Abstract
Mycobacterium tuberculosis resides within the phagocytes of its host. It ensures its continued survival through arresting the normal maturation of its phagosome, which is retained within the early endosomal system of the macrophage. Although individual bacterial components have been shown to modulate phagosome biogenesis, the mechanism(s) active in live, intact bacteria remain elusive. We have developed a genetic screen that facilitates the isolation of mutants defective in arresting the maturation of their phagosomes. Macrophages were incubated with iron-dextran that was chased into lysosomes. The cells were subsequently infected with M. tuberculosis from a library of transposon-mutagenized bacteria. After four rounds of enrichment, the majority of mutants isolated were unable to prevent acidification of their phagosomes and were attenuated for intracellular survival. The genes affected range in function from those with no known homologues to putative transporters and lipid synthesis enzymes. Further characterization of these bacteria is needed. In addition to clarifying the processes active in modulation of phagosome biogenesis by M. tuberculosis, this screen may be applicable to other pathogens that restrict the maturation of their phagosome.
Pathogenic Mycobacterium spp. are known to reside in vacuoles that fail to exhibit the normal progression of phagosomes to phagolysosomes. Studies from several groups (1–3) have demonstrated that the phagosomes containing these bacilli retain many of the characteristics of early endosomes and remain accessible to material internalized by means of the rapid recycling endosomal system.
The normal progression of phagosomes after internalization of inert particles by phagocytes involves the transient acquisition of the GTPase rab5, phosphorylation of phosphatidylinositol (PI) to generate PI-3-phosphate (PI3P) by the PI kinase VPS34, and accumulation of the PI3P-binding protein early endosome autoantigen 1 (EEA1) (4–7). As the association with EEA1 diminishes, phagosomes show increased accumulation of rab7 and increased fusion with lysosomes. Within the lumen of phagosomes, one observes increased acidification and the accumulation of lysosomal hydrolases that are processed into their active forms as the environment within phagosomes becomes increasingly hydrolytic. In contrast, the phagosomes containing pathogenic Mycobacterium spp. fail to acidify below pH 6.2 (8, 9), remain positive for the early endosomal GTPase rab5 (10–12), and do not acquire EEA1 (13, 14). Despite the extensive documentation of the aberrant retention of a range of host proteins, these studies do not provide an explanation of how the bacterium achieves this process of arrest.
Several mechanisms have been proposed for this phenomenon, including the effects of ammonia production (15), the close apposition between the bacterium and its vacuole membrane (16), the ability of surface lipids such as lipoarabinomannan (LAM) and cord factor to inhibit vesicular fusion (13, 14, 17), and the activity of a bacterial serine/threonine kinase pknG (18). The disparity between data from different groups suggests that there may be more than one mechanism responsible for the arrest of phagosome maturation by Mycobacterium.
The availability of the Mycobacterium tuberculosis genome has facilitated several genetic screens designed to identify genes implicated in the infection process (19–21). Although it is likely that the resulting lists of genes contain the mediators of the arrest of phagosome maturation, the lists are too complex to identify such components directly. The lysosomal environment is clearly bacteriostatic for M. tuberculosis, but it seems to be only weakly bactericidal (22, 23). Therefore, using bacterial death to identify mutants defective in arresting phagosome maturation will be an inefficient process and will be masked by other deleterious mutations.
To overcome these problems, we developed a mechanical screen to enrich for mutants that were unable to block the progression of their phagosomes and prevent their fusion with preformed lysosomes. In brief, macrophages were fed with iron-dextran that was chased into the lysosomes. The cells were then infected with pools of transposon-mutagenized M. tuberculosis and lysed, and the iron-dextran-loaded lysosomes were isolated by magnetic selection. The bacteria in this fraction were recovered, and the selection process was repeated several times before isolation of colonies that were analyzed by Southern blot to identify putative siblings. These mutants were then characterized for phenotype and genotype. The range of mutants isolated and verified suggests that the process may be multifactorial and that alteration in the efficiency of cell division, the surface of the bacilli, and several export systems all may influence the capacity of M. tuberculosis to arrest maturation of its phagosome.
Materials and Methods
Bacterial Strains and Cells. CDC1551, a clinical isolate of M. tuberculosis, was used as the host for the generation of the random transposon-mutagenized library. All of the liquid cultures of M. tuberculosis were performed in 7H9-OADC medium. Bone marrow macrophages (BMMO) were derived from BALB/c mice and maintained in DMEM supplemented with 10% FCS, 5% horse serum, and 20% L cell-conditioned medium.
Generation of the Transposon Library. The M. tuberculosis CDC1551 transposon mutant library was constructed as described (24), with the following modifications. Briefly, M. tuberculosis was electroporated with plasmid pPR30 and plated on 7H11-ADC plates with kanamycin (25 μg/ml) at 30°C for 4–6 weeks to select transformants. Bacteria were cultured for 48 h in 100 μl of 7H9 medium before counterselection on 7H9 plates with kanamycin (25 μg/ml) and 2% sucrose at 39°C. This short-term growth step was included to minimize the presence of siblings in the final library. Individual mutant colonies (8,000) were picked from multiple plates and, after culture, were frozen in 96-well plates. The random nature of the library was confirmed by Southern blot hybridization of 100 independent clones. The library was screened in eight pools of 1,000 mutants generated from the archived stocks.
Preparation of Iron-Dextran. Ten milliliters of 1.2 M FeCl2 was mixed with 10 ml of 1.8 M FeCl3 and agitated extensively while 10 ml of 30% NH3 was added dropwise. A further 100 ml of 5% NH3 was added to the flask, which was placed on a magnet until the precipitate had gathered on the bottom. The supernatant was decanted, and the precipitate was washed with three sequential changes of H2O. The sediment was suspended in 80 ml of 0.3 M HCl and stirred with a magnetic stirrer for 30 min. Four grams of dextran (40 kDa) was added and stirred for a further 30 min. The solution was dialyzed against H2O (6 liters) for four to five changes over a 2-day period. This suspension was centrifuged for 20 min at 20,000 × g to pellet aggregates and filtered through a sterile filter (0.22 μm). The sterile solution was stored at 4°C.
Development of the Screen for Isolation of Mutants Defective in the Arrest of Phagosome Maturation. Eight T75 flasks confluent with BMMO (5 × 106 cells) were incubated with 5 ml of iron-dextran in H2O (≈40 mg/ml iron-dextran) mixed 1:1 with 2× Optimem medium supplemented with 20% FCS. Cells were incubated for 2 h, rinsed in warmed medium, and chased overnight. Each flask was then infected with a recombined pool of 1,000 transposon-mutagenized M. tuberculosis CDC1551 at a multiplicity of infection of 10:1 in 1 ml of uptake buffer (PBS with 4.5 mg/ml glucose, 5 mg/ml defatted BSA, and 1.0 mg/ml gelatin) and 4 ml DMEM with 5% FCS preequilibrated at 37°C. Bacteria were dispersed by 10 passages through a 25-gauge needle before mixing with the DMEM. BMMO were infected for 60 min, rinsed with warm medium, and incubated for a further 60 min. Cells were rinsed with cold homogenization buffer (HB) (250 mM sucrose/0.5 mM EGTA/0.1% gelatin/20 mM Tris, pH 7.0) and scraped into 4 ml of buffer. The cell suspension was centrifuged at 700 × g for 10 min, resuspended in 1 ml of cold HB, and lysed by multiple passage (≈12 times) through a 25-gauge syringe. The lysate was subjected to low-speed centrifugation to remove nuclei and unlysed cells (300 × g for 10 min at 4°C). The supernatant was then applied to a Miltenyi Biotec (Auburn, CA) MiniMACS column placed in the magnetic selector. The homogenate was run through under gravity and retained for analysis. The column was washed with 2 ml of HB and then removed from the magnetic selector, and bound material was eluted with 1 ml of 0.5% Tween 20 plus 0.05% SDS in H2O. The eluant was centrifuged (2,000 × g for 10 min), the supernatant was removed, and the pellet was resuspended in 500 μl of 7H9 medium. Two 150-μl aliquots were plated on 7H11 agar plates supplemented with kanamycin (20 μg/ml), and the remainder was frozen at –80°C. After 3–5 days of culture on plates, the bacteria were scraped from the surface of the plates and placed in 7H9 medium for 2–5 days before repeating the selection procedure. This selection was performed on each of the eight pools and was repeated either three or four times.
Southern Blot Analysis. The extraction of the M. tuberculosis chromosomal DNA was performed as described (25). For the Southern blotting, 2 μg of chromosomal DNA was digested with SalI, electrophoresed, and transferred to a nylon membrane. A probe specific for the aph gene was amplified by PCR from the pUC4K (Amersham Pharmacia Biotech) by using the primers Kan-up (5′-att gaa gga gaa aac tca ccg-3′) and Kan-down (5′-tca gac taa act ggc tga cg-3′), and was labeled with the digoxygenin-11-dUTP. Prehybridization and hybridization were carried out at 65°C by using the DIG Easy Hyb solution (Roche Molecular). The membrane was then washed and developed by using the DIG Luminescent Detection Kit (Roche).
Identification of the Transposon Insertion Sites. The transposon insertion sites were identified by ligation-mediated PCR as described (26). The primers used for amplification were Salgd (5′-tag ctt att cct caa ggc acg agc) and either IS1 (5′-ctt ctg cag caa cgc cag gtc cac act-3′) or IS2 (5′-gag gcg gca gaa agt cgtc agg tcag-3′). The products were sequenced on an Applied Biosystems Automated 3730 DNA Analyzer.
Macrophage Survival Assays. Before we performed the survival assays, the mutants were assayed for growth defects by measuring OD in 7H9 liquid medium over a period of 25 days, and by comparison of colony size on 7H11 plates. No differences were observed between any of the isolates. Fresh BMMO grown in Petri dishes were transferred to 24-well plates, at a density of 3 × 105 cells per well, in 1 ml of medium. Forty-eight hours later, the confluent monolayers were washed once with medium without antibiotics, and infected with mycobacteria at a multiplicity of infection of 1:1. After 1 h of infection, extracellular mycobacteria were removed by washing, and 1 ml of medium without antibiotic was added to each well. The number of intracellular bacteria was determined by lysing the macrophages with 1 ml of H2O containing 0.05% Tween 20 (four wells per strain and time point) at 0, 2, 4, 6, and 8 days postinfection. The lysate was serial diluted in 7H9 medium and plated onto 7H11 plates. Colony-forming units were scored after 3–4 weeks of incubation at 37°C.
Measurement of Intravacuolar pH. Fully confluent monolayers of macrophages were incubated with M. tuberculosis labeled with N-hydroxysuccinimide (NHS)-carboxyfluorescein at a multiplicity of infection of 25:1 for 10 min at ambient temperature (8). The level of substitution with NHS-carboxyfluorescein used does not affect bacterial viability. Cells were washed in PBS at ambient temperature and placed in a cuvette at 37°C in a water-jacketed holder in a PTI QM4SE spectrofluorometer. The pH was calculated from fluorescence ratio analysis after excitation at 450/490 nm and measurement at 520 nm. A standard pH curve was generated by measuring the emission levels from fluorescent M. tuberculosis after phagocytosis by BMMO. These cells were treated with nigericin (10 μM) in 120 mM KCl to collapse the proton gradient artificially, and the infected cell monolayers were placed in a standard series of buffers from pH 4.0–7.5.
Internalization of bound bacteria was confirmed by probing fixed, nonpermeabilized coverslips with rabbit anti-fluorescein and Texas red anti-rabbit IgG (The Jackson Laboratory). Although at an early time point (20 min) we could observe extracellular bacteria, we could detect no extracellular bacteria after 60 min of incubation.
Quantification of Fusion with Lysosomes by Electron Microscopy. Macrophages in T25 flasks were incubated with albumin-coated colloidal gold (EM Science) for 120 min, rinsed, and chased for 120 min. These cells were then infected with M. tuberculosis at 20:1 or fed 1-μm-diameter IgG-coated latex beads for 60 min, rinsed, and replaced in warm medium for a further 60 min. At that time, the cells were fixed in 1% glutaraldehyde and processed for conventional transmission electron microscopy as detailed (27).
Micrographs were taken from each sample, and the percentage of bacteria-containing vacuoles that colocalized with colloidal gold was calculated. Fifty vacuoles were scored for each mutant.
Results and Discussion
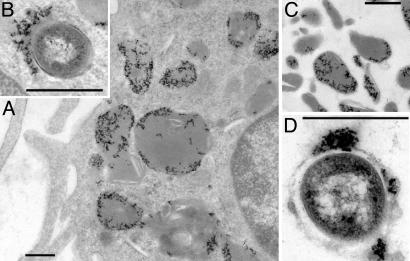
Development of the Screen and Isolation of Mutants. The mechanical screen relies on data from preliminary experiments that demonstrated that M. tuberculosis survived transient exposure (2 h) to the lysosomal milieu (data not shown) and was based on the premise that mutants defective in arresting maturation of the phagosome will be enriched in late endosomal/lysosomal compartments. BMMO, preloaded with iron-dextran, were infected with transposon-mutagenized M. tuberculosis CDC1551 as detailed. Examination of the cells by electron microscopy revealed iron-dextran in dense, lysosome-like compartments and the occasional colocalization of iron-dextran and bacteria (Fig. 1 A and B). Cells were lysed, and the homogenates were applied to Miltenyi Biotec MiniMACS columns in their magnetic holder. The flow-through fractions contained nuclei, mitochondria, and other cellular organelles (data not shown). The columns were removed from their holder, and the bound material was eluted. Electron microscopy of these fractions revealed a highly enriched preparation of dense lysosome-like vacuoles containing iron-dextran, and a few bacteria enclosed in iron-dextran-containing vacuoles (Fig. 1 C and D). The lysosomal fraction was lysed and plated onto 7H11 plates for 3–5 days before scraping the plates into liquid medium and repeating the selection for further enrichment.
Fig. 1.
Electron microscopical analysis of fractions from the phagosome isolation screen. (A) Macrophages after internalization of iron-dextran and overnight chase illustrating that the majority of iron-dextran (the dense flocculent material) is localized to dense lysosome-like compartments. (B) After infection with the pools of transposon-mutagenized M. tuberculosis, a small number of bacteria can be observed in dense compartments that also contain iron-dextran. (C) The fraction bound by the magnetic column eluted with PBS after removal of the column from the magnetic holder. The fraction is rich in dense lysosome-like vacuoles containing iron-dextran. In the mutant isolation method, this fraction is eluted in the presence of detergent to lyse these compartments. (D) Among the dense, iron-dextran-containing vacuoles, there are a few vesicles that also contain bacteria. This fraction should be enriched for mutants defective in arresting the process of phagosome maturation. (Scale bar, 0.5 μm.)
In the preliminary screen, this selection was repeated three times. After the final isolation step, 48 individual colonies from each of the eight pools were picked from the agar plates, expanded in liquid culture, and processed for Southern blot analysis to identify potential families of siblings (data not shown). In the preliminary screen, the largest family of putative siblings had 12 members. To confirm the reproducibility of the selection and to enlarge the sibling families, we decided to return to the original pools of mutants and repeat the screen with four rounds of selection. Analysis of the DNA from the second screen revealed a higher degree of enrichment. From the second screen, the most abundant family of possible siblings had 38 members from a single pool; moreover, this mutant was isolated independently in the same two pools in both the first and second screen. The sites of insertion of the transposons were identified by sequencing products generated by ligation-mediated PCR amplification across the insertion junctions. The validated siblings isolated in both screens are shown in Table 1.
Table 1. The mutants isolated in both the first and second screen.
| Pool no. | Rv no. | MT no. | Annotated function | Hits (Southern blot) | Confirmation (LM-PCR) |
|---|---|---|---|---|---|
| First screen | |||||
| 1 | 0918 | 0944 | Unknown | 4 | 2/3 |
| 2 | 3527 | 3627 | Unknown | 12 | 4/4 |
| 4 | 3527 | 3627 | Unknown | 7 | 4/5 |
| 4 | 0249c | 0263 | Succinate dehydrogenase | 4 | 3/4 |
| 5 | 2930c | 2999 | fadD26 | 5 | 2/2 |
| 5 | 1522c | 1573 | mmpL12 | 4 | 2/2 |
| 8 | 1426c | 1469 | Esterase | 6 | 2/6 |
| 8 | 1361 | 1406 | PPE19 | 5 | 3/3 |
| Second screen | |||||
| 1 | 2941 | 3011 | fadD28 | 6 | 3/4 |
| 2 | 3527 | 3627 | Unknown | 38 | 5/5 |
| 3 | 1819c | 1867 | ABC transporter | 5 | 4/5 |
| 3 | Intergenic | 3491.1 | Unknown | 7 | 3/3 |
| 4 | 3527 | 3627 | Unknown | 14 | 3/3 |
| 5 | 2107 | 2166 | PE22 | 33 | 4/4 |
| 6 | 2336 | 2399 | Unknown | 13 | 3/3 |
| 6 | 0986 | 1014 | ABC transporter | 17 | 7/7 |
| 7 | 0986 | 1014 | ABC transporter | 9 | 7/7 |
| 7 | 3378c | 3488 | Unknown | 7 | 6/6 |
| 8 | 1522c | 1573 | mmpl12 | 8 | 4/4 |
| 8 | 3377c | 3487 | Terpenoid cyclase | 14 | 9/12 |
| 8 | 3156 | 3244 | NuoL | 5 | 2/3 |
The ORF numbers are from the H37Rv (Rv) or CDC1551 (MT) genomic databases. The putative siblings (Hits) identified by Southern blot were confirmed by ligation-mediated (LM)-PCR. Those mutants that are underlined were isolated multiple times in either independent pools or in independent screens.
Phenotypic Analysis of the Isolated Mutants: Intraphagosomal pH. The screen was designed to select mutants on the basis of their inability to avoid fusion with lysosomes. However, as with any such screen, the validity of the method can be determined only by demonstrating that at least some of the mutants have the predicted phenotype. Clearly, from the range of genes enriched in the screen, we cannot identify a single bacterial effector that regulates phagosome maturation. This finding is not surprising because it is likely that many factors can influence modulation of the vacuole. Furthermore, the phenotypes of the selected mutants have to be validated, and, ultimately, their genotypes must be confirmed. Nonetheless, there are several “themes” apparent in the list of annotated genes isolated, including several putative transporters, several lipid processing enzymes, and unknown genes (Table 1).
To evaluate the maturation status of the phagosomes, we decided first to measure the pH of the vacuoles containing the different mutants. We have shown that the pH of vacuoles containing pathogenic Mycobacterium spp. in resting BMMO was ≈6.3–6.4 (8), dropping to pH 5.2 on activation of the macrophage with IFN-γ (28). To examine the pH of the vacuoles containing mutant M. tuberculosis, the bacteria were labeled with N-hydroxysuccinimide (NHS)-carboxyf luorescein, a pH-sensitive fluorochrome. The bacteria were bound to a monolayer of macrophages at room temperature, washed, and placed in a cuvette of medium at 37°C inside a QM4SE spectrofluorometer. The fluorescence emission was recorded at 520 nm after excitation at 450/490 nm. The ratios were converted into pH from a standard curve determined experimentally. This assay measures the average phagosomal pH across the monolayer of cells; therefore, it is important to note that all bound bacteria were internalized within 60 min.
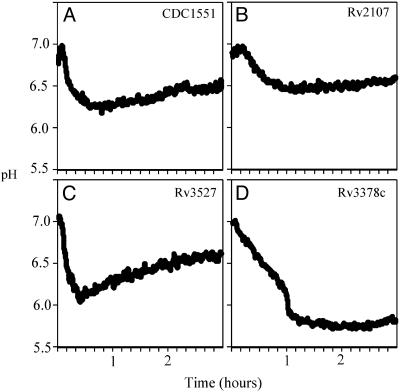
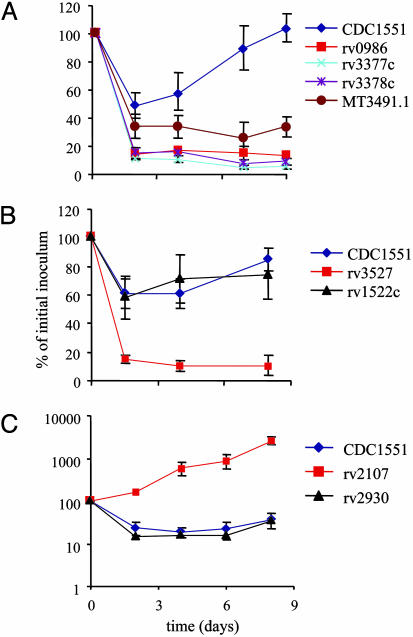
Consistent with our belief that the screen-isolated mutants delivered preferentially to later endosomal compartments, the majority of mutants studied demonstrated a lowered phagosomal pH (Fig. 2). The mutants fell into three basic classes: those that generated acidification profiles that remained similar to wild-type (Fig. 2 A), such as Rv2107 (Fig. 2B); those that showed transient acidification of their phagosome followed by rebound to close to wild-type pH, like Rv3527 (Fig. 2C); and, finally, those that allowed vacuole acidification to fall and remain close to pH 5.5, such as Rv3378c (Fig. 2D). Table 2 displays the three classes of mutants detailed in Table 1. Table 2 summarizes the pH profiles for the mutants by providing the pH measurements for each mutant 30 min and 3 h postinfection.
Fig. 2.
Analysis of the kinetics of acidification of phagosomes containing M. tuberculosis mutants. Graphs illustrating the pH of the phagosomes containing bacteria labeled with N-hydroxysuccinimide (NHS)-carboxyfluorescein after uptake by macrophages. (A) Wild-type CDC1551 goes into vacuoles that drop to pH 6.2 over the course of 40 min. There is a slight “rebound” over time before the vacuole pH settles to around pH 6.4. (B) The mutation in Rv2107 has little effect on the pH of the phagosome. Mutants showing this phenotype are grouped as “like-wt” mutants because their pH profile fails to reveal a phenotype. (C) The mutation in Rv3527 leads to the phagosomes acidifying close to pH 6.0 within 20–30 min before rebounding to pH levels comparable to those of wild-type bacteria by 3 h. Mutants that fall into this group are referred to as “transient acidification” mutants. (D) Finally, the mutation of Rv3378c leads to a severe inability to halt the acidification of the phagosome, and the bacilli end up in vacuoles that equilibrate to pH 5.7. Clearly this is the more extreme phenotype that we wish to select for in the screen. These mutants are grouped as “acidification” mutants. The grouping of all mutants characterized is given in Table 2.
Table 2. The acidification profile of the different mutants studied by fluorescence ratio spectrofluorometry.
| pH profile | ORF | pH at 30 min | pH at 3 h |
|---|---|---|---|
| Wild type | Rv0406c | 6.4 | 6.5 |
| Rv1819c | 6.2 | 6.4 | |
| Rv2107c | 6.5 | 6.6 | |
| Transient acidification | Rv1522c | 6 | 6.4 |
| Rv3527 | 6.1 | 6.5 | |
| Acidification | Rv0986c | 6.1 | 5.9 |
| Rv1426c | 6.2 | 6 | |
| Rv2930c | 6.3 | 5.8 | |
| Rv3377c | 6.2 | 5.9 | |
| Rv3378c | 6.3 | 5.7 | |
| Mt3491.1 | 6.1 | 5.8 |
The mutants were assigned to a group dependent on whether they exhibited a wild-type phenotype, a transient acidification phenotype, or an acidification phenotype. The table shows the pH values at 30 min and 3 h postinternalization.
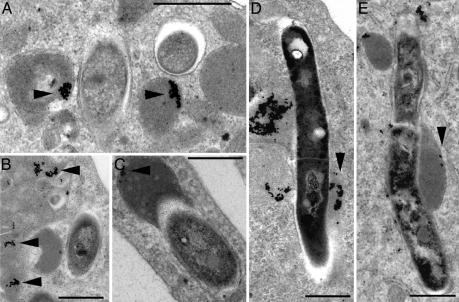
Electron Microscopy of Macrophages Loaded with Colloidal Gold Before Infection. To provide an independent measurement of the trafficking of the mutant and wild-type bacteria, we performed electron microscopy on BMMO that were preloaded with BSA-coated colloidal gold before infection with a subset of mutants (Fig. 3). We scored the percentage of bacteria-containing phagosomes that also contained colloidal gold and recorded these data in a histogram (Fig. 4). The increased accessibility of the vacuoles containing mutants Rv3377c, Rv3378c, Rv0986, and Rv3527 in comparison with both wild-type bacteria and Rv2107c mutants is consistent with the lower pH profiles obtained in the previous experiment.
Fig. 3.
Electron microscopy of BMMO preloaded with BSA-colloidal gold before infection with bacterial mutants. (A) CDC1551 resides in membrane-bound compartments that show minimal-fusion gold-containing vesicles (arrowheads). (B) The strain mutated in Rv2107 shows a similar distribution. (C) In contrast, the strain mutated in Rv3377c shows an enhanced access to colloidal gold-containing vacuoles (arrowheads). Finally, those bacteria containing a transposon in Rv3527 have a preponderance of elongated bacteria that colocalize with colloidal gold (D) or with iron-dextran (E) (arrowheads). The number of bacterial vacuoles containing collodal gold was scored and is shown in Fig. 4. (Scale bar, 0.5 μm.)
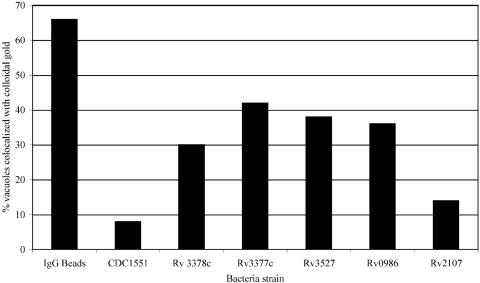
Fig. 4.
The percentage of bacteria-containing vacuoles that colocalize with colloidal gold. The relative accessibility and fusion of bacteria-containing vacuoles with late endosomal/lysosomal compartments labeled with colloidal gold mirrors closely the degree of acidification observed for all of the bacteria strains examined here. The mutants in Rv2107 and wild-type CDC1551 show a similar degree of loading, whereas Rv3377c, Rv3378c, Rv0986, and Rv3527 all demonstrate an increased acquisition of colloidal gold. However, none of the bacteria-containing vacuoles acquire as much colloidal gold label as the IgG bead-containing compartments.
Electron microscopical analysis revealed that all bacilli were retained within membrane-bound compartments (Fig. 3). Interestingly, we observed that the Rv3527 mutant produced elongated bacteria that resided in vacuoles that tended to fuse with both colloidal gold- and iron-dextran-containing vacuoles.
Phenotypic Analysis of the Isolated Mutants: Intramacrophage Survival. The isolation of M. tuberculosis mutants defective in the arrest of phagosome maturation facilitated the testing of the long-held assumption that the inability to regulate maturation of the vacuole will reduce the bacteria's survival in macrophages. Until now, this contention had been tested only through activation of the host cell by cytokines (28, 29) or through stimulation with ATP (28–31), both of which have pleotropic effects on the host cell.
The survival profiles of all mutants determined to have an altered intramacrophage phenotype are illustrated in Fig. 5. In brief, the majority of mutants shown to have a lowered vacuolar pH and increased localization with colloidal gold-loaded lysosomes demonstrated reduced viability in macrophages. Many of the mutants were attenuated severely and exhibited over a log fewer bacteria than the wild-type controls. None of these bacteria showed a growth defect either in liquid medium or on agar plates (data not shown). These data support the contention that modulation of the vacuole impacts directly on bacterial survival. We were, however, surprised to note that one of the mutants (Rv2107) was hypervirulent and outgrew the wild-type control by 100× over the infection period. Electron microscopy revealed that the bacterium was retained within phagosomes comparable to those of the wild-type bacilli (Fig. 3).
Fig. 5.
Graphs illustrating the survival curves of mutants after infection of macrophages. There is a reasonable consensus between acidification of the phagosomes and the survival of mutants in macrophages. (A) The survival profile of the mutants Rv0986, Rv3377c, Rv3378c, and MT3491.1 in comparison with wild-type CDC1551. These mutants all showed an acidification profile and were assigned to the “acidification” group. (B) The colony-forming units of Rv3527 and Rv1522c that had a “transient acidification” pH profile. Rv3527 is clearly attenuated for intramacrophage survival whereas Rv1522c shows no obvious survival defect. (C) The survival profile of Rv2930c and Rv2107. Rv2930 generated an acidified phagosome profile yet shows little attenuation, whereas Rv2107, which did not acidify, is actually hypervirulent in macrophages.
Interpretation of the Genotypes/Phenotypes. Clearly a great deal of work is now needed to determine the molecular basis of the phenotypes observed. For some of the mutants, the explanation seems likely to be trivial, whereas, in the others, the underlying explanation may be an indirect effect either through an alteration of cell wall structure or through the export of an effector molecule.
Of the apparently trivial mutants, Rv3527 is the most obvious. The transposon has inserted in ORF Rv3527, which has no identifiable homologues. However, the bacterium tends to form elongated threads in culture. Electron microscopy of macrophages infected with ΔRv3527 reveal elongated bacteria in vacuoles containing colloidal gold or iron-dextran (Fig. 3). These bacilli have incomplete septae between daughter bacteria, suggesting that this gene is involved in bacterial septation. It has been shown that bacterial clumps tend to go to the lysosome after uptake (32); therefore, it is likely that this mutant has difficulty modulating phagosome maturation. Interestingly, although the phagosomes containing these mutants do acidify early, the pH of the vacuoles is close to that observed for wild-type bacteria 3 h postinfection.
Three of the mutants had insertions in genes that have been proposed, through homology, to have transporter function. Rv0986 is the first ORF in a set of three genes that encode a putative ABC transporter that is closely homologous to the AttFGH locus of Agrobacterium tumefaciens, a locus required for efficient infection of plants (33). Rv1819c is similar to another ABC transporter system and is annotated as the ATP-binding protein and possible drug-transport ABC transporter (discussed in ref. 34). It is interesting to note that an increasing number of MDR-type ABC transporters have been shown to transport lipids (35), so it is feasible that Rv0986 or Rv1819c might transport a lipid to the exterior of the bacterium. Finally, we observed enrichment of mutants in mmpL12, one of a family of membrane proteins with multiple membrane-spanning domains. Another member of this family, mmpL7, has been shown to export the lipid phthiocerol dimycocerosate, which is important for the continuity of the bacterial cell wall and is implicated in differential colonization of tissues in the mouse (36–38). Of the putative transporter mutants isolated, the insertion in Rv0986 has the strongest phenotype with respect to survival and vacuole acidification.
The majority of other mutants characterized fall into the “metabolism” group. This group includes mutants in Rv2930 (FadD26), a fatty acid CoA ligase that is involved in synthesis of phthiocerol dimycocerosate (36, 38); in Rv1426c (LipO), a possible esterase or lipase; in Rv3156 (NuoL), a NADH dehydrogenase; and in Rv0249c, a succinate dehydrogenase. However, these mutants were not enriched strongly (Table 1) and show minimal alteration in intracellular survival. The most intriguing set of transposon insertions is three independent insertions within an operon of five genes that encode putative enzymes involved in the biosynthesis or modification of isoprenol-based compounds. Among the genes in this operon are lytb1 and dxs2, which belong to the deoxyxylulose 5-phosphate/methylerythritol 4-phosphate (DOXP/MEP) pathway of isoprenol biosynthesis. The first insertion is at the start of the operon in a region of the chromosome designated as an ORF in CDC1551 (Mt3491.1) and as an intergenic region upstream of idsB in H37Rv. Transcriptional analysis by RT-PCR revealed markedly reduced transcript from this operon in this mutant (data not shown). The other two genes disrupted are Rv3377c, which is homologous to terpenoid cyclases responsible for cyclization of geranylgeranylpyrophosphate or farnylpyrophosphate to generate a range of products such as sterol, antibiotics, or cyclic isoprenols such as the anti-microtubule agent Taxol (39–41), and Rv3378c, which possesses the D81DXXD motif diagnostic of prenyltransferase enzymes. Rv3377c and Rv3378c have a lower guanine/cytosine (GC) content (54.35% and 48.36%, respectively) than the surrounding chromosome (65%), implying that the locus was acquired by horizontal gene transfer. The operon is also present in Mycobacterium bovis, indicating that it invaded these bacteria before the divergence of the two species.
Concluding Remarks. The ability to modulate the maturation of the phagosome after uptake by macrophages is often presented as key to the success of M. tuberculosis. The screen described here tends to support this contention because the majority of mutants that are defective in arrest of phagosome maturation are attenuated for intracellular survival. The more extreme of those mutants reside in vacuoles of pH 5.7–5.8. The explanations behind these phenotypes are likely diverse, and not all will relate to a specific bacterial effector. Nonetheless, the results imply that modulation of phagosome maturation is likely a multifactorial process that is mediated by other components in addition to cell wall lipids such as lipoarabinomannan and cord factor (14, 17). Further research is needed to elucidate these mechanisms.
The usefulness of this genetic screen may not be restricted to M. tuberculosis. The screen could be applied to any intracellular pathogen that modulates the progression of its phagosome, survives transient exposure to the lysosomal environment, and for which a mutational approach is feasible. The limitation likely lies in the ability of the bacterium to survive the lysosomal milieu for the duration of the screen; however, the use of cell lines instead of primary macrophages and the addition of proteinase inhibitors to reduce lysosomal activity might widen this window of opportunity.
Acknowledgments
This work was supported by U.S. Public Health Service Grants AI33348, AI057086, and HL55936 (to D.G.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: BMMO, bone marrow macrophage.
References
- 1.Clemens, D. L. (1996) Trends Microbiol. 4, 113–118. [DOI] [PubMed] [Google Scholar]
- 2.Fratti, R. A., Vergne, I., Chua, J., Skidmore, J. & Deretic, V. (2000) Electrophoresis 21, 3378–3385. [DOI] [PubMed] [Google Scholar]
- 3.Russell, D. G. (2001) Nat. Rev. Mol. Cell. Biol. 2, 569–577. [DOI] [PubMed] [Google Scholar]
- 4.Vitale, G., Alexandrov, K., Ullrich, O., Horiuchi, H., Giner, A., Dobson, C., Baykova, O., Gournier, H., Stenmark, H. & Zerial, M. (1995) Cold Spring Harbor Symp. Quant. Biol. 60, 211–220. [DOI] [PubMed] [Google Scholar]
- 5.Vieira, O. V., Botelho, R. J., Rameh, L., Brachmann, S. M., Matsuo, T., Davidson, H. W., Schreiber, A., Backer, J. M., Cantley, L. C. & Grinstein, S. (2001) J. Cell Biol. 155, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenmark, H. & Olkkonen, V. M. (2001) Genome Biol. 2, 3007.1–3007.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenmark, H., Aasland, R. & Driscoll, P. C. (2002) FEBS Lett. 513, 77–84. [DOI] [PubMed] [Google Scholar]
- 8.Sturgill-Koszycki, S., Schlesinger, P. H., Chakraborty, P., Haddix, P. L., Collins, H. L., Fok, A. K., Allen, R. D., Gluck, S. L., Heuser, J. & Russell, D. G. (1994) Science 263, 678–681. [DOI] [PubMed] [Google Scholar]
- 9.Oh, Y. K. & Straubinger, R. M. (1996) Infect. Immun. 64, 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Via, L. E., Deretic, D., Ulmer, R. J., Hibler, N. S., Huber, L. A. & Deretic, V. (1997) J. Biol. Chem. 272, 13326–13331. [DOI] [PubMed] [Google Scholar]
- 11.Clemens, D. L., Lee, B. Y. & Horwitz, M. A. (2000) Infect. Immun. 68, 2671–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley, V. A. & Schorey, J. S. (2003) Mol. Biol. Cell 14, 3366–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fratti, R. A., Backer, J. M., Gruenberg, J., Corvera, S. & Deretic, V. (2001) J. Cell Biol. 154, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergne, I., Chua, J. & Deretic, V. (2003) J. Exp. Med. 198, 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, A. H., Hart, P. D. & Young, M. R. (1980) Nature 286, 79–80. [DOI] [PubMed] [Google Scholar]
- 16.de Chastellier, C. & Thilo, L. (1998) Res. Immunol. 149, 699–702. [DOI] [PubMed] [Google Scholar]
- 17.Indrigo, J., Hunter, R. L., Jr., & Actor, J. K. (2003) Microbiology 149, 2049–2059. [DOI] [PubMed] [Google Scholar]
- 18.Walburger, A., Koul, A., Ferrari, G., Nguyen, L., Prescianotto-Baschong, C., Huygen, K., Klebl, B., Thompson, C., Bacher, G. & Pieters, J. (2004) Science 304, 1800–1804. [DOI] [PubMed] [Google Scholar]
- 19.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2003) Mol. Microbiol. 48, 77–84. [DOI] [PubMed] [Google Scholar]
- 20.Sassetti, C. M. & Rubin, E. J. (2003) Proc. Natl. Acad. Sci. USA 100, 12989–12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamichhane, G., Zignol, M., Blades, N. J., Geiman, D. E., Dougherty, A., Grosset, J., Broman, K. W. & Bishai, W. R. (2003) Proc. Natl. Acad. Sci. USA 100, 7213–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong, J. A. & Hart, P. D. (1975) J. Exp. Med. 142, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes, M. S., Paul, S., Moreira, A. L., Appelberg, R., Rabinovitch, M. & Kaplan, G. (1999) Infect. Immun. 67, 3199–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelicic, V., Jackson, M., Reyrat, J. M., Jacobs, W. R., Jr., Gicquel, B. & Guilhot, C. (1997) Proc. Natl. Acad. Sci. USA 94, 10955–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baulard, A., Kremer, L. & Locht, C. (1996) J. Bacteriol. 178, 3091–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prod'hom, G., Lagier, B., Pelicic, V., Hance, A. J., Gicquel, B. & Guilhot, C. (1998) FEMS Microbiol. Lett. 158, 75–81. [DOI] [PubMed] [Google Scholar]
- 27.Mwandumba, H. C., Russell, D. G., Nyirenda, M. H., Anderson, J., White, S. A., Molyneux, M. E. & Squire, S. B. (2004) J. Immunol. 172, 4592–4598. [DOI] [PubMed] [Google Scholar]
- 28.Schaible, U. E., Sturgill-Koszycki, S., Schlesinger, P. H. & Russell, D. G. (1998) J. Immunol. 160, 1290–1296. [PubMed] [Google Scholar]
- 29.Via, L. E., Fratti, R. A., McFalone, M., Pagan-Ramos, E., Deretic, D. & Deretic, V. (1998) J. Cell Sci. 111, 897–905. [DOI] [PubMed] [Google Scholar]
- 30.Kusner, D. J. & Adams, J. (2000) J. Immunol. 164, 379–388. [DOI] [PubMed] [Google Scholar]
- 31.Kusner, D. J. & Barton, J. A. (2001) J. Immunol. 167, 3308–3315. [DOI] [PubMed] [Google Scholar]
- 32.Schuller, S., Neefjes, J., Ottenhoff, T., Thole, J. & Young, D. (2001) Cell Microbiol. 3, 785–793. [DOI] [PubMed] [Google Scholar]
- 33.Matthysse, A. G., Yarnall, H. A. & Young, N. (1996) J. Bacteriol. 178, 5302–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braibant, M., Gilot, P. & Content, J. (2000) FEMS Microbiol. Rev. 24, 449–467. [DOI] [PubMed] [Google Scholar]
- 35.Borst, P., Zelcer, N. & van Helvoort, A. (2000) Biochim. Biophys. Acta 1486, 128–144. [DOI] [PubMed] [Google Scholar]
- 36.Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. (1999) Mol. Microbiol. 34, 257–267. [DOI] [PubMed] [Google Scholar]
- 37.Camacho, L. R., Constant, P., Raynaud, C., Laneelle, M. A., Triccas, J. A., Gicquel, B., Daffe, M. & Guilhot, C. (2001) J. Biol. Chem. 13, 13. [DOI] [PubMed] [Google Scholar]
- 38.Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R., Jr. (1999) Nature 402, 79–83. [DOI] [PubMed] [Google Scholar]
- 39.Dairi, T., Hamano, Y., Kuzuyama, T., Itoh, N., Furihata, K. & Seto, H. (2001) J. Bacteriol. 183, 6085–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamano, Y., Kuzuyama, T., Itoh, N., Furihata, K., Seto, H. & Dairi, T. (2002) J. Biol. Chem. 277, 37098–37104. [DOI] [PubMed] [Google Scholar]
- 41.Lin, X., Hezari, M., Koepp, A. E., Floss, H. G. & Croteau, R. (1996) Biochemistry 35, 2968–2977. [DOI] [PubMed] [Google Scholar]