Abstract
The actin cytoskeleton is a key component in the machinery of eukaryotic cells, and it self-assembles out of equilibrium into a wide variety of biologically crucial structures. Although the molecular mechanisms involved are well characterized, the physical principles governing the spatial arrangement of actin filaments are not understood. Here we propose that the dynamics of actin network assembly from growing filaments results from a competition between diffusion, bundling and steric hindrance, and is responsible for the range of observed morphologies. Our model and simulations thus predict an abrupt dynamical transition between homogeneous and strongly bundled networks as a function of the actin polymerization rate. This suggests that cells may effect dramatic changes to their internal architecture through minute modifications of their nonequilibrium dynamics. Our results are consistent with available experimental data.
The dynamics of actin cytoskeleton is essential to the function of living cells. Here, Foffano et al. describe a nonequilibrium filament model to mimic the formation of cytoskeleton and pinpoint the key role played by the actin entanglement during the transition from homogeneous to bundled networks.
The cytoskeleton of living cells is an extremely dynamic system, of which actin is a vital component. Actin monomers are continuously assembled during polymerization; at the same time, actin filaments are bundled together by crosslinkers to form a large variety of structures. Tight bundles thus appear in filopodia, in stress fibres and in the contractile ring involved in cell division, whereas homogeneous networks are found in the cell cortex and in the lamella1. Understanding the fundamental mechanisms that determine the formation and the morphology of the actin cytoskeleton is thus necessary to explain how the cell regulates its own shape, internal structure and motility.
A tool of choice to characterize these structures is to isolate a few essential ingredients and study the result of their interactions. Bottom-up experiments thus put one type of crosslinker in solution together with purified actin, resulting in in vitro reconstituted actin networks2,3. As filaments grow and become crosslinked into bundles, the morphology of the resulting networks strongly depends on their assembly kinetics: different protocols leading to the same filament number and lengths through different kinetic pathways thus result in different structures, demonstrating that the observed phases are out of equilibrium4,5,6. The structure of keratin networks similarly results from the competition between filament elongation and bundle formation7. Despite the highly dynamic nature of these experiments, theoretical attempts to describe such networks have largely relied on equilibrium physics8,9,10, modelling morphological transitions as the result of the competition between crosslinker binding and thermal fluctuations. In many cases, however, the bundling of two or more filaments by hundreds of crosslinkers can involve energies of the order of thousands of kBT11,12, indicating that equilibrium thermal fluctuations alone cannot account for the presence of structural disorder in the final networks.
Here we propose a theoretical framework to account for the architecture of actin networks from the dynamics of their assembly. We study the simplest situation of actin structures assembling de novo from a fixed number of initially short filaments, mirroring existing in vitro experiments4,5,7, as well as, for example, cytoskeletal reassembly in a newly formed bleb13, and actin recovery following drug treatment14. We consider a system of polymerizing and diffusing filaments that tend to bundle irreversibly when they come into contact (Fig. 1a), as observed experimentally15. Bundling can however be sterically blocked by the presence of other filaments (Fig. 1b) that becomes increasingly likely as the filaments elongate. At early times, the filaments are very short and diffusion is fast. Bundling thus proceeds unimpeded by steric constraints, and the number of bundles in the solution decreases over time as thicker bundles are formed through the merging of thinner ones. As a result, blocking becomes less likely and further bundling events are facilitated in a positive feedback mechanism. As the filaments grow, diffusion slows down and bundles come into contact more rarely. As a consequence, blocking finally outpaces reaction and the system becomes kinetically arrested. This basic mechanism allows us to formulate simple, experimentally testable scaling predictions for the bundle size and concentration, including an abrupt change in system behaviour upon kinetic trapping. We numerically validate these predictions in simulations of rod-like bundles over five orders of magnitude in concentration and four orders of magnitude in filament growth velocity, a much broader range than is accessible to existing detailed simulations16. We thus develop a robust, easily extendable framework to describe the nonequilibrium physics of cytoskeletal network assembly.
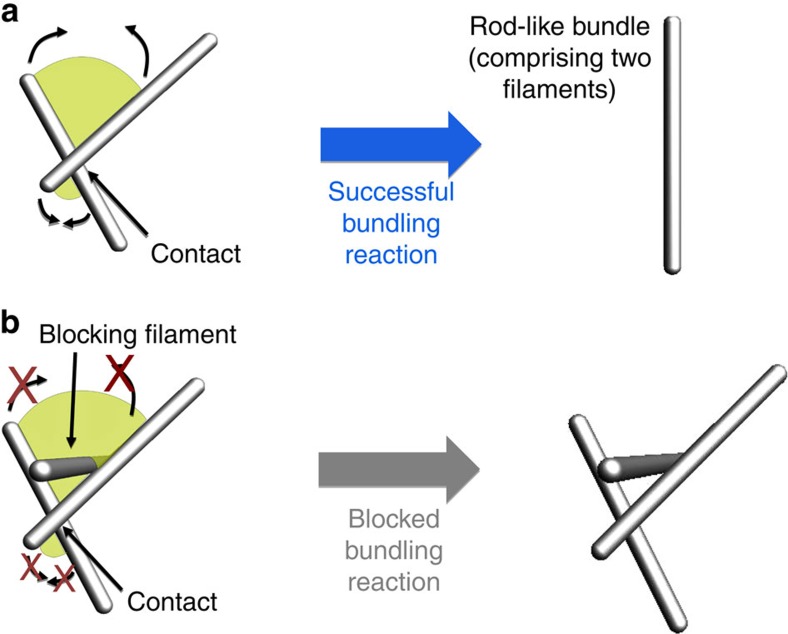
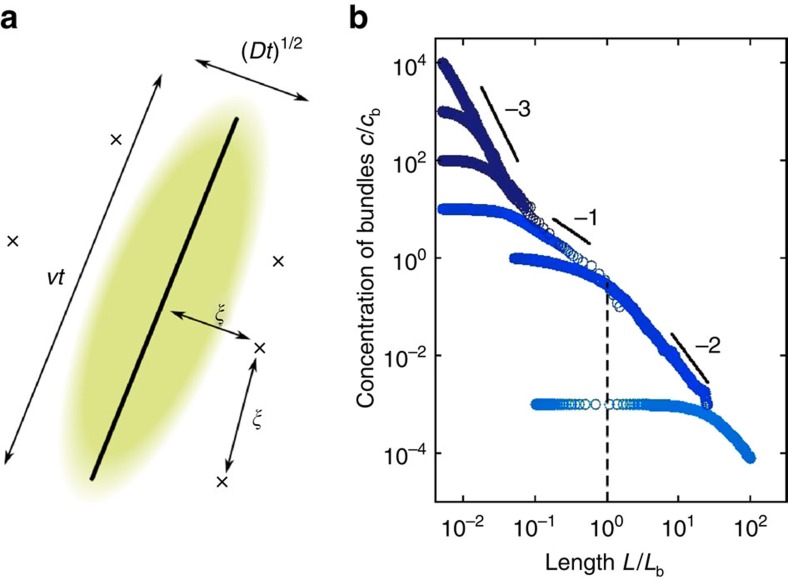
Figure 1. Basic mechanisms of filament network assembly.
(a) When two actin filaments come into contact, they attempt a bundling reaction (thin arrows). If there are no filaments in the surroundings, the attempt results in a single bundle. (b) If other filaments are found on the path of the bundling reaction (green surface), bundling is blocked because of steric interaction (red crosses).
Results
A model for kinetic arrest based on filament entanglement
We model actin bundles as impenetrable, infinitely thin, rigid17 rods in a homogeneous solution of crosslinkers. These rods grow at a constant velocity v, and their diffusion coefficient is given by 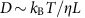 in the Rouse approximation18, where L=vt is the length of a filament and η is the viscosity of the surrounding solution. When two rods come within a distance b of the order of the size of a crosslinker, they react with a rate k to merge into a rod-like bundle as in Fig. 1a. Note that the chemical rate constant k is associated with the crosslinker binding rate and not the filament merging time. Indeed, the latter is much shorter than the typical filament reaction time, as further detailed in the discussion. Although nonmerged bundles may be connected by a few crosslinkers, such connections are short-lived (∼1 s for α-actinin12) and we neglect them over the timescale of minutes involved in network formation. As a result, kinetic trapping in our model arises from steric entanglement between densely packed rods. This scenario is consistent with experimental evidence that entanglement can induce kinetic trapping in actin networks even in the absence of crosslinkers19.
in the Rouse approximation18, where L=vt is the length of a filament and η is the viscosity of the surrounding solution. When two rods come within a distance b of the order of the size of a crosslinker, they react with a rate k to merge into a rod-like bundle as in Fig. 1a. Note that the chemical rate constant k is associated with the crosslinker binding rate and not the filament merging time. Indeed, the latter is much shorter than the typical filament reaction time, as further detailed in the discussion. Although nonmerged bundles may be connected by a few crosslinkers, such connections are short-lived (∼1 s for α-actinin12) and we neglect them over the timescale of minutes involved in network formation. As a result, kinetic trapping in our model arises from steric entanglement between densely packed rods. This scenario is consistent with experimental evidence that entanglement can induce kinetic trapping in actin networks even in the absence of crosslinkers19.
Filament interactions involve several dynamical regimes
We first develop a mean-field approach considering a homogeneous solution of isotropically oriented rods of concentration c. This concentration accounts for bundles of any thickness, including single filaments, which we see as ‘one-filament bundles', and evolves according to
 |
where L=vt and r(c, L) is the rate with which one rod bundles with any other.
In dilute systems, where the average distance between two rods is much larger than their length (cL3<<1), r(c, L) is effectively due to a two-body interaction. We denote it by r(2) and estimate it separately in the case of reaction-limited and diffusion-limited systems. In a reaction-limited system, two rods within interaction range b bundle at a rate k. As the probability for a rod to be within interaction range of another is ∼cbL2, the total two-body rate of bundling is:
 |
where Ar is a dimensionless prefactor of order one. In the diffusion-limited case, the orientation of the rods can rotationally diffuse over the whole sphere in a much shorter time than it takes them to come into contact with one another. The rate with which one rod encounters any other through diffusion is thus given by 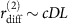 (ref. 20). Therefore:
(ref. 20). Therefore:
 |
where Ad is another dimensionless prefactor. As evidenced by the different L-dependences of r(2) in equations (2 and 3), while the rods grow, diffusion slows down relative to reaction and the dynamics transition from reaction limited to diffusion limited. This happens at a critical length 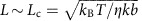 .
.
In concentrated systems (cL3≳1) bundling is affected by the presence of surrounding rods, yielding a rate:
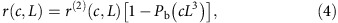 |
where the rate r(2) at which bundling is attempted is given by equations (2 and 3) and Pb is the probability for the attempt to be blocked as in Fig. 1b. To determine the blocking probability, we note that 1−Pb=(1−pb)N−2, where pb is the probability for an individual, randomly placed rod to block the attempt, and N is the total number of rods in the system. To estimate pb, we consider two rods with tangent vectors 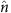 and
and 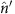 coming into contact at their midpoints. Denoting with V the volume of the system, with
coming into contact at their midpoints. Denoting with V the volume of the system, with 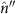 the orientation of the third, potentially blocking rod and letting
the orientation of the third, potentially blocking rod and letting 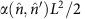 be the area of the bundling path pictured in Fig. 1b, the probability that the third rod intersects this path reads
be the area of the bundling path pictured in Fig. 1b, the probability that the third rod intersects this path reads 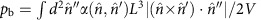 . In the thermodynamic limit (N, V→∞) this yields
. In the thermodynamic limit (N, V→∞) this yields 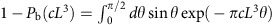 , which we plot in Fig. 2b. The blocking probability Pb becomes large at large concentration c, accounting for the experimental observation that although bundling speeds up with increasing c at low c (because of binary collisions), the opposite trend is observed at higher concentrations (when three or more body blocking becomes predominant)4.
, which we plot in Fig. 2b. The blocking probability Pb becomes large at large concentration c, accounting for the experimental observation that although bundling speeds up with increasing c at low c (because of binary collisions), the opposite trend is observed at higher concentrations (when three or more body blocking becomes predominant)4.
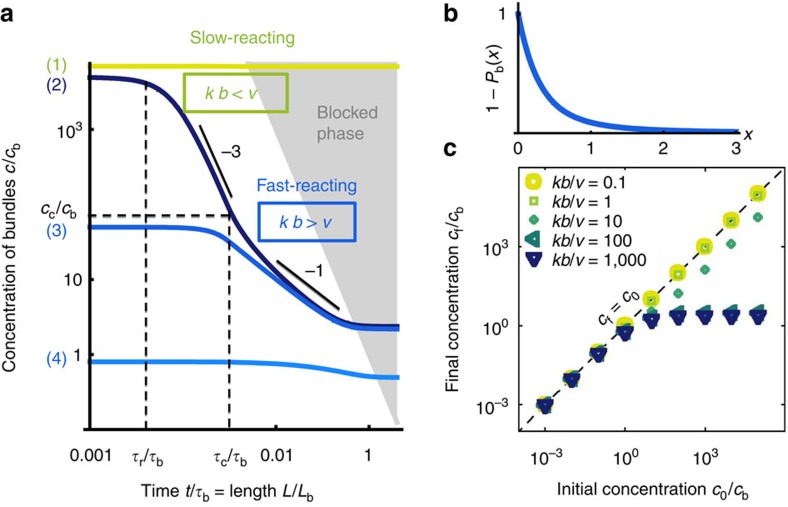
Figure 2. Dynamical evolution of our mean-field model.
(a) Numerical solutions of equations (1, 2, 3, 4) for the four different scenarios (1) through (4) described in the main text. Here the system transitions from reaction limited to diffusion limited through r(2)=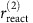 for L<Lc and r(2)=
for L<Lc and r(2)=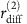 for L≥Lc. An alternative, smoother interpolation 1/r(2)=1/
for L≥Lc. An alternative, smoother interpolation 1/r(2)=1/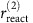 +1/
+1/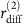 yields similar curves. Here kb/v=0.1 for slow-reacting filaments (light grey (green) curve), and kb/v=1,000 for fast-reacting filaments (dark grey (blue) curves). The grey region materializes the condition cL3>1, where blocking is observed. (b) Probability 1−Pb(cL3) that an attempted bundling event does not get blocked. (c) Final bundle concentration cf as a function of c0. The dashed line materializes the final concentration of a homogeneous network of single filaments (cf=c0).
yields similar curves. Here kb/v=0.1 for slow-reacting filaments (light grey (green) curve), and kb/v=1,000 for fast-reacting filaments (dark grey (blue) curves). The grey region materializes the condition cL3>1, where blocking is observed. (b) Probability 1−Pb(cL3) that an attempted bundling event does not get blocked. (c) Final bundle concentration cf as a function of c0. The dashed line materializes the final concentration of a homogeneous network of single filaments (cf=c0).
Mean-field dynamical scenarios and final morphologies
We now use our model to predict the final structure of a system of filaments. As shown in Fig. 2a, four different scenarios can develop, depending on the initial bundle concentration c0, on the reaction rate k and on the polymerization velocity v. As L(t=0)=0, the system always starts off in the cL3<<1 reaction-limited regime, implying, through equations (1 and 2):
 |
with 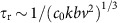 . This solution predicts a crossover from c=c0 for
. This solution predicts a crossover from c=c0 for 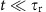 to c∝t−3 for
to c∝t−3 for  . Scenario (1) (Fig. 2a, topmost line) applies to slow-reacting filaments (kb<v), for which blocking happens before this first transition. The concentration c thus never departs from its initial value c0: a homogeneous network of single filaments of concentration c0 is formed.
. Scenario (1) (Fig. 2a, topmost line) applies to slow-reacting filaments (kb<v), for which blocking happens before this first transition. The concentration c thus never departs from its initial value c0: a homogeneous network of single filaments of concentration c0 is formed.
For fast-reacting filaments (kb>v) blocking takes over at a time larger than 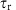 . Bundles thus form, and three possible scenarios can develop depending on c0. Scenario (2) (Fig. 2a, second line from the top) describes cases where substantial bundling takes place before the system transitions from reaction limited to diffusion limited, that is,
. Bundles thus form, and three possible scenarios can develop depending on c0. Scenario (2) (Fig. 2a, second line from the top) describes cases where substantial bundling takes place before the system transitions from reaction limited to diffusion limited, that is, 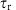 <
<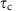 =Lc/v, or equivalently
=Lc/v, or equivalently 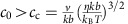 . Equation (5) is thus valid for t<
. Equation (5) is thus valid for t<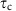 , and the c∝t−3 decay is valid for
, and the c∝t−3 decay is valid for 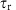 <t<
<t<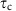 . As a result cL3 remains constant while the rods grow, thus staving off blocking as long as t<
. As a result cL3 remains constant while the rods grow, thus staving off blocking as long as t<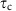 . At
. At 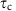 , the system becomes diffusion limited, and equations (1 and 3) imply that the concentration decays as
, the system becomes diffusion limited, and equations (1 and 3) imply that the concentration decays as 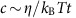 as long as
as long as 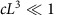 . Blocking then induces kinetic arrest for cL3∼1, implying
. Blocking then induces kinetic arrest for cL3∼1, implying 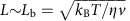 , or equivalently
, or equivalently 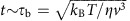 , yielding a final concentration
, yielding a final concentration 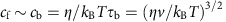 independent of the initial concentration c0. Scenario (3) (Fig. 2a, third line from the top) is relevant for cb<c0<cc. In this regime
independent of the initial concentration c0. Scenario (3) (Fig. 2a, third line from the top) is relevant for cb<c0<cc. In this regime 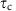 <
<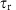 and the bundle concentration thus transitions from its initial plateau directly to the c∝t−1 diffusive regime. As in the previous scenario, blocking occurs at t=
and the bundle concentration thus transitions from its initial plateau directly to the c∝t−1 diffusive regime. As in the previous scenario, blocking occurs at t=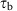 , resulting in a final concentration of the order of cb. Finally, scenario (4) (Fig. 2a, bottommost line) is relevant for c0<cb. In that case, by the time the filaments get into contact through diffusion at
, resulting in a final concentration of the order of cb. Finally, scenario (4) (Fig. 2a, bottommost line) is relevant for c0<cb. In that case, by the time the filaments get into contact through diffusion at 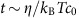 the system is already concentrated, and most bundling attempts are therefore blocked, yielding cf∼c0.
the system is already concentrated, and most bundling attempts are therefore blocked, yielding cf∼c0.
Our results regarding the final morphology of the networks are summarized in Fig. 2c: in low-concentration (c0<cb) and/or slow-reacting (kb≲v) systems, no bundling takes place. The final state is a homogeneous network of single filaments of concentration cf∼c0. In contrast, in fast-reacting, high-concentration systems (c0>cb and kb>v), the system evolves to a network of bundles with concentration cf∼cb independent of c0 and a characteristic number of filaments per bundle equal to c0/cf∼c0/cb. For c0≥cb, going from a slow- to a fast-reacting system produces an abrupt shift from a homogeneous network of filaments to a network of bundles with a concentration lower by several orders of magnitude.
Brownian dynamics simulations
To assess the validity of our homogeneous, isotropic, mean-field dynamical scenarios, we conduct numerical simulations of our model. We simulate a solution of initially very short, randomly oriented, impenetrable rods and implement their growth as well as their standard Brownian dynamics with diffusion coefficients D||=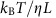 , D⊥=D||/2 for their longitudinal and transverse translation, respectively, and Dr=6D||/L2 for their rotation18. For each time step, the algorithm assesses the probability for each rod to react with its closest neighbour (the ‘target' rod) that is assumed to be fixed (the fixed rod is itself moved in a separate step). To estimate this probability, we write the Fokker–Planck equation describing the stochastic dynamics of the distance d between the two rods in the limit where d<<L (cases that violate this condition are benign in practice, as they yield a negligible reaction probability anyway):
, D⊥=D||/2 for their longitudinal and transverse translation, respectively, and Dr=6D||/L2 for their rotation18. For each time step, the algorithm assesses the probability for each rod to react with its closest neighbour (the ‘target' rod) that is assumed to be fixed (the fixed rod is itself moved in a separate step). To estimate this probability, we write the Fokker–Planck equation describing the stochastic dynamics of the distance d between the two rods in the limit where d<<L (cases that violate this condition are benign in practice, as they yield a negligible reaction probability anyway):
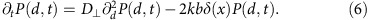 |
Here P(d, t) is the probability distribution of d∈[0, +∞), and the right-hand side includes a sink term representing reactions between the two rods in the limit of a very short interaction range b. Equation (6) assumes the convention 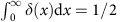 . This yields a probability of reaction between the two rods initially separated by d0 over one time step dt of the simulation:
. This yields a probability of reaction between the two rods initially separated by d0 over one time step dt of the simulation:
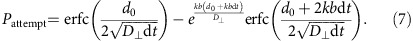 |
Note that equation (7) is and must be fully valid even in cases where d0 is of the order of the typical diffusion and reaction length scales 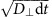 and kbdt. The algorithm implements a bundling attempt with a probability Pattempt.
and kbdt. The algorithm implements a bundling attempt with a probability Pattempt.
In the case where bundling is indeed attempted, the algorithm determines if any blocking rod is present in the bundling path (Fig. 1). If there is one, bundling is aborted and the attempting filament is moved in close proximity to the closest blocking rod. If bundling is successful, the attempting rod is deleted, representing its merging with the target rod. In the case where bundling is not attempted, diffusion proceeds as in normal Brownian dynamics, although with a reflecting boundary between rods to ensure their impenetrability21. Note that a single blocking rod can never derail bundling if it is not itself entangled with the rest of the network. Indeed, if the blocking rod is free to move and bundle with the attempting rod, they will do so within a few diffusion steps and the bundling of the two first rods will then be allowed to proceed. Thus, the transition to kinetic arrest is a true many-body effect in our simulations.
Snapshots of the resulting dynamics are shown in Fig. 3 at times corresponding to the four successive regimes of scenario (2). The evolution of the concentration in our simulations confirms the four predicted scenarios for the evolution of the rod concentration (Fig. 4a). Consistent with Fig. 2c, they also show that the final morphologies are either homogeneous networks of single filaments or strongly bundled phases, with an abrupt transition from one to the other (Fig. 4b) as a high-concentration system goes from slow reacting to fast reacting. The final structures do not display significant overall orientational order (Fig. 4c), consistent with both our model and in vitro observations4,5,7.
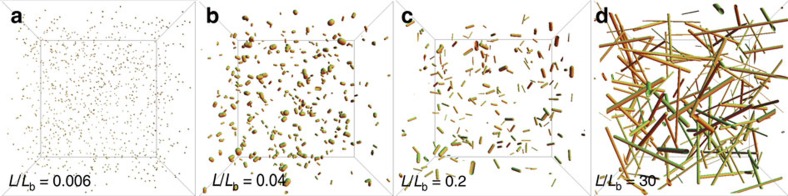
Figure 3. Snapshots of our Brownian dynamics simulations.
The state of the system in each of the four different regimes identified in our mathematical model are illustrated: (a) initial plateau c∝t0, (b) reaction-limited regime c∝t−3, (c) diffusion-limited regime c∝t−1 and (d) blocked regime c∝t0. To facilitate visualization despite order of magnitude changes in filament concentrations, each of the four panels is taken from a different simulation with a different box size, ensuring that between 100 and 1,000 rods are visible in each picture. The corresponding parameters and absolute concentrations can be read off from the white-filled symbols in Fig. 4a. In this figure the cross-sectional area of each rod is proportional to the number of filaments within the bundle.
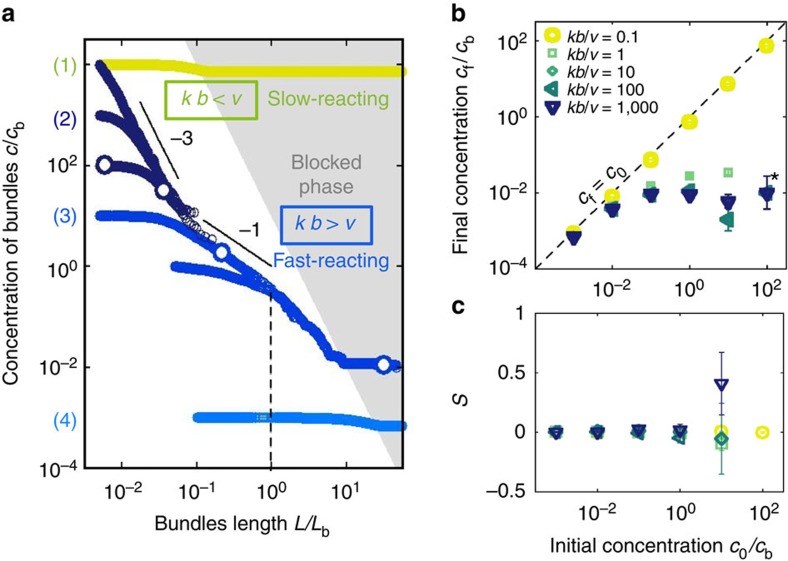
Figure 4. Dynamical evolution of our Brownian dynamics simulations.
(a) Concentration of solutions of N=103 rods as a function of their length for kb/v=0.1 (light grey (green)) and kb/v=1,000 (dark (blue) symbols). The initial concentration (c0) and filament length (L0) for each simulation can be read on the graph, and the white-filled symbols indicate the data points used for the snapshots of Fig. 3. The grey region materializes the condition cL3>10, where blocking is observed. (b) Final bundle concentration as a function of c0 for N=104 rods of initial length L0/Lb=0.1. Error bars give an estimate of the relative uncertainty on the number of remaining filaments at the end of the simulation 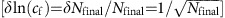 . For the most highly concentrated, fast-reacting conditions investigated (c0/cb=102 and kb/v≥1, marked with an asterisk), bundling is so strong that all rods in our simulations collapse into one, terminating the dynamics for reasons independent of blocking. (c) Scalar nematic order parameter
. For the most highly concentrated, fast-reacting conditions investigated (c0/cb=102 and kb/v≥1, marked with an asterisk), bundling is so strong that all rods in our simulations collapse into one, terminating the dynamics for reasons independent of blocking. (c) Scalar nematic order parameter 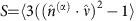 for the data of (b) following blocking. In the definition of S the index α refers to the α-th rod, and
for the data of (b) following blocking. In the definition of S the index α refers to the α-th rod, and 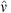 is the eigenvector corresponding to the largest eigenvalue of the tensorial order parameter
is the eigenvector corresponding to the largest eigenvalue of the tensorial order parameter 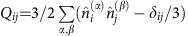 (ref. 32). Error bars show the s.e.m. associated with the determination of the average involved in the computation of S, again indicating the error associated with small filament numbers. The nematic order parameter is not computed for final states where only one rod is present (namely, for the data points with c0/cb=102, kb/v≥1).
(ref. 32). Error bars show the s.e.m. associated with the determination of the average involved in the computation of S, again indicating the error associated with small filament numbers. The nematic order parameter is not computed for final states where only one rod is present (namely, for the data points with c0/cb=102, kb/v≥1).
The good agreement between our theory and simulations comes with one quantitative difference. Because of the more complex geometry of the simulations, kinetic arrest there sets in for a relatively large value of cL3 (≃10), allowing more time for bundling and thus driving the final concentration down. As seen in Fig. 4a, this delayed blocking reveals an additional dynamical regime with a slope steeper than −1. In this regime, the rod crosses over to a faster-than-diffusive exploration of space thanks to its ballistic polymerization, leading to a speed-up of bundling before blocking (see Methods and Fig. 6). Note however that this regime can never fully develop if blocking is present, and thus that the scaling scenarios described above remain valid in our simulations, although with modified prefactors.
Figure 6. Characterization of the L>Lb scaling regime in a system without blocking.
(a) Schematic of the situation considered for our scaling argument, showing the rod of interest (solid line), the area explored by it in a time t (coloured region) and the other rods intersecting the plane of the figure (crosses). (b) Evolution of the concentration in a Brownian dynamics simulation identical to that of Fig. 4a with blocking turned off. The predicted −2 slope regime is clearly visible for L>Lb. The absence of blocking induces fast, unhindered decay of the filament concentrations towards zero, attesting to the importance of blocking for the stabilization of the cytoskeletal morphologies discussed above.
Discussion
The cytoskeleton of living cells is fundamentally out of equilibrium, and is constantly shaped by two major active processes: the operation of embedded molecular motors, and the constant self-assembly of its components. Although the statistical mechanics of the former is the subject of a substantial experimental and theoretical literature22, our understanding of the collective dynamics induced by the latter is very limited. Inspired by recent experiments, we introduce a versatile theoretical framework to investigate this problem, based on rate equations supplemented with a mean-field, entanglement-induced kinetic trapping term. Brownian dynamics simulations validate our theoretical assumptions, and show that our results are robust to changes in the detailed interactions between bundles.
We analyse our model in a simple situation consistent with existing in vitro experiments4,5,7. Although quantitative comparisons are impeded by technical challenges in resolving single filaments and thin bundles in these specific studies, our main qualitative predictions are all paralleled by the data. Bundle densities thus vary over orders of magnitude upon changes in the initial filament concentration c0, and the timescale required for their formation decreases sharply upon an increase of c0 (ref. 4). This is reminiscent of our predicted transition from the slowly relaxing (c∝t−1 at early times) scenario (3) at low c0 to the faster (c∝t−3) scenario (2) at larger c0. An increasing crosslinker concentration (analogous to an increase in k in the model) further induces a sharp transition from a homogeneous (scenario (1)) to a bundled network (scenario (2) or (3)). An additional 10-fold increase in crosslinker concentration however hardly modifies the mesh size of the network, strongly reminiscent of our abrupt transition from a slow-reacting to a fast-reacting system of fixed concentration cb for c0>cb (ref. 4). Our model also predicts that an increased reaction rate is equivalent to a decreased polymerization velocity through the dimensionless parameter kb/v. Consistent with this, in ref. 5 an increase in v through the use of the formin mDia1 causes the final bundle concentration to rapidly increase, then plateau out. More quantitatively, refs 4, 5 use crosslinker α-actinin at concentrations of the order of cα≈2 μM. Given the α-actinin–actin binding rate kon=5 μM−1 s−1 (ref. 23), we estimate that two actin filaments within an interaction distance b≈30 nm (the size of an α-actinin molecule) bind with a rate k=koncα=10 s−1. For v=10−2 μm s−1, this yields kb/v≈30 for the typical initial actin filament concentration c0≈0.1 μM. This is consistent with the formation of bundles observed under the aforementioned experimental conditions, and suggests that those in vitro assays can indeed transition between scenarios (1), (2) and (3) as their parameters are varied. We moreover predict 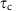 ≈370 s and
≈370 s and 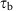 ≈2,000 s, comparable to the observed gelation time t≈600 s.
≈2,000 s, comparable to the observed gelation time t≈600 s.
These quantitative estimates further allow a discussion of the domain of validity of our model's main assumptions. We first discuss our approximation that the merging between two bundles is instantaneous. In general, the time required to merge two filaments is the sum of the time for the two filaments to find each other and form their first crosslink, plus a time 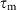 required to complete their merging. Direct measurements15 indicate that the latter timescale is of the order of a few hundred milliseconds at most. (This number is measured in the presence of large beads that slow down the merging dynamics because of hydrodynamic friction; the merging timescale
required to complete their merging. Direct measurements15 indicate that the latter timescale is of the order of a few hundred milliseconds at most. (This number is measured in the presence of large beads that slow down the merging dynamics because of hydrodynamic friction; the merging timescale 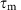 is probably significantly smaller in the situation considered here, where such beads are not present.) This timescale is much shorter than the typical evolution timescales
is probably significantly smaller in the situation considered here, where such beads are not present.) This timescale is much shorter than the typical evolution timescales 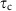 and
and 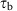 evaluated above. More quantitatively, we estimate in Methods that the delay
evaluated above. More quantitatively, we estimate in Methods that the delay 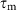 to merging will have negligible effects on the final concentration provided that
to merging will have negligible effects on the final concentration provided that 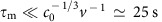 , confirming the merging time can indeed be neglected.
, confirming the merging time can indeed be neglected.
Our model also neglects actin bending and crosslinking, treating bundles as rigid rods throughout their dynamics. To assess the domain of validity of this approximation24, we compare the energetic incentive for two bundles entangled with the rest of the network to merge over a fraction of their length and compare it with the bending cost of doing so (Fig. 5). Guided by the detailed simulations of ref. 25, we consider bundles of N≃10 filaments with persistence length 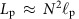 , where
, where 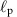 ≃10 μm is the actin persistence length, and assume that merging the two bundles brings a free energy bonus
≃10 μm is the actin persistence length, and assume that merging the two bundles brings a free energy bonus 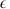 ≃4kBT per crosslinker with a typical spacing between crosslinks of
≃4kBT per crosslinker with a typical spacing between crosslinks of 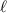 ≃b≃30 nm. As shown in Fig. 5, these parameters imply that filament bending will become prevalent for network mesh sizes of the order of
≃b≃30 nm. As shown in Fig. 5, these parameters imply that filament bending will become prevalent for network mesh sizes of the order of 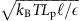 ≃3 μm. This estimate is in line with the final morphologies observed in ref. 25, where bundles are bent on length scales of the order of one to two μm, comparable to the network mesh size. This estimate suggests that networks with smaller mesh sizes, which include most cellular structures as well as the structure-defining early stages of our simulations, will undergo only moderate bundle bending, compatible with our approach. In contrast, networks with larger mesh sizes, including those studied in some in vitro assays, will undergo significant bending and deformation, and may therefore not be well described by our model. These deformations could facilitate the collapse of entangled structures towards a more energetically favourable (that is, more crosslinked) state. This could compromise the mechanical integrity of these networks and account for the formation of inhomogeneous structures with large gaps as observed in refs 26, 27.
≃3 μm. This estimate is in line with the final morphologies observed in ref. 25, where bundles are bent on length scales of the order of one to two μm, comparable to the network mesh size. This estimate suggests that networks with smaller mesh sizes, which include most cellular structures as well as the structure-defining early stages of our simulations, will undergo only moderate bundle bending, compatible with our approach. In contrast, networks with larger mesh sizes, including those studied in some in vitro assays, will undergo significant bending and deformation, and may therefore not be well described by our model. These deformations could facilitate the collapse of entangled structures towards a more energetically favourable (that is, more crosslinked) state. This could compromise the mechanical integrity of these networks and account for the formation of inhomogeneous structures with large gaps as observed in refs 26, 27.
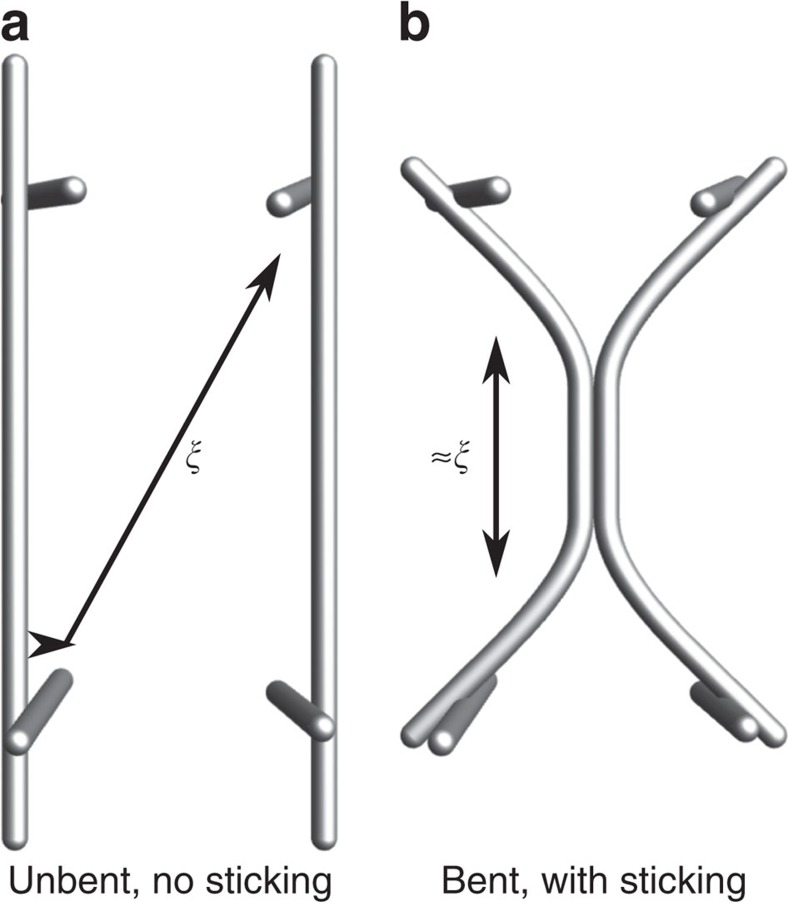
Figure 5. Assessment of the bundles' propensity to bend.
We consider two vertical bundles each blocked by the rest the network, with typical mesh size ξ (a). Thermal fluctuations or internal network stresses may cause the two filaments to deform and come into contact (b). Denoting by 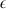 the binding free energy associated with the binding of a crosslinker and by
the binding free energy associated with the binding of a crosslinker and by 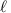 the typical distance between crosslinks, we assess whether such configurations are energetically favourable by comparing the energy bonus
the typical distance between crosslinks, we assess whether such configurations are energetically favourable by comparing the energy bonus 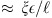 due to crosslinker binding to the energy penalty ≈kBTLp/ξ because of filament bending, where Lp is the persistence length of the bundle. The latter exceeds the former for mesh sizes smaller than
due to crosslinker binding to the energy penalty ≈kBTLp/ξ because of filament bending, where Lp is the persistence length of the bundle. The latter exceeds the former for mesh sizes smaller than 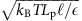 , (≃3 μm with the parameters of the main text). Our treatment of bundles as rigid is thus justified in these networks.
, (≃3 μm with the parameters of the main text). Our treatment of bundles as rigid is thus justified in these networks.
The good agreement between our predictions and the experiments of refs 4, 5 suggests that topological entanglement between filaments could be the major driver of kinetic arrest in cytoskeletal systems. Depending on the system considered, its action in regulating the thickness of actin bundles could be complemented by other mechanisms. For instance, the build-up of elastic strains28 has been proposed to regulate the width of bundles crosslinked by the very short crosslinker fascin29, although the importance of this mechanism is less clear in the α-actinin bundles used in refs 4, 5. Other effects ignored here, for example, transient sticking between unbundled filaments or the effective increase in length incurred by a bundle upon coalescence with another, may thus not be essential to gain a first understanding of the resulting network structures. Such effects could however easily be included in our framework if warranted by more precise experimental comparisons, as will the physiologically important effects of spontaneous filament nucleation or the coexistence of several crosslinker types with different bundling behaviours30. Detailed simulations will also be useful in assessing the influence of the addition of these and other experimentally relevant features to our model. Although our current solution-like model does not explicitly describe the network's mechanical properties, it does predict its typical mesh size and bundle thickness, whose relationship to the network's mechanical response has been the subject of substantial modelling efforts31. Finally, further experimental and theoretical work is needed to elucidate the network structure in the biologically relevant presence of depolymerization/severing that could give rise to fundamentally nonequilibrium steady states.
Overall, our study provides a first theoretical account of the nonequilibrium mechanisms responsible for the actin structures observed in vivo and in vitro. It further illustrates that these dynamical processes can lead to sharp transitions between dramatically different network structures, hinting that cells need only harness relatively modest changes in their internal composition to generate the large variety of morphologies that characterize the cytoskeleton.
Methods
Speed-up of bundling before blocking
Here we rationalize the speed-up of bundling observed in our Brownian dynamics simulations just before the system becomes blocked in Fig. 4a. This speed-up is a signature of the system crossing over to a new scaling regime for r(2) as L becomes larger than Lb, that is, as the longitudinal growth of the rod becomes faster than its longitudinal diffusion. In practice, this regime has little incidence on our model as bundling becomes hindered by blocking precisely at L=Lb (in our mean-field discussion) or shortly thereafter (in our simulations). Here we present a scaling argument showing that c∝t−2 in this regime, and display numerical evidence to that effect.
Let us consider a rod of interest lying in the plane of Fig. 6a. As the rod diffuses and grows, it encounters other rods that intersect the plane of the figure, and attempts to bundle with them. In an homogeneous, isotropic solution, the typical distance between two rods is 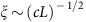 (ref. 31). The typical number of other rods encountered by the rod of interest after a time t is
(ref. 31). The typical number of other rods encountered by the rod of interest after a time t is 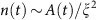 , with A(t) the typical area of the plane of the figure visited by the rod of interest within a time t. The width of this area is of the order of
, with A(t) the typical area of the plane of the figure visited by the rod of interest within a time t. The width of this area is of the order of 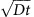 , with
, with 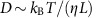 the typical diffusion coefficient of the rod (representing a combination of transverse and rotational diffusion). Here we consider a diffusing and growing rod with length
the typical diffusion coefficient of the rod (representing a combination of transverse and rotational diffusion). Here we consider a diffusing and growing rod with length  , implying that the rate of longitudinal diffusion of the rod (inducing a displacement
, implying that the rate of longitudinal diffusion of the rod (inducing a displacement 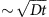 ) is negligible in front of its growth rate (inducing a displacement ∼vt). As a result the length of the area grows as vt and
) is negligible in front of its growth rate (inducing a displacement ∼vt). As a result the length of the area grows as vt and 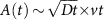 . Combining these expressions and using L=vt we find that
. Combining these expressions and using L=vt we find that 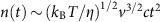 .
.
The typical reaction rate r(2) is the rate at which the rod of interest encounters other rods, yielding 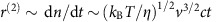 . Applying equation (1) in the absence of blocking we thus find
. Applying equation (1) in the absence of blocking we thus find
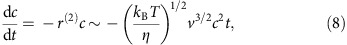 |
which yields in the long-time asymptotic limit
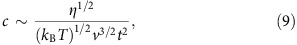 |
This c∝t−2 scaling is indeed observed in our simulations in the absence of blocking, as shown in Fig. 6b.
Incidence of delayed bundling on the final concentration
Here we assess the effect of a finite bundle merging time 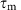 on the final rod concentration. We place ourselves in the diffusion-limited regime that, as described in Results, is the important one when considering the transition to blocking. The rate of bundling attempts is thus
on the final rod concentration. We place ourselves in the diffusion-limited regime that, as described in Results, is the important one when considering the transition to blocking. The rate of bundling attempts is thus
 |
To account for the additional hindrance to bundling, we assume that two rods that come into contact at a time t only complete their bundling at time t+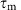 . During this time interval, the two unbundled rods are linked together and thus diffuse as a single object, implying that they count as a single rod in estimating the concentration that enters the attempt rate of equation (10). However, as they are not yet bundled they retain their full blocking power towards other bundling events until t+
. During this time interval, the two unbundled rods are linked together and thus diffuse as a single object, implying that they count as a single rod in estimating the concentration that enters the attempt rate of equation (10). However, as they are not yet bundled they retain their full blocking power towards other bundling events until t+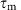 . Denoting by c(t) the number of independently diffusing objects at time t, the full mean-field bundling rate thus reads:
. Denoting by c(t) the number of independently diffusing objects at time t, the full mean-field bundling rate thus reads:
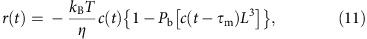 |
as the concentration of rods relevant for blocking at time t is identical to the concentration of free rods at time t−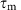 . Finally, here we assume that the bundling dynamics becomes abruptly blocked as cL3 exceeds one (note that this approximation preserves all the scaling results derived in the Results section).
. Finally, here we assume that the bundling dynamics becomes abruptly blocked as cL3 exceeds one (note that this approximation preserves all the scaling results derived in the Results section).
Rescaling time by 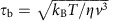 and concentration by
and concentration by 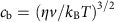 , the full dimensionless concentration equation becomes
, the full dimensionless concentration equation becomes
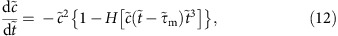 |
where 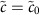 for
for 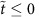 and H is the Heaviside step function. The final solution of this equation displays two regimes, depending on whether kinetic arrest takes over before or after the first bundling event is completed:
and H is the Heaviside step function. The final solution of this equation displays two regimes, depending on whether kinetic arrest takes over before or after the first bundling event is completed:
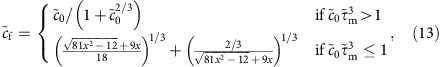 |
with 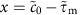 .
.
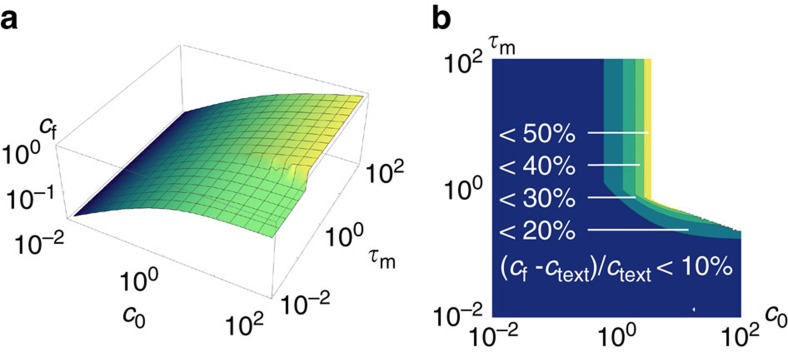
These final concentrations are plotted in Fig. 7, along with their relative deviation from the result at 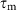 =0. In practice, our results are insensitive to the value of
=0. In practice, our results are insensitive to the value of 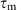 as long as
as long as 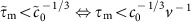 , as discussed in the Results section.
, as discussed in the Results section.
Figure 7. Quantitative effect of delayed bundling on cf.
(a) Final concentrations as given by equation (13) in absolute value. (b) Corresponding deviations relative to the 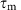 =0 prediction (denoted as ctext). Such violations are substantial only in the regime where
=0 prediction (denoted as ctext). Such violations are substantial only in the regime where 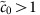 and
and 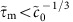 .
.
Data availability
The computer code used for this study as well as the data generated and analysed are available from the corresponding author on request.
Additional information
How to cite this article: Foffano, G. et al. The dynamics of filament assembly define cytoskeletal network morphology. Nat. Commun. 7, 13827 doi: 10.1038/ncomms13827 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Emmanuel Trizac for stimulating discussions. This work was supported by grants from Université Paris-Sud's Attractivité and CNRS' PEPS-PTI programs, Marie Curie Integration Grant PCIG12-GA-2012-334053, ‘Investissements d'Avenir' LabEx PALM (ANR-10-LABX-0039-PALM), ANR Grant ANR-15-CE13-0004-03 and ERC Starting Grant 677532. The group of M.L. belongs to the CNRS consortium CellTiss.
Footnotes
Author contributions M.L. designed the research. G.F., N.L. and M.L. carried out the analytical calculations. G.F. carried out the numerical simulations. G.F. and M.L. wrote the paper.
References
- Fletcher D. A. & Mullins R. D. Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch A. R. & Kroy K. A bottom-up approach to cell mechanics. Nat. Phys. 12, 231 (2006). [Google Scholar]
- Pelletier O. et al. Structure of actin cross-linked with α-actinin: a network of bundles. Phys. Rev. Lett. 91, 148102 (2003). [DOI] [PubMed] [Google Scholar]
- Falzone T. T., Lenz M., Kovar D. R. & Gardel M. L. Assembly kinetics determine the architecture of α-actinin crosslinked F-actin networks. Nat. Commun. 3, 1861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone T. T., Oakes P. W., Sees J., Kovar D. R. & Gardel M. L. Actin assembly factors regulate the gelation kinetics and architecture of F-actin networks. Biophys. J. 104, 1709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieleg O., Kayser J., Brambille G., Cipelletti L. & Bausch A. R. Slow dynamics and internal stress relaxation in bundled cytoskeletal networks. Nat. Mat. 10, 236–242 (2011). [DOI] [PubMed] [Google Scholar]
- Kayser J., Grabmayr H., Harasim M., Herrmann H. & Bausch A. Assembly kinetics determine the structure of keratin networks. Soft Matter 8, 8873 (2012). [Google Scholar]
- Zilman A. G. & Safran S. A. Role of cross-links in bundle formation, phase separation and gelation of long filaments. Europhys. Lett. 63, 139 (2003). [Google Scholar]
- Borukhov I., Bruinsma R., Gelbart W. & Liu A. J. Structural polymorphism of the cytoskeleton: a model of linker-assisted filament aggregation. Proc. Natl Acad. Sci. USA 102, 3673 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyron C. J. et al. Equilibrium phase diagram of semi-flexible polymer networks with linkers. Europhys. Lett. 102, 38003 (2013). [Google Scholar]
- Ferrer J. M. et al. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc. Natl Acad. Sci. USA 105, 9221–9226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H., Yasuda R. & Kinosita K. Jr Strength and lifetime of the bond between actin and skeletal muscle alpha-actinin studied with an optical trapping technique. Biochim. Biophys. Acta 1290, 83–88 (1996). [DOI] [PubMed] [Google Scholar]
- Charras G. T., Hu C.-H., Coughlin M. & Mitchison T. J. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175, 477 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotsch C. & Radmachr M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys. J. 78, 520–535 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streichfuss M. et al. Measuring forces between two single actin filaments during bundle formation. Nano Lett. 11, 3676 (2011). [DOI] [PubMed] [Google Scholar]
- Kim T., Hwang W. & Kamm R. D. Computational analysis of a cross-linked actin-like network. Exp. Mech. 49, 91–104 (2009). [Google Scholar]
- Takatsuki H., Bengtsson E. & Mansson A. Persistence length of fascin-cross-linked actin filament bundles in solution and the in vitro motility assay. Biochim. Biophys. Acta 1840, 1933–1942 (2014). [DOI] [PubMed] [Google Scholar]
- Doi M. & Edwards S. F. The Theory of Polymer Dynamics Clarendon Press (1986). [Google Scholar]
- Viamontes J., Oakes P. W. & Tang J. X. Isotropic to nematic liquid crystalline phase transition of F-actin varies from continuous to first order. Phys. Rev. Lett. 97, 118103 (2006). [DOI] [PubMed] [Google Scholar]
- Van Kampen N. Stochastic Processes in Physics and Chemistry North-Holland Personal Library (1992). [Google Scholar]
- Tse Y.-L. S. & Andersen H. C. Modified scaling principle for rotational relaxation in a model for suspensions of rigid rods. J. Chem. Phys. 139, 044905 (2013). [DOI] [PubMed] [Google Scholar]
- Prost J., Jülicher F. & Joanny J.-F. Active gel physics. Nat. Phys. 11, 111–117 (2015). [Google Scholar]
- Kuhlman P. A., Ellis J., Critchley D. R. & Bagshaw C. R. The kinetics of the interaction between the actin-binding domain of α-actinin and F-actin. FEBS Lett. 339, 297–301 (1994). [DOI] [PubMed] [Google Scholar]
- Pandolfi R. J., Edwards L., Johnston D., Becich P. & Hirst L. S. Designing highly tunable semiflexible filament networks. Phys. Rev. E 89, 062602 (2014). [DOI] [PubMed] [Google Scholar]
- Müller K. W. et al. Rheology of semiflexible bundle networks with transient linkers. Phys. Rev. Lett. 112, 238102 (2014). [DOI] [PubMed] [Google Scholar]
- Lieleg O. et al. Structural polymorphism in heterogeneous cytoskeletal networks. Soft Matter 5, 1796 (2009). [Google Scholar]
- Nguyen L. T. & Hirst L. S. Polymorphism of highly cross-linked F-actin networks: Probing multiple length scales. Phys. Rev. E 83, 031910 (2011). [DOI] [PubMed] [Google Scholar]
- Grason G. M. Colloquium: geometry and optimal packing of twisted columns and filaments. Rev. Mod. Phys. 87, 401 (2015). [Google Scholar]
- Claessens M. M. A. E., Semmrich C., Ramos L. & Bausch A. R. Helical twist controls the thickness of F-actin bundles. Proc. Natl Acad. Sci. USA 105, 8819 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller K. M., Lieleg O. & Bausch A. R. Structural and viscoelastic properties of actin/filamin networks: cross-linked versus bundled networks. Biophys. J. 97, 83–89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broedersz C. P. & MacKintosh F. C. Modeling semiflexible polymer networks. Rev. Mod. Phys. 86, 995 (2014). [Google Scholar]
- Chaikin P. M. & Lubensky T. C. Principles of Condensed Matter Physics Cambridge University Press (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computer code used for this study as well as the data generated and analysed are available from the corresponding author on request.