Abstract
Duchenne's muscular dystrophy (DMD) is a fatal neuromuscular disease caused by absence of dystrophin. Utrophin is a chromosome 6-encoded dystrophin-related protein (DRP), sharing functional motifs with dystrophin. Utrophin's ability to compensate for dystrophin during development and when transgenically overexpressed has provided an important impetus for identifying activators of utrophin expression. The utrophin promoter A is transcriptionally regulated in part by heregulin-mediated, extracellular signal-related kinase-dependent activation of the GABPα/β transcription factor complex. Therefore, this pathway offers a potential mechanism to modulate utrophin expression in muscle. We tested the ability of heregulin to improve the dystrophic phenotype in the mdx mouse model of DMD. Intraperitoneal injections of a small peptide encoding the epidermal growth factor-like region of heregulin ectodomain for 3 months in vivo resulted in up-regulation of utrophin, a marked improvement in the mechanical properties of muscle as evidenced by resistance to eccentric contraction mediated damage, and a reduction of muscle pathology. The amelioration of dystrophic phenotype by heregulin-mediated utrophin up-regulation offers a pharmacological therapeutic modality and obviates many of the toxicity and delivery issues associated with viral vector-based gene therapy for DMD.
Duchenne's muscular dystrophy (DMD) is the most common X-linked neuromuscular disease and affects 1 in 3,500 newborn males. The disease is caused by mutations in the DMD gene (1–3) resulting in quantitative and/or qualitative disturbances in expression of dystrophin, the gene product of the DMD locus (4). Dystrophin is associated with the membrane-bound dystroglycan/sarcoglycan complex (DGC) that forms an important link with laminin, a constituent of the extracellular matrix (5).
Dystrophin is member of the spectrin superfamily of proteins that includes the spectrins, α-actinins, and dystrophin and its closely related proteins. The dystrophin-related family consists of dystrophin, dystrophin-related protein (DRP) or utrophin (encoded on chromosome 6), DRP2 (chromosome X), and dystrobrevin (chromosome 18). Utrophin is considered the autosomal homolog of dystrophin because it shares structural and functional motifs throughout the length of the molecule (6–10). Utrophin is capable of associating with the dystroglycan/sarcoglycan complex at the sarcolemma and binds F-actin with similar affinity to dystrophin as well (11, 12). Prenatally, utrophin is thought to carry out dystrophin's function because it is expressed at high levels throughout the sarcolemma (13). Indeed, necrosis in dystrophin-deficient mdx mice occurs only once the high perinatal utrophin levels have declined to the lower levels noted in adults (14). Postnatally, utrophin is replaced by dystrophin in extrasynaptic compartments of normal muscle fiber; remaining utrophin expression is restricted to the neuromuscular (NMJ) and myotendinous junctions in mature myofibers (8, 9, 14). Direct evidence for the ability of utrophin to functionally substitute for dystrophin comes from experiments demonstrating that transgene-driven utrophin overexpression can effectively rescue dystrophin-deficient muscle in mdx mice (15–17).
Thus, a promising therapeutic strategy for DMD consists of identifying molecules that modulate utrophin expression by activation of its promoter (18). Previously, we and others had identified heregulin (HRG) as a molecule capable of transactivating utrophin promoter A via extracellular signal-related kinase-dependent activation of the ets-related transcription factor complex GABPα/β in cultured muscle (19, 20); these findings suggest the therapeutic potential of these molecules if they are delivered to dystrophin-deficient muscle in vivo. In this study, we have tested the ability of HRG to improve the dystrophic phenotype in the mdx mouse model of DMD.
Materials and Methods
Mice. We obtained cohorts of 4-week-old male dystrophin-deficient C57BL/10ScSn-Dmdmdx/J (mdx) and C57BL/10ScSn (controls) from The Jackson Laboratory. The mdx:utrn–/– dystrophin–utrophin double mutants were bred from mdx and utrn–/– (utrophin-null mutant) mice, a kind gift from Josh Sanes and Mark Grady (Washington University, St. Louis). All animal experiments were performed according to Danish and U.S. laws and approved by the Danish Animal Inspectorate and Institutional Animal Care and Use Committee at the University of Pennsylvania.
Physiological Studies. Physiological evaluation of muscle was performed as described (21, 22). Briefly, extensor digitorum longus (EDL) muscle was dissected from 16-week-old mdx mice that had been injected three times a week for 3 months with either a peptide encoding the epidermal growth factor-like, 71-aa region of the human HRG β1 molecule (GenBank accession no. M94166, amino acid residues 176–246: TSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNE FTGDRCQNYVMASFYKHLGIEFMEAEELYQK) in PBS (R & D Systems) or PBS alone (controls). The dose used was 8 μg of peptide per kg of body weight, and it was delivered via the i.p. route. The injection material was freshly prepared by diluting in PBS a stock solution of 1 μM HRG in PBS on the day of the injection. The recombinant peptide was expressed in Escherichia coli and had a purity of >97% as determined by silver staining and endotoxin level of <1.0 endotoxin unit per μg of peptide. The freshly dissected mdx EDL were weighed and placed in a custom-built organ bath containing a 1.8° step-motor driven micrometer (length controller; precision, 2.5 μm) and a Grass FT-03 force transducer (Astro-Med, West Warwick, RI). Muscle was attached at either end by using 5-0 nylon and maintained in oxygenated Ringer's solution (pH 7.4) at 24°C for the duration of the experiment. Stimulation was performed by using platinum field electrodes connected to a Grass S48 stimulator. Eccentric contraction (ECC) force drop was calculated by using the difference of isometric force generation during the 1st and 10th tetanus of the standard ECC protocol [supramaximal stimulus of 700 msec (500-msec isometric phase, 200-msec eccentric phase); lengthening L0/10; lengthening velocity, 0.5 L0/sec]. Data were digitized and acquired by using a PowerLab/8SP A/D converter and software (AD Instruments, Colorado Springs, CO). At the end of physiological studies, muscles were immersed in 0.5% procion orange dye (Sigma) in oxygenated Ringer's solution for 5 min, flash-frozen in isopentane cooled in liquid nitrogen, and stored at –80°C before histological analysis.
Morphological Studies. Previously described methods were used for morphological studies (14). Briefly, 7- to 12-μm serial cryosections of EDL and tibialis anterior (TA) from the previously described 16-week-old mdx mice and TA from treated/untreated mdx:utrn–/– were fixed in a 1:1 mixture of acetone/methanol for 5 min at 4°C. Sections were processed with hematoxylin and eosin (H&E), anti-laminin antibody (1 μg/ml, Sigma), and Hoechst dye 33342 (Sigma) for morphological studies. Indirect immunofluorescence was performed by using affinity-purified BH11 antibody against utrophin (7, 14) (1 μg/ml), rhodamine-conjugated bungarotoxin (4 μg/ml, Molecular Probes), and Alexa Fluor 488 goat anti-rabbit antibody (Molecular Probes). Slides were examined by using an Olympus BX51 microscope equipped with epifluorescence optics and a Nikon CoolPix 995 digital camera. Area measurements of necrosis were done by using image 4.0.2 (Scion, Frederick, MD) with necrotic focus defined as a necrotic area >3,200 μm2.
Molecular and Biochemical Studies. Molecular and biochemical evaluation of utrophin used previously described methods and reagents (7, 19). Briefly, RNA was extracted from frozen aliquots of muscle. Five micrograms of RNA was reverse transcribed into cDNA by using the Superscript Choice system (Invitrogen Life Technologies), and 10% (vol/vol) purified cDNA was used as template for semiquantitative PCR using primers for murine utrophin (19). As an internal control for efficiency of reverse transcription, we simultaneously amplified glyceraldehyde 3-phosphate dehydrogenase. PCR products were resolved on 2% agarose gels, and quantification was performed on aliquots that were within the linear amplification range for each reaction by using a Typhoon 8600 scanner and imagequant 1.2 software (Amersham Pharmacia). For Western blotting, 50-mg pieces of frozen TA muscle were weighed, homogenized in sample buffer, and resolved on 4–12% PAGEr Gold gradient SDS polyacrylamide gels (BioWhittaker). Proteins were electrotransferred to nitrocellulose membrane (Bio-Rad). Efficiency of transfer and even loading of lanes was verified by using Ponceau S staining. Membranes were probed with the affinity-purified BH 11 utrophin antibody (7) (1 μg/ml), detected on a Typhoon 8600 scanner by using a goat anti-rabbit horseradish peroxidase antibody (Pierce) and the SuperSignal Western Dura ECL kit (Pierce). imagequant 1.2 software was used for quantification. Serum creatine kinase (CK) was measured by using an indirect CK colorimetric assay kit and standards (Sigma).
Statistical Analysis. Student's t test was used throughout this study to calculate P values for determination of statistical significance. All results are shown as average ± SEM; HRG-treated mdx is shown in red, and control mdx is shown in blue. Dashed lines represent data from age-matched (normal) C57BL/10 mice.
Results
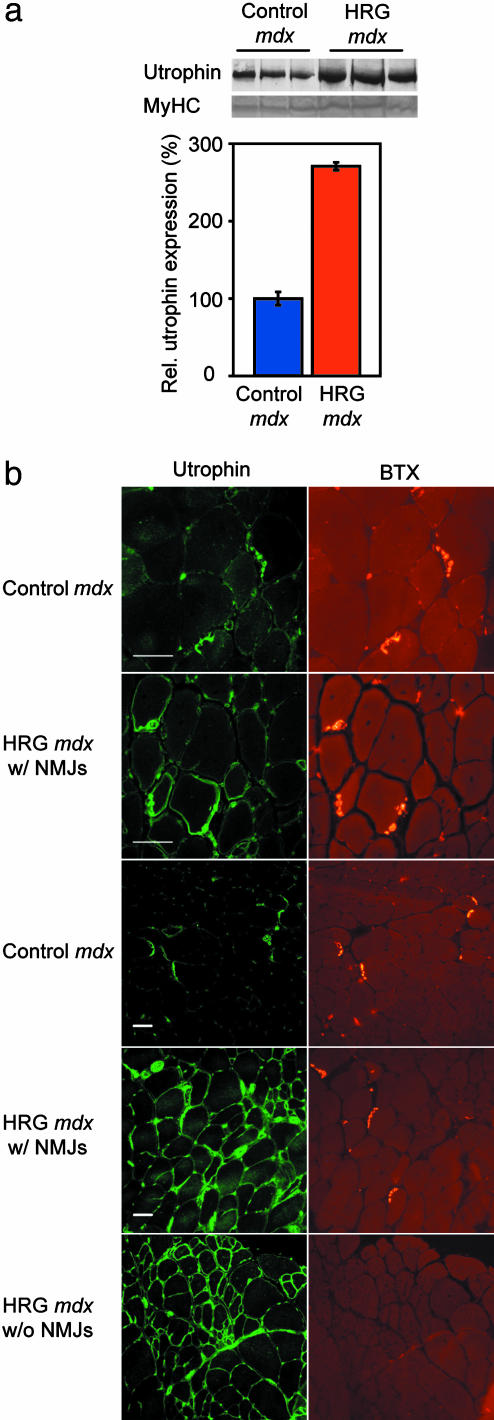
Four-week-old male mdx mice were injected on alternate days with a small ≈8-kDa peptide containing the epidermal growth factor-like region of HRG ectodomain (dose of 8 μg/kg in PBS-treated mdx group) or PBS alone (control mdx group) for 3 months and then killed, and various muscle groups were dissected and processed. We performed Western blots and immunofluorescence to determine the extent and distribution of utrophin up-regulation achieved in vivo. Utrophin expression was increased ≈2.7-fold at the protein level in treated compared to control mdx mice (Fig. 1a); a similar increase was seen in HRG-treated cultured muscle cells (19, 20). In mdx muscle, the ubiquitously transcribed utrophin protein is enriched at nerves and vessels; within myofibers, it is enriched at NMJs. In the myofibers of treated mdx mice, up-regulated utrophin was seen to spread throughout the sarcolemma rather than remaining confined to the NMJs in synapse-rich regions (14) (Fig. 1b). Increased sarcolemmal labeling was also noted in regions of muscle with a paucity of synapses, as validated by a lack of bungarotoxin labeling (Fig. 1b Lower). Semiquantitative RT-PCR revealed that utrophin mRNA was up-regulated ≈1.8-fold (n = 24; P < 0.005, data not shown). These observations are consistent with levels and distribution of utrophin predicted to be potentially beneficial (23), based on the time course of developmental down-regulation (14) and transgenic studies (16).
Fig. 1.
Increased utrophin expression in HRG-treated mdx mice. (a) Utrophin levels in blots of three control and three HRG-treated mdx mice (Upper). (Lower) Myosin heavy chain (MyHC) controls for even loading. Quantification revealed an ≈2.7-fold increase in utrophin in HRG-treated muscle (n = 6; P < 0.005). (b) First and second row from top show a synaptic region from control mdx and HRG-treated TA, respectively, at a ×40 magnification. Third and fourth rows are similar to the first and second rows, but at a ×20 magnification. The fifth row shows an extrasynaptic region of HRG-treated mdx.(Left) Utrophin labeling (BH11 Ab) for NMJs. (Right) Bungarotoxin (BTX) labeling for NMJs. Muscle shows increased sarcolemmal utrophin in the HRG-treated mdx compared to the untreated mdx, where utrophin, within the myofiber, remains restricted to the NMJ region. Labeling of peripheral nerve and vascular utrophin is visible in all panels as well. (Scale bar, 50 μm.)
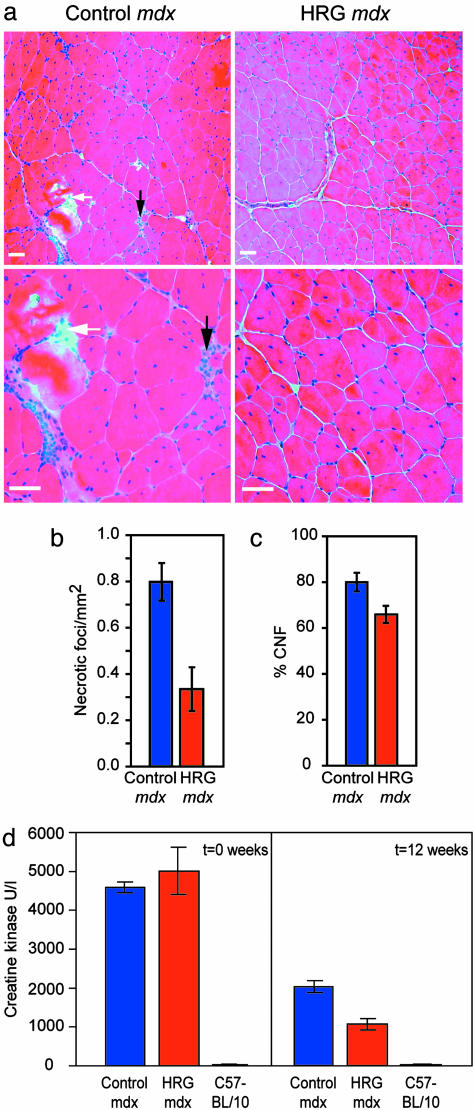
To determine whether the utrophin up-regulation achieved in vivo was associated with reduction of muscle pathology, we analyzed the EDL and TA muscles from treated and control mdx mice. The TA was used in addition to the EDL because the mdx mice have only occasional foci of degeneration at 16 weeks of age in limb muscles (24). Representative H&E-stained cryosections show that the treated mdx mice had less degeneration and necrosis compared to control mdx (Fig. 2a), corresponding to a >2-fold reduction of the number of necrotic foci in treated mdx mice (Fig. 2b). No significant changes were noted in single fiber area, cross-sectional area, or total myofiber numbers in treated mdx compared to control mdx mice (data not shown). Because mdx muscle constantly regenerates in response to chronic inflammation and muscle damage, it has a much larger percentage of centrally nucleated fibers (CNF) compared to normal mice (24). Indeed, CNF proportion is commonly used as an index to monitor the efficiency of gene therapy trials in mdx mice (16). There was a significant reduction in the CNF proportion in treated mdx compared to control mdx EDL (Fig. 2c, 65.9 ± 3.8% vs. 80.0 ± 0.1%; nmice = 20; nfibers = 19,787; P < 0.01). Consistent with the reduction of muscle pathology, a significant reduction (≈50%) was noted in the level of serum CK compared to levels seen in age-matched mdx controls (Fig. 2d). The reduction in muscle degeneration, proportion of CNF, and CK levels offer morphological and biochemical evidence of reduced muscle pathology in HRG-treated mdx mice. To address whether the benefit was utrophin-dependent, double mutant mdx:utrn–/– (25–28) were treated with HRG or PBS. The double mutant mice have a very short lifespan, are underweight, and have severe muscle pathology (27, 28). No benefit was noted with respect to lifespan, body weight, and muscle pathology (Fig. 3), suggesting utrophin dependence of the HRG-mediated improvement noted in mdx mice.
Fig. 2.
Reduction of degeneration and chronic inflammation in HRG-treated mdx mice. (a) H&E staining of low- and high-power fields (Upper and Lower) of control mdx TA muscle (from two representative mice) showing multiple foci of muscle degeneration and infiltration with cellular exudate (black arrows) and phagocytosis (white arrows) compared with HRG mdx TA muscle (from two representative mice), consistent with reduction of necrotic foci (b), CNFs (c), and serum CK (d) in treated mdx mice. (Scale bar, 50 μm.)
Fig. 3.
HRG-treated double mutant mdx:utrn–/– mice show no overall improvement. There was no significant difference in body weight (a); average time in therapy before death for age-matched mice (b); and representative muscle pathology as visualized by H&E staining (c) between untreated and HRG-treated double mutant mdx:utrn–/– respectively.
To quantify the degree of functional improvement in muscle resulting from utrophin up-regulation, we analyzed the physiological properties of skeletal muscle. Freshly dissected EDL muscles were tested ex vivo in a muscle organ bath, equipped with a force transducer and stepper motor-driven length controller, to determine their mechanical properties. The mdx EDL has a well characterized susceptibility to damage by lengthening contractions resulting in an inability to generate adequate force after a series of ECCs (22, 29, 30). Post-ECC drop in force production (force drop) occurs because of increased sarcolemmal stresses as a consequence of dystrophin deficiency. We saw a significant reduction in post-ECC force drop (29) in treated mdx mice (Fig. 4 a and b, 54.3 ± 3.8% vs. 67.9 ± 3.8%, respectively; n = 20; P < 0.02). Additionally, staining of EDL in procion orange dye after ECC revealed that treated mdx mice were less susceptible to membrane damage during ECC as evidenced by their ability to exclude the dye molecules (21) (Fig. 4 a Insets and c, 7.8 ± 1.4% vs. 20.4 ± 3.8%, respectively; nmice = 20, nfibers = 6,095; P < 0.03). Although there was a marked improvement in the post-ECC isometric force generated in the treated group, specific force per se was not increased. Interestingly, the nature of physiological improvement reported here is similar to the improvement (increased resilience to damage by lengthening contractions coupled with no increase in specific force) recently reported by using the internally truncated R4-R23 microdystrophin construct in a viral-vector-based gene therapy trial of mdx mice (31). This decrease in post-ECC force drop, along with decreased post-ECC dye uptake by damaged muscle, provides physiological evidence for improvement in sarcolemmal integrity and muscle function in HRG-treated mdx mice.
Fig. 4.
Functional improvement of muscle in HRG-treated mdx mice. (a) Physiological studies demonstrate greater force drop between the 1st (solid) and 10th (stippled) ECC in control mdx (Left) compared to HRG-treated mdx (Right) mice. (Insets) Decreased ability of control mdx to exclude procion orange dye from ECC-damaged fibers. (b) Quantification reveals a functional improvement in post-ECC force generation in treated mdx vs. control mdx mice. (c) Similarly, a lesser proportion of fibers in treated mdx was unable to exclude dye. Dashes show force drop and dye-exclusion in C57BL/10 adult mice (29).
Discussion
In this study, we have shown that administration of a small HRG peptide by simple i.p. injections led to utrophin up-regulation and a significant functional improvement of the dystrophic phenotype in mdx mice in vivo. Utrophin was likely required for mediating the benefit, because no improvement was seen in double mutant mdx:utrn–/– mice treated with HRG. The strategy of administering HRG need not be used in isolation; it could also be used in combination with other therapeutic interventions such as stem cells (32), vectorless oligonucleotide (33, 34), viral-vector-based gene therapy (31, 35), or myostatin blockade (36) in DMD management. Indeed, a recent study used muscle progenitor cells to systemically deliver human microdystrophin to dystrophic muscle, demonstrating the benefits of combinatorial strategies for the muscular dystrophies (37). Based on the nature of physiological benefit noted in this study and those noted previously by using myostatin blockade (36), HRG-mediated utrophin up-regulation would be predicted to synergistically potentiate the benefits of concomitant myostatin blockade in mdx mice; however, experiments need to be performed to test this hypothesis.
Conceptually, the postnatal activation of the utrophin promoter to increase its expression is similar to “reactivation” strategies used to augment fetal hemoglobin production after birth, as an adjunct to other therapies in sickle cell anemia (38). Akin to drugs used for fetal hemoglobin reactivation, HRG is likely to have pleiotrophic effects and activate genes that share similar promoter motifs and/or signaling pathways as utrophin. The amelioration of phenotype reported here is not as dramatic as that reported by using transgenic means to drive utrophin expression in mdx mice (15–17). This difference may be related to the levels of utrophin up-regulation achieved or the limited period that HRG was administered in this series of experiments. However, in contrast to studies where transgenic mice were created (15–17) or viral vectors used (35) to achieve utrophin up-regulation, utrophin promoter activation via a small peptide has a fundamental advantage of obviating the immune and delivery problems associated with germ-line modification and/or somatic gene therapy in DMD patients.
Acknowledgments
We thank Drs. Josh Sanes and Mark Grady (Washington University, St. Louis) for the kind gift of utrophin-null mutant mice, and Drs. Carsten Bönnemann and Kelly Perkins (University of Pennsylvania) for helpfuldiscussions and insightful comments. This study was supported in part by grants from Association Française contre les Myopathies (France), Duchenne Parents Project (The Netherlands), and Lundbeckfonden (Denmark), as well as National Institute of Arthritis and Musculoskeletal and Skin Diseases and National Eye Institute (National Institutes of Health) Grants AR048871 and EY013862. T.O.B.K. was the recipient of a predoctoral fellowship from the Faculty of Medicine, Copenhagen University (Denmark).
Abbreviations: DMD, Duchenne's muscular dystrophy; NMJ, neuromuscular junction; HRG, heregulin; EDL, extensor digitorum longus; ECC, eccentric contraction; TA, tibialis anterior; H&E, hematoxylin and eosin; CK, creatine kinase; CNF, centrally nucleated fibers.
References
- 1.Monaco, A. P., Bertelson, C. J., Middlesworth, W., Colletti, C. A., Aldridge, J., Fischbeck, K. H., Bartlett, R., Pericak-Vance, M. A., Roses, A. D. & Kunkel, L. M. (1985) Nature 316, 842–845. [DOI] [PubMed] [Google Scholar]
- 2.Monaco, A. P., Neve, R. L., Colletti-Feener, C., Bertelson, C. J., Kurnit, D. M. & Kunkel, L. M. (1986) Nature 323, 646–650. [DOI] [PubMed] [Google Scholar]
- 3.Koenig, M., Hoffman, E. P., Bertelson, C. J., Monaco, A. P., Feener, C. & Kunkel, L. M. (1987) Cell 50, 509–517. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman, E. P., Brown, R. H., Jr., & Kunkel, L. M. (1987) Cell 51, 919–928. [DOI] [PubMed] [Google Scholar]
- 5.Ibraghimov-Beskrovnaya, O., Ervasti, J. M., Leveille, C. J., Slaughter, C. A., Sernett, S. W. & Campbell, K. P. (1992) Nature 355, 696–702. [DOI] [PubMed] [Google Scholar]
- 6.Love, D. R., Hill, D. F., Dickson, G., Spurr, N. K., Byth, B. C., Marsden, R. F., Walsh, F. S., Edwards, Y. H. & Davies, K. E. (1989) Nature 339, 55–58. [DOI] [PubMed] [Google Scholar]
- 7.Khurana, T. S., Hoffman, E. P. & Kunkel, L. M. (1990) J. Biol. Chem. 265, 16717–16720. [PubMed] [Google Scholar]
- 8.Nguyen, T. M., Ellis, J. M., Love, D. R., Davies, K. E., Gatter, K. C., Dickson, G. & Morris, G. E. (1991) J. Cell Biol. 115, 1695–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlendieck, K., Ervasti, J. M., Matsumura, K., Kahl, S. D., Leveille, C. J. & Campbell, K. P. (1991) Neuron 7, 499–508. [DOI] [PubMed] [Google Scholar]
- 10.Tinsley, J. M., Blake, D. J., Roche, A., Fairbrother, U., Riss, J., Byth, B. C., Knight, A. E., Kendrick-Jones, J., Suthers, G. K., Love, D. R., et al. (1992) Nature 360, 591–593. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura, K., Ervasti, J. M., Ohlendieck, K., Kahl, S. D. & Campbell, K. P. (1992) Nature 360, 588–591. [DOI] [PubMed] [Google Scholar]
- 12.Winder, S. J., Hemmings, L., Maciver, S. K., Bolton, S. J., Tinsley, J. M., Davies, K. E., Critchley, D. R. & Kendrick-Jones, J. (1995) J. Cell Sci. 108, 63–71. [DOI] [PubMed] [Google Scholar]
- 13.Clerk, A., Morris, G. E., Dubowitz, V., Davies, K. E. & Sewry, C. A. (1993) Histochem. J. 25, 554–561. [PubMed] [Google Scholar]
- 14.Khurana, T. S., Watkins, S. C., Chafey, P., Chelly, J., Tome, F. M., Fardeau, M., Kaplan, J. C. & Kunkel, L. M. (1991) Neuromuscul. Disord. 1, 185–194. [DOI] [PubMed] [Google Scholar]
- 15.Tinsley, J. M., Potter, A. C., Phelps, S. R., Fisher, R., Trickett, J. I. & Davies, K. E. (1996) Nature 384, 349–353. [DOI] [PubMed] [Google Scholar]
- 16.Tinsley, J., Deconinck, N., Fisher, R., Kahn, D., Phelps, S., Gillis, J. M. & Davies, K. (1998) Nat. Med. 4, 1441–1444. [DOI] [PubMed] [Google Scholar]
- 17.Deconinck, N., Tinsley, J., De Backer, F., Fisher, R., Kahn, D., Phelps, S., Davies, K. & Gillis, J. M. (1997) Nat. Med. 3, 1216–1221. [DOI] [PubMed] [Google Scholar]
- 18.Dennis, C. L., Tinsley, J. M., Deconinck, A. E. & Davies, K. E. (1996) Nucleic Acids Res. 24, 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khurana, T. S., Rosmarin, A. G., Shang, J., Krag, T. O., Das, S. & Gammeltoft, S. (1999) Mol. Biol. Cell 10, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gramolini, A. O., Angus, L. M., Schaeffer, L., Burton, E. A., Tinsley, J. M., Davies, K. E., Changeux, J. P. & Jasmin, B. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3223–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrof, B. J., Shrager, J. B., Stedman, H. H., Kelly, A. M. & Sweeney, H. L. (1993) Proc. Natl. Acad. Sci. USA 90, 3710–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moens, P., Baatsen, P. H. & Marechal, G. (1993) J. Muscle Res. Cell Motil. 14, 446–451. [DOI] [PubMed] [Google Scholar]
- 23.Khurana, T. S. & Davies, K. E. (2003) Nat. Rev. Drug Discovery 2, 379–390. [DOI] [PubMed] [Google Scholar]
- 24.Coulton, G. R., Morgan, J. E., Partridge, T. A. & Sloper, J. C. (1988) Neuropathol. Appl. Neurobiol. 14, 53–70. [DOI] [PubMed] [Google Scholar]
- 25.Grady, R. M., Merlie, J. P. & Sanes, J. R. (1997) J. Cell Biol. 136, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deconinck, A. E., Potter, A. C., Tinsley, J. M., Wood, S. J., Vater, R., Young, C., Metzinger, L., Vincent, A., Slater, C. R. & Davies, K. E. (1997) J. Cell Biol. 136, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grady, R. M., Teng, H., Nichol, M. C., Cunningham, J. C., Wilkinson, R. S. & Sanes, J. R. (1997) Cell 90, 729–738. [DOI] [PubMed] [Google Scholar]
- 28.Deconinck, A. E., Rafael, J. A., Skinner, J. A., Brown, S. C., Potter, A. C., Metzinger, L., Watt, D. J., Dickson, J. G., Tinsley, J. M. & Davies, K. E. (1997) Cell 90, 717–727. [DOI] [PubMed] [Google Scholar]
- 29.Brooks, S. V. (1998) J. Muscle Res. Cell Motil. 19, 179–187. [DOI] [PubMed] [Google Scholar]
- 30.Gillis, J. M. (1999) J. Muscle Res. Cell Motil. 20, 605–625. [DOI] [PubMed] [Google Scholar]
- 31.Harper, S. Q., Hauser, M. A., DelloRusso, C., Duan, D., Crawford, R. W., Phelps, S. F., Harper, H. A., Robinson, A. S., Engelhardt, J. F., Brooks, S. V., et al. (2002) Nat. Med. 8, 253–261. [DOI] [PubMed] [Google Scholar]
- 32.Gussoni, E., Soneoka, Y., Strickland, C. D., Buzney, E. A., Khan, M. K., Flint, A. F., Kunkel, L. M. & Mulligan, R. C. (1999) Nature 401, 390–394. [DOI] [PubMed] [Google Scholar]
- 33.Lu, Q. L., Mann, C. J., Lou, F., Bou-Gharios, G., Morris, G. E., Xue, S. A., Fletcher, S., Partridge, T. A. & Wilton, S. D. (2003) Nat. Med. 9, 1009–1014. [DOI] [PubMed] [Google Scholar]
- 34.Bertoni, C., Lau, C. & Rando, T. A. (2003) Hum. Mol. Genet. 12, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert, R., Nalbantoglu, J., Petrof, B. J., Ebihara, S., Guibinga, G. H., Tinsley, J. M., Kamen, A., Massie, B., Davies, K. E. & Karpati, G. (1999) Hum. Gene Ther. 10, 1299–1310. [DOI] [PubMed] [Google Scholar]
- 36.Bogdanovich, S., Krag, T. O., Barton-Davis, E. R., Morris, L. D., Whittemore, L., Ahima, R. & Khurana, T. S. (2002) Nature 420, 418–421. [DOI] [PubMed] [Google Scholar]
- 37.Bachrach, E., Li, S., Perez, A. L., Schienda, J., Liadaki, K., Volinski, J., Flint, A., Chamberlain, J. & Kunkel, L. M. (2004) Proc. Natl. Acad. Sci. USA 101, 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olivieri, N. F. & Weatherall, D. J. (1998) Hum. Mol. Genet. 7, 1655–1658. [DOI] [PubMed] [Google Scholar]