Abstract
The constituents of seminal fluid are a complex mixture of proteins and other molecules, most of whose functions have yet to be determined and many of which are rapidly evolving. As a step in elucidating the roles of these proteins and exposing potential functional similarities hidden by their rapid evolution, we performed comparative structural modeling on 28 of 52 predicted seminal proteins produced in the Drosophila melanogaster male accessory gland. Each model was characterized by defining residues likely to be important for structure and function. Comparisons of known protein structures with predicted accessory gland proteins (Acps) revealed similarities undetectable by primary sequence alignments. The structures predict that Acps fall into several categories: regulators of proteolysis, lipid modifiers, immunity/protection, sperm-binding proteins, and peptide hormones. The comparative structural modeling approach indicates that major functional classes of mammalian and Drosophila seminal fluid proteins are conserved, despite differences in reproductive strategies. This is particularly striking in the face of the rapid protein sequence evolution that characterizes many reproductive proteins, including Drosophila and mammalian seminal proteins.
In most internally fertilizing animals, sperm are exposed to a variety of seminal plasma proteins in both the male and female reproductive tracts. In insects and mammals, males have developed multiple sex accessory tissues that secrete seminal proteins that are transferred to the female during mating. In Drosophila, the accessory gland, ejaculatory bulb, and ejaculatory duct serve as the male reproductive tract accessory tissues. The male accessory glands of Drosophila melanogaster produce ≈80 seminal fluid proteins (1). Receipt of male accessory gland proteins (Acps) by females induces a variety of physiological, behavioral, and reproductive changes, including an increase in egg-production and in the female's egg-laying and ovulation rates, a decrease in her propensity to remate, an increase in her ability to store sperm, and a decrease in the mated female's lifespan (reviewed in ref. 2).
Eighteen Acp genes had previously been reported (reviewed in ref. 2) and a comprehensive EST screen (1) identified an additional 57 candidate Acp genes, bringing the total identified to 75. Subsequent gene annotation and expression analysis (J.L.M., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. Siggia, C.F.A., and M.F.W., unpublished results) led us to consider 52 of the 75 as particularly robust candidates for Acps (male-biased expression and encoding secreted proteins). Of these 52, the 15 tested thus far are known to be accessory gland-specific in their expression (3–5), and the 15 tested thus far have been confirmed to encode seminal proteins that are transferred to the female during mating (refs. 4 and 6–8, and K. Ravi Ram, S. Ji, and M.F.W., unpublished results; see www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm). As a group, Acps are roughly 2-fold more divergent than proteins in nonreproductive tissues (1, 9); indeed one, Acp26Aa, is one of the most rapidly evolving genes in the D. melanogaster genome (10, 11). This rapid divergence has been suggested to result from positive Darwinian selection (12, 13) and to be due to the participation of seminal fluid proteins in the evolutionary dynamics of antagonistic evolution (14), sperm competition (15), and/or sexual selection (16).
In male mammals, the prostate, epididymis, seminal vesicles, bulbourethral glands, and ampullae secrete seminal proteins that are then transferred to females during mating. Among mammals, each of the sex accessory tissues, except for the epididymis, exhibits great variation in biology, anatomy, and function. Nonetheless, in combination, these tissues produce a general set of protein classes common among mammals. These classes include protease inhibitors, proteases, lipid modifying enzymes, lectins, CRISPs (cysteine-rich secreted proteins), antimicrobial peptides, and antioxidants (reviewed in refs. 17–19). These protein classes have proposed involvements in semen coagulation, sperm motility, sperm maturation, sperm acrosome reaction, sperm viability, sperm capacitation, and control of immune activity within the female reproductive tract of mammals (reviewed in refs. 17–19).
Primary sequence comparisons can sometimes miss conserved elements of a protein, which can be seen only at the structural level. A comparative structural modeling (CM) approach can reveal these structural/functional similarities undetectable at the sequence level. Reproductive proteins, such as Acps, serve as good candidates for a CM approach, because their rapid evolution generates high levels of sequence diversity (20) yet their structures are likely to remain conserved (21). Here, we used CM to investigate whether there are essential molecular processes in the seminal fluid that are conserved between mammals and flies, despite the differences in reproductive strategies and the fact that many specific Drosophila and mammalian seminal fluid proteins are rapidly evolving (1, 11–13, 22–25). Our data illustrate how CM can provide insight into possible functional relationships undetectable by primary sequence alignments. The CM approach also predicts candidate-active sites, surface-ligand-binding regions, structurally important residues, residues predictive of novel functions, and insight into the patterns of rapid evolution at the structural level. The utility of this approach was previously validated with a single test case: CM-based prediction that Acp62F is a serine protease inhibitor was verified biochemically (7). By extending this approach to the remaining 51 robust candidate Acps, we report here that 28 encoded proteins show predicted structural similarities to proteins in the Protein Data Bank (PDB). Interestingly, many of the D. melanogaster Acp structures fell into the same general protein classes as in the mammalian seminal fluid. This conservation suggests that D. melanogaster may be a good general model in which to study the mechanisms of essential reproductive strategies. The 23 remaining Acps whose structures are unrecognizable by CM searches are primarily short peptides with no domains with identifiable protein structures; they are thus good candidates for future NMR or crystallographic studies.
Materials and Methods
Sequence and Structure Analysis. The Acp protein sequences used in our modeling were taken from an annotated and validated list of D. melanogaster Acp gene sequences (J.L.M., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. Siggia, C.F.A., and M.F.W., unpublished results; see www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm). GenBank accession numbers for CRISP structural homologs are as follows: AEG-1 Mus musculus, Q03401; AEG-1 Homo sapiens, P54107; TPX-1 Homo sapiens, P16562; allurin Xenopus laevis, AAL12003; and Vespula vulgaris venom allergen, 1_QNXA. The CG10284 Drosophila simulans EST sequence was obtained from GenBank under accession number BG642232 (1). PDB files were downloaded from the protein databank (www.rcsb.org/pdb) (26).
Generating 3D Models. 3D templates were identified by means of a combination of three different protein-threading programs, because each program emphasizes different features of a protein's predicted structure. The use of multiple programs [superfamily, 3d-pssm, and fugue3 (27–29)] allowed us to (i) compare alignments to find the optimal one, (ii) ensure that candidate hits were not missed by using a single program, and (iii) validate that the significance of a hit from one program is supported by other programs. Of the 51 Acps tested, 28 display significant alignments in multiple programs. The 28 Acps that aligned are Acp29AB (CG17797), Acp76A (CG3801), CG1462, CG1652, CG1656, CG4847, CG6069, CG6168, CG6289, CG8093, CG8137, CG8194, CG9334, CG9997, CG10284, CG10363 (TepIV) (30), CG10433, CG10956, CG11598, CG11664, CG11864, CG14034, CG17097, CG17575, CG17843, CG18284, CG31872, and BG642378. Acp62F (7) was previously structurally characterized and was not included in our analysis. The remaining 23 were also tested, but they did not disclose any structural similarities.
Each of the 28 Acps with predicted structural similarities also gave significant PDB blast hits, but we used the threading servers to generate multiple alternative alignments from which to choose the most structurally congruent alignment. The selection steps to identify the most accurate alignment include the following: (i) ranking, based upon their significance values, only those structure/sequence alignments with >95% confidence of a match between an Acp's sequence and a known structure (% identities between template structures and Acps, structural alignments, fugue Z-scores, and 3d-pssm and superfamily E-values and the completed Acp models can be found at www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm); (ii) determining the most congruent alignment by searching for similar gap sizes and matching of conserved residues; and (iii) refining the alignment to remove gaps within α-helices and β-strands and to ensure that no structurally conserved residues within a family had been shifted. Once an accurate alignment was determined, generation of a 3D model was performed with the program modeller6 version 2 (31). Each model generated was inspected for the following features: (i) surface exposure, primarily consisting of hydrophilic residues; (ii) spatial orientation of structural residues, disulfide bridges and secondary structure hydrophobic interactions; (iii) consistency of stereochemical properties, root mean square deviations (rmsd) of α-carbons being calculated by superimposing the two structures by means of an iterative fit with swisspdbview 3.7 (32) to ensure no significant structural deviations (rmsd < 1) were present; and (iv) active site integrity, i.e., the set of active site residues have to be intact and show the correct spatial orientation. Models were ranked depending upon these steps, with threading of a given Acp with the fewest differences to a known structure ranked the highest. All models generated were quality-checked with the what_check (33) check program (www.cmbi.kun.nl/gv/servers/WIWWWI), and all major structural errors (not found in the template structure) were corrected by refining the existing alignment or modeling the Acp to another structure in the same structural class. Acps without significant hits from the above structural server outputs also did not yield significant structural similarities when submitted to the Structure Prediction Meta Server (34) (http://BioInfo.PL/Meta), which utilizes 13 servers to generate a consensus structure prediction.
Although these models could still potentially contain errors, because the modeling was performed by means of CM, the accuracy of CM Acp models is probably higher than models generated by fold recognition and/or ab initio methods (35). Additionally, many of our predicted protein classes for the Acps overlap with protein classes considered “reliable models” by MODBASE (36), which uses other methods to check the models.
Results
Thirteen Acps Resemble Regulators of Proteolysis. Seven Acps thread onto members of the protease inhibitor class. Seven D. melanogaster seminal fluid proteins are suggested to be structural members of the serine protease inhibitor class (serpins: Acp62F (7), Acp76A (3), CG6289, CG8137, CG9334, CG10956, and BG642378). Six of the seven predicted serpin Acps are composed of the typical nine α-helices and three β-sheets, whereas Acp62F had previously been found to match a small class of structurally unique serine protease inhibitors (7). Many serpins inhibit serine proteases: the protease attacks the serpin-active site and becomes irreversibly covalently bound in a serpin–protease complex (37). The typical serpin contains an ≈18-aa reactive center loop (RCL). Protease inhibition by serpins depends upon this RCL, although proteins with an RCL can also be noninhibitory. The serpin-type Acps' loop regions vary from 14 to 19 residues (Acp76A-19, BG642378-14, CG6289-15, CG8137-18, CG9334-18, and CG10956-17), which is within or close to the canonical loop length (15–22 residues).
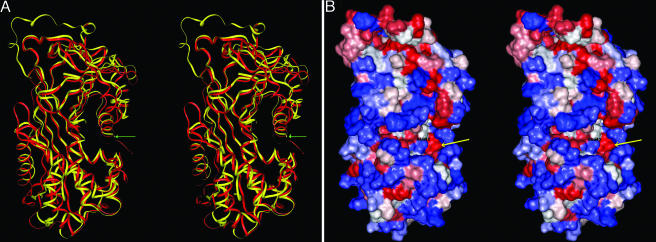
Some serpins also perform additional roles such as hormone transport (38). For example, the overall structure of protein C inhibitor (PCI) is similar to that of most other cleaved serpins, except for an N-terminal shortening of the first α-helix (Fig. 1A). This N-terminal shortening results in the formation of a large hydrophobic pocket (Fig. 4, which is published as supporting information on the PNAS web site). It is predicted that this site in PCI binds hormones; it is also found in two other hormone-binding serpins (39). Based on our structural modeling, two Acps, Acp76A and CG8137, also have N-terminal truncations of their first α-helix (see www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm for alignments and Fig. 1 A for the CG8137 model). These truncations form a hydrophobic pocket along their surfaces (Fig. 1B for CG8137 pocket), similar to the one in PCI (Fig. 4). This finding suggests that these two Acps may also be involved in the binding or transport of hydrophobic hormones. Acp76A and CG8137's hydrophobic pockets differ in electrostatic potential between the surfaces at the centers of these pockets and their rims from that observed in PCI (Fig. 1B), which suggests a difference in specificity (electrostatic surfaces not shown).
Fig. 1.
Candidate hormone-binding pocket of CG8137. (A) Superimposed stereoview ribbon diagrams of CG8137 and PCI (cleavage fragment not shown), in the same orientation as the surface potential figures, show the conserved serpin family secondary structures. PCI ribbons are in red and CG8137 ribbons are in yellow, with a green arrow indicating the position of the truncated N-terminal helices forming the hydrophobic pocket. (B) Molecular surface of CG8137 is shown in stereoview with hydrophobic surface areas shown in red and hydrophilic surfaces shown in blue. The yellow arrow indicates the position of the candidate hydrophobic pocket. Glu-42 in CG8137 is primarily responsible for the difference in electrostatic surface potential charge (not shown) between CG8137 and PCI.
Six Acps thread onto members of three protease classes. Members of three of the four protease types (aspartic, cysteine, metallo-, and serine proteases) are found among predicted Acps (Table 1); only aspartic proteases have not been identified. CG6168 and CG11864 seem to be members of the metalloprotease class and CG4847 a member of the cysteine protease class. More specifically, the structures of CG6168, CG11864, and CG4847 thread to members of the aminopeptidase, astacin metalloprotease, and papain-like cysteine protease classes, respectively. CG11864, CG6168, and CG4847 do not display major structural differences in their models (Table 1), when compared with their respective known structures.
Table 1. Acp predicted active sites.
| Proteins in predicted classes | Predicted active site residues |
|---|---|
| Proteases | |
| CG4847 | Cys-197, His-338 |
| CG6069 | Unknown |
| CG6168 | His-136, Asp-152, Glu-187, Asp-224, His-302 |
| CG9997 | Unknown |
| CG11664 | His-62, Asp-108, Ser-198 |
| CG11864 | His-144, Glu-145, His-148, His-154, Tyr-204 |
| Lipases | |
| CG8093 | Ser-167, Asp-341, His-376 |
| CG11598 | Ser-157, Asp-325, His-355 |
| CG14034 | Gly-150, Asp-175, His-243 |
| CG17097 | Ser-855, His-1033, His-1063 |
| CG18284 | Ser-228, Asp-401, His-432 |
| CG31872 | Ser-845, Asp-1018, His-1049 |
| Lectins | |
| Acp29AB | Gln-199, Thr-201, Asn-205, Gln-206, Glu-222, Lys-223 |
| CG1652 | Leu-120, Leu-122, Asp-128, Asp-129, Ser-143, Ala-144 |
| CG1656 | Glu-125, Thr-127, Asp-133, Asp-134, Leu-148, Asp-149 |
| Thioredoxin | |
| CG17843 (N-term.) | Cys-69, Cys-72 |
| CG17843 (C-term.) | Cys-451, Cys-454, Cys-512, Cys-515 |
| RNase | Catalytic sites: His-114, Lys-170, His-171; Substrate binding: Trp-117 |
| CG8194 |
Amino acid positions are given for each predicted active site residue within a given structure. The CRISPs and the Defensin do not contain characterized active sites and are therefore not included in this table.
A number of structural differences from consensus exist in the predicted active sites of some Acp members of the serine protease class (CG6069, CG9997, and CG11664), suggesting a function other than proteolysis. The typical serine catalytic triad consists of a His, Asp, and Ser. However, in CG6069, the His is replaced by an Asn residue, and the serine is not present (see www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm for alignments). Neither difference is due to an error in structural alignment, because adjacent conserved structural residues align. Loss of two active site residues suggests that CG6069 may not function as a protease; however, the active site protease pocket is still present and is positively charged by an Arg. The most extreme structural differences from canonical serine proteases occur in CG9997, which does not maintain any of the active site residues (see www.mbg.cornell.edu/wolfner/tables/AcpCModels.htm for alignments) yet shares structural similarities with 34 different eukaryotic and bacterial serine proteases.
There Are Six Lipase-Related Acps. Lipases are lipid-modifying enzymes belonging to the α/β hydrolase fold family. Five of the six lipase family members detected as Acps (CG8093, CG11598, CG17097, CG18284, and CG31872) model most accurately to the acid lipase class (Table 1), whereas CG14034 models best to members of the phospholipase class. Acid lipases are able to withstand acidic conditions, unlike most other lipases that function around pH 7.0–8.0. Previous work has shown the presence of lipolytic activity within the male accessory gland (40). Accessory gland triacylglycerol lipase activity was optimal at a pH of ≈5.5 (40), which is consistent with the majority of our threaded lipases being of the acid lipase class.
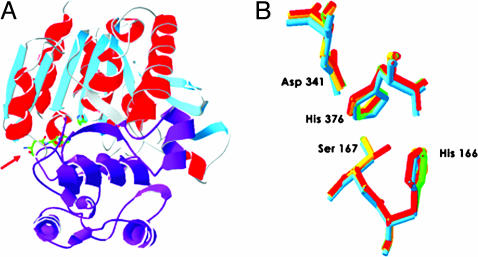
All five Acps that are similar to acid lipases contain a cap-domain predicted to cover the active site serine (Fig. 2A). This domain, found in canonical acid lipases, is thought to serve as a lid to regulate when lipid substrates can access the active site (Fig. 5) (41). When the active site is made accessible, lipase activity depends on a catalytic triad composed of the conserved consensus sequence (Gly-X1-Ser-X2-Gly) and two other residues (His and Asp). Four of the five predicted acid-type lipase Acps contain this triad (Table 1) in the correct spatial arrangement of canonical catalytic residues (Fig. 2B). The two exceptions within the catalytic triad are CG17097, where a His replaces the Asp, and CG14034, where a Gly replaces the Ser.
Fig. 2.
Acid-lipase structure characteristics of CG8093 and active-site comparison of CG8093, CG18284, CG11598, and 1hlg. (A) Complete predicted structure of CG8093 and its predicted CAP-domain in purple, which when removed (Fig. 5, which is published as supporting information on the PNAS web site) exposes the active site (red arrow). (B) Overlay of CG8093 (red), CG18284 (blue), CG11598 (green), and 1hlg (orange) active sites showing similar spatial orientation. The lipase catalytic triad residues Ser, His, and Asp are superimposed to show active site spatial integrity across multiple acid lipase structures. The additional His, adjacent to the Ser, is required for active site stability and is a component of the lipase signature motif, GHSXG. Residue positions are given for CG8093 only.
Three Predicted Acps Have Structural Features of Lectins. Three Acps identified fall into the C-type lectin (C-TL) class. Lectins possess a carbohydrate recognition domain (CRD), where interaction occurs with a given carbohydrate chain in the target. Acps CG1652, CG1656, and Acp29AB possess a domain with characteristics of a CRD (Table 1). The binding of sugars by C-TLs is calcium-dependent; the calcium ion serves as the nucleus of the binding site.
Carbohydrate specificity of C-TLs is determined by a set of six key residues. CG1652, CG1656, and Acp29AB have been modeled onto members of the C-TL class. Given C-TLs' specificity of binding to sugars, we can predict the residues important for binding for these three Acps (Table 1). For each Acp, the spatial orientation of the critical residues within its binding pocket is similar to that of structures onto which it was threaded (Fig. 6, which is published as supporting information on the PNAS web site). However, the residues predicted to bind sugars differ between each Acp and the structure onto which it threads. Thus, if these Acps bind sugars as predicted by their structures, it is likely that they will bind to different carbohydrate moieties from those bound by the known C-TLs.
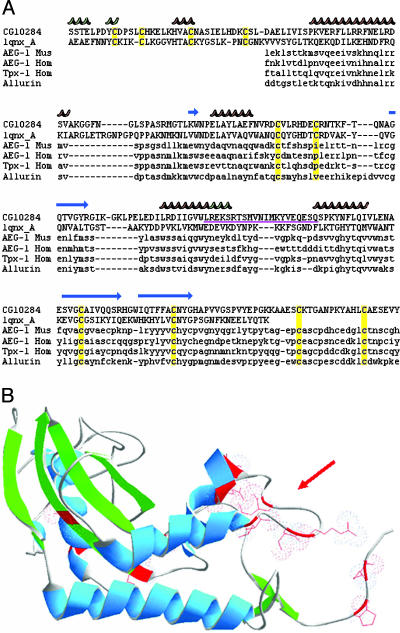
Two Acps Resemble CRISPs. It is particularly intriguing that structures of two Acps resemble CRISPs, because cysteine-rich proteins in the CRISP family have been associated with sperm-related functions in several vertebrates (42, 43). CG10284 and CG17575 both align to the CRISP structural family of proteins, which contains multiple disulfide bridges (CG10284 alignment is shown in Fig. 3A). The structural similarities between these family members are only evident by structural comparison, not by primary sequence comparisons (blastp).
Fig. 3.
Sequence and structural characteristics of the CRISP domain of CG10284. (A) Sequence alignment of the CRISP domains (entire protein coding regions are not shown) of CG10284 (D. melanogaster), and members of the CRISP family 1qnx_A(Vespula vulgaris), AEG-1 (Mus musculus), AEG-1 (Homo sapiens), TPX-1 (Homo sapiens), and allurin (Xenopus laevis). Residues of known (1qnx_A) and threaded (CG10284) structure are capitalized. CG10284 residue positions 48–283 are shown. Overlapping predicted and known secondary structural regions are depicted above each alignment. α-Helices are colored red, β-strands blue, and 310 helices green. Conserved cysteines are highlighted in yellow. CG10284's region with the excessive amino acid replacements between D. simulans and D. melanogaster is underlined in purple. (B) Overall structure and mapping of amino acid replacements of CG10284 when compared between D. simulans and D. melanogaster. The overall structure of CG10284 threaded onto 1qnx_A (venom antigen) is shown in ribbon representation with α-helices shown in blue and β-strands shown in green. Amino acid replacement sites are shown in red and have been marked by exposing their side chains and electron density clouds. The red arrow points to the loop region where a high frequency of amino acid replacements exists between D. simulans and D. melanogaster.
Active sites or residues important for sperm-binding are not yet known for any of the CRISPs in vertebrates or Drosophila. However, one of the two Drosophila CRISPs, CG10284, has a nonsynonymous substitution rate/synonymous substitution rate (dN/dS) of 1.8 (1) between a D. simulans EST sequence and D. melanogaster, and thus seems likely to have been subject to positive selection. Mapping the amino acid changes between the D. simulans EST and D. melanogaster onto the CRISP structure indicates a clustering of variable sites around a single loop (Fig. 3B) of the predicted structure. This clustering suggests the loop might be important for CG10284's function, and subject to strong evolutionary pressure for amino acid diversification.
Five Acps Seem to Perform Protective Functions. CG17843 threads to an antioxidant class of proteins. Thioredoxin-like proteins catalyze dithiol-disulfide oxidoreduction reactions, which can be involved in protection from oxidative stress (44) and chemokine-like activities (45), among other processes. CG17843 contains two structural domains, each of which is predicted to contain the characteristic thioredoxin-like fold (Cys-X1-X2-Cys) (Table 1). Each fold consists of either a Pro or Gly at position X1, which is predicted to reduce the disulfide bridge conformational strain and to evade steric crowding. The N-terminal and C-terminal domains both match disulfide oxidoreductases. The C-terminal domain structure contains two thioredoxin folds within the same structure (Fig. 7, which is published as supporting information on the PNAS web site). The two folds are on opposite ends of the structure, but appear to use the same groove for substrate binding (Fig. 7).
Four Acps are predicted antimicrobial peptides. β-defensins are antimicrobial peptides that typically inhibit Gram-positive bacteria and contribute to the host immune defense system. We have identified an Acp, CG10433, that contains structurally predicted β-defensin domains at both its N-terminal and C-terminal ends. Additionally, three peptide-sized Acps (CG9029, CG9074, and CG14560) have a high content of proline residues (25.8%, 16.7%, and 24.6%, respectively). Models for these three peptides could not be generated, but their proline richness is consistent with an antimicrobial role because proline-rich peptides are found in many organisms' innate immune responses (46).
CG8194 Is a Predicted RNase. CG8194 threads best to a secreted RNase, suggesting that it might function in RNA hydrolysis. RNA hydrolysis by T2-type-secreted RNases requires a conserved active site composed of His, Lys, and His, which make up the catalytic site, and a Trp as part of the substrate-binding site (47). These active site residues are conserved in CG8194 (Table 1). Not only does CG8194 fall into this class of RNases, it threads best to S-RNases from Solanaceae. S-RNases are proteins secreted by pistil cells, which are later taken up into the pollen tubes, where they are involved in self/non-self recognition to mediate self-incompatibility (48). The threading of CG8194 to a protein involved in gamete recognition suggests the possibility, by analogy, that CG8194's RNase activity could participate in assortative fertilization in Drosophila (49).
Discussion
Many reproductive proteins evolve rapidly (20), which may in part relate to the varied reproductive strategies found across taxa. To address whether conservation of reproductive protein structure and function exists amidst their rapid sequence evolution, we used a CM approach to reveal similarities undetectable at the primary sequence level. Proteins with sequence similarities as low as 30% can still allow medium accuracy comparative structural models to be generated (35). CM has many advantages over sequence alignment, including the ability to predict protein partners, identify antibody epitopes, characterize functional active sites, assist in site-directed mutagenesis, and map rapidly evolving regions. A CM approach was previously used to predict the active site specificity and structurally critical residues of one test-case Acp (Acp62F) (7). Therefore, we analyzed a defined set of 51 additional Acp genes (J.L.M., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. Siggia, C.F.A., and M.F.W., unpublished results), identified from previous screens (1, 2), by CM. Our analyses demonstrate that using a CM-based approach not only provides better insight into the possible functions of new collections of as yet uncharacterized proteins, but also allows comparisons between the predicted functional relationships of reproductive proteins between highly divergent organisms.
In mammals, multiple regulators of proteolysis, particularly metalloand serine proteases and serpins, are present in seminal fluid (17–19) and have been suggested to function in capacitation and fertilization. For example, the prostate-specific antigen (PSA) is a serine protease that is involved in sperm motility, probably by processing of semenogelins (50). In Drosophila, proteins predicted to be involved in proteolysis represent close to half (13 of 29) of the modelable Acps. These Acps may serve as coagulation factors (51) and hence may participate in mating plug assembly (52), or in regulation of the proteolytic cascades that trigger the immune response (53). Acp regulation of proteolytic cleavage could also protect sperm, cleave active peptide hormones from a prohormone precursor, protect prohormones from premature proteolysis (4), or ensure proteolysis at correct sites (54); some of these processes are known to regulate a male seminal fluid peptide hormone, Acp26Aa (54). In addition to regulating proteolysis, some serpin class members, at least in mammals, are able to bind hormones. One example of this is PCI, which is a major component of the mammalian seminal plasma that binds hydrophobic hormones and is required for male fertility (55). Intriguingly, two predicted Acp serpins, CG8137 and Acp76A, contain predicted hydrophobic pockets similar to PCIs. Given the importance of the hydrophobic hormone juvenile hormone in Drosophila reproduction (56) and Acp response (57), perhaps these Acps are involved in its binding or transport (or of another hydrophobic molecule).
D. melanogaster accessory gland proteins assist in the storage and utilization of sperm (58, 59); a similar role has been associated with seminal fluid proteins in mammals (reviewed in refs. 17–19 and 60). Proteins in several classes are known to interact with sperm in mammals; these classes are also found among Drosophila Acps: lipases, lectins, and CRISPs. In mammals, secreted lipid carriers and modifiers in the seminal plasma HE1, clusterin, lipocalin-type prostaglandin D synthase, and cholesterol-binding protein, comprising up to 30% of the total seminal fluid secretions, function in lipid binding/modification in mammalian seminal fluid (61, 62). It is possible that the six Acp lipases may also play a role in lipid modification or in providing energy to the sperm, which may affect sperm motility and/or viability. In vertebrates, sperm–egg interactions have been shown to be in part mediated by members of the lectin (63) and CRISP (42, 43) protein classes. Although little is known about the carbohydrate moieties on the sperm surface of Drosophila, they seem important for sperm–egg interaction (64); thus, lectin-like Acps Acp29AB, CG1652, and CG1656 may perform an analogous function to lectin-like spermadhesins in mammals. Vertebrate sperm-binding CRISPs have been identified in a number of organisms including rats (DE) (65), humans (ARP/hCRISP-1) (43), and in Xenopus egg jelly (allurin) (42). It is therefore intriguing to have identified Acp members of the CRISP family, CG10284 and CG17575, which may also mediate sperm–egg interactions.
Protective roles of seminal fluid proteins against microbes and oxidative stress have been studied for the most part only in mammals. Microbes introduced into the female reproductive tract during mating could result in infections that negatively impact reproductive success. Many antimicrobial proteins have been identified in the mammalian seminal fluid, such as Bin1b (66) and mBD-12 (67), which are of the same β-defensin protein class as Acp CG10433. Male flies are known to transfer at least four antimicrobial peptides or proteins to females (68, 69). Mammalian seminal fluid antioxidant proteins have been shown to protect sperm from oxidative stress (70). This finding may be attributed to secreted forms of superoxide dismutase and peroxidases present in the mammalian seminal fluid of rodents (71, 72); therefore, the presence of a thioredoxin-like Acp, CG17843, may also protect sperm from oxidative stress.
Conclusions
The use of CM on this collection of Acps provides a holistic view of Acp roles in the seminal fluid and provides a comprehensive look at seminal fluid proteins. The 3D structures of 29 Acps (one from ref. 7 plus 28 from this study) have elucidated their predicted roles. Recent studies have revealed an astonishingly rapid evolution of several important reproductive proteins (20). Our results are thus intriguing in showing an overall conservation of structural classes among seminal fluid proteins in organisms as different as insects and mammals. These conserved protein classes likely govern common essential reproductive processes in fly and mammalian seminal fluid. Even with these protein class similarities, some of the 29 modelable Acps are rapidly evolving at the primary sequence level [e.g., CG10284, Acp29AB (13), and Acp62F (12)]. Thus, our results show that modelability is not restricted to slowly evolving proteins. The putative structures for these Acps may aid in understanding the functions and selective pressures associated with their rapid evolution, as has been the case for mammalian MHC molecules (73). The remaining 23 Acps could not be comparatively modeled possibly because they are too short or because they are evolving rapidly enough that their structures are not conserved. These 23 Acps may include good candidates for physiological modifiers whose mechanisms or actions may be more taxon-specific.
Supplementary Material
Acknowledgments
We thank Drs. A. Clark, A. Fiumera, M. Goldberg, J. Lis, S. Suarez, members of the Wolfner laboratory, and anonymous reviewers for helpful comments. This work was supported by National Institutes of Health (NIH) Grant HD38921 (to M.F.W.). Part of this research was conducted by using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York State, federal agencies, foundations, and corporate partners. J.L.M. was supported by an NIH Genetics and Development Training Grant and subsequently by a Department of Education Graduate Assistance in Areas of National Need Fellowship.
Abbreviations: Acp, accessory gland protein; CRISP, cysteine-rich secretory protein; PCI, protein C inhibitor; C-TL, C-type lectin; CM, comparative structural modeling.
References
- 1.Swanson, W. J., Clark, A. G., Waldrip-Dail, H. M., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfner, M. F. (2002) Heredity 88, 85–93. [DOI] [PubMed] [Google Scholar]
- 3.Wolfner, M. F., Harada, H. A., Bertram, M. J., Stelick, T. J., Kraus, K. W., Kalb, J. M., Lung, Y. O., Neubaum, D. M., Park, M. & Tram, U. (1997) Insect Biochem. Mol. Biol. 27, 825–834. [DOI] [PubMed] [Google Scholar]
- 4.Monsma, S. A., Harada, H. A. & Wolfner, M. F. (1990) Dev. Biol. 142, 465–475. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P. S., Stumm-Zollinger, E., Aigaki, T., Balmer, J., Bienz, M. & Bohlen, P. (1988) Cell 54, 291–298. [DOI] [PubMed] [Google Scholar]
- 6.Coleman, S., Drahn, B., Petersen, G., Stolorov, J. & Kraus, K. (1995) Insect Biochem. Mol. Biol. 25, 203–207. [DOI] [PubMed] [Google Scholar]
- 7.Lung, O., Tram, U., Finnerty, C. M., Eipper-Mains, M. A., Kalb, J. M. & Wolfner, M. F. (2002) Genetics 160, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertram, M. J., Neubaum, D. M. & Wolfner, M. F. (1996) Insect Biochem. Mol. Biol. 26, 971–980. [DOI] [PubMed] [Google Scholar]
- 9.Civetta, A. & Singh, R. S. (1995) J. Mol. Evol. 41, 1085–1095. [DOI] [PubMed] [Google Scholar]
- 10.Aguadé, M., Miyashita, N. & Langley, C. H. (1992) Genetics 132, 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsaur, S. C. & Wu, C. I. (1997) Mol. Biol. Evol. 14, 544–549. [DOI] [PubMed] [Google Scholar]
- 12.Begun, D. J., Whitley, P., Todd, B. L., Waldrip-Dail, H. M. & Clark, A. G. (2000) Genetics 156, 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguadé, M. (1999) Genetics 152, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice, W. R. (1996) Nature 381, 232–234. [DOI] [PubMed] [Google Scholar]
- 15.Clark, A. G., Aguadé, M., Prout, T., Harshman, L. G. & Langley, C. H. (1995) Genetics 139, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberhard, W. G. & Cordero, C. (1995) Trends Ecol. Evol. 10, 493–496. [DOI] [PubMed] [Google Scholar]
- 17.Dacheux, J. L., Gatti, J. L. & Dacheux, F. (2003) Microsc. Res. Tech. 61, 7–17. [DOI] [PubMed] [Google Scholar]
- 18.Kravets, F. G., Lee, J., Singh, B., Trocchia, A., Pentyala, S. N. & Khan, S. A. (2000) Prostate 43, 169–174. [DOI] [PubMed] [Google Scholar]
- 19.Kirchhoff, C., Osterhoff, C., Pera, I. & Schroter, S. (1998) Andrologia 30, 225–232. [DOI] [PubMed] [Google Scholar]
- 20.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- 21.Chothia, C. & Lesk, A. M. (1986) EMBO J. 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyckoff, G. J., Wang, W. & Wu, C. I. (2000) Nature 403, 304–309. [DOI] [PubMed] [Google Scholar]
- 23.Kingan, S. B., Tatar, M. & Rand, D. M. (2003) J. Mol. Evol. 57, 159–169. [DOI] [PubMed] [Google Scholar]
- 24.Jensen-Seaman, M. I. & Li, W. H. (2003) J. Mol. Evol. 57, 261–270. [DOI] [PubMed] [Google Scholar]
- 25.Aguadé, M. (1998) Genetics 150, 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, J., Blundell, T. L. & Mizuguchi, K. (2001) J. Mol. Biol. 310, 243–257. [DOI] [PubMed] [Google Scholar]
- 28.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 29.Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001) J. Mol. Biol. 313, 903–919. [DOI] [PubMed] [Google Scholar]
- 30.Lagueux, M., Perrodou, E., Levashina, E. A., Capovilla, M. & Hoffmann, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 11427–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marti-Renom, M. A., Stuart, A. C., Fiser, A., Sanchez, R., Melo, F. & Sali, A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325. [DOI] [PubMed] [Google Scholar]
- 32.Guex, N., Diemand, A. & Peitsch, M. C. (1999) Trends Biochem. Sci. 24, 364–367. [DOI] [PubMed] [Google Scholar]
- 33.Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 34.Ginalski, K., Elofsson, A., Fischer, D. & Rychlewski, L. (2003) Bioinformatics 19, 1015–1018. [DOI] [PubMed] [Google Scholar]
- 35.Baker, D. & Sali, A. (2001) Science 294, 93–96. [DOI] [PubMed] [Google Scholar]
- 36.Pieper, U., Eswar, N., Stuart, A. C., Ilyin, V. A. & Sali, A. (2002) Nucleic Acids Res. 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye, S. & Goldsmith, E. J. (2001) Curr. Opin. Struct. Biol. 11, 740–745. [DOI] [PubMed] [Google Scholar]
- 38.Jerabek, I., Zechmeister-Machhart, M., Binder, B. R. & Geiger, M. (2001) Eur. J. Biochem. 268, 5989–5996. [DOI] [PubMed] [Google Scholar]
- 39.Huntington, J. A., Kjellberg, M. & Stenflo, J. (2003) Structure (Cambridge) 11, 205–215. [DOI] [PubMed] [Google Scholar]
- 40.Smith, G. M., Rothwell, K., Wood, S. L., Yeaman, S. J. & Bownes, M. (1994) Biochem. J. 304, 775–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roussel, A., Canaan, S., Egloff, M. P., Riviere, M., Dupuis, L., Verger, R. & Cambillau, C. (1999) J. Biol. Chem. 274, 16995–7002. [DOI] [PubMed] [Google Scholar]
- 42.Olson, J. H., Xiang, X., Ziegert, T., Kittelson, A., Rawls, A., Bieber, A. L. & Chandler, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen, D. J., Ellerman, D. A., Busso, D., Morgenfeld, M. M., Piazza, A. D., Hayashi, M., Young, E. T., Kasahara, M. & Cuasnicu, P. S. (2001) Biol. Reprod. 65, 1000–1005. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, H., Herzenberg, L. A., Bai, J., Araya, S., Kondo, N., Nishinaka, Y. & Yodoi, J. (2001) Proc. Natl. Acad. Sci. USA 98, 15143–15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertini, R., Howard, O. M., Dong, H. F., Oppenheim, J. J., Bizzarri, C., Sergi, R., Caselli, G., Pagliei, S., Romines, B., Wilshire, J. A., et al. (1999) J. Exp. Med. 189, 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gennaro, R., Zanetti, M., Benincasa, M., Podda, E. & Miani, M. (2002) Curr. Pharm. Des. 8, 763–778. [DOI] [PubMed] [Google Scholar]
- 47.Ohgi, K., Iwama, M., Tada, K., Takizawa, R. & Irie, M. (1995) J. Biochem. (Tokyo) 117, 27–33. [DOI] [PubMed] [Google Scholar]
- 48.Ida, K., Norioka, S., Yamamoto, M., Kumasaka, T., Yamashita, E., Newbigin, E., Clarke, A. E., Sakiyama, F. & Sato, M. (2001) J. Mol. Biol. 314, 103–112. [DOI] [PubMed] [Google Scholar]
- 49.Markow, T. A. (1997) Proc. Natl. Acad. Sci. USA 94, 7756–7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kise, H., Nishioka, J., Kawamura, J. & Suzuki, K. (1996) Eur. J. Biochem. 238, 88–96. [DOI] [PubMed] [Google Scholar]
- 51.Leadley, R. J., Jr. (2001) Curr. Top. Med. Chem. 1, 151–159. [DOI] [PubMed] [Google Scholar]
- 52.Lung, O. & Wolfner, M. F. (2001) Insect Biochem. Mol. Biol. 31, 543–551. [DOI] [PubMed] [Google Scholar]
- 53.Khush, R. S. & Lemaitre, B. (2000) Trends Genet. 16, 442–449. [DOI] [PubMed] [Google Scholar]
- 54.Park, M. & Wolfner, M. F. (1995) Dev. Biol. 171, 694–702. [DOI] [PubMed] [Google Scholar]
- 55.Uhrin, P., Dewerchin, M., Hilpert, M., Chrenek, P., Schofer, C., Zechmeister-Machhart, M., Kronke, G., Vales, A., Carmeliet, P., Binder, B. R. & Geiger, M. (2000) J. Clin. Invest. 106, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubrovsky, E. B., Dubrovskaya, V. A. & Berger, E. M. (2002) Insect Biochem. Mol. Biol. 32, 1555–1565. [DOI] [PubMed] [Google Scholar]
- 57.Moshitzky, P., Fleischmann, I., Chaimov, N., Saudan, P., Klauser, S., Kubli, E. & Applebaum, S. W. (1996) Arch. Insect Biochem. Physiol. 32, 363–374. [DOI] [PubMed] [Google Scholar]
- 58.Tram, U. & Wolfner, M. F. (1999) Genetics 153, 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue, L. & Noll, M. (2000) Proc. Natl. Acad. Sci. USA 97, 3272–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suarez, S. S. (2002) Reprod. Domest. Anim. 37, 140–143. [DOI] [PubMed] [Google Scholar]
- 61.Fouchecourt, S., Charpigny, G., Reinaud, P., Dumont, P. & Dacheux, J. L. (2002) Biol. Reprod. 66, 458–467. [DOI] [PubMed] [Google Scholar]
- 62.Okamura, N., Kiuchi, S., Tamba, M., Kashima, T., Hiramoto, S., Baba, T., Dacheux, F., Dacheux, J. L., Sugita, Y. & Jin, Y. Z. (1999) Biochim. Biophys. Acta 1438, 377–387. [DOI] [PubMed] [Google Scholar]
- 63.Romero, A., Romao, M. J., Varela, P. F., Kolln, I., Dias, J. M., Carvalho, A. L., Sanz, L., Topfer-Petersen, E. & Calvete, J. J. (1997) Nat. Struct. Biol 4, 783–788. [DOI] [PubMed] [Google Scholar]
- 64.Perotti, M. E., Cattaneo, F., Pasini, M. E., Verni, F. & Hackstein, J. H. (2001) Mol. Reprod. Dev. 60, 248–259. [DOI] [PubMed] [Google Scholar]
- 65.Klemme, L. M., Roberts, K. P., Hoffman, L. B., Ensrud, K. M., Siiteri, J. E. & Hamilton, D. W. (1999) Gene 240, 279–288. [DOI] [PubMed] [Google Scholar]
- 66.Li, P., Chan, H. C., He, B., So, S. C., Chung, Y. W., Shang, Q., Zhang, Y. D. & Zhang, Y. L. (2001) Science 291, 1783–1785. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, Y., Nagase, T., Makita, R., Fukuhara, S., Tomita, T., Tominaga, T., Kurihara, H. & Ouchi, Y. (2002) J. Immunol. 169, 2516–2523. [DOI] [PubMed] [Google Scholar]
- 68.Lung, O., Kuo, L. & Wolfner, M. F. (2001) J. Insect Physiol. 47, 617–622. [DOI] [PubMed] [Google Scholar]
- 69.Samakovlis, C., Kylsten, P., Kimbrell, D. A., Engstrom, A. & Hultmark, D. (1991) EMBO J. 10, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen, H., Cheung, M. P., Chow, P. H., Cheung, A. L., Liu, W. & O, W. S. (2002) Reproduction 124, 491–499. [DOI] [PubMed] [Google Scholar]
- 71.Perry, A. C., Jones, R. & Hall, L. (1993) Biochem J 293, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vernet, P., Rigaudiere, N., Ghyselinck, N., Dufaure, J. P. & Drevet, J. R. (1996) Biochem. Cell Biol. 74, 125–131. [DOI] [PubMed] [Google Scholar]
- 73.Hughes, A. L. & Nei, M. (1988) Nature 335, 167–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.