Abstract
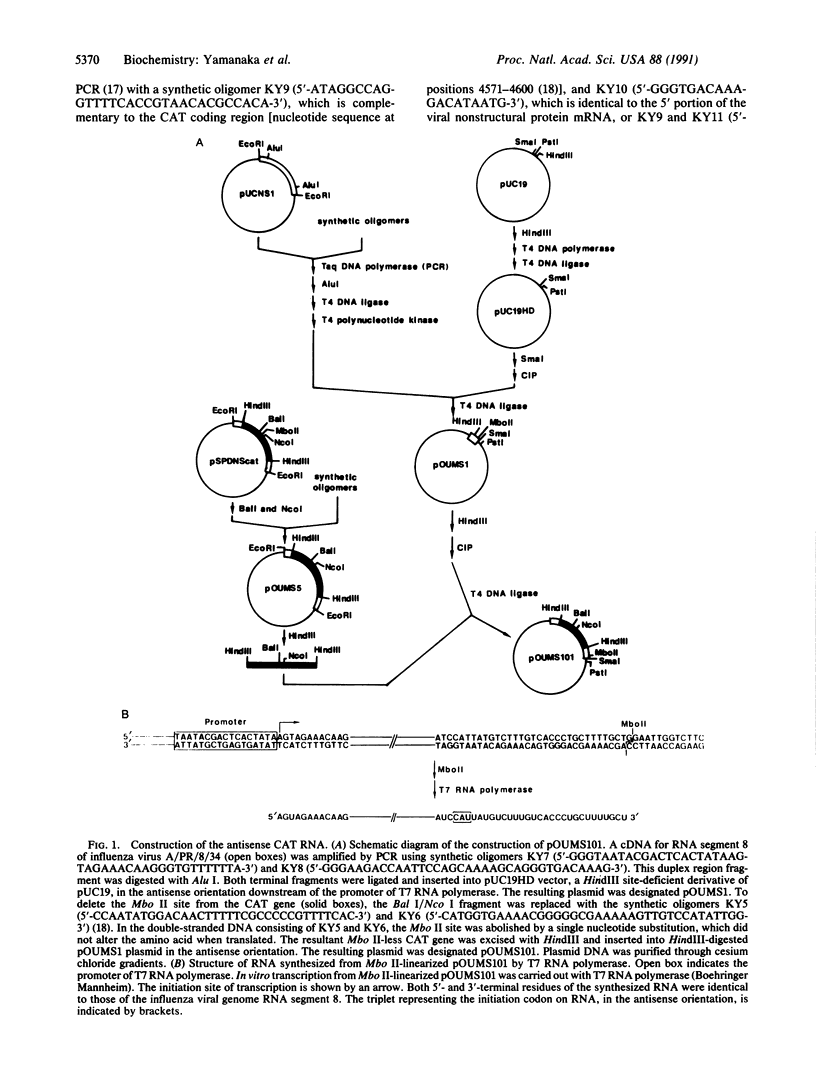
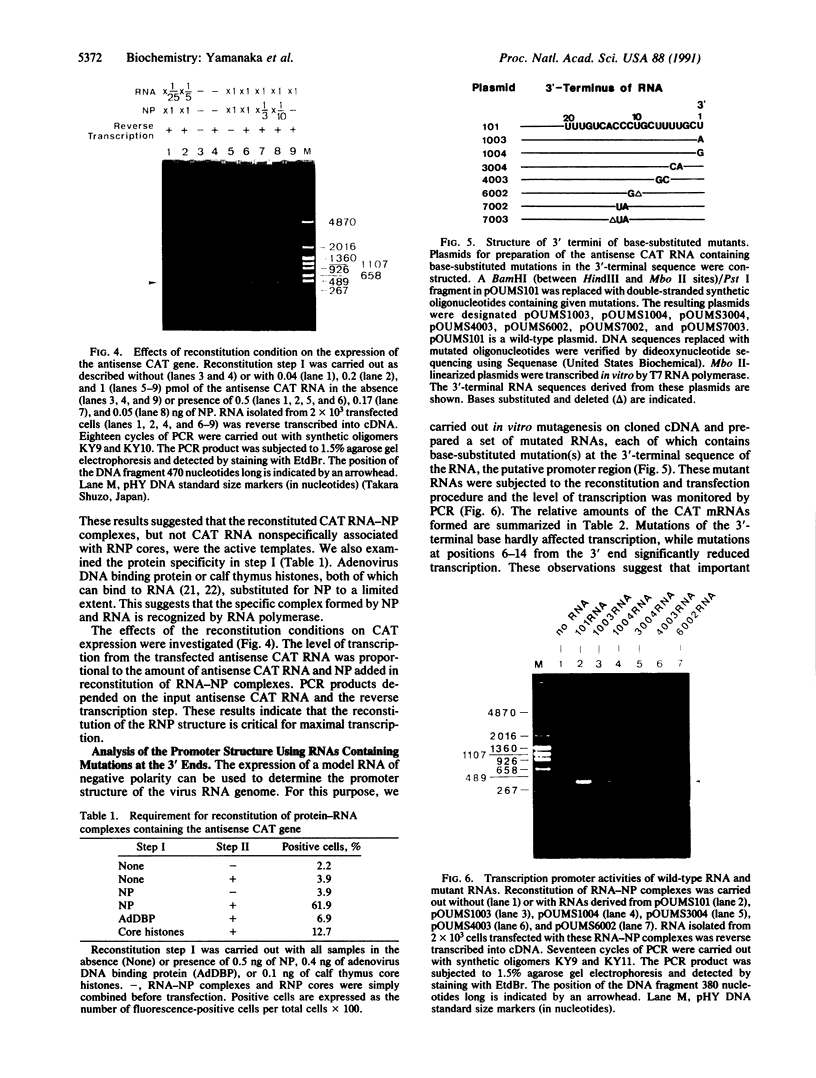
A system for the expression of a foreign gene derived from negative polarity RNA was developed using influenza virus, a negative-stranded RNA virus. From cDNA for the influenza virus RNA genome segment 8, the region coding for the nonstructural protein was deleted and replaced by the chloramphenicol acetyltransferase (CAT) gene. The resulting DNA sequence was placed under the control of the promoter of T7 RNA polymerase such that the antisense RNA to CAT mRNA was produced when transcribed by T7 RNA polymerase. Transfection of HeLa cells with this antisense CAT RNA in the presence of the helper ribonucleoprotein cores led to no significant production of the CAT. In contrast, when the RNA was covered with purified nucleoprotein prior to transfection, the CAT gene was efficiently expressed. This indicated that the viral RNA polymerase transcribed the RNA transfected as the RNA-nucleoprotein complexes. In addition, this system was used for analysis of the cis-acting region in transcription and the promoter structure of the viral RNA genome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., French R., Janda M., Loesch-Fries L. S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleghon V. G., Klessig D. F. Association of the adenovirus DNA-binding protein with RNA both in vitro and in vivo. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8947–8951. doi: 10.1073/pnas.83.23.8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans R. W., Content J., Duesberg P. H. Structure of the ribonucleoprotein of influenza virus. J Virol. 1972 Oct;10(4):795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R., Janda M., Ahlquist P. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science. 1986 Mar 14;231(4743):1294–1297. doi: 10.1126/science.231.4743.1294. [DOI] [PubMed] [Google Scholar]
- Honda A., Mukaigawa J., Yokoiyama A., Kato A., Ueda S., Nagata K., Krystal M., Nayak D. P., Ishihama A. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J Biochem. 1990 Apr;107(4):624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- Honda A., Uéda K., Nagata K., Ishihama A. Identification of the RNA polymerase-binding site on genome RNA of influenza virus. J Biochem. 1987 Nov;102(5):1241–1249. doi: 10.1093/oxfordjournals.jbchem.a122163. [DOI] [PubMed] [Google Scholar]
- Honda A., Uéda K., Nagata K., Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988 Dec;104(6):1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A., Nagata K. Viral RNA polymerases. CRC Crit Rev Biochem. 1988;23(1):27–76. doi: 10.3109/10409238809103119. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Finch J. T., Winter G., Robertson J. S. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983 Sep;34(2):619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- Levis R., Huang H., Schlesinger S. Engineered defective interfering RNAs of Sindbis virus express bacterial chloramphenicol acetyltransferase in avian cells. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4811–4815. doi: 10.1073/pnas.84.14.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Krystal M., Enami M., Parvin J. D., Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989 Dec 22;59(6):1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- Malone R. W., Felgner P. L., Verma I. M. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T., Ishikawa M., Motoyoshi F., Semba K., Okada Y. In vitro transcription of infectious RNAs from full-length cDNAs of tobacco mosaic virus. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5043–5047. doi: 10.1073/pnas.83.14.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani S., Colonno R. J. In vitro synthesis of an infectious RNA from cDNA clones of human rhinovirus type 14. J Virol. 1985 Nov;56(2):628–632. doi: 10.1128/jvi.56.2.628-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak D. P., Sivasubramanian N., Davis A. R., Cortini R., Sung J. Complete sequence analyses show that two defective interfering influenza viral RNAs contain a single internal deletion of a polymerase gene. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2216–2220. doi: 10.1073/pnas.79.7.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P. The genes of influenza virus. Cell. 1977 Jan;10(1):1–10. doi: 10.1016/0092-8674(77)90133-7. [DOI] [PubMed] [Google Scholar]
- Palter K. B., Alberts B. M. The use of DNA-cellulose for analyzing histone-DNA interactions. Discovery of nucleosome-like histone binding to single-stranded DNA. J Biol Chem. 1979 Nov 10;254(21):11160–11169. [PubMed] [Google Scholar]
- Parvin J. D., Palese P., Honda A., Ishihama A., Krystal M. Promoter analysis of influenza virus RNA polymerase. J Virol. 1989 Dec;63(12):5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons M. W., Schulze I. T., Hirst G. K., Hauser R. Isolation and characterization of the ribonucleoprotein of influenza virus. Virology. 1969 Oct;39(2):250–259. doi: 10.1016/0042-6822(69)90045-2. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. M., Doyle M. V., Mark D. F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Ishihama A., Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990 Jul 5;265(19):11151–11155. [PubMed] [Google Scholar]
- Yamanaka K., Ishihama A., Nagata K. Translational regulation of influenza virus mRNAs. Virus Genes. 1988 Oct;2(1):19–30. doi: 10.1007/BF00569734. [DOI] [PubMed] [Google Scholar]
- van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]