Abstract
The opium poppy, Papaver somniferum, is one of mankind's oldest medicinal plants. Opium poppy today is the commercial source of the narcotic analgesics morphine and codeine. Along with these two morphinans, opium poppy produces approximately eighty alkaloids belonging to various tetrahydrobenzylisoquinoline-derived classes. It has been known for over a century that morphinan alkaloids accumulate in the latex of opium poppy. With identification of many of the enzymes of alkaloid biosynthesis in this plant, biochemical data suggested involvement of multiple cell types in alkaloid biosynthesis in poppy. Herein the immunolocalization of five enzymes of alkaloid formation in opium poppy is reported: (R,S)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase central to the biosynthesis of tetrahydroisoquinoline-derived alkaloids, the berberine bridge enzyme of the sanguinarine pathway, (R,S)-reticuline 7-O-methyltransferase specific to laudanosine formation, and salutaridinol 7-O-acetyltransferase and codeinone reductase, which lead to morphine. In capsule and stem, both O-methyltransferases and the O-acetyltransferase are found predominantly in parenchyma cells within the vascular bundle, and codeinone reductase is localized to laticifers, the site of morphinan alkaloid accumulation. In developing root tip, both O-methyltransferases and the O-acetyltransferase are found in the pericycle of the stele, and the berberine bridge enzyme is localized to parenchyma cells of the root cortex. Laticifers are not found in developing root tip, and, likewise, codeinone reductase was not detected. These results provide cell-specific localization that gives a coherent picture of the spatial distribution of alkaloid biosynthesis in opium poppy.
The opium poppy, Papaver somniferum, is the source of the narcotic analgesics codeine and morphine, which accumulate in specialized internal secretory cells called laticifers (1). In the aerial parts of the plant, the laticifer cells are anastomosed, forming an articulated network (2, 3). Laticifers are found associated with the vascular bundle in all plant parts. The morphinan alkaloids morphine, codeine, and thebaine are found both in roots and in aerial plant parts and specifically accumulate in vesicles within laticifers (4, 5). The benzo[c]phenanthridine alkaloid sanguinarine is found in root tissue. The syntheses of sanguinarine and of the tetrahydrobenzylisoquinoline alkaloid laudanine are completely understood at the enzyme level (6). Nearly all enzymes of morphine biosynthesis have also been described (6). In more recent years, cDNAs encoding 10 enzymes of alkaloid biosynthesis in P. somniferum have been isolated and characterized as follows. On the pathway leading from l-tyrosine to the first tetrahydrobenzylisoquinoline alkaloidal intermediate (S)-norcoclaurine, tydc, a cDNA encoding tyrosine/dopa decarboxylases, has been isolated (7). Transformation of (S)-norcoclaurine to the central isoquinoline alkaloid biosynthetic intermediate (S)-reticuline is understood at both the enzyme and gene level. (S)-Norcoclaurine is O-methylated by (R,S)-norcoclaurine 6-O-methyltransferase (8, 9). (S)-Coclaurine is next N-methylated by (R,S)-coclaurine N-methyltransferase (10) and the cDNA encoding this enzyme has been characterized (S. Haase, J.Z., S. Frick, and T.M.K., unpublished data). (S)-N-methylcoclaurine is then hydroxylated by the cytochrome P450-dependent monooxygenase CYP80B1 [(S)-N-methylcoclaurine 3′-hydroxlyase] (11). The cDNA encoding the corresponding cytochrome P450 reductase has been isolated as well (12). (S)-3′-hydroxy-N-methylcoclaurine is methylated to (S)-reticuline by (R,S)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT) (13). The cDNA 4′omt has been isolated and characterized from P. somniferum (J.Z., M. L. Diaz Chavez, and T.M.K., unpublished data).
(S)-Reticuline is a central intermediate of isoquinoline alkaloid biosynthesis, which leads to a plethora of alkaloidal structures. In P. somniferum, (R,S)-reticuline can be methylated by (R,S)-reticuline 7-O-methyltransferase (7OMT) (for which the cDNA 7omt has been described) to the tetrahydrobenzylisoquinoline laudanine (9). The N-methyl group of (S)-reticuline can alternatively be oxidatively cyclized by the berberine bridge enzyme (BBE) to C-8 of (S)-scoulerine (11, 14, 15). (S)-Scoulerine is then further converted in these plants to the antimicrobial benzo[c]phenanthridine alkaloid sanguinarine. Along the pathway on which (S)-reticuline is specifically converted to morphine, cDNAs encoding two biosynthetic enzymes have been identified. Salutaridinol 7-O-acetyltransferase (SalAT), encoded by salat, transfers an acetyl moiety from acetyl-CoA to the 7-hydroxyl group of salutaridinol (16, 17). Codeinone reductase (COR) is encoded by cor and catalyzes the penultimate step in morphine biosynthesis, the NADPH-dependent reduction of the keto moiety of codeinone to the 6-hydroxyl group of codeine (18, 19).
The combined results of the enzymological and molecular genetic work suggested a spatial distribution of alkaloid biosynthesis in opium poppy. It has also been suggested, based on the failure to produce morphine in undifferentiated P. somniferum cell cultures, that morphine accumulation is related to cytodifferentiation, noting that laticifers are absent from these cultures but present in tissue cultures (20, 21). We now address the question of cell-specific localization of the enzymes of alkaloid biosynthesis and the site of gene transcript accumulation. The results of these localization experiments provide us with an insight into the multiple levels of regulation involved in P. somniferum alkaloid biosynthesis. The necessity of understanding the cell-specific expression of alkaloid biosynthetic genes is also central to choosing proper gene transcriptional promoters for the design of meaningful metabolic engineering experiments with P. somniferum.
Herein, fluorescence immunocytological localization is carried out with the 4′OMT (reticuline pathway), the 7OMT (laudanine pathway), BBE (sanguinarine pathway), SalAT, and- COR (morphine pathway) (Fig. 1) with P. somniferum capsule, stem, and root tissue sections. In this manner, the spatial organization of enzymes occurring before and after a central branch point is analyzed. In addition, in situ localization of 7omt and cor1 was performed to correlate the site of gene transcription to enzyme accumulation.
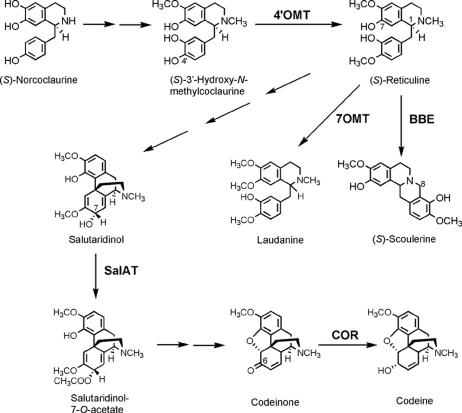
Fig. 1.
Schematic presentation of the biosynthetic pathway from (S)-norcoclaurine to codeine, laudanine, and (S)-scoulerine in P. somniferum. The positions of the enzymes localized in this study are indicated in bold print.
Materials and Methods
Plant Material. P. somniferum plants were either grown outdoors from April to July in Saxony-Anhalt, Germany, or grown throughout the year in a green house at the Leibniz Institute of Plant Biochemistry at 24°C with 18 h of light and 50% humidity. Plant tissue was harvested for immunolocalization and in situ hybridization experiments within 1–2 days after flower petal fall.
Protein Expression and Antibody Production. The following prokaryotic expression vectors were used for heterologous expression of the various P. somniferum cDNAs used in this study: 4′omt in pQE (Qiagen, Hilden, Germany), salat and cor in pCR T7NT/TOPO (Invitrogen), and mlp 15 and 7omt in pHIS8 [modified pET 28b vector (22)]. Overexpression in Escherichia coli (strain BL21 for 7omt, salat, cor, and mlp 15; strain M15 for 4′omt) was carried out according to the manufacturer's instructions (Invitrogen) for pCR T7NT/TOPO. Recombinant protein synthesis was induced by addition of isopropyl β-d-thiogalactopyranoside (1 mM final concentration) to the medium, and cells were harvested after incubation for 10–12 h at 28°C. Purification of the proteins containing an N-terminal histidine extension was achieved by cobalt affinity chromatography by using a Talon resin (Clontech) according to the manufacturer's instructions. The buffer system chosen was 50 mM Tris·HCl, pH 7.0, containing 500 mM NaCl, 25 mM imidazole, 10% glycerol, and 10 mM 2-mercaptoethanol. Because overexpression of major latex protein (MLP) 15 resulted in inclusion body formation, it was purified after denaturation according to the Clontech manual instructions. The bbe cDNA from Eschscholzia californica was expressed in Spodoptera frugiperda Sf9 cells and purified exactly according to refs. 15 and 23. Protein solutions in a final concentration of 1 mg/ml were used for antibody production by Biogenes (Berlin). Antibodies against the 4′OMT, 7OMT, SalAT, COR, and BBE were each produced in two rabbits. Animals were inoculated three times at 4-week intervals according to the company's standard protocol. Two weeks after the final immunization, the animals were bled. For coimmunolocalization experiments, two guinea pigs were inoculated three times with MLP 15; the second immunization followed the first by 14 days, and the third was 4 weeks later. After an additional 2 weeks, the animals were bled.
Western Blot Analysis. For the preparation of protein gel blots, plant material of P. somniferum was frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. The powder was extracted with 1.5 volumes (wt/vol) 50 mM Tris·HCl, pH 7.5, containing 20 mM DTT, 0.1% Triton X-100, and 2% polyvinylpyrrolidone. Cellular debris was removed by centrifugation at 10,000 × g for 10 min at 4°C. For electrophoretic resolution, 10–25 μg of protein was subjected to SDS/PAGE in a 12% acrylamide gel (24). The resolved proteins were transferred to a nitrocellulose filter according to ref. 25. The filter was incubated at room temperature for 1 h in 20 mM Tris·HCl, pH 7.5, containing 150 mM NaCl, 0.5% Tween 20, and 3% powdered milk. Antibody-containing serum was diluted 1:200 to 1:500 in fresh 20 mM Tris·HCl, pH 7.5, containing 150 mM NaCl, 0.5% Tween 20, and 3% powdered milk and added to the filter for 1 h at room temperature. The filter was then washed with the same buffer four times each for 10 min at room temperature. Secondary antibody (anti-rabbit IgG alkaline phosphatase conjugate or anti-guinea pig alkaline phosphatase conjugate, Sigma) was diluted 1:5,000 in fresh 20 mM Tris·HCl, pH 7.5, containing 150 mM NaCl, 0.5% Tween 20, and 3% powdered milk and added to the filter for 1–2 h at room temperature. The filter was afterward washed with the same buffer four times each for 10 min at room temperature. Sites of secondary antibody binding were visualized after addition of 0.4 mM nitro blue tetrazolium chloride and 0.4 mM 5-bromo-4-chloro-3-indolyl phosphate in 20 mM Tris·HCl, pH 9.5, containing 150 mM NaCl and 5 mM MgCl2 to the filter. Color development times were typically 20 min.
Embedding of Plant Material. Capsule, stem, and root tissue of P. somniferum were embedded for immunolocalization and in situ hybridization experiments. Approximately 3- × 3-mm explants were fixed for 2 h at room temperature under reduced pressure in PBS (1.5 mM KH2PO4/8mMNa2HPO4/135 mM NaCl/3mM KCl, pH 7.0–7.2) containing 3% paraformaldehyde and 0.1% Triton X-100. The fixed tissue was dehydrated in a graded ethanol series and embedded in polyethylene glycol (PEG) (PEG 1500/PEG 4000, 1:2). For longitudinal sections of stem and for in situ hybridization, ethanol was substituted with Rotihistol (Roth, Karlsruhe, Germany). Within 48 h at 60°C, the Rotihistol was replaced with Paraplast (Sigma), and the tissue was subsequently embedded in Paraplast. Embedded tissue was sectioned with a rotation microtome HM 335E (Microm, Hei-ldelberg, Germany). The PEG-embedded sections were ≈1–3 μm thick and the Paraplast-embedded sections were ≈6–10 μm thick. The paraffin sections were floated on distilled water droplets on poly-l-lysine coated slides, which were then baked at 45°C for 2–16 h. The paraffin was removed from the Paraplast-embedded sections with Roticlear (Roth), and the sections were rehydrated with a graded ethanol series and finally transferred to PBS. The PEG-embedded sections were floated on 45% PEG 6000 in PBS. The PEG 6000 was subsequently removed with PBS.
Immunolocalization. Immunolocalization of the P. somniferum alkaloid biosynthetic enzymes and of MLP 15 was carried out according to ref. 26, with the exception that 3–5% BSA in PBS was used as the blocking and antibody-containing solution. Anti-MLP 15, anti-COR, anti-SalAT, anti-4′OMT, and anti-BBE antibodies were diluted 1:500, and anti-7OMT antibody was diluted 1:1,000. Anti-callose antibody (Biosupplies, Parkville, Australia) was used at a final concentration of 10 μg/ml. The following secondary antibodies were used in a 1:500 dilution: Alexa 488 goat anti-rabbit IgG, Alexa 568 goat anti-guinea pig IgG, and Alexa 350 goat anti-mouse IgG (Molecular Probes). The immunostained sections were sealed in a citifluor antifadent mounting medium (Citifluor, Leicester, U.K.). Staining with aniline blue was carried out according to ref. 27.
In Situ Hybridization. In situ hybridization was carried out according to ref. 28. For the generation of sense and antisense RNA, 7omt and cor were subcloned into pGEM-T Easy (Promega). In vitro transcription was performed with digoxigenin-UTP (Roche Molecular Biochemicals) for 7omt and with fluroescein-UTP (Roche Molecular Biochemicals) for cor initiated from either the T7 or SP6 promoter according to the manufacturer's instructions. Hybridizing transcript was visualized either with anti-digoxigenin antibody alkaline phosphatase conjugate or with anti-fluorescein antibody alkaline phosphatase conjugate diluted 1:1,000 in 1% blocking reagent (Roche Molecular Biochemicals) and 1% acetylated BSA (Biotrend, Cologne, Germany) in Tris-buffered saline (100 mM Tris·HCl, pH 7.4, containing 150 mM NaCl).
Microscopy. Bright field microscopy and fluorescence microscopy were performed by using an Axioskop microscope (Zeiss). For fluorescence detection, the appropriate filter combination was used. Photographs were made with a Fuji video camera HZ300, and for fluorescence microscopy a Sony charge-coupled device camera was used.
Results and Discussion
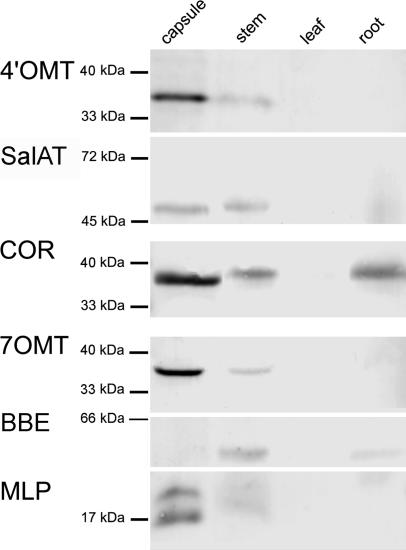
Organ-Specific Localization of Enzymes. Polyclonal antibodies raised against 4′OMT, SalAT, 7OMT, COR, BBE, and MLP 15 were initially evaluated by hybridization to protein gel blots that contained crude protein extracts from P. somniferum capsule, stem, leaf, and root. In general, the antibody preparations crossreacted with a single band of appropriate size blotted from a SDS/PAGE gel (Fig. 2). The MLP family of P. somniferum contains at least eight genes divided into two subfamilies (29). It was therefore not surprising that anti-MLP 15 antibody reacted with more than one low-molecular-weight protein. With the exception of BBE, the richest source of alkaloid biosynthetic enzymes and MLP 15 was capsule tissue followed by stem tissue. Anti-BBE antibody reacted with protein from stem and root. Benzo[c]phenanthridine and low levels of morphinan alkaloids were found in root tissue (data not shown) and the enzyme levels detected on the protein gel blot were correspondingly low. Based on this Western blot analysis and previous knowledge of distribution of enzyme activities in the plant (6, 16–18), capsule, stem, and root material was used for subsequent immunolocalization experiments.
Fig. 2.
Western blot of P. somniferum protein probed with anti-4′OMT, anti-SalAT, anti-COR, anti-7OMT, anti-BBE, or anti-MLP 15. Crude protein extracts were prepared from capsule, stem, leaf, and root tissue of mature plants. Sites of secondary antibody binding were visualized with nitro blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate.
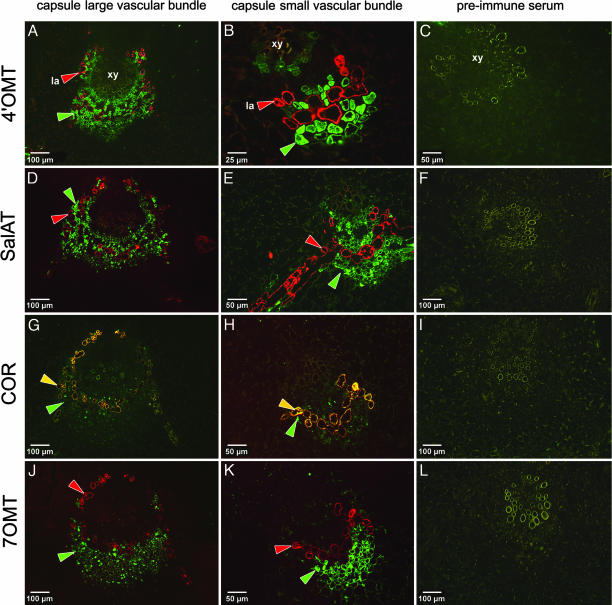
Tissue-Specific Localization of Enzymes. The capsule (fruit) of opium poppy is a rich source of latex and, therefore, of morphinan alkaloids. The capsule dissected in cross section revealed the parietal placenta with attached ovules. Tissue containing the large- and small-vascular bundle was excised and fixed (Fig. 5 A and B, which is published as supporting information on the PNAS web site). The reddish-brown discoloration that forms upon excision is a typical indicator of the oxidation (polymerization of phenolics) of exuded latex. Immunolocalization of the alkaloid biosynthetic enzymes and MLP 15 in cross sections prepared from this area of capsule indicate that two cell types are involved (Fig. 3). Localized (green fluorescence) to parenchyma cells that surround laticifers within the large- and small-vascular bundle (Fig. 3 A and B), was 4′OMT, which participates in the central isoquinoline alkaloid biosynthetic pathway leading to (S)-reticuline (Fig. 1). The red fluorescence shows the position of the latex-specific MLP 15 and is indicative of a laticifer. Likewise, SalAT, an enzyme specific to morphine biosynthesis, is localized to the same cell type as 4′OMT (Fig. 3 D and E, green fluorescence) within the vascular bundle. In contrast, COR, which is the penultimate enzyme of morphine biosynthesis occurring downstream of SalAT, is clearly colocalized with MLP 15 to laticifers, demonstrated by the yellow fluorescence, which results from an overlay of red and green fluorescence (Fig. 3 G and H). In capsule, COR was also found to a small extent in parenchyma cells, a phenomenon that was not observed in stem (Fig. 6, which is published as supporting information on the PNAS web site). Shown by green fluorescence in Fig. 3 J–L is 7OMT, which lies on the biosynthetic pathway to laudanine (Fig. 1) but not to morphine and is localized to parenchyma cells within the vascular bundle but, in contrast to 4′OMT and SalAT, only to those cells distal to laticifers Fig. 3 J and K, red fluorescence). The preimmune serum controls for each anti-biosynthetic enzyme antibody are shown in Fig. 3 C, F, I, and L. Fig. 3 E and G nicely demonstrates the presence of fused laticifer cells (fluorescing red in E and yellow in G) that form an articulated network in capsule.
Fig. 3.
Immunolabeling of alkaloid biosynthetic enzymes in cross sections of capsule of P. somniferum. Shown are cross sections through the large vascular bundle as indicated by the fusion of several vascular bundles (A, D, G, and J) and through a small vascular bundle of the capsule wall (B, E, H, and K); also shown are the preimmune serum controls (C, F, I, and L). The green arrows indicate the position of anti-biosynthetic enzyme antibody; the red arrows indicate the position of anti-MLP 15 antibody and is indicative of laticifer cells; the yellow arrows indicate colocalization of a biosynthetic enzyme with MLP 15. (A–C) Localization of 4′OMT. xy, xylem; la, laticifer. (D–F) Localization of SalAT. (G–I) Localization of COR (yellow is indicative of colocalization of COR and MLP 15). (J–L) Localization of 7OMT. Single sections were probed with two primary antibodies (anti-MLP and anti-biosynthetic enzyme) and two secondary antibodies. Red (excitation, 568 nm; emission long-pass filter, 590 nm) and green (excitation, 488 nm; emission long-pass filter, 520 nm) fluorescence images were overlaid in all micrographs.
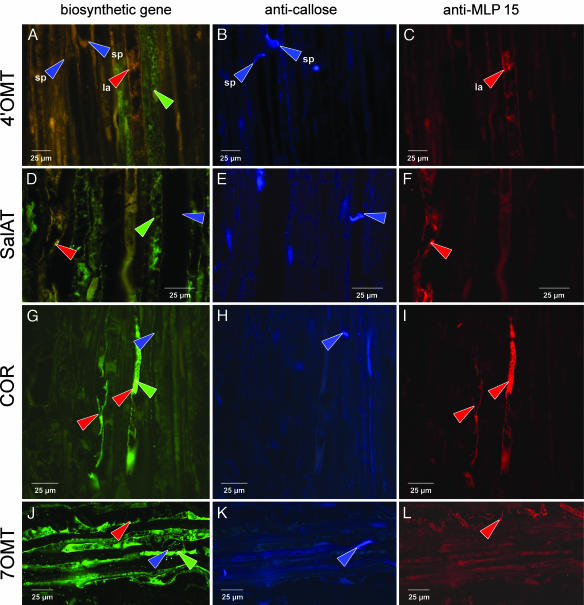
Stem tissue that lies within 2–4 cm of the capsule is also a rich source of latex. In longitudinal-sections of upper stem, sieve elements with their sieve plates and vesiculated laticifers can be readily identified (Fig. 5C). Immunolocalization of the alkaloid biosynthetic enzymes and MLP 15 in longitudinal-sections prepared from this area of stem also indicate that two cell types are involved in alkaloid biosynthesis in P. somniferum (Fig. 4). In these experiments, single sections were irradiated with different excitation wavelengths to visualize alkaloid biosynthetic enzyme, callose of sieve plates, or MLP 15 of laticifer cells. In Fig. 4A, 4′OMT (green arrow, green fluorescence) has been localized to cells adjacent to a laticifer (Fig. 4C, red arrow and red fluorescence). The sieve element identified by the presence of a sieve plate (Fig. 4B, blue arrow and blue fluorescence) is spatially separated from the cells containing biosynthetic enzyme and also shows no green fluorescence. SalAT, as in capsule, is localized similarly to 4′OMT (Fig. 4D). Again, the distinctly different position of a laticifer and sieve element are indicated by red and blue arrows and shown respectively also in Fig. 4 F and E. COR, on the other hand, is localized only to laticifers (Fig. 4G, green fluorescence). The same cells in Fig. 4 G and I are labeled with anti-COR and anti-MPL 15 antibody. The sieve element indicated by blue arrows in Fig. 4 G and H is devoid of green or red fluorescence. Also consistent with those results obtained in capsule, 7OMT is localized to the vascular bundle but not to laticifers or sieve elements (Fig. 4 J–L).
Fig. 4.
Immunolabeling of alkaloid biosynthetic enzymes, callose, and MLP 15 in longitudinal sections of stem of P. somniferum. Each row of micrographs represents a single section irradiated with a different excitation wavelength to visualize alkaloid biosynthetic enzyme (A, D, G, and J), callose of sieve plates (B, E, H, and K), or MLP 15 of laticifer cells (C, F, I, and L). The green arrows indicate the position of anti-biosynthetic enzyme antibody; the red arrows indicate the position of anti-MLP 15 antibody and is indicative of laticifer cells; the blue arrows indicate the position of anti-callose antibody and is indicative of sieve plates. (A) Localization of 4′OMT. (D) Localization of SalAT. (G) Localization of COR. (J) Localization of 7OMT. (B, E, H, and K) Callose of sieve elements. (C, F, I, and L) MLP 15 in laticifers. Single sections were probed with three primary antibodies (anti-biosynthetic enzyme, anti-MLP, and anti-callose) and three secondary antibodies. Green (excitation, 488 nm; emission long-pass filter, 520 nm), blue (excitation, 568 nm; emission long-pass filter, 590 nm), and red (excitation, 365 nm; emission long-pass filter, 420 nm) fluorescence images are presented individually. la, laticifer; sp, sieve plate.
Finally, developing root tips were sectioned, and the immunolocalization of 7OMT, BBE, and SalAT was carried out (Figs. 7A and 8, which are published as supporting information on the PNAS web site). The 7OMT and SalAT were found in the pericycle within the stele. Laticifers were not found in root tip, and likewise, COR was not immunologically detected. BBE localized to the largest part of the root cortex that consists of thin-walled parenchyma cells, which often have large intercellular spaces. This tissue proved to be quite fragile and severed when treated with anti-BBE antibody. BBE was not detected in aerial parts of the plant by fluorescence immunocytology. In the P. somniferum variety used in this study, (S)-scoulerine, the product of the reaction catalyzed by BBE, was below the limits of detection by liquid chromatography MS, suggesting that too little of this vesicular enzyme may be present for detection by fluorescence immunocytological methods in single cells. The vesicular localization of BBE has recently been demonstrated by ImmunoGold labeling of shoot tissue of E. californica and P. somniferum (30).
Roles of the Vascular Bundle and Laticifers in Alkaloid Biosynthesis. Tyrosine/dopa decarboxylase participates in the very early stages of tetrahydrobenzylisoquinoline alkaloid biosynthesis, acting at the interface between primary and secondary metabolism (6). Tydc transcript has been reportedly detected in phloem in P. somniferum aerial plant parts (31). This finding is consistent with the results reported herein. Transcripts of 7omt and cor were detected in phloem, and cor transcript was localized to laticifer cells (Fig. 7B and Fig. 9, which is published as supporting information on the PNAS web site). Based on the results obtained herein, early stages of isoquinoline alkaloid biosynthesis in P. somniferum occur in parenchyma cells associated with the vascular bundle and surrounding laticifers in aerial plant parts. Although cDNAs encoding each biosynthetic enzyme have not yet been identified, it appears that beginning from the primary metabolite l-tyrosine, enzymatic transformations through to the branch point intermediate (S)-reticuline occur in parenchyma associated with laticifers. Diversification into laudanine then takes place only in those parenchyma cells that lie distal to laticifers. Further transformation of (S)-reticuline into morphine is probably spatially located in the parenchyma that surround laticifers at least up until the 7-O-acetylation of salutaridinol. Thebaine, the subsequent intermediate in codeinone synthesis, is found in large quantities in the vesicles of latex (5), which implies a transport of either salutaridinol-7-O-acetate or thebaine from the surrounding parenchyma into the laticifer. The localization of the enzyme catalyzing demethylation of thebaine into neopinone is not known because an in vitro enzyme assay has not yet been established. Codeinone is then reduced in the laticifer into codeine, consistent with both the immunolocalization of COR herein and the known site of codeine accumulation in latex (5). The demethylation of codeine into morphine, as for the demethylation of thebaine into neopinone, still requires more study before a cell-specific localization can be determined. The third branch in alkaloid biosynthesis occurring after (S)-reticuline, which has been investigated herein, that leads to benzo[c]phenanthridines appears to proceed in thin-walled parenchyma of root cortex, based on the cellular immunolocalization of BBE.
Two cell types, laticifers and phloem parenchyma, have been identified herein for the biosynthesis of isoquinoline alkaloids in P. somniferum. It has been recently reported that sieve elements are the alkaloid biosynthetic cell of P. somniferum, based on immunolocalization of CYP80B1, BBE, and COR (32), an observation that we never made in our own studies, whether enzymatic, molecular genetic, or immunocytological. This finding is also inconsistent with the endoplasmic reticulum membrane localization of the cytochrome P450-dependent CYP80B1 and the necessity for an endoplasmic reticulum membrane-bound cytochrome P450 reductase for enzyme activity that such an enzyme likely be located in the cytosol of a sieve element (33). In addition, BBE is enzymatically active at pH 9–10 and completely inactive at neutral or acidic pH. BBE is located in planta in smooth vesicles with a basic pH as demonstrated both biochemically and by ImmunoGold labeling, in which experiments BBE was immunolocalized to smooth vesicles in idioblasts of P. somniferum (14, 30). Likewise, it is unlikely to be functional in the cytosol of a sieve element. Based on the low quality of the fluorescence micrographs presented in that work and the general inability to clearly discern cells types in those pictures, we can only conclude that the localization of all three investigated alkaloid biosynthetic enzymes to sieve elements represents a misinterpretation of the micrographs. This misinterpretation may also have resulted from the use of a commercial low-morphine variety (P. somniferum cv. Marianne) in the Bird et al. (32) study that further hampered specific detection of signals from low-level biosynthetic enzymes.
In conclusion, two cell types, parenchyma within the vascular bundle and laticifers, are sites of the biosynthesis of isoquinoline alkaloids in P. somniferum. The early stages of morphine biosynthesis occur in parenchyma cells surrounding laticifers and then at late stages, possibly at the level of either salutaridinol-7-O-acetate or thebaine, moves into the laticifer, which is the storage site of the morphinans thebaine, codeine, and morphine. The role of intercellular transporters of alkaloidal intermediates as well as intracellular transport into vesicles within laticifers adds a potential level of regulation to morphine biosynthesis that still needs to be investigated.
Supplementary Material
Acknowledgments
This paper is dedicated to Professor Detlef Gröger (Leibniz-Institut für Pflanzenbiochemie), on the occasion of his 75th birthday. We thank Dr. Bettina Hause (Leibniz-Institut für Pflanzenbiochemie) and Professor Gerhard Wanner (Ludwig-Maximilians-Univerität, Munich) for helpful discussions that allowed us to develop the techniques necessary to carry this project through to fruition. This work was supported by the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie.
Abbreviations: BBE, berberine bridge enzyme; COR, codeinone reductase; 4′OMT, (R,S)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; 7OMT, (R,S)-reticuline 7-O-methyltransferase; MLP, major latex protein; SalAT, salutaridinol 7-O-acetyltransferase; PEG, polyethylene glycol.
References
- 1.Kutchan, T. M., Ayabe, S. & Coscia, C. J. (1985) in The Chemistry and Biology of Isoquinoline Alkaloids, eds. Phillipson, J. D., Roberts, M. F. & Zenk, M. H. (Springer, Berlin), pp. 281–294.
- 2.Felkova, M. & Babkova, K. (1958) Pharmazie 13, 220. [PubMed] [Google Scholar]
- 3.Fairbairn, J. W. & Kapoor, L. D. (1960) Planta Med. 8, 49–61. [Google Scholar]
- 4.Nessler, C. L. & Mahlberg, P. G. (1978) Am. J. Bot. 65, 978–983. [Google Scholar]
- 5.Roberts, M. F., McCarthy, D., Kutchan, T. M. & Coscia, C. J. (1983) Arch. Biochem. Biophys. 222, 599–609. [DOI] [PubMed] [Google Scholar]
- 6.Kutchan, T. M. (1998) in The Alkaloids, ed. Cordell, G. A. (Academic, San Diego), Vol. 50, pp. 257–316. [Google Scholar]
- 7.Facchini, P. J. & De Luca, V. (1994) J. Biol. Chem. 269, 26684–26690. [PubMed] [Google Scholar]
- 8.Rüffer, M., Nagakura, N. & Zenk, M. H. (1983) Planta Med. 49, 131–137. [DOI] [PubMed] [Google Scholar]
- 9.Ounaroon, A., Decker, G., Schmidt, J., Lottspeich, F. & Kutchan, T. M. (2003) Plant J. 36, 808–819. [DOI] [PubMed] [Google Scholar]
- 10.Frenzel, T. & Zenk, M. H. (1990) Phytochemistry 29, 3491–3497. [Google Scholar]
- 11.Huang, F.-C. & Kutchan, T. M. (2000) Phytochemistry 53, 555–564. [DOI] [PubMed] [Google Scholar]
- 12.Rosco, A., Pauli, H. H., Priesner, W. & Kutchan, T. M. (1997) Arch. Biochem. Biophys. 348, 369–377. [DOI] [PubMed] [Google Scholar]
- 13.Frenzel, T. & Zenk, M. H. (1990) Phytochemistry 29, 3505–3511. [Google Scholar]
- 14.Steffens, P., Nagakura, N. & Zenk, M. H. (1985) Phytochemistry 24, 2577–2583. [Google Scholar]
- 15.Dittrich, H. & Kutchan, T. M. (1991) Proc. Natl. Acad. Sci. USA 88, 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenz, R. & Zenk, M. H. (1995) J. Biol. Chem. 270, 31091–31096. [DOI] [PubMed] [Google Scholar]
- 17.Grothe, T., Lenz, R. & Kutchan, T. M. (2001) J. Biol. Chem. 276, 30717–30723. [DOI] [PubMed] [Google Scholar]
- 18.Lenz, R. & Zenk, M. H. (1995) Eur. J. Biochem. 233, 132–139. [DOI] [PubMed] [Google Scholar]
- 19.Unterlinner, B., Lenz, R. & Kutchan, T. M. (1999) Plant J. 18, 465–475. [DOI] [PubMed] [Google Scholar]
- 20.Kutchan, T. M., Ayabe, S., Krueger, R. J., Coscia, E. M. & Coscia, C. J. (1983) Plant Cell Rep. 2, 281–284. [DOI] [PubMed] [Google Scholar]
- 21.Rush, M. D., Kutchan, T. M. & Coscia, C. J. (1985) Plant Cell Rep. 4, 237–240. [DOI] [PubMed] [Google Scholar]
- 22.Jez, J. M., Ferrer, J. L., Bowman, M. E., Dixon, R. A. & Noel, J. P. (2000) Biochemistry 39, 890–902. [DOI] [PubMed] [Google Scholar]
- 23.Kutchan, T. M., Bock, A. & Dittrich, H. (1994) Phytochemistry 35, 353–360. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 25.Kyhse-Andersen, J. (1984) J. Biochem. Biophys. Meth. 10, 203–209. [DOI] [PubMed] [Google Scholar]
- 26.Hause, B., Demus, U., Teichmann, C., Parthier, B. & Wasternack, C. (1996) Plant Cell Physiol. 37, 641–649. [DOI] [PubMed] [Google Scholar]
- 27.Gerlach, D. (1984) Botanische Mikrotechnik (Thieme, Stuttgart), 3rd Ed.
- 28.St-Pierre, B., Vazquez-Flota, F. A. & De Luca, V. (1999) Plant Cell 11, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nessler, C. L. & Burnett, R. J. (1992) Plant Mol. Biol. 20, 749–752. [DOI] [PubMed] [Google Scholar]
- 30.Bock, A., Wanner, G. & Zenk, M. H. (2002) Planta 216, 57–63. [DOI] [PubMed] [Google Scholar]
- 31.Facchini, P. J. & De Luca, V. (1995) Plant Cell 11, 1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird, D. A., Franceschi, V. R. & Facchini, P. J. (2003) Plant Cell 15, 2626–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauli, H. H. & Kutchan, T. M. (1998) Plant J. 13, 793–801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.