vassiliadis enrichment broth. Appl. Environ. Microbiol. 42 615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassiliadis P., Trichopoulos D., Kalandidi A., Xirouchaki E. (1978). Isolation of Salmonellae from sewage with a new procedure of enrichment. J. Appl. Bacteriol. 44 233–239. 10.1111/j.1365-2672.1978.tb00795.x [DOI] [PubMed] [Google Scholar]
- Wang H., Gill V. S., Cheng C. M., Gonzalez-Escalona N., Irvin K. A., Zheng J., et al. (2015). Evaluation and comparison of rapid methods for the detection of Salmonella in naturally contaminated pine nuts using different pre enrichment media. Food Microbiol. 46 58–65. 10.1016/j.fm.2014.06.028 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. R., Brown E., Keys C., Strain E., Luo Y., Muruvanda T., et al. (2016). Whole Genome DNA sequence analysis of Salmonella subspecies enterica serotype Tennessee obtained from related peanut butter foodborne outbreaks. PLoS ONE 11:e0146929 10.1371/journal.pone.0146929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 426–429. 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Performance of Resphera Insight and other tools on novel isolates of S. enterica. Y-axis shows the Diagnostic True Positive Rate (DTP), i.e., the percentage of sequences assigned unambiguously to S. enterica. Lines show the average DTP for all reads covering each gene position for a given read length range. DTP rates over 99.5% are achieved for Resphera Insight with 300bp sequences that span the first 100bp of the 16S rRNA gene.