Significance
An old enigma in insect toxicology, the mode of action (MoA) of selective chitin biosynthesis inhibitors in arthropods, is resolved. Benzoylureas, buprofezin, and etoxazole share a MoA by directly interacting with chitin synthase 1. The finding that a single mutation confers striking levels of insecticide resistance against three putative different MoAs has important ramifications on resistance management strategies and rational use of insecticides against major agricultural pests and vectors of human diseases. Our results also show that CRISPR/Cas9-mediated gain-of-function mutations in single-copy genes of highly conserved target sites in arthropods provide opportunities for comprehensive insecticide resistance investigations across species boundaries and against several insecticide classes.
Keywords: insecticide resistance, benzoylureas, CRISPR/Cas9, resistance management, mosquito control
Abstract
Despite the major role of chitin biosynthesis inhibitors such as benzoylureas (BPUs) in the control of pests in agricultural and public health for almost four decades, their molecular mode of action (MoA) has in most cases remained elusive. BPUs interfere with chitin biosynthesis and were thought to interact with sulfonylurea receptors that mediate chitin vesicle transport. Here, we uncover a mutation (I1042M) in the chitin synthase 1 (CHS1) gene of BPU-resistant Plutella xylostella at the same position as the I1017F mutation reported in spider mites that confers etoxazole resistance. Using a genome-editing CRISPR/Cas9 approach coupled with homology-directed repair (HDR) in Drosophila melanogaster, we introduced both substitutions (I1056M/F) in the corresponding fly CHS1 gene (kkv). Homozygous lines bearing either of these mutations were highly resistant to etoxazole and all tested BPUs, as well as buprofezin—an important hemipteran chitin biosynthesis inhibitor. This provides compelling evidence that BPUs, etoxazole, and buprofezin share in fact the same molecular MoA and directly interact with CHS. This finding has immediate effects on resistance management strategies of major agricultural pests but also on mosquito vectors of serious human diseases such as Dengue and Zika, as diflubenzuron, the standard BPU, is one of the few effective larvicides in use. The study elaborates on how genome editing can directly, rapidly, and convincingly elucidate the MoA of bioactive molecules, especially when target sites are complex and hard to reconstitute in vitro.
Insects pose tremendous threats to humans in two main areas. Pathogens causing diseases such as malaria, dengue fever, and more recent problems caused by the Zika virus, are vectored by mosquitos, such as the Anopheles gambiae and Aedes aegypti, and cause severe global health problems (1). Furthermore, the sustainability of agricultural yields, which need to meet predicted population growth (2), is seriously threatened by pest insects and mites. The diamondback moth Plutella xylostella, a global lepidopterous pest of brassicaceous vegetables, is one of the economically most important agricultural pests in the world, particularly due to it having developed resistance to almost all chemical classes of insecticides applied for its control under continuous insecticide pressure (3).
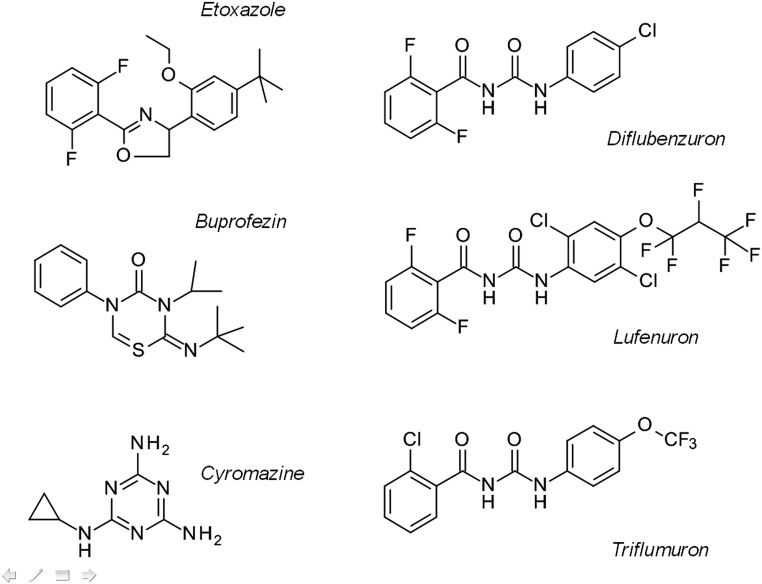
Protection of food sources and human health from invertebrate pests is critically reliant on insecticides (4, 5). Insecticides are classified according to mode of action (MoA) and chemistry into several groups through the IRAC (Insecticide Resistance Action Committee) insecticide grouping system, which is the basis for their rational use and resistance management strategies (4). The vast majority of current insecticides have neurotoxic and muscle action (>80%), whereas only a relatively small proportion interfere with growth and development (insect growth regulators, IGRs) and thus are highly selective to targeted arthropod pests as there are often no physiologically related processes or target sites present in vertebrates. IGRs are a group of chemically diverse compounds including the microbial-derived pyrimidine-nucleoside peptides, benzoylureas (BPUs), oxazolines, and thiadiazines (6) that all interfere with chitin biosynthesis or transport and deposition pathways. The MoA of the antifungal pyrimidine-nucleoside antibiotics is by their function as substrate analogs of UDP-N-acetylglucosamine at the catalytic site of chitin synthase (CHS) and are thus considered competitive inhibitors (7–9). BPUs (10), such as the major mosquito larvicide diflubenzuron and the agriculturally widely used insecticides triflumuron and lufenuron, represent a group of compounds (group 15 with regard to the IRAC grouping system; see also Fig. S1) that inhibit chitin biosynthesis by a unique yet elusive mechanism of action independent of the catalytic reaction of CHS itself (6, 10, 11). Although the sulfonylurea receptor (SUR) has been suggested as the direct target of BPUs (12) by affecting chitin biosynthesis indirectly by altering vesicle trafficking, its role in chitin biosynthesis inhibition remains controversial (13, 14). Furthermore, it was recently shown that SUR is dispensable for cuticle formation and chitin biosynthesis in Drosophila melanogaster (15).
Fig. S1.
Chemical structures of insecticides used in this study. According to the IRAC classification system (www.irac-online.org/modes-of-action/), Etoxazole belongs to group 10B (mite growth inhibitors), BPUs (Diflubenzuron, Lufenuron, Triflumuron) belong to group 15 (inhibitors of chitin biosynthesis, type 0), whereas Buprofezin belongs to group 16 (inhibitors of chitin biosynthesis, type 1). Cyromazine belongs to group 17 (moulting disruptor, dipteran).
Buprofezin (group 16) and etoxazole (group 10B) are two other, chemically different compounds (Fig. S1) highly selective to sucking agricultural pests that have also been proposed to interfere with chitin biosynthesis or cuticle formation (16, 17). Etoxazole is an oxazoline acaricide widely used against pest mite species but with limited activity on insects (18). Genomic mapping of a recessive monogenic etoxazole resistance locus in the two-spotted spider mite Tetranychus urticae, together with additional genetic and biochemical evidence, suggests that a single mutation in CHS1 is associated with etoxazole resistance; this mutation, I1017F (T. urticae numbering), is located in the C-terminal transmembrane domain. Therefore, it is likely that CHS1 is the molecular target of etoxazole as well as the chemically different acaricides clofentezine and hexythiazox (19, 20). Based on the similarity of symptoms for poisoning observed following exposure to both BPUs and etoxazole, as well as their inhibitory potential on chitin biosynthesis in isolated integuments of lepidopteran larvae, it has been hypothesized that they share the same MoA (18). The same direct MoA of BPU on CHS1, but not SUR, was later also postulated (19). However, no molecular evidence for such a possible association exists; there have been reports of BPU resistance in the diamondback moth in subtropical areas with intensive use of BPUs (21), but the molecular mechanism remains unknown. Furthermore, functional evidence of the involvement of the I1017F mutation in resistance could not be provided, given that in vitro approaches using recombinant protein expression are not feasible for large oligomeric integral protein complexes, especially when interactions are pre- or postcatalytic or involve the oligomerization of the complex (6, 19). As functional evidence is missing, the MoA through which chitin biosynthesis inhibitors exert their insecticidal activity remains uncertain.
Recent advances in genome modification technology, and especially the emergence of CRISPR/Cas9 (22), allow the application of “reverse” genetics approaches to provide in vivo evidence of the linkage between genotypes with phenotypes, including the study of insecticide MoA via generation of gain-of-function/loss-of-function mutations.
Here, we study and further select BPU resistance in P. xylostella and analyze the genetics of resistance as well as the possible association of identified point mutations in its CHS1 gene with the phenotype. We use CRISPR to generate the corresponding single mutations associated with BPU (and etoxazole) resistance in D. melanogaster, a model organism that is equipped with an efficient genetic “toolbox” enabling the fast and reliable study of the contribution of individual mutations to resistance. Toxicity bioassays with genome-modified flies are used to reveal insensitivity to BPUs and buprofezin, thus attempting to provide compelling evidence for the functional interaction with CHS1 as the molecular target site.
Results
Selection and Characterization of BPU Resistance in P. xylostella.
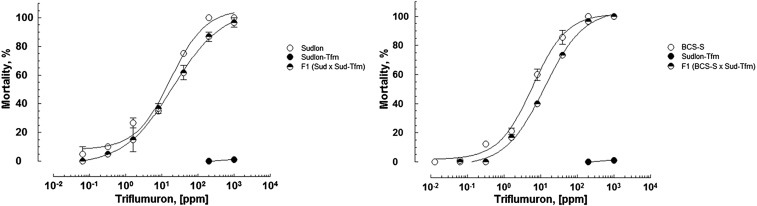
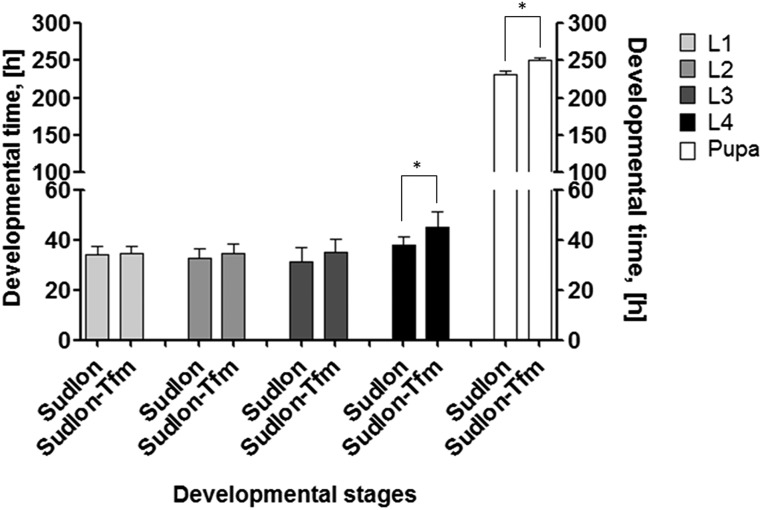
Low but significant resistance levels against diflubenzuron and triflumuron were detected in a P. xylostella strain (Sudlon) recently sampled in a Philippine cabbage field. The strain was maintained under laboratory conditions since 2011 to investigate target-site mutations in ryanodine receptors conferring resistance to diamide insecticides (23, 24). BPU insecticides have been used for diamondback moth control in Philippine cabbage in the past and were recently abandoned due to development of resistance. The Sudlon strain was reselected with triflumuron under laboratory conditions, resulting in the strain Sudlon-Tfm. Selection for 10 generations resulted in high BPU cross-resistance compared with the parental strain and reference strains BCS-S and Japan (Table 1). The selected strain Sudlon-Tfm was not only resistant to chemically diverse BPUs but also etoxazole (>178-fold), a chitin biosynthesis inhibitor of a different chemical class. Reciprocal crosses between Sudlon-Tmf and BCS and Sudlon revealed that the resistance was inherited autosomal recessive (Fig. 1) with a degree of dominance ranging from –0.73 to –0.88 in all reciprocal crosses (Table S1). Comparison of the postembryonic developmental time of strains Sudlon and Sudlon-Tfm showed that Sudlon-Tfm had a significantly longer larval (fourth instar) and pupal development time (Fig. S2), which could be indicative of possible fitness costs associated with the selected BPU resistance trait in Plutella.
Table 1.
Log-dose probit mortality for commercial BPU insecticides and etoxazole tested against third instar larvae of different strains of diamondback moth in leaf-dip bioassays (96 h)
| Compound | Strain | n | LC50, ppm | 95% CL* | Slope | RR† |
| Diflubenzuron | BCS-S | 300 | 36 | 21.0–60.3 | 1.3 | |
| Japan | 300 | 45 | 24–85 | 1.2 | 1 | |
| Sudlon | 300 | 317 | 118–855 | 1.2 | 9 | |
| Sudlon-Tfm | 300 | >1,000 | >28 | |||
| Triflumuron | BCS-S | 420 | 5.3 | 4.2–6.9 | 1 | |
| Japan | 420 | 11.6 | 7.8–17.3 | 0.89 | 2 | |
| Sudlon | 420 | 17.6 | 10.5–29.5 | 0.88 | 3 | |
| Sudlon-Tfm | 180 | >1,000 | >188 | |||
| Lufenuron | BCS-S | 450 | 1.8 | 0.96–3.5 | 1.3 | |
| Japan | 420 | 1.2 | 0.28–4.7 | 0.47 | 1 | |
| Sudlon | 390 | 0.63 | 0.2591–1.510 | 0.86 | 1 | |
| Sudlon-Tfm | 330 | 354 | 57–2189 | 0.94 | 196 | |
| Flucycloxuron | BCS-S | 240 | 0.16 | 0.15–0.18 | 1.3 | |
| Japan | 240 | 0.36 | 0.21–0.63 | 1.6 | 2 | |
| Sudlon | 240 | 0.091 | 0.068–0.12 | 1.1 | 1 | |
| Sudlon-Tfm | 540 | 179 | 27–1183 | 0.50 | 1,119 | |
| Etoxazole | BCS-S | 120 | 2.8 | 1.7–4.4 | 0.99 | |
| Sudlon | 120 | 5.3 | 3.2–8.7 | 0.99 | 2 | |
| Sudlon-Tfm | 120 | >500 | >178 |
95% confidence limits.
Resistance ratio (based on strain BCS-S).
Fig. 1.
Log-dose mortality data for triflumuron tested against third instar larvae of diamondback moth strains BCS-S, Sudlon, and Sudlon-Tfm as well as combined reciprocal crosses (F1). Error bars represent SEM.
Table S1.
Log-dose probit-mortality data for triflumuron tested against instar larvae of diamondback moth strains BCS-S, Sudlon, and Sudlon-Tfm as well as their respective reciprocal crosses (F1)
| Strain | n | LC50, ppm | 95% CL* | SLOPE | RR† | D‡ |
| BCS-S | 630 | 6.1 | 4.6–8.2 | 0.95 | ||
| Sudlon | 420 | 18 | 11–30 | 0.89 | 3 | |
| Sudlon-Tfm | 180 | >1,000 | >164 | |||
| Reciprocal crosses | ||||||
| F1§ | 210 | 12 | 9.2–16 | 0.81 | 2 | −0.73 |
| F1¶ | 240 | 23 | 17–31 | 0.78 | 1 | −0.88 |
95% confidence limits.
Resistance ratio.
Degree of dominance.
BCS-S × Sud-Tfm.
Sudlon × Sud-Tfm.
Fig. S2.
Comparison of the postembryonic developmental time (±SD) of strains Sudlon and Sudlon-Tfm. Sudlon-Tfm shows a significant longer larval (L4) and pupal development (P < 0.0005, R2 = 0.3, n = 50).
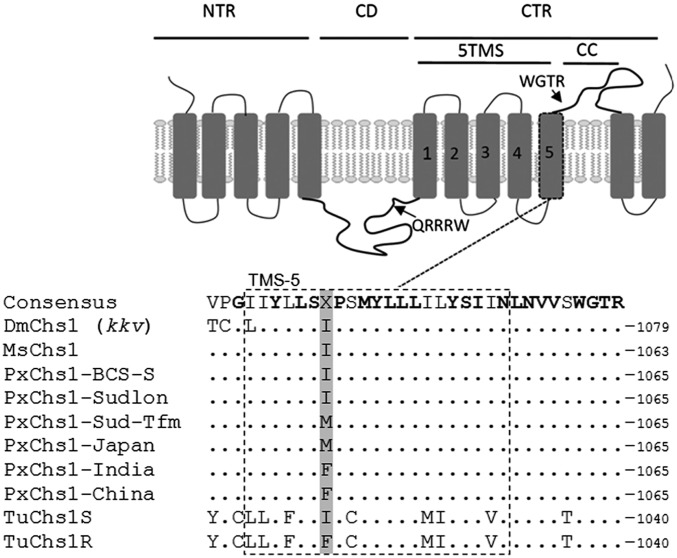
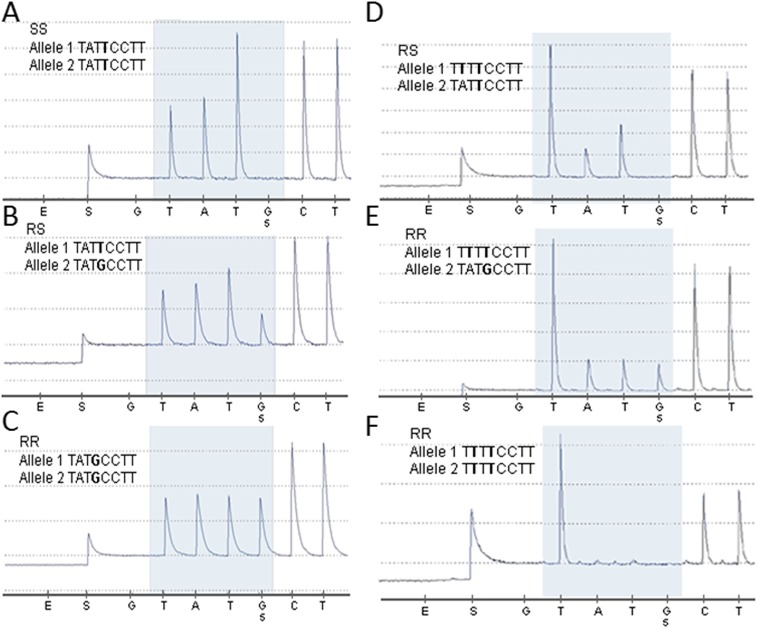
Based on (i) the identical symptoms of poisoning observed following exposure to both BPUs and etoxazole, (ii) the inheritance of resistance in an autosomal and recessive way in line to the etoxazole resistance phenotype previously reported in spider mites (19), and (iii) the strong genetically based evidence that etoxazole likely acts on CHS1 but not SUR (19), we subsequently cloned and sequenced the full-length CHS1 gene of P. xylostella strains BCS-S (GenBank accession no. KX420688), Sudlon (GenBank accession no. KX420689), and Sudlon-Tfm (GenBank accession no. KX420690) to compare the sequences between BPU-resistant and -susceptible strains. Compared with the CHS1, cDNA sequence of both susceptible strains BCS-S and Sudlon, a single nonsynonymous SNP resulting in a isoleucine (I)-to-methionine (M) amino acid change at position 1042 (P. xylostella numbering) in the C-terminal region of CHS1 of strain Sudlon-Tfm was found (Fig. 2). Genotyping of individual larvae by pyrosequencing of amplified CHS1 fragments covering that region revealed that the I-to-M amino acid substitution at position 1042 (I1042M), which was completely absent in the BCS-S strain, was present at low frequency in the Sudlon strain and fixed (100%) in the resistant Sudlon-Tfm strain after selection with triflumuron (Table 2 and Fig. S3).
Fig. 2.
Location of the two mutations conferring resistance. (Top) Schematic representation of domain architecture of CHS1, redrafted from ref. 19. 5TMS, cluster of five transmembrane segments; CC, coiled-coil motif; CD, catalytic domain; CTR, C-terminal region; NTR, N-terminal region. Rectangular boxes represent transmembrane domains. Arrows point to signature sequences QRRRW (catalytic domain) and WGTR (N-terminal region). (Bottom) Aligned amino acid sequences of helix 5 in the 5TMS clusters of CHS1 of D. melanogaster (Dm), M. sexta (Ms), six strains of P. xylostella (Px), and T. urticae (Tu; S, etoxazole susceptible; R, etoxazole resistant). Conserved residues are shown in bold. The position of the I1042M/F substitution in resistant P. xylostella (I1017F in etoxazole-resistant mites) is indicated in gray.
Table 2.
Genotyping (individual larvae) by pyrosequencing for a CHS1 target-site mutation (I1042M) in different strains of diamondback moth
| Strain | N | Frequency of genotype, % | |||||
| SS I1042, ATT/ATT | RS I/M1042, ATT/ATG | RR M1042, ATG/ATG | RR L1042, TTG | RR M/L1042, ATG/TTG | RR M/F1042, ATG/TTT | ||
| BCS-S | 30 | 100 | 0 | 0 | |||
| Sudlon | 30 | 97 | 0 | 3 | |||
| Sudlon-Tfm | 40 | 0 | 0 | 100 | |||
| Japan | 30 | 50 | 43 | 7 | |||
| Japan* | 6 | 0 | 0 | 100 | |||
| Px-China | 59 | 27 | 20 | 25 | 3 | 2 | 22 |
| Px-India | 23 | 52 | 30 | 17 | |||
| Reciprocal crosses | |||||||
| F1-A (BCS-S × Sudlon-Tfm) | 40 | 0 | 100 | 0 | |||
| F1-B (Sudlon × Sudlon-Tfm) | 63 | 0 | 89 | 11 | |||
Survivors of BPU treatment (>100 ppm).
Fig. S3.
SNP pyrosequencing assay results for the CHS I1042M mutation (ATT/ATG, printed in bold letters) in amplified cDNA and gDNA fragments of P. xylostella. (A) Homozygous ATT, genotype SS. (B) Heterozygous ATT/ATG, genotype RS. (C) Homozygous ATG, genotype RR. (D) Heterozygous TTT/ATT, genotype RS. (E) Heterozygous TTT/ATG, genotype RR. (F) Homozygous TTT, genotype RR.
The frequency of the 1042M/1042M alleles was 7% in a population from Japan, whereas the frequency of the 1042M/1042M in survivors of BPU treatment (>100 ppm) of the same population was 100% (Table 2). The correlation between mutation and resistance is significant (R2 = 0.9779, P = 0.0002). The I1042M mutation was also present at relatively high frequencies in field populations of P. xylostella sampled from cabbage fields in China and India with known BPU control failures (Table 2). Furthermore, genotyping of amplified CHS1 fragments of individual larvae of the Chinese field strain revealed another mutation, I1042F, which has been associated (19) with etoxazole resistance in T. urticae (Fig. 2, corresponding position I1017F).
Drosophila Flies Bearing the Mutations Corresponding to I1042M and I1017F Are Resistant to BPUs and Other Chitin Biosynthesis Inhibitors.
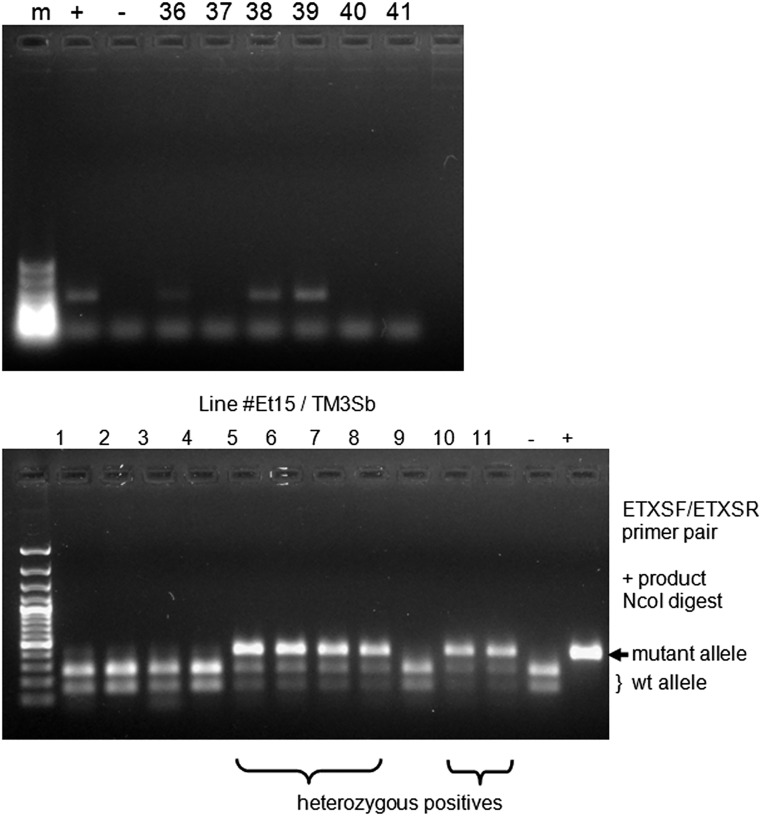
We identified the ortholog CHS gene in Drosophila (krotzkopf verkehrt or kkv; SI Results and Fig. S4), and to generate in kkv the I1056F/M mutations corresponding to I1017F in T. urticae and I1042M in P. xylostella, respectively (Fig. 2), we injected strain y1 M{nos-Cas9.P}ZH-2A w* (referred to as nos.Cas9 below) embryos with the appropriate gRNAs/donor plasmid mixes (SI Materials and Methods and Fig. S5) and screened progeny for genome-modified alleles. For the I1056F mutation, there were indications for the presence of homology-directed repair (HDR)-derived alleles within the sample at 16 different lines—that is, ∼20% of the total number (i.e., 77) of lines that gave G1 progeny. G1 individuals from each of three different original (G0) lines were crossed to balancer flies and screened to identify positive heterozygotes (Fig. S6). Several independent lines were established, and at least one became readily homozygous after balancing (line Et15); this line was verified by sequencing the relevant genomic region and shown to be genome-modified as expected, carrying the I1056F mutation at the kkv gene. Similarly, for the I1056M mutation, HDR-derived alleles were found (Fig. S6) in pools from 16 lines out of the 48 screened (∼33%), and individuals from three lines were crossed to balancers and screened. Several lines were sequence-verified as homozygous; line Px39 was selected for conducting toxicity bioassays.
Fig. S4.
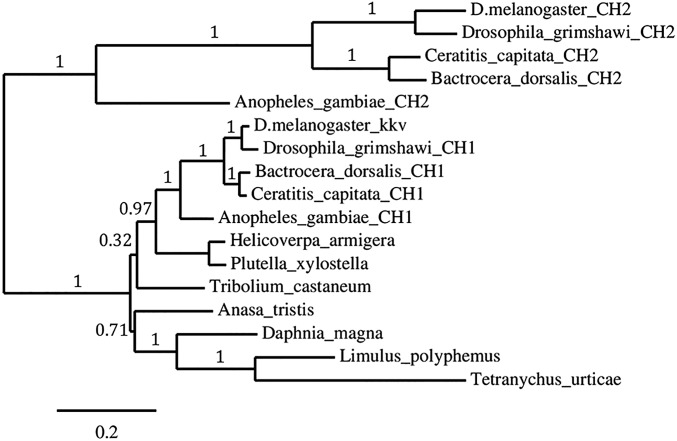
Phylogenetic analysis of several arthropod CHS protein sequences. Dipteran CHSs: A. gambiae (accession no. CH1: XP_321336.5, CH2: XP_321951.2), Bactrocera dorsalis (CHS1: XP_011203784.1, CHS2: AGC38392.1), Ceratitis capitata (CHS1: XP_012157009.1, CHS2: XP_012161954.1), Drosophila grimshawi (CHS1: XP_001994028.1, CHS2: XP_001985562.1), D. melanogaster (kkv: NP_524233.1, CHS2: NP_524209.3). Other insect CHSs: P. xylostella (Lepidoptera, BAF47974.1), Helicoverpa armigera (Lepidoptera, AKJ54482.1), Tribolium castaneum (Coleoptera, NP_001034491.1), Anasa tristis (Hemiptera, AFM38193.1). Other arthropod CHSs include spider mite T. urticae (XP_015781017.1), horseshoe crab Limulus polyphemus (XP_013790798.1), and crustacean Daphnia magna (KZS08010.1). Phylogenetic analysis was performed as described in ref. 40 using the one-click mode (www.phylogeny.fr/index.cgi). Numbers on branches indicate the approximate likelihood ratio test confidence index.
Fig. S5.
Nucleotide and deduced amino acid sequence of an 800-bp fragment of kkv exon 6 (corresponding to 3R:5381406:5382206 at the BDGP6 genome assembly), flanking position 1056 of the D. melanogaster amino acid sequence (I, shown in black background), equivalent to 1042 in Plutela xylostella and 1017 in T. urticae. Light gray areas indicate the CRISPR/Cas9 targets selected (gRNA444, gRNA658), whereas dark gray areas indicate the corresponding PAM (–NGG) triplets. Vertical arrows denote break points for CRISPR/Cas9-induced double stranded breaks. Ovals mark differences between target (wild-type) and donor (genome modified) sequences (red for I1056M and blue for I1056F). A C→T synonymous transition common for both designs at position 407 abolishes an NcoI cleavage site (CCATGG, underlined); an A→T transversion at position 492 generates a codon alteration (ATC→TTC) that results in the I1056F mutation, whereas a C→G transversion at position 494 generates a different codon alteration (ATC→ATG) that results in the I1056M mutation. Six extra synonymous mutations present at the CRISPR targets in the I1056M design are shown in red letters. Horizontal arrows indicate the relative positions of the primers used for diagnostic screening [ETXSF, ETXSR, ETXSM (blue, specific for I1056F), and PxM (red, specific for I1056M); see Table S2].
Fig. S6.
Screening for genome-modified flies. (Top) PCR screening after digestion with NcoI of template DNA from pools of G1 flies derived from different G0 (injected) individuals using a specific primer pair (ETXS/PxM) for I1056M mutation [–, nos.Cas9 DNA (negative control); +, donor plasmid template (positive control)]. (Bottom) Screening of G2 flies from line Et15 crossed to balancer TM3Sb, PCR amplification with a generic primer pair (ETXSF/ETXSR), and digestion of 395-bp product with NcoI. The modified allele remains uncut, whereas the wild-type allele is cut in two smaller bands; positives are heterozygous at this stage.
Toxicity assays with Drosophila larvae of strains nos.Cas9 and yw (both of which contribute to the genetic background of genome-modified flies) indicated that the strains carrying the wild-type kkv allele were sensitive to etoxazole at concentrations around 10 mg/L, without any significant differences observed between the two strains. Larvae did not manage to pupate or even grow to third instar. On the contrary, larvae from the genome-modified strains Et15 and Px39 bearing either the I1056F or I1056M homozygous mutation managed to grow and undergo molting without any visible problem, virtually all pupated, and adults eclosed normally when exposed to etoxazole concentrations as high as 10,000 mg/L, although at the highest concentrations (>1,000 mg/L) adults were dying just after eclosion. The LC50 values (with their corresponding 95% fiducial limits) for the susceptible (nos.Cas9) and resistant (Et15, Px39) lines and the associated resistance ratios are shown in Table 3.
Table 3.
Bioassay results (LC50 values and associated resistance ratios) of genome-modified flies (Et15, Px39) versus relevant unmodified controls (nos.Cas9) for five different insecticides
| Insecticides | Strains | LC50, ppm (95% CL) | Resistance ratio |
| Etoxazole | Et15 (I1017F) | >10,000 | >1,077 |
| Px39 (I1042M) | >10,000 | >1,077 | |
| nos.Cas9 | 9.28 (0.73–14.00) | 1 | |
| Diflubenzuron | Et15 (I1017F) | >5,000 | >15,625 |
| Px39 (I1042M) | >5,000 | >15,625 | |
| nos.Cas9 | 0.32 (0.24–0.42) | 1 | |
| Lufenuron | Et15 (I1017F) | 16.66 (8.70–66.47) | 111.06 |
| Px39 (I1042M) | >20 | >133 | |
| nos.Cas9 | 0.15 (0.11–0.18) | 1 | |
| Cyromazine | Et15 (I1017F) | 0.23 (0.21–0.25) | 0.74 |
| Px39 (I1042M) | 0.30 (0.21–0.41) | 1 | |
| nos.Cas9 | 0.31 (0.25–0.34) | 1 | |
| Buprofezin | Et15 (I1017F) | >1,000 | >18.79 |
| Px39 (I1042M) | 1,276.65 (1,110.36–1,554.15) | 24.07 | |
| nos.Cas9 | 53.20 (41.24–65.72) | 1 |
Bioassay screens indicated a gross difference in the toxicity between both Px39 (I1056M) and Et15 (I1056F) Drosophila lines for diflubenzuron (LC50 nos.Cas9, 0.322 mg/L vs. LC50 Et15 and LC50 Px39, >5,000 mg/L), lufenuron (LC50 nos.Cas9, 0.148 mg/L vs. LC50 Et15, 16.659 mg/L and LC50 Px39, >20 mg/L), and buprofezin (LC50 nos.Cas9, 53.2 mg/L vs. LC50 Et15, >1,000mg/L and LC50 Px39, 1,276.654 mg/L). Such levels of at least partial cross-resistance support a common MoA between etoxazole, BPUs, and buprofezin. However, cyromazine toxicity is not affected either by the I1056M or the I1056F mutation, indicating either a different binding mode or another MoA.
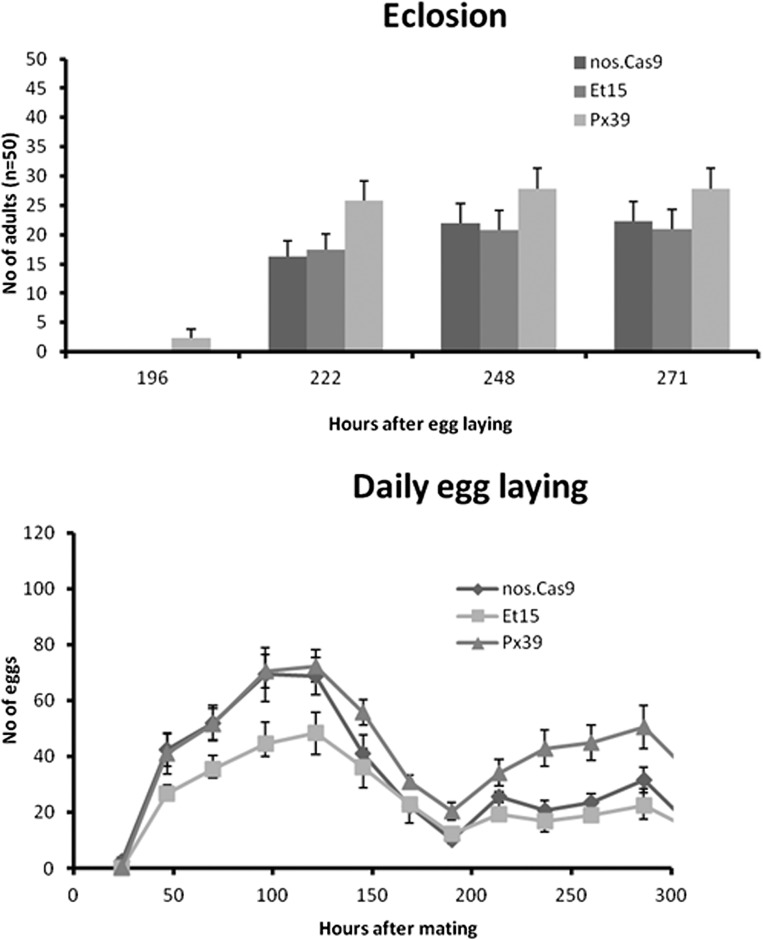
The genome-modified fly lines used for bioassays were examined for certain life table parameters (SI Materials and Methods and Fig. S7), but no significant difference was observed in the flies bearing the I1042M mutation, contrary to the result from Plutella (Fig. S2).
Fig. S7.
(Top) Comparison of developmental timing and number of eclosed adults out of 50 original eggs among different Drosophila lines used in the bioassays of this study. No significant difference was found between Px39 (I1042M) or Et15 (I1017F) versus nos.Cas9 wild-type controls (P = 0.151 and P = 0.4, respectively). (Bottom) Comparison of average daily fecundity among lines (n = 8 for Et15 and nos.Cas9, n = 7 for Px39). No evidence for reduced fecundity is found for line Px39 (I1042M). Error bars represent SEM.
Discussion
Resistance against the major chitin biosynthesis inhibitor class of insecticide chemistry (i.e., BPUs) was detected and subsequently selected in a recently collected Philippine field population of the diamondback moth P. xylostella, one of the most important agricultural pests in brassicaceous crops worldwide. The presence and frequency of the amino acid substitution I1042M was highly correlated with cross-resistance against several BPUs such as diflubenzuron, triflumuron, lufenuron, and flucycloxuron. Surprisingly, the P. xylostella mutation in CHS1 gene lies at the same location of a previously documented mutation (I1017F) conferring etoxazole resistance (19). Introduction of either mutation in D. melanogaster by a CRISPR/Cas9 coupled with HDR genome modification approach showed a similar resistance phenotype across different chemical classes of IGRs, such as BPUs, etoxazole, and buprofezin, but not cyromazine. This is compelling evidence that BPUs, buprofezin, and etoxazole share the same MoA and directly interact with CHS1.
Our chosen genetic validation approach is further supported by a contemporary study showing that the introduction of a single point mutation in an alpha6 subunit of the nicotinic acetylcholine receptor of Drosophila by CRISPR/Cas9 genome editing copying a mutation associated with spinosad resistance in thrips resulted in a spinosad-resistant phenotype in genome-modified flies (25). Our results show that a reverse genetics strategy is exceptionally suitable for the elucidation of the MoA of insecticides and/or functional validation of mutations associated with insecticide resistance in a wide array of targets that are otherwise difficult to study. CRISPR/Cas9 has already been used in Drosophila for resistance research before (25, 26). However, in this study, we have generated lines bearing homozygous recessive gain-of-function mutations in a single-copy gene, thus enabling comprehensive investigation—that is, comparative bioassays for these particular mutations against several insecticide classes. The fact that most target sites between arthropods are highly conserved allows screening of different mutations across species boundaries. This strategy has several potential valuable ramifications, as it can be used in a large number of molecular targets and a wide array of chemical classes of insecticides.
Procedures toward the investigation of insecticide MoA and resistance mechanisms typically involve in vitro screening systems (27), electrophysiology (28), direct ligand/receptor–insecticide interactions either in vivo (24) or in silico (29), functional expression of enzymes (30, 31), or genetic mapping linkage analysis (19, 32). However, there are cases where in vitro screening is not applicable because the native proteins or protein complexes cannot be reconstituted or recombinantly expressed. One such example is CHS1 because of its structure as a large oligomeric integral membrane protein that catalyzes both polymerization of sugars and translocation of the nascent chitin fiber across the plasma membrane. No structural information is available on CHS1 complexes, and even the quaternary structure is not known (although trimeric complexes have been purified from Manduca sexta, they could be building blocks of higher order complexes) (33), thus rendering impossible any effort to model interactions. Attempts in recombinant expression have failed to generate active complexes. In this and other cases, the interaction between target site and insecticides can be more complex than simply inhibiting natural substrate or ligand binding, making it even harder to develop a functional screening assay.
The elucidation of the MoA of the chitin biosynthesis inhibitor classes BPU and buprofezin (i.e., IGR insecticides) that have been used against major agricultural pests and disease vectors for many years, directly acting on CHS, as well as the identification of BPU target-site resistance mutations has important implications and impact for the rational use of insecticides and insecticide resistance management. It will directly affect the IRAC classification (4) of those molecules, which are currently assigned to a MoA group (MoA group 15) different from etoxazole (MoA group 10) and buprofezin (MoA 16). Our study provides compelling evidence that both classes affect the same target protein, CHS1, thus justifying their subgrouping in a single MoA class. The finding that a single mutation confers high levels of insecticide resistance against three putative different MoAs has important effects on resistance management strategies, which are largely based on rotation of insecticide MoA groups, to avoid selection for target-site resistance by repeatedly applying chemistries addressing the same binding site.
The presence of the CHS1 resistance mutation in diamondback moth populations from different countries, in particular, is an important consideration for rational use and management of insecticides against this major pest. The slightly but significantly extended development time of fourth instar larvae and pupae in strain Sudlon-Tfm indicated a putative fitness cost in Plutella, possibly associated with this mutation. However, this was not confirmed in Drosophila lines, where the mutation was isolated in an isogenic background. It is possible that unrelated genetic loci in the multiresistant Sudlon-Tfm laboratory strain (24) might have contributed to the fitness cost observed.
The developed pyrosequencing diagnostic as well as possible additional field-applicable technologies to detect the presence of CHS1 target-site mutations provides a tool allowing us to screen rapidly for the presence of resistant genotypes to adjust resistance management strategies based on MoA rotation accordingly.
The findings may also have implications for public health insecticide-based vector control interventions. The larvicide diflubenzuron is one of the most important insecticides that have been used against mosquitoes, particularly in regions such as Europe, where neurotoxic insecticides are banned from use in mosquito breeding sites. Screening of A. aegypti and Aedes albopictus populations, the major vectors of arbovirus including Dengue and Zika, from several geographical regions for possible resistant CHS1 alleles will guide appropriate resistance management strategies to ensure the sustainability of control interventions. This discovery will also potentially have a bearing on the choice of insecticide for new human pathogen vector control, such as against the malaria mosquito A. gambiae s.s (34).
Materials and Methods
Chemicals.
Insecticides (diflubenzuron, triflumuron, lufenuron, and flucycloxuron) used for P. xylostella bioassays were of technical grade (purity >98%) and provided in-house (Bayer CropScience). Commercial insecticide formulations were used for Drosophila bioassays, namely Borneo [11% (wt/vol) etoxazole; Hellapharm], Dimilin [48% (wt/vol) diflubenzuron; Syngenta], Match [50% (wt/vol) lufenuron; Syngenta], Trigard [75% (wt/vol) cyromazine; Syngenta], and Applaud [25% (wt/vol) buprofezin; Syngenta]. All other chemicals were purchased from Sigma-Aldrich.
Insects.
The susceptible reference strain (BCS-S) of P. xylostella L. (Lepidoptera: Plutellidae) has been maintained under laboratory conditions for more than 20 y without exposure to insecticides. Strain Sudlon was collected in a cabbage field located in Sudlon, Cebu Island, in the Philippines in 2011 as described elsewhere (24). The BPU-resistant strain Sudlon-Tfm was obtained by selecting strain Sudlon for 10 generations with triflumuron by incrementally increasing its concentration to 1,000 mg·L−1. The Japan strain was collected in Mizobe, Japan in 2010. Finally, the strains from China and India were collected from cabbage in 2014. All strains were maintained on cabbage plants (Brassica oleracea) as recently described (24). Strain Sudlon-Tfm was maintained on triflumuron- (1,000 mg·L−1) treated cabbage plants. The Drosophila strain y1 M{nos-Cas9.P}ZH-2A w* (nos.Cas9; stock no. 54591 at Bloomington Stock Center) (35) as well as yw strain and the strain yw; TM3 Sb e/TM6B Tb Hu e (containing third chromosome balancers, provided by Christos Delidakis, Institute of Molecular Biology and Biotechnology/ Foundation for Research and Technology Hellas and University of Crete, Heraklion, Crete, Greece) were used in this study. Drosophila strains were typically cultured at 25 °C temperature, 60–70% humidity, and 12/12-h photoperiod on standard fly diet.
Bioassays.
Leaf dip bioassays with third instar diamondback moth larvae were conducted after IRAC method no. 7 (www.irac-online.org) as described recently (24). Control mortality was less than 10%. LC50 values and their corresponding 95% fiducial limits were calculated using Prism 5.03 (GraphPad Software, Inc.). For Drosophila bioassays, second instar larvae were collected and transferred in batches of 20 into new vials containing fly food supplemented with different insecticide concentrations. Larval development, molting, pupal eclosion, and adult survival were monitored for a period of 10–12 d. Five to six insecticide concentrations that cause 5–95% mortality (when applicable) were tested in triplicate, together with relevant negative (no insecticide) controls, in genome-modified flies and wild-type (nos.Cas9 and/or yw) controls. Dose-dependent molting and/or mortality curves were constructed from dose–response data, and LC50 values were calculated with PoloPlus (LeOra Software). A χ2 test was used to assess how well the individual LC50 values agreed with the calculated linear regression lines.
Crossing Experiments.
Pupae of strains BCS-S, Sudlon, and Sudlon-Tfm were collected and kept in Petri dishes individually until they hatched. After sex determination, 50 virgin females of Sudlon-Tfm were crossed with 50 males of Sudlon strain or BCS-S strain and vice versa. Because there was no difference obtained between the two reciprocal crosses, the F1 generation was pooled for further studies. The F1 generation was backcrossed with the respective parental strains. The backcross was conducted following the same approach as the reciprocal crosses; there was no difference obtained among the offspring, so samples were pooled. Third instar larvae were used for leaf dip bioassays to obtain the individual LC50 values for triflumuron. The degree of dominance (D) was calculated using Stone’s equation. (36). Larvae of the different strains were preserved in RNAlater (Ambion) and analyzed for the I1042M/F mutation by pyrosequencing.
Pyrosequencing.
Individual P. xylostella larvae were ground in lysis buffer, and total genomic DNA (approximately 400 ng per larvae) was extracted using DNAdvance Tissue Kit (Agencourt) according to the to the supplier’s recommended protocol. A gene fragment of 210 bp was amplified by PCR from 50-ng aliquots of gDNA using the primer pair PxCHS1-forward and PxCHS1-reverse (Table S2), designed with Assay Design Software (PSQ-Biotage AB, now Qiagen). The primer pair is based on a ClustalW aligned consensus sequence of CHS1 of diamondback moth found in GenBank (accession number AB271784) as well as internally sequenced CHS1 of strains BCS-S, Japan, and Sudlon. The pyrosequencing protocol comprised 35 PCR cycles with 0.5 μM forward and biotinylated reverse primer in 30 μL reaction mixtures containing 1× Taq enzyme reaction mix (RedTaq Jumpstart Master Mix, Sigma-Aldrich) and cycling conditions of 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min and a final elongation step at 72 °C for 5 min. The single-strand DNA required for pyrosequencing was prepared as described in ref. 23. The pyrosequencing reactions were carried out according to the manufacturer’s instructions using the PSQ 96 Gold Reagent Kit (Qiagen), and the sequence-PxCHS1-seq (Table S2) for genotyping. The pyrograms were analyzed using the SNP Software (Qiagen).
Table S2.
List of primers used in this study
| Primer name | Sequence |
| 444F | 5′-CTTCGTAATGCACCAGAACTCTTG-3′ |
| 444R | 5′-AAACCAAGAGTTCTGGTGCATTAC-3′ |
| 658F | 5′-CTTCGCTTCCTTCAGAGTGGAATC-3′ |
| 658R | 5′-AAACGATTCCACTCTGAAGGAAGC-3′ |
| ETXSF | 5′-CCTGTAAGTCGAACATCCAG-3′ |
| ETXSR | 5′-AGCTCAGCATGCTCTTCTGC-3′ |
| ETXSM | 5′-AGCAGGTACATGGACGGGAA-3′ |
| PxM | 5′-GCAGCAGGTACATGGACGGC-3′ |
| CHS11F | 5′-TCCTCCGACAACATCACACG-3′ |
| CHS11R | 5′-AACATGAAGGCCAGGATCGG-3′ |
| CHS12F | 5′-GGACCCGGAACCATTTTCCT-3′ |
| CHS12R | 5′-TGCTCCACGGGATCATTGTC-3′ |
| CHS20F | 5′-GTCGGACTCAGACACTTCGG-3′ |
| CHS20R | 5′-ACCGCGAAAATGCCAAGATG-3′ |
| PxCHS1-forward | 5′-CGCTTGTTTGCATCCTCAAGAG-3′ |
| PxCHS1-reverse | 5′-[btn]TTCGCACTCTTCTTTGCGTCTTC-3′ |
| PxCHS1-seq | 5′-GGTATCATTTACCTTCTGTC-3′ |
Genomic Engineering Strategy.
An ad hoc CRISPR/Cas9 genomic engineering strategy was devised to generate the I1056M/F mutations (equivalent to the I1042M and I1017F mutation in P. xylostella and T. urticae CHS1, respectively; SI Materials and Methods, Fig. 2, and Fig. S5) at the kkv gene in D. melanogaster. Potential CRISPR targets in the region of interest were identified using the online tool Optimal Target Finder (37) (tools.flycrispr.molbio.wisc.edu/targetFinder/), and two targets with no predicted off-target hits were selected to generate RNA expressing plasmids gRNA444 and gRNA658, respectively, targeting the relevant genomic regions (SI Materials and Methods and Fig. S5). We constructed de novo (Genscript) two donor plasmids for HDR, encompassing genomic region 3R:5380538:5383542 but with certain modifications compared with the wild-type genomic sequence (Fig. S5).
Drosophila DNA Purification and Amplification.
DNA was purified from Drosophila tissues by DNAzol (MRC) according to the manufacturer’s instructions. PCR amplification with relevant primer pairs (Table S2) was typically performed with Kapa Taq DNA Polymerase (Kapa Biosystems). The conditions used were 95 °C for 2 min, followed by 30–35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 1 min followed by a final extension step for 2 min.
Generation and Selection of Genome-Modified Flies.
We used transgenic flies with the genotype y1 M{nos-Cas9.P}ZH-2A w* that carry a transgene expressing Cas9 protein during oogenesis under control of nanos regulatory sequences (35) and injected embryos as described in SI Materials and Methods. Screening was performed by isolating DNA from sets of ∼30 individuals per vial (mostly pupae, but also adults and third instar larvae, depending on availability). In case the presence of genome-modified alleles was indicated in the pool (Fig. S6), several individual G1 flies from the same original cross were first crossed again with nos.Cas9 flies to generate G2 progeny and then individually screened to positively identify which of these G1 flies indeed carried genome-modified alleles. Lines originating from positive G1 flies were established, and individual G2 flies (expected to be heterozygous for the mutant allele at a 50% ratio) were balanced against a strain containing third chromosome balancers (yw; TM3 Sb e/TM6B Tb Hu e). Flies potentially containing modified alleles were screened as before after being back-crossed to the original balancer stock (Fig. S6); the progeny of positives (bearing the modified allele opposite to one of the balancer chromosomes) was used to generate homozygous lines by crossing between siblings and selecting against the marker phenotype (Sb or Tb Hu) for the relevant balancer. All lines used were sequence-verified.
SI Materials and Methods
Genome Modification Strategy.
Genomic region sequencing in nos.Cas9 flies.
We used three primer pairs (CHS11F/CHS11R, CHS12F/CHS12R, and CHS20F/CHS20R; Table S2) to generate three overlapping DNA fragments encompassing genomic region 3R: 5380505:5383684 (numbering according to BDGP6 genome assembly) by PCR amplification (see main text), using DNA from y1 M{nos-Cas9.P}ZH-2A w* flies. This region was sequenced to identify possible variations from the published genome sequence (none detected).
Generation of gRNA plasmids.
We used gRNA vector pU6-BbsI-chiRNA (38), after digestion with BbsI, and we ligated specific double-stranded DNA oligos generated by annealing of single-stranded DNA oligos 444F/444R and 658F/658R (Table S2) to generate RNA expressing plasmids gRNA444 and gRNA658, respectively, targeting the relevant genomic regions (Fig. S5).
Donor plasmids for HDR.
We constructed de novo (Genscript) two donor plasmids for HDR encompassing genomic region 3R:5380538:5383542 but with certain modifications compared with the wild-type genomic sequence (Fig. S5): for the I1056F mutation (equivalent to position 1042 in P. xylostella and 1017 in T. utrticae), a C→T synonymous transition that abolishes an NcoI restriction site in the donor sequence and an A→T transversion that generates the I1056F mutation (codon alteration ATC→TTC) and abolishes a FokI restriction site in the donor sequence (Fig. S5). For the I1056M mutation, apart from the C→T synonymous transition abolishing the NcoI site and the C→G transversion that generates the I1056M mutation (codon alteration ATC→ATG), we added six more synonymous changes, three on each of the gRNA CRISPR targets (Figs. S5), to avoid cleavage of the donor plasmid by the CRISPR/Cas9 procedure used to target the genome.
Drosophila embryo injection and G1 screening.
Embryos were injected with a plasmid mixture containing 75 ng/μL of each gRNA plasmid and 100 ng/μL of donor plasmid in injection buffer (2 mM sodium phosphate, pH 6.8–7.8, 100 mM KCl), according to optimal concentrations defined in ref. 39. Injected G0 adults were back-crossed with nos.Cas9 flies, and G1 progeny was initially screened en masse to identify crosses that had produced G1 flies that underwent HDR events.
Screening was performed by isolating DNA from sets of ∼30 individuals per vial (mostly pupae, but also adults and third instar larvae, depending on availability), digesting ∼2 μg of total DNA with NcoI (to preferentially digest wild-type alleles but not the modified alleles that contain the donor sequence where NcoI is absent) and using ∼30 ng of the digested DNA as template for amplification either with “mutant-specific” primer pairs (ETXSF/ETXSM yielding a 247-bp product for I1056F or ETXSF/PxM yielding a 249-bp product for I1056M) or a “generic” (ETXSF/ETXSR) primer pair (Table S2) yielding a 395-bp product (Fig. S6). In case the generic primer pair was used, the product was further digested with NcoI and/or FokI for allele evaluation. For individual flies, screening was performed as described above but with template DNA derived from individual fly DNA preps (2–5 ng per reaction).
Life Table Parameters.
P. xylostella life tables.
Strains Sudlon and Sudlon-Tfm were placed in plastic cages in a climate chamber and kept under standard conditions at 25 ± 1 °C, relative humidity 50 ± 10%, and LD 16:8 for one generation to adapt to the conditions. To investigate postembryonic developmental stages, 50 adults of each strain were transferred to a cage, and females were allowed to lay eggs for 6 h; afterward the cabbage plants were removed, and egg development was monitored. Fifty newly hatched first instar larvae were transferred to a six-well plate containing a wetted filter paper and a cabbage leaf disk (2-cm diameter). Development was recorded every 6 h until pupation. Data were analyzed for significant differences between strains by Student’s t test.
Drosophila life tables.
To determine the developmental time from egg to pupa/adult, cages with 20 adult females and 20 adult males were covered with cherry-agar plates layered with yeast. The flies were left to adapt for several hours, and after plate replacement, they were left to lay eggs for ∼5 h. Fifty eggs were transferred in small vials with fly diet (five replicates for each line) and left to grow in standard conditions (25 ± 1 °C, relative humidity 70 ± 10%, LD 12:12). Pupation and adult eclosion timing and number of pupae/adults were monitored. For determination of fecundity, 20 crosses (one virgin female × one male) per line were set into vials covered with a cherry-agar plate. The number of eggs was counted, and the plate was replaced daily. Data were analyzed for significant differences between strains by Student’s t test.
SI Results
Insect genomes typically contain two genes for CHS with different developmental roles: CHS1 is only expressed at molting, whereas CHS2 is continuously expressed in midgut. To verify which of the two Drosophila candidate genes (kkv or CHS2) is more related to the Plutella CHS, the percentage of sequence similarity was calculated and it was found that the P. xylostella CHS1 is significantly more related to kkv (sequence identity 68.3%) than to Drosophila CHS-2 (37.6%). The T. urticae CHS1 shares 48.6% identity with kkv and 34% with Drosophila CHS2. Furthermore, a phylogenetic analysis of several arthropod CHSs (Fig. S4) clearly indicates that kkv is grouped together with P. xylostella and other insect CHS1 proteins in a clade with strong support, whereas CHS2 belongs to a different clade. Thus, kkv was considered the appropriate target for genome modification.
Acknowledgments
We thank Athanasia Zampouka (University of Crete) for her help with Drosophila bioassays and cloning, Dr. Maria Riga (University of Crete) for valuable discussions regarding Drosophila life table experiments, and Prof. Christos Delidakis (Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology and University of Crete) for providing fly stocks and very helpful advice. T.V.L. is supported by Fund for Scientific Research Flanders (FWO) Grants G009312N and G053815N. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618258113/-/DCSupplemental.
References
- 1.World Health Organization 2016 World Malaria Report, 2015 (WHO, Geneva, Switzerland). Available at www.who.int/mediacentre/factsheets/fs387/en/. Accessed October 31, 2016.
- 2.Food and Agriculture Organization of the United Nations 2010 How to Feed the World in 2050. Available at www.fao.org/fileadmin/templates/wsfs/docs/expert_paper/How_to_Feed_the_World_in_2050.pdf. Accessed October 31, 2016.
- 3.Talekar NS, Shelton AM. Biology, ecology and management of the diamondback moth. Annu Rev Entomol. 1993;38:275–301. doi: 10.1146/annurev-ento-010715-023622. [DOI] [PubMed] [Google Scholar]
- 4.Sparks TC, Nauen R. IRAC: Mode of action classification and insecticide resistance management. Pestic Biochem Physiol. 2015;121:122–128. doi: 10.1016/j.pestbp.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt S, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526(7572):207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merzendorfer H. Chitin synthesis inhibitors: Old molecules and new developments. Insect Sci. 2013;20(2):121–138. doi: 10.1111/j.1744-7917.2012.01535.x. [DOI] [PubMed] [Google Scholar]
- 7.Cohen E, Casida JE. Properties of Tribolium gut chitin synthetase. Pestic Biochem Physiol. 1980;13:121–128. [Google Scholar]
- 8.Zhang D, Miller MJ. Polyoxins and nikkomycins: Progress in synthetic and biological studies. Curr Pharm Des. 1999;5(2):73–99. [PubMed] [Google Scholar]
- 9.Ruiz-Herrera J, San-Blas G. Chitin synthesis as target for antifungal drugs. Curr Drug Targets Infect Disord. 2003;3(1):77–91. doi: 10.2174/1568005033342064. [DOI] [PubMed] [Google Scholar]
- 10.Sun R, Liu C, Zhang H, Wang Q. Benzoylurea chitin synthesis inhibitors. J Agric Food Chem. 2015;63(31):6847–6865. doi: 10.1021/acs.jafc.5b02460. [DOI] [PubMed] [Google Scholar]
- 11.Cohen E. 2010. Chitin biochemistry: Synthesis, hydrolysis and inhibition. Advances in Insect Physiology, eds Jérôme C, Stephen JS (Academic, London), Vol 38, pp 5–74.
- 12.Matsumura F. Studies on the action mechanism of benzoylurea insecticides to inhibit the process of chitin synthesis in insects: A review on the status of research activities in the past, the present and the future prospects. Pestic Biochem Physiol. 2010;97:133–139. [Google Scholar]
- 13.Akasaka T, et al. The ATP-sensitive potassium (KATP) channel-encoded dSUR gene is required for Drosophila heart function and is regulated by tinman. Proc Natl Acad Sci USA. 2006;103(32):11999–12004. doi: 10.1073/pnas.0603098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangishetti U, et al. Effects of benzoylphenylurea on chitin synthesis and orientation in the cuticle of the Drosophila larva. Eur J Cell Biol. 2009;88(3):167–180. doi: 10.1016/j.ejcb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Meyer F, Flötenmeyer M, Moussian B. The sulfonylurea receptor Sur is dispensable for chitin synthesis in Drosophila melanogaster embryos. Pest Manag Sci. 2013;69(10):1136–1140. doi: 10.1002/ps.3476. [DOI] [PubMed] [Google Scholar]
- 16.Uchida M, Asai T, Sugimoto T. Inhibition of cuticle deposition and chitin biosynthesis by a new insect growth regulator, buprofezin in Nilaparvata lugens Stål. Agric Biol Chem. 1985;49:1233–1234. [Google Scholar]
- 17.De Cock A, Degheele D. Effects of buprofezin on the ultrastructure of the third instar cuticle of the insect Trialeurodes vaporariorum. Tissue Cell. 1991;23(5):755–762. doi: 10.1016/0040-8166(91)90028-r. [DOI] [PubMed] [Google Scholar]
- 18.Nauen R, Smagghe G. Mode of action of etoxazole. Pest Manag Sci. 2006;62(5):379–382. doi: 10.1002/ps.1192. [DOI] [PubMed] [Google Scholar]
- 19.Van Leeuwen T, et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc Natl Acad Sci USA. 2012;109(12):4407–4412. doi: 10.1073/pnas.1200068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demaeght P, et al. High resolution genetic mapping uncovers chitin synthase-1 as the target-site of the structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae. Insect Biochem Mol Biol. 2014;51:52–61. doi: 10.1016/j.ibmb.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teixeira LA, Andaloro JT. Diamide insecticides: Global efforts to address insect resistance stewardship challenges. Pestic Biochem Physiol. 2013;106:76–78. [Google Scholar]
- 22.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troczka B, et al. Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem Mol Biol. 2012;42(11):873–880. doi: 10.1016/j.ibmb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Steinbach D, et al. Geographic spread, genetics and functional characteristics of ryanodine receptor based target-site resistance to diamide insecticides in diamondback moth, Plutella xylostella. Insect Biochem Mol Biol. 2015;63:14–22. doi: 10.1016/j.ibmb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Zimmer CT, et al. A CRISPR/Cas9 mediated point mutation in the alpha 6 subunit of the nicotinic acetylcholine receptor confers resistance to spinosad in Drosophila melanogaster. Insect Biochem Mol Biol. 2016;73:62–69. doi: 10.1016/j.ibmb.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somers J, Nguyen J, Lumb C, Batterham P, Perry T. In vivo functional analysis of the Drosophila melanogaster nicotinic acetylcholine receptor Dα6 using the insecticide spinosad. Insect Biochem Mol Biol. 2015;64:116–127. doi: 10.1016/j.ibmb.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Troczka BJ, et al. Stable expression and functional characterisation of the diamondback moth ryanodine receptor G4946E variant conferring resistance to diamide insecticides. Sci Rep. 2015;5:14680. doi: 10.1038/srep14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong K, et al. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem Mol Biol. 2014;50:1–17. doi: 10.1016/j.ibmb.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Reilly AO, et al. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem J. 2006;396(2):255–263. doi: 10.1042/BJ20051925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nauen R, et al. Development of a lateral flow test to detect metabolic resistance in Bemisia tabaci mediated by CYP6CM1, a cytochrome P450 with broad spectrum catalytic efficiency. Pestic Biochem Physiol. 2015;121:3–11. doi: 10.1016/j.pestbp.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 31.Riga M, et al. Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochem Mol Biol. 2015;65:91–99. doi: 10.1016/j.ibmb.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Heckel DG. Genomics in pure and applied entomology. Annu Rev Entomol. 2003;48:235–260. doi: 10.1146/annurev.ento.48.091801.112624. [DOI] [PubMed] [Google Scholar]
- 33.Maue L, Meissner D, Merzendorfer H. Purification of an active, oligomeric chitin synthase complex from the midgut of the tobacco hornworm. Insect Biochem Mol Biol. 2009;39(9):654–659. doi: 10.1016/j.ibmb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Hemingway J, et al. Averting a malaria disaster: Will insecticide resistance derail malaria control? Lancet. 2016;387(10029):1785–1788. doi: 10.1016/S0140-6736(15)00417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111(29):E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone BF. A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. Bull World Health Organ. 1968;38(2):325–326. [PMC free article] [PubMed] [Google Scholar]
- 37.Gratz SJ, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren X, et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Reports. 2014;9(3):1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]