Significance
In the vertebrate eye, a monolayer of cells, called the retinal pigment epithelium (RPE), is between the choroidal blood supply and the retina. The RPE provides metabolic support for the retina, including delivery of glucose and other nutrients. Here, we show that reductive carboxylation of α-ketoglutarate, a type of metabolism that supports growth and survival of cancer cells, is a prominent feature of RPE cells. We show that extreme oxidative stress can overwhelm the reductive carboxylation pathway. However, we also found that the RPE can be protected from extreme oxidative stress by supplementation with an NAD+ precursor or α-ketoglutarate.
Keywords: reductive carboxylation, RPE, metabolism, age-related macular degeneration, oxidative stress
Abstract
The retinal pigment epithelium (RPE) is a monolayer of pigmented cells that requires an active metabolism to maintain outer retinal homeostasis and compensate for oxidative stress. Using 13C metabolic flux analysis in human RPE cells, we found that RPE has an exceptionally high capacity for reductive carboxylation, a metabolic pathway that has recently garnered significant interest because of its role in cancer cell survival. The capacity for reductive carboxylation in RPE exceeds that of all other cells tested, including retina, neural tissue, glial cells, and a cancer cell line. Loss of reductive carboxylation disrupts redox balance and increases RPE sensitivity to oxidative damage, suggesting that deficiencies of reductive carboxylation may contribute to RPE cell death. Supporting reductive carboxylation by supplementation with an NAD+ precursor or its substrate α-ketoglutarate or treatment with a poly(ADP ribose) polymerase inhibitor protects reductive carboxylation and RPE viability from excessive oxidative stress. The ability of these treatments to rescue RPE could be the basis for an effective strategy to treat blinding diseases caused by RPE dysfunction.
The retinal pigment epithelium (RPE) is a monolayer of postmitotic cells situated between the photoreceptors of the retina and the choroidal blood supply. The interaction of the RPE and photoreceptors is critical to maintaining vision. Functions of the RPE include phagocytosis of shed photoreceptor outer segments, recycling of retinoids, production and secretion of cytokines and chemokines, and mediating the exchange of nutrients and metabolites between the choroid and photoreceptors (1, 2). RPE cells provide crucial metabolic support for the retina.
RPE dysfunction can lead to photoreceptor death and retinal degenerative disease, such as age-related macular degeneration (AMD), which is the leading cause of irreversible vision loss in the elderly human population (3–5). RPE cells are exposed to ongoing oxidative stress from the combined effects of light, choroidal O2, polyunsaturated fatty acids, and retinoids (6). The resulting impairment in RPE energy metabolism and function by oxidative stress is one likely mechanism for the pathogenesis of AMD (3, 7–11).
Mitochondria support the active energy metabolism of the RPE (10–12). A recent report showed that RPE is less stable and less able to support the retina when it is forced to rely on glycolytic rather than mitochondrial metabolism (13). Another recent report supports the importance of mitochondria in RPE by showing that bolstering mitochondrial activity makes these cells more resilient to oxidative damage (14).
In mitochondria, citrate can be generated from acetyl CoA and oxaloacetate as part of the TCA cycle. However, under hypoxic conditions, some cells also produce citrate via reductive carboxylation of α-ketoglutarate (αKG) through the action of NADPH-dependent isocitrate dehydrogenases (IDH) (15–17). Reductive carboxylation occurs in a small cohort of cells from liver, heart, brown adipocytes, and quiescent fibroblasts (18–20), where it supports redox homeostasis and synthesis of lipids, nucleotides, and urea (16, 18).
Reductive carboxylation of αKG to isocitrate by NADPH and CO2 can be catalyzed by IDH1 and IDH2, two of three isoforms of IDH. Overexpression of IDH1 or IDH2 can enhance protection from oxidative damage, whereas interference with their activities causes oxidation of glutathione and accumulation of reactive oxygen species (21–24). Loss of IDH1 and IDH2 activity also slows lipid synthesis and cell growth (25). IDH2 KO mice have dysfunctional mitochondria, poor redox homeostasis, and accelerated heart failure (26). Other studies have highlighted the importance of pyridine nucleotides, NAD+/H, and NADP+/H in reductive carboxylation (15, 16). Pyridine nucleotides have diverse metabolic roles in differentiation, survival, and protection from oxidative stress (27).
We report here that RPE cells have an exceptionally high capacity for reductive carboxylation. They acquire it with maturation and use it for viability, redox balance, mitochondrial function, and response to oxidative stress. Reductive carboxylation in RPE is compromised when oxidative stress becomes excessive. Remarkably, supplementing with either an NAD+ precursor or an inhibitor of poly(ADP ribose) polymerase (PARP) protects reductive carboxylation and RPE cell viability from the effects of oxidative stress.
Results
There Is an Unusually High Metabolic Flux Through Reductive Carboxylation in RPE.
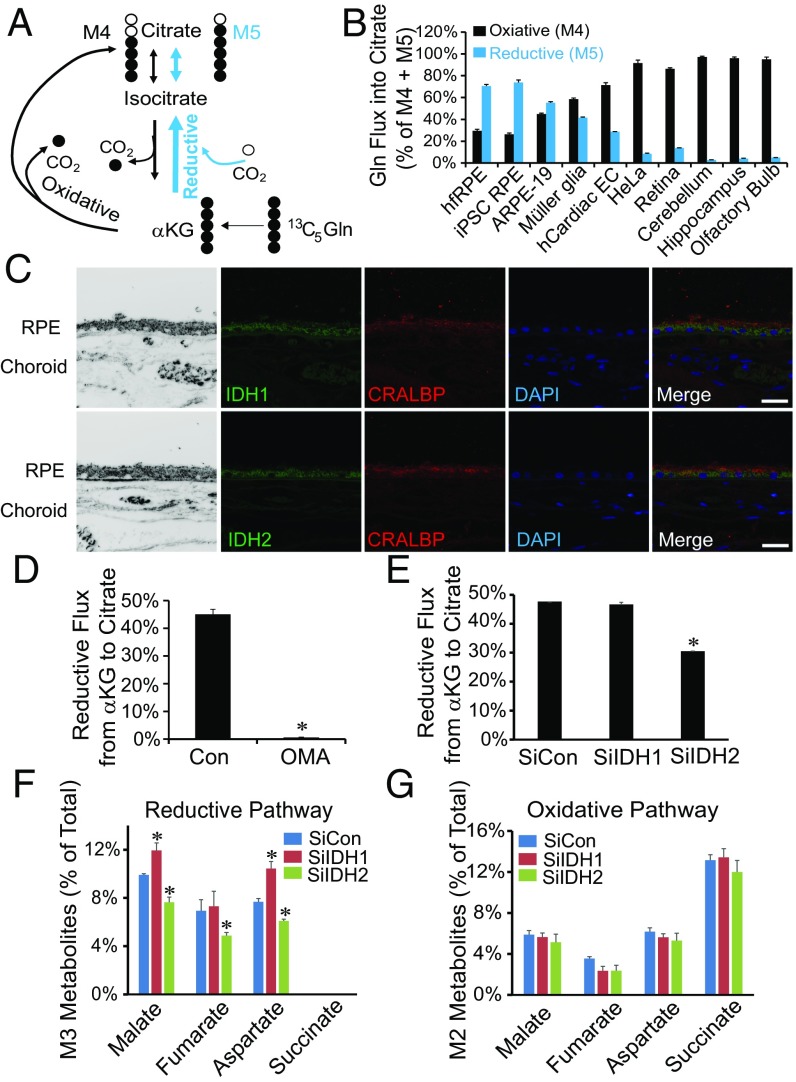
We quantified 13C-labeled metabolites from RPE cells that were fed U-13C glutamine for 1 h. The labeled glutamine can be made into M5 αKG, which then is either (i) oxidized in the TCA cycle (curved black line in Fig. 1A) into M4 intermediates (labeled with four 13C) or (ii) carboxylated (blue line in Fig. 1A) into M5 isocitrate and citrate (Fig. S1). M4 and M5 are the two most abundant species of labeled citrate produced in these experiments (Fig. S2A). We performed this analysis with human fetal retinal pigment epithelium (hfRPE) cells, human induced pluripotent stem cell (iPSC)-derived RPE cells, ARPE-19 cells (a human RPE cell line), cultured murine Müller glia, human cardiac endothelial cells (hCardiac ECs), HeLa cells (a cancer cell line), isolated mouse retina, and slices of murine neuronal tissues (cerebellum, hippocampus, and olfactory bulb). M4 is the major form of citrate made in the retina, other neuronal tissues, hCardiac ECs, and cancer cells (HeLa). In striking contrast, M5 is the major form in RPE (Fig. 1B).
Fig. 1.
Human RPE cells have an unusually high rate of reductive carboxylation from αKG to citrate through IDH2. (A) Schematic illustrating reductive carboxylation. U-13C glutamine produces either four carbon-labeled (M4) citrate through the classical oxidative pathway or five carbon-labeled (M5) citrate through reductive carboxylation of αKG. (B) Human fetal and control iPSC RPEs have much higher reductive (M5) and less oxidative (M4) activity compared with the other tissues or cell lines tested. Mean ± SD (n = 3). To emphasize the metabolic specialization, the data are shown as percentage of total M5 (reductive) and M4 (oxidative) incorporation of 13C into citrate from U-13C glutamine. A more detailed presentation of the data is provided in Table S1. (C) Expression of IDH1 and IDH2 in human adult RPE by immunofluorescence. Cellular retinaldehyde binding protein (CRALBP) is a marker of RPE. (Scale bar: 50 μm.) (D and E) Reductive flux of hfRPE cells is inhibited by either 5 mM OMA or siRNA silencing of IDH2 but not IDH1 (n = 3). Reductive flux was calculated as percentage of total flux from U-13C glutamine that went to M5 citrate (reductive). (F and G) Effect of IDH1 or IDH2 knockdown on M3 metabolites from the reductive pathway and M2 metabolites from the oxidative pathway. Mean ± SD (n = 3). *P < 0.05 vs. Con (control) or SiCon (nontarget siRNA control).
Fig. S1.
A schematic of metabolic intermediate labeling from 13C glutamine. 13C glutamine is either oxidized through the TCA cycle generating M4 or M2 carbons (black circles), or it undergoes reductive carboxylation into M5 citrate, producing M3 oxaloacetate (OAA), malate, aspartate, and fumarate (red circles).
Fig. S2.
Reductive carboxylation in human RPE is unusually active. (A) Isotopologue analysis of labeled citrate in hfRPE and mouse retina by GC-MS with U-13C glutamine for 1 h. (B and C) Rates of incorporation of 13C from U-13C glutamine or 1,2 13C-glucose into metabolites in hfRPE and mouse retina (mRetina). The total abundances of metabolites for hfRPE were citrate = 400 ± 54 pmol/μg protein and pyruvate = 86 ± 8 pmol/μg protein (SEM; n = 15), and the total abundances for retina were citrate = 134 ± 29 pmol/μg protein and pyruvate = 69 ± 7 pmol/μg protein (SEM; n = 10). ME, malic enzyme. (D) Human fetal and iPSC RPEs have much higher reductive flux than the other tissue or cells tested. Reductive flux is estimated as the fraction of citrate that is M5 divided by the fraction of αKG that is M5 after labeling with U-13C glutamine for 1 h. Data are mean ± SD (n = 4). We also compared the ratios of total picomoles of M5 citrate to total picomoles of M5 αKG in the steady state from 1 to 2 h for hfRPE [2.75 ± 0.64 (SD; n = 9)] and mRetina [1.17 ± 0.21 (SD; n = 4)]. (E) Human fetal RPE has more labeled M3 metabolites from 13C glutamine than murine retina (n = 4).
Table S1.
Reductive flux to citrate in different cell lines and tissues
| Cell line/tissue | Total CIT, μmol/μg protein | % CIT with 13C label (M1–M6) | Reductive path, % M5 CIT | Oxidative path, % M4 CIT |
| hfRPE | 17.5 ± 0.5 | 37.5 ± 5.3 | 17.7 ± 1.5 | 8.1 ± 1.4 |
| iPSC RPE | 27.2 ± 2.6 | 47.3 ± 4.5 | 25.7 ± 1.2 | 10.0 ± 1.1 |
| ARPE-19 | 20.0 ± 2.8 | 20.9 ± 0.0 | 9.9 ± 0.7 | 8.1 ± 0.6 |
| Müller glia | 20.0 ± 3.3 | 55.0 ± 0.9 | 5.1 ± 0.2 | 30.4 ± 0.9 |
| hCardiac EC | 12.3 ± 2.5 | 17.9 ± 0.8 | 4.5 ± 0.3 | 11.2 ± 0.3 |
| HeLa | 26.7 ± 0.6 | 65.4 ± 0.9 | 4.2 ± 0.4 | 45.2 ± 0.2 |
| Retina | 6.9 ± 0.9 | 27.6 ± 0.8 | 2.7 ± 0.2 | 17.1 ± 1.0 |
| Cerebellum | 7.6 ± 1.0 | 18.9 ± 0.6 | 0.3 ± 0.1 | 11.3 ± 0.7 |
| Hippocampus | 7.6 ± 0.8 | 29.0 ± 1.4 | 0.9 ± 0.3 | 20.5 ± 1.2 |
| Olfactory bulb | 5.1 ± 0.6 | 15.5 ± 2.1 | 0.5 ± 0.0 | 9.7 ± 1.4 |
CIT, citrate.
The rate at which labeled citrate accumulates (Fig. S2B) shows that reductive carboxylation is more active than oxidation in hfRPE, whereas oxidation is more active than reductive carboxylation in mouse retina. We also evaluated two other pathways that can generate cytosolic NADPH. Both malic enzyme activity (formation of M3 pyruvate from U-13C glutamine) (Fig. S2B) and pentose phosphate pathway (PPP) activity [formation of M1 pyruvate from 1,2 13C-glucose (28)] (Fig. S2C) are minor compared with reductive carboxylation in hfRPE cells.
Comparison of M5 citrate and M5 αKG in cells and tissues incubated with 13C-glutamine shows that reductive carboxylation is more active in RPE than in other cells and tissues (Fig. S2D). M3 malate, fumarate, and aspartate are made from M5 citrate produced by reductive carboxylation in the matrix and exported to the cytosol (Fig. S1). These indicators of reductive carboxylation are sixfold more abundant in RPE than in retina (Fig. S2E).
NADP-Dependent IDH2 Catalyzes Reductive Carboxylation in RPE Cells.
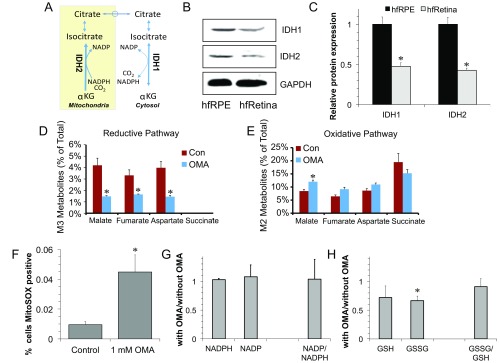
Reductive carboxylation can be catalyzed by the NADP-dependent enzymes IDH1 and IDH2, which are distributed in the cytosol and mitochondria, respectively (Fig. S3A). IDH1 and IDH2 are approximately twofold more abundant in hfRPE than in human fetal retina (Fig. S3 B and C). In adult human tissue, IDH1 and IDH2 are abundant in RPE but nearly undetectable in the choroid (Fig. 1C). Oxalomalate (OMA), a competitive inhibitor of NADP-dependent IDH (20), inhibits reductive carboxylation of αKG to citrate in hfRPE (Fig. 1D). M3 malate, M3 fumarate, and M3 aspartate are approximately threefold less abundant when OMA is present, and metabolites from the oxidative pathway increase slightly (Fig. S3 D and E). Inhibition of IDH activity by OMA increases the number of cells positive for MitoSOX red staining (Fig. S3F). However, OMA treatment has only minor effects on the steady-state levels of NADP+, NADPH, oxidized glutathione (GSSG), and glutathione (GSH) (Fig. S3 G and H). Interpretation of these findings is complicated by the absence of information about flux through the NADP(H) redox cycle and the distributions of pools of NADP+ and NADPH within the cell. Additional studies will be required to determine whether inhibition of IDH1 and IDH2 influences redox flux through NADP(H), whether it causes compensatory changes from other NADPH pathways, and whether it affects cytosolic and mitochondrial distributions of NADPH and reduced glutathione.
Fig. S3.
Expression of IDH1 and IDH2 and effects of OMA. (A) Schematic that shows reductive carboxylation mediated by cytosolic IDH1 and mitochondrial IDH2. (B) Immunoblot showing IDH1 and IDH2 protein expression in hfRPE cells and human fetal retina (hfRetina). (C) Quantification of B normalized to GAPDH. *P < 0.05 vs. hfRPE (n = 3). (D and E) Inhibition of reductive carboxylation OMA decreases labeled metabolites from 13C glutamine in the reductive pathway, and labeled metabolites were slightly increased in the oxidative pathway. Con, control. Data are mean ± SD (n = 3). (F) Quantification of the effect of OMA treatment on mitochondrial reactive oxygen species (ROS) formation based on the number of MitoSOX-positive hfRPE cells. (G) Quantification of the effects of OMA on NADPH and NADP levels. (H) Quantification of effect of OMA on oxidized and reduced forms of glutathione. *P < 0.05 between with vs. without OMA.
We used siRNAs to diminish expression of IDH1 or IDH2 in hfRPE cells (Fig. S4 A–C). Knocking down IDH2 restricts reductive flux to citrate (Fig. 1E), and it decreases formation of M3 malate, M3 fumarate, and M3 aspartate (Fig. 1F). There is no effect on the M2 isotopologues, indicating that IDH knockdown does not affect oxidative flux in the mitochondria (Fig. 1G). There is no effect of knocking down IDH1 on reductive flux from αKG to citrate (Fig. 1E). We suggest that this is because IDH1 does not catalyze the reductive reaction in the scheme in Fig. 2A and Fig. S1. Knocking down IDH1 makes more cytosolic M5 citrate available to form M3 malate and M3 aspartate (Fig. 1F and upper right of Fig. S1).
Fig. S4.
NADP-dependent IDH1 and IDH2 in hfRPE regulates NAD/NADH, mitochondrial bioenergetics, and fatty acid synthesis. (A–C) hfRPE cells were transfected with siGENOME human IDH1 and IDH2 siRNA on the initial and third day after plating. Cells transfected with nontarget siRNA were used as controls. The silencing efficiency was (A) determined by Western blot analysis of IDH1 and IDH2 expression and (B and C) quantified. Data are mean ± SD (n = 3). *P < 0.05 vs. SiCon (siRNA control). (D) Knockdown of IDH1 or IDH2 increases NADH in hfRPE. (Data are normalized to the control value in D–F.) (E) Inhibition of NADP-dependent IDHs by 5 mM OMA increases NAD/NADH (n = 3). Con, control. (F) Knockdown of either IDH1 or IDH2 increases the ADP/ATP ratio relative to control (n = 3). (G) Enrichment of palmitate after knockdown of IDH1 or IDH2 in hfRPE cells (n = 3). Mean ± SD (n = 3). *P < 0.05 vs. Con or SiCon (nontarget siRNA control).
Fig. 2.
NADP-dependent IDH activity influences NAD/NADH, mitochondrial bioenergetics, glycolysis, and lipid synthesis in hfRPE. (A) Schematic illustrating NAD metabolism by NADP-dependent IDH (blue arrows) in mitochondria. Production of NAD+ (red) comes from reductive carboxylation in mitochondria and separately in the cytosol. (B and C) The effect of IDH1 or IDH2 knockdown on NAD+ and NAD+/NADH levels relative to control (n = 3). (D) Knockdown of both IDH1 and IDH2 increases the AMP/ATP ratio. (E) Schematic illustration of U-13C glucose metabolism. Black circles represent the labeled carbons. (F and G) Knockdown of IDH2 inhibits glucose metabolism in hfRPE. Decreased levels of M3 pyruvate and M2 citrate were determined by isotopologue analysis after incubating hfRPE with 5 mM U-13C glucose for 15 min. (H) Schematic illustrating the role of IDH in palmitate synthesis from U-13C glutamine. Black circles represent the labeled carbons. ACL, ATP citrate lyase. (I) The effect of IDH1 or IDH2 knockdown on the relative abundance of palmitate isotopologues in hfRPE cells (arbitrary units) derived from U-13C glutamine incubated with hfRPE for 48 h. (J and K) Knockdown of IDH1 increases labeled malate and aspartate from U-13C glutamine, whereas IDH2 knockdown decreases labeled malate and aspartate. M3 and M1 isotopologues include carbons that complete more than one TCA cycle. Mean ± SD (n = 3). *P < 0.05 vs. SiCon (nontarget siRNA control).
Reductive Carboxylation Contributes to Redox Homeostasis in RPE.
Loss of redox homeostasis can perturb mitochondrial bioenergetics and glucose metabolism. Together with nicotinamide nucleotide transhydrogenase (NNT), IDH2 helps convert mitochondrial NADH into NAD+ (29) (Fig. 2A). Knocking down IDH2 increases [NADH] and decreases [NAD+], whereas knocking down IDH1 only increases [NADH] (Fig. 2 B and C and Fig. S4D). Inhibiting both IDH1 and IDH2 by OMA substantially lowers the NAD+/NADH ratio (Fig. S4E). Consistent with the lowering of NAD+/NADH, we found that inhibiting IDH increases AMP/ATP and ADP/ATP (Fig. 2D and Fig. S4F). To examine whether loss of IDH activity influences glycolysis, we treated RPE cells with U-13C glucose for 15 min and quantified incorporation of 13C into pyruvate and citrate (Fig. 2E). Knockdown of IDH2 but not IDH1 causes a small but significant inhibition of M3 pyruvate and M2 citrate formation (Fig. 2 F and G). This result is consistent with inhibition of glycolysis caused by the diminished NAD+/NADH ratio. These findings show that IDH1 and IDH2 influence RPE redox metabolism. A more detailed interpretation of the effects of IDH suppression on redox metabolism is complicated by the absence of information about the distributions of the metabolites in mitochondrial vs. cytosolic pools.
Reductive Carboxylation Contributes to Fatty Acid Synthesis in RPE.
Fatty acids are made from acetyl-CoA produced by ATP citrate lyase (Fig. 2H). We incubated RPE cells with 2 mM U-13C glutamine in DMEM with 1% FBS for 48 h and then, quantified 13C in palmitate. The RPE culture medium was replaced with DMEM, because standard RPE culture medium contains unlabeled glutamate and aspartate. Cells carboxylate acetyl-CoA into malonyl-CoA, which then contributes two carbons to each elongation cycle of fatty acid synthesis. Knockdown of IDH2 inhibits M2, M4, and M6 palmitate formation, whereas knockdown of IDH1 increases M2 palmitate (Fig. 2I and Fig. S4G). Inhibition of IDH1 causes accumulation of cytosolic citrate, which is broken down by citrate lyase into acetyl CoA and oxaloacetate. Knockdown of IDH1 also causes malate and aspartate to accumulate (Fig. 2 J and K). These findings support the model shown in Fig. 2H.
Reductive Carboxylation Increases as RPE Cells Mature.
When RPE cells mature and differentiate, they shift to mitochondrial oxidative metabolism to support their specialized functions (30). We analyzed reductive carboxylation in hfRPE cells at different stages of maturity in culture. RPE cells were either plated at varying densities and cultured for 1 wk or plated at a fixed density and cultured for up to 8 wk. Mature RPE cells have hexagonal morphology, tight junctions, and melanin pigment granules. These characteristics start to appear in hfRPE cells seeded at 169,000 in a 35-mm dish after 1 wk in culture, although they are most pronounced when the cells are seeded at a density of 327,000 (Fig. S5 A–F). Similar hexagonal morphology and pigmentation appear in hfRPE cells seeded at 100,000 and cultured for 4 wk and become more pronounced after 8 wk in culture (Fig. S5 G–J). Along with increasing cellular maturity, reductive flux from αKG to citrate increased by approximately twofold (Fig. S5 K and L) as did the M3 intermediates from the reductive pathway (Fig. S4M). M2 metabolites from the oxidative pathway did not increase, except at the very highest cell density (Fig. S4N).
Fig. S5.
Reductive carboxylation increases with RPE cell maturity. Bright-field images of hfRPE either (A–F) plated at varying cell densities (k indicates thousands of cells per 35 mm) and cultured for 1 wk or (G–J) plated at 100,000 per 35 mm and cultured for 1–8 wk (W1–W8). RPE cells with higher initial plating densities or ≥4 wk in culture show greater hexagonal morphology and pigmentation, consistent with increased RPE maturity. (K and L) Reductive flux increases with RPE cell plating density and time in culture. Mean ± SD (n = 3). *P < 0.05 vs. cells at 28,000 or cells at W1. (M and N) Levels of intermediates derived from reductive and oxidative pathways with increasing RPE plating density. Reductive flux increases with an increased RPE maturity, whereas oxidative flux is not affected, except at the highest plating density. *P < 0.05 vs. 28,000 cells (n = 3).
Reductive Carboxylation in RPE Is Compromised by Excessive Oxidative Stress.
An important feature of RPE cells is their ability to withstand oxidative damage. A widely used method to inflict oxidative damage on RPE cells is exposure to millimolar concentrations of H2O2. Although such a high level of stress would not occur in a normal physiological setting, H2O2 treatment provides a controllable and reproducible technique for an initial assessment of the biochemical response of RPE metabolism to oxidative damage.
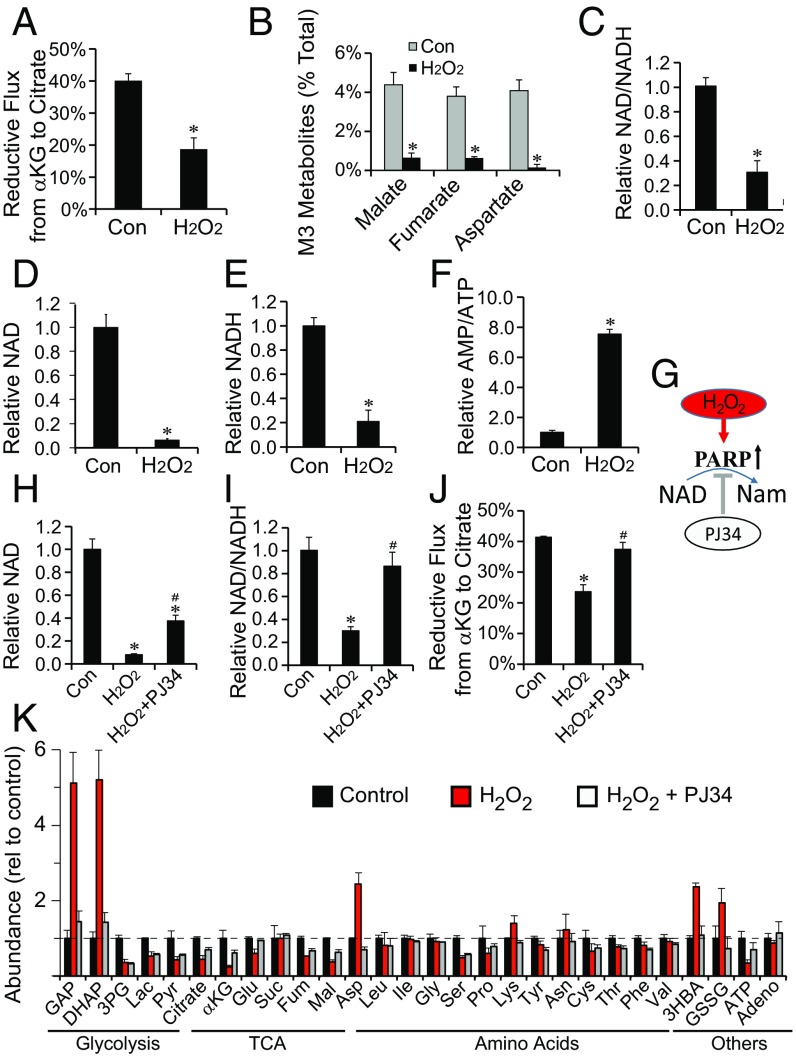
H2O2 does not affect reductive carboxylation at concentrations up to 0.25 mM. At 1 mM, it does not kill the cells, but it inhibits more than 50% of reductive flux (Fig. 3A), and it blocks formation of M3 intermediates (Fig. 3B). It also substantially decreases the NAD+/NADH ratio (Fig. 3C) and the total NAD+ and NADH levels (Fig. 3 D and E). NAD+ is required for glycolysis and mitochondrial bioenergetics, and therefore, flux through glycolysis decreases by ∼70% (Fig. S6A), and the AMP/ATP ratio increases approximately fivefold after H2O2 exposure (Fig. 3F). H2O2 increases flux through the PPP by 15-fold, but even under these conditions, the total flux through the PPP is only ∼10% of the flux through glycolysis (Fig. S6A).
Fig. 3.
Oxidative stress inhibits reductive carboxylation in hfRPE. (A) Reductive flux is significantly diminished in hfRPE exposed to 1 mM H2O2. hfRPE were labeled for 1 h with U-13C glutamine. (B) M3 metabolites derived from the reductive pathway are diminished by H2O2 exposure. *P < 0.05 vs. control (n = 3). (C–F) H2O2 treatment decreases NAD+/NADH, NAD+, and NADH, and it increases AMP/ATP in hfRPE. (G) A schematic illustrating how H2O2 stimulation of PARP activity leads to the depletion of NAD. PJ34 is a potent PARP inhibitor. (H–J) PJ34 (10 μM) counteracts the effects of H2O2 on NAD+ and restores NAD+/NADH and reductive flux. (K) Quantification of metabolites in hfRPE after exposure to H2O2 reveals significant changes in the abundance of glycolytic intermediates and amino acids. PJ34 counteracts the effects of H2O2. Mean ± SD (n = 3). Adeno, adenosine; Nam, nicotinamide. *P < 0.05 vs. Con (control); #P < 0.05 vs. H2O2.
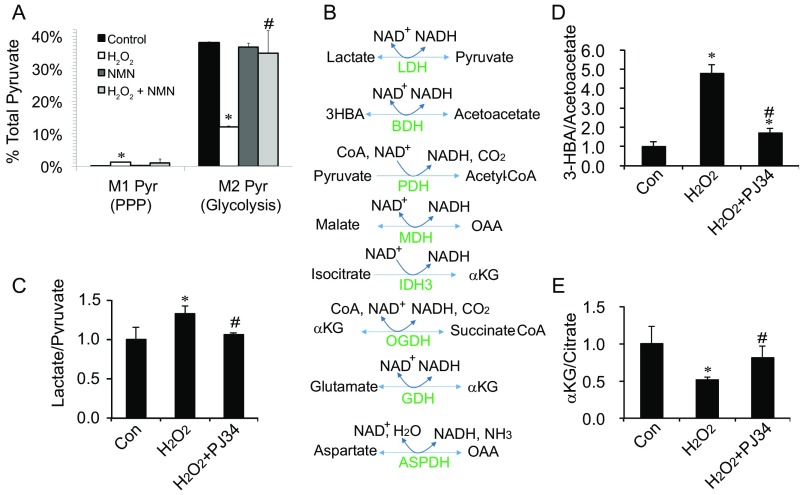
Fig. S6.
NMN and PARP inhibition can counteract metabolic deficiencies caused by H2O2. (A) 1 mM H2O2 inhibits glycolysis and stimulates PPP activity. hfRPE cells were incubated with 1,2 13C glucose with vs. without 1 mM H2O2 and with vs. without 5 mM NMN. (B) NAD+-dependent dehydrogenases involved in glucose metabolism. ASPDH, aspartate dehydrogenase; BDH, 3HBA dehydrogenase; GDH, glutamate dehydrogenase; MDH, malate dehydrogenase; OGDH, αKG dehydrogenase; PDH, pyruvate dehydrogenase. (C–E) PJ34 (10 μM) normalized the ratios of lactate/pyruvate, 3HBA/acetoacetate, and αKG/citrate after treatment with H2O2 for 1 h. Data are presented as mean ± SD (n = 3). *P < 0.05 vs. Con (control); #P < 0.05 vs. H2O2.
Oxidative Stress Depletes NAD+ and Adversely Affects RPE Survival.
H2O2 causes DNA damage (31), which stimulates PARP in RPE cells (32). Excessive PARP activity can deplete NAD+ by using it as a substrate to synthesize poly(ADP ribose) (PAR) (Fig. 3G) (33, 34). To determine if PARP is responsible for the effect of H2O2 on NAD+ in RPE cells, we treated them with 10 µM PJ34, a potent cell-permeable PARP inhibitor (EC50 = 20 nM). PJ34 restored NAD+ to five times its level in H2O2-treated RPE cells, and it restored NAD+/NADH and reductive carboxylation (Fig. 3 H–J). These findings show that PARP-mediated depletion of NAD+ can inhibit reductive carboxylation in RPE cells. We also quantified metabolites from hfRPE cells treated with H2O2 either with or without PJ34. H2O2 causes massive accumulation of glyceraldehyde 3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP), and it increases levels of aspartate, β-hydroxybutyrate (3HBA), and GSSG. Remarkably, PJ34 completely blocks these effects (Fig. 3K).
GAP and DHAP accumulate, because GAPDH requires NAD+. Depletion of NAD+ by PARP also might affect other NAD+-dependent dehydrogenases (Fig. S6B). PJ34 counteracts the effect of H2O2 on lactate/pyruvate and 3HBA/acetoacetate ratios (Fig. S6 C and D). It also counteracts the effect of H2O2 on αKG/citrate (Fig. S6E), which influences reductive flux in cancer cells (35).
We treated hfRPE with 1 mM H2O2 in the absence or presence of PJ34 and monitored cell death by lactate dehydrogenase (LDH) in the culture medium (Fig. S7A) and by cell density (Fig. S7 B–E). Cells exposed to H2O2 died within 48 h, but concurrent treatment with 100 nM PJ34 completely blocked H2O2-induced cell death. This finding indicates that RPE cell death from H2O2 is caused by depletion of NAD+ by PARP.
Fig. S7.
PARP inhibition blocks H2O2-induced hfRPE cell death. (A) LDH release into the medium at 24, 48, and 72 h in hfRPE cells with or without 1 mM H2O2 in the presence or absence of the PARP inhibitor, PJ34, at either 10 or 100 nM. Data are mean ± SD (n = 3). *P < 0.05 vs. Con (control) without treatment; #P < 0.05 vs. H2O2. (B–E) Bright-field images of hfRPE cells at the 72-h time point from A.
Oxidative Damage in RPE Is Enhanced When Reductive Carboxylation Is Inhibited.
Because reductive carboxylation can produce NADPH needed to reduce glutathione, we asked whether impairment of the reductive pathway makes RPE cells more vulnerable to oxidative stress. We cultured hfRPE cells for 10 d with or without knockdown of IDH1 and IDH2. We then treated them with 1 mM H2O2 for 24 h and stained them with ethidium homodimer to quantify cell death (Fig. S8 A–F). IDH2 siRNA alone induces cell death threefold more than control siRNA. H2O2 alone increases cell death 65-fold (Fig. S8G). H2O2 with IDH1 knockdown did not cause additional death, but together with IDH2 knockdown, it increased death 95-fold. The findings suggest that IDH2 can help to protect RPE from oxidative stress.
Fig. S8.
Inhibition of reductive carboxylation enhances the toxic effects of H2O2, and supplementation with αKG suppresses them. (A–F) Knockdown of IDH2 but not IDH1 increases hfRPE cell death as assessed by ethidium homodimer (EthD) staining of dead cells. SiCon, siRNA control. (G) Quantification of dead cells per field from A–F. *P < 0.05 vs. SiCon (nontarget siRNA control); #P < 0.05 vs. SiCon (nontarget siRNA control) treated with H2O2. (H) dmαKG, a methyl-esterified, cell-permeable form of αKG, fully suppresses cell death caused by H2O2 as assessed by EthD staining. (I) dmαKG fully suppresses cell death caused by H2O2 as assessed by EthD staining (n = 4). *P < 0.05 vs. control; #P < 0.05 vs. H2O2 alone.
αKG Protects RPE from Oxidative Stress.
We reasoned that enhancing reductive carboxylation by providing excess αKG as a substrate for IDH2 could enhance resistance of RPE cells to H2O2. Fig. S8 H and I shows that 1 mM dimethyl α-ketoglutarate (dmαKG) protects hfRPE cells completely from the damaging effects of 1 mM H2O2.
An NAD+ Precursor Protects RPE from Oxidative Stress.
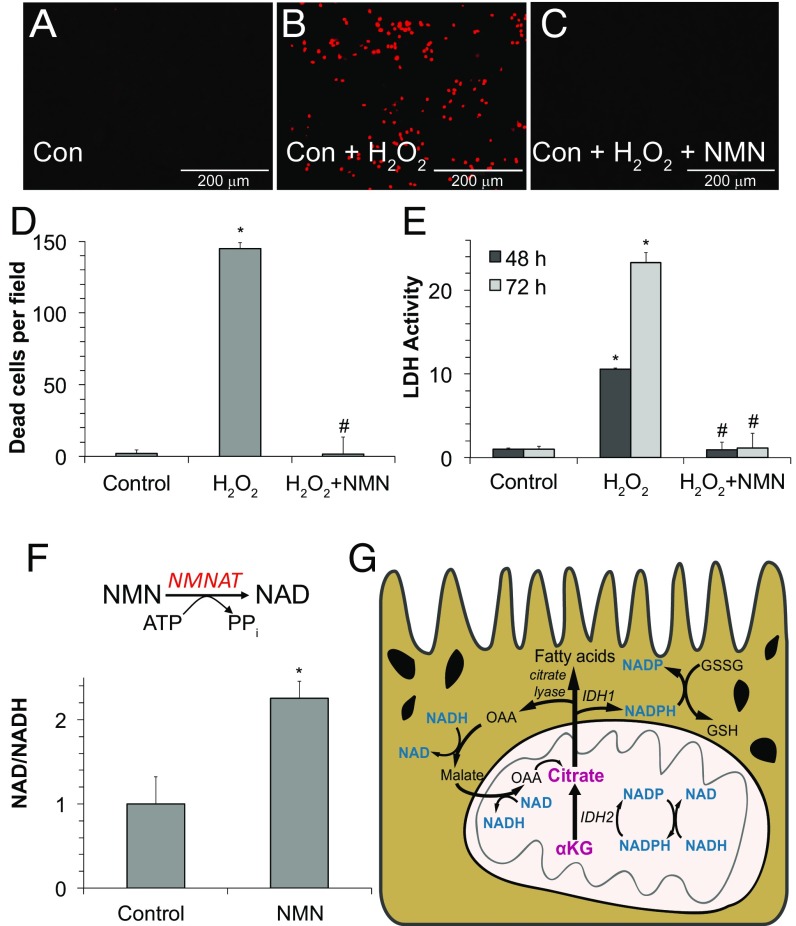
Blocking PARP activity restores NAD+, and it overrides the effect of H2O2 on reductive carboxylation (Fig. 3). Another way to restore NAD+ is to supplement the culture medium with nicotinamide mononucleotide (NMN), an NAD+ precursor. Remarkably, supplementation with NMN completely prevents RPE cell death induced by H2O2 (Fig. 4 A–E), and it doubles the ratio of NAD+/NADH (Fig. 4F). NMN also counteracts the effect of H2O2 on glycolysis (Fig. S6A). However, it does not counteract the stimulatory effect of H2O2 on the low level of PPP activity present in RPE (Fig. S6A). Taken altogether, our findings indicate that IDH2-mediated reductive carboxylation is a pathway enriched in RPE cells that might enhance their resistance to oxidative stress (Fig. 4G).
Fig. 4.
NMN protects RPE cells from oxidative damage. (A–C) iPSC RPE cells cultured for 8 wk were treated with 1 mM H2O2 in either the presence or absence (Con, control) of 1 mM NMN for 48 h. Cell death was quantified by fluorescence imaging of ethidium homodimer (EthD) staining. (D) Quantification of EthD staining from A–C. (E) LDH activity released into the medium under the same conditions as in experiments in A–C. (F) NMN, which is a precursor to NAD+, increases cellular NAD+/NADH in hfRPE. NMNAT, nicotinamide mononucleotide adenylyltransferase. All data are mean ± SD (n = 3). *P < 0.05 vs. control; #P < 0.05 vs. H2O2. (G) Model for the role of reductive carboxylation in RPE. IDH2 converts αKG into isocitrate and then, citrate by reductive carboxylation. Citrate exits the mitochondria into the cytoplasm to make either cytosolic NADPH (via IDH1) or acetyl CoA (via citrate lyase) for fatty acid synthesis. The pyridine nucleotides are highlighted in blue to illustrate some of the vital reactions in RPE metabolism that would be compromised by depletion of NAD+.
Discussion
We report here that reductive carboxylation plays a major role in the energy metabolism of RPE cells. Reducing power stored in NADH in the mitochondrial matrix can be transferred to citrate by NNT and IDH2 (29). Citrate produced in the matrix can be transported to the cytoplasm, where IDH1 can use it to reduce cytosolic NADP+ to NADPH (Fig. 4G). The RPE can use cytosolic NADPH to protect itself from oxidative stress. A direct test of this role for reductive carboxylation in RPE cells will require quantification of redox flux through NADP(H) and evaluation of the intracellular distributions of pyridine nucleotides. We favor the idea of the IDH2-mediated pathway shown in Fig. 4G, because knockdown of IDH2 inhibits incorporation of 13C from glutamine into fatty acids. However, additional studies will be required to evaluate the contribution from an IDH1-mediated pathway (16) that can generate mitochondrial NADPH, such as occurs in tumor cell spheroids (17) and pyruvate dehydrogenase-deficient cells (36).
The high level of activity of reductive carboxylation in RPE may help to explain the importance of mitochondria in supporting RPE function and enhancing protection from oxidative stress (13, 14). The RPE provides glucose to the retina by transporting it from the choroid, and therefore, this process would be most efficient if the RPE consumes as little glucose for itself as possible. We suggest that reductive carboxylation is a way for RPE cells to generate NADPH without consuming glucose (Fig. 4G). Consistent with this suggestion, other pathways that produce cytosolic NADPH, the PPP and malic enzyme, appear to be less active than reductive carboxylation in RPE cells.
RPE cannot survive long-term deficiency of NAD+ (37, 38), and a mutation in nicotinamide nucleotide adenylyltransferase 1 (NMNAT1), the enzyme that converts NMN into NAD+, causes severe retinal degeneration in humans (39–41). We have shown that, when oxidative stress is excessive, PARP depletes NAD+. This depletion blocks glycolysis and reductive carboxylation, because they require pyridine nucleotides.
An important implication of our findings is that failure of reductive carboxylation in the RPE could be a key factor in the pathogenesis of RPE-related diseases, such as AMD. Dysfunction of RPE mitochondria and increasing susceptibility to oxidative stress with age are linked to AMD (8, 11). Oral antioxidant supplements have been shown to have only modest effects on AMD disease progression (42). In our studies, NMN, PJ34, and dmαKG restore reductive carboxylation and protect RPE cells from oxidative damage very effectively, and therefore, they may represent promising therapeutic targets for AMD.
Our findings about RPE metabolism suggest a hypothesis to explain RPE dysfunction in diseases, like AMD (8). DNA damage in RPE mitochondria accumulates with age (8), and mismatches between respiratory complex subunits encoded by the mitochondrial and nuclear genomes can increase oxidative stress (43). Single-strand breaks in DNA can stimulate PARP to make PAR (44), and PARP activity is required for DNA repair in RPE cells (32). The negatively charged PAR facilitates DNA repair by loosening the structure of chromatin (45). NAD+ is consumed by PARP, but NAD+ also is a required substrate for Sirt1, a nuclear enzyme that deacetylates histones (46). Sirt1 activity slows when NAD+ is depleted by PARP (47), leaving histones more acetylated, which loosens chromatin structure even further.
Loosening of chromatin by accumulation of PAR and acetylated histones is important for DNA repair, but it also can allow genes that normally are inaccessible within heterochromatin to become more generally accessible to polymerases and transcription factors (48). We suggest that photoreceptors, specialized for aerobic glycolysis (49, 50), become less glycolytic, whereas RPE cells that rely on mitochondria for reductive carboxylation develop a more conventional metabolism. Loss of the unique and complementary metabolic features of these tissues may compromise the essential symbiotic relationship between the RPE and the retina. An attractive feature of this hypothesis is that each of its predictions will be testable with currently available technology.
Inhibiting PARP rescues RPE from oxidative stress. Inhibiting PARP also blocks DNA repair (32), and therefore, it is unlikely that PARP inhibitors could be an effective long-term therapy. However, we found that supplementation with an NAD+ precursor also is a very effective way to protect RPE cells from oxidative stress. This approach to therapy is likely to be more successful than PARP inhibition, because it can restore glycolysis, reductive carboxylation, and sirtuin activity without compromising DNA repair.
NAD+ declines with age (47, 51), and supplementation with NAD+ precursors can delay aging, liver disease, and vascular dysfunction (52–54). NAD+ precursors, such as nicotinamide riboside, can be used safely in humans (55, 56). In the absence of animal models for AMD, our findings highlight the need for clinical trials that evaluate NAD+ precursors for their ability to prevent and treat AMD.
Our study focused on identification and quantification of reductive carboxylation in RPE. Follow-up studies on RPE metabolism should investigate relative contributions of reductive carboxylation, malic enzyme activity, and the PPP to NADPH production. It also is important to determine how these pathways are regulated and how they are influenced by stress, aging, and disease states.
In summary, we found that reductive carboxylation is a prominent feature of healthy RPE cells. This metabolic pathway enhances the ability of RPE to maintain its cellular redox potential, and it contributes to lipid synthesis and possibly, resistance to oxidative stress. We found that excessive oxidative stress depletes NAD+ in RPE cells, and supplementation with NAD+ precursors restores pyridine nucleotides and rescues RPE from cell death.
Materials and Methods
Cell Culture.
Dissections of fetal tissue (16–18 wk gestation) to isolate RPE were performed following a previously published protocol (57). Use of fetal RPE cells as described did not qualify as human subjects research, as determined by the University of Washington Institutional Review Board. Isolated fetal RPE formed a confluent, pigmented monolayer of hexagonal cells after ∼4 wk in culture. ARPE-19 cells were cultured in the RPE medium. hCardiac ECs were obtained from Lonza and cultured according to the manufacturer’s protocol. Müller glial cells were isolated from mice as reported (58) and cultured in Neurobasal medium with 10% (vol/vol) FBS.
Stable Isotope-Labeled Metabolite Analysis.
RPE cells and all other cells or tissues (Fig. 1B) were changed into Krebs–Ringer Bicarbonate medium (59) with 5 mM glucose and 2 mM U-13C glutamine for 1 h or 5 mM U-13C glucose for 15 min. The decrease in glucose concentration in the media during the course of those experiments was negligible. For fatty acid analysis, RPE cells were labeled with 2 mM U-13C glutamine in DMEM with 1% FBS for 48 h. RPE cells were homogenized, and the metabolites were extracted, dried, derived with N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (TBDMS), and analyzed by GC-MS (Agilent 7890/5975C) as described in detail (60–62).
Additional details and other materials and methods are in SI Materials and Methods. Research involving human subjects described in SI Materials and Methods was approved by the University of Washington Institutional Review Board, and informed consent was obtained.
SI Materials and Methods
Reagents.
OMA was obtained from Cayman Chemical; uniformly labeled 13C glucose was from Cambridge Isotope Laboratories, Inc. 13C glutamine and reagents were purchased from Sigma-Aldrich unless otherwise specified.
Cell Culture.
Human fetal eyes, with a typical gestational age of 16–20 wk, were harvested and shipped overnight on ice in RPMI media containing antibiotics from Advanced Bioscience Resources Inc. Dissections of fetal tissue to isolate RPE were performed within 24 h of procurement following a previously published protocol (57). The resulting fetal RPE sheets were incubated at 37 °C with 5% (50,000 ppm by volume) CO2 and cultured in an RPE medium [MEMα; Gibco supplemented with 5% (vol/vol) FBS; N1 Medium Supplement (Sigma-Aldrich); a stock solution of hydrocortisone, triiodothyronine, and taurine; nonessential amino acids (NEAAs); and a penicillin-streptomycin solution]. Isolated fetal RPE formed a confluent, pigmented monolayer of hexagonal cells after about 3–4 wk in culture, was passaged using a 0.25% trypsin-EDTA solution (Gibco), and was passed through a 40-µm nylon cell strainer (BD Falcon) to collect a suspension of single cells. The collected fetal RPE cells were plated onto 35-mm tissue culture plates (Corning) precoated with Growth Factor Reduced (GFR) Matrigel Matrix (Corning) with the addition of 10 µM Y-27632 dihydrochloride (Tocris Bioscience). When cells reached confluency, typically in 1–2 wk, RPE cells were cultured in new RPE medium without Y-27632 and decreased FBS concentration (1%). ARPE-19 cells were cultured in the RPE medium. hCardiac ECs were obtained from Lonza and cultured according to the manufacturer’s protocol. Müller glia cells were isolated from mice as reported (58) and cultured in Neurobasal medium with 10% (vol/vol) FBS. All cell lines were periodically tested for Mycoplasma contamination, and they were negative.
iPSC Generation and Differentiation into RPE.
Peripheral blood mononuclear cells (PBMCs) were obtained from normal control participants and made into iPSCs in a University of Washington Institutional Review Board-approved protocol (HSD 43143). Informed consent was obtained from all participants before inclusion in the study, and all experiments were conducted according to the principles expressed in the Declaration of Helsinki. PBMCs were separated from fresh blood samples using a Ficoll-Hypaque (GE Healthcare) gradient. Erythroblasts were generated by expanding 2 million PBMCs in culture for 9 d in a QBSF-60 Serum-Free Medium (Fisher Scientific) supplemented with stem cell factor (50 ng/mL), IL-3 (10 ng/mL), erythropoietin (2 U/mL), insulin-like growth factor 1 (IGF1; 40 ng/mL), dexamethasone (1 μM), ascorbic acid (50 ng/mL), and a penicillin-streptomycin solution. Episomal vectors encoding the reprogramming factors OCT4, SOX2, KLF4, LIN28A, LMYC, and an shRNA targeting p53 were obtained from Addgene (catalog nos. 27077, 27078, 27080, and 27082). The episomal vectors were introduced into the in vitro expanded erythroblasts using the Nucleofector II device (Kit V, Program T-019; Lonza). After 48–72 h, transfected erythroblasts were plated onto a layer of irradiated mouse embryonic fibroblasts (iMEFs) in a reprogramming medium. Approximately 12 d after nucleofection, colonies with the morphological characteristics of human ES cells (hESCs) began to emerge, and the medium was then switched to hESC medium [DMEM:F12 1:1, Knockout Serum Replacement (Life Technologies), NEAA, l-glutamine, sodium pyruvate, sodium bicarbonate, β-mercaptoethanol, basic fibroblast growth factor (bFGF) (10 ng/mL), penicillin-streptomycin solution]. After colonies with ES cell-like morphology had grown to a sufficient size, they were manually transferred to separate iMEF-coated culture dishes for expansion in hESC medium. Eventually, iPSC cultures were moved to feeder-free plates and grown in E8 medium (Life Technologies).
RPE Differentiation and Culture.
iPSC-derived RPE was differentiated as reported (63). Briefly, human iPSCs cultured in Essential 8 Media (Gibco) on GFR Matrigel Matrix (Corning) were passaged using Dispase (Gibco) and plated onto GFR Matrigel-coated six-well plates in DMEM F12 basal media containing N2 Medium Supplement (Gibco), sodium pyruvate (Gibco), a 10% (vol/vol) BSA solution (Sigma-Aldrich), NEAA, Hepes (Gibco), sodium bicarbonate, and a penicillin-streptomycin solution and supplemented with 10 µM Y-27632, 50 ng/mL Recombinant Human Noggin (R&D Systems), 10 ng/mL Recombinant Human DKK-1 (R&D Systems), 10 ng/mL Recombinant Human IGF1 (Sigma-Aldrich), and 10 mM nicotinamide (Sigma-Aldrich). On day 1 after passage, Y-27632 was excluded from the medium; on day 3, the media were changed to the N2-supplemented media described previously containing 10 ng/mL Recombinant Human Noggin, DKK, IGF 5 ng/mL Basic Fibroblast Growth Factor (Invitrogen), and 10 mM nicotinamide. On day 5, N2-supplemented media containing 10 ng/mL DKK and IGF and 100 ng/mL Recombinant Human/Murine/Rat Activin A (Peprotech) were added to the cell cultures. From day 7 to day 14, cells were treated with N2-supplemented media containing 100 ng/mL Activin, 10 µM SU 5402 (Tocris Bioscience), and 1 µM Vasoactive Intestinal Peptide (Sigma-Aldrich). From day 14 onward, cells were cultured in N2-supplemented media until RPE began to appear (∼4 wk). RPE cells were either manually picked or trypsinized depending on the quality of differentiation. The RPE was then plated on Matrigel Matrix and cultured as described previously in an RPE medium containing 5% (vol/vol) FBS and 10 µM Y-27632. After reaching confluency, the FBS concentration in the media was decreased to 1%, and Y-27632 was no longer added to the media.
Stable Isotope-Labeled Metabolite Analysis.
RPE cells and all other cells or tissue were changed into Krebs–Ringer Bicarbonate medium (59) with 5 mM glucose and 2 mM U-13C glutamine or 5 mM U-13C glucose with or without OMA for varying times. The cells were then quickly rinsed with ice-cold 0.9% NaCl and scraped with cold 80% (vol/vol) methanol in water. RPE cells were homogenized, and the metabolites were extracted, dried, derived with TBDMS, and analyzed by GC-MS (Agilent 7890/5975C) as described in detail (60–62). For palmitate labeling, metabolites were extracted with a mixture of chloroform:methanol (2:1 vol/vol), derived by Methyl-8 reagent (Pierce), and analyzed by GC-MS. The measured distribution of mass isotopologues was corrected for natural abundance of 13C metabolite intensity.
Liquid Chromatography/MS Analysis of Metabolites.
NAD+, NADH, NADP, NADPH, ATP, ADP, AMP, GSH, and GSSG were measured by a Waters Xevo TQ Tandem Mass Spectrometer with ACQUITY UPLC as we reported previously (62). The metabolites from RPE cells were separated by the UPLC BEH Amide Analytic Column (Waters). The mobile phase was (A) water with 10 mM ammonium acetate (pH 8.9) and (B) acetonitrile:water (95:5) with 10 mM ammonium acetate (pH 8.9). (All solvents are LC-MS Optima Grade from Fisher Scientific.) The gradient elution was (i) 95–61% B at 6 min, (ii) 61–44% B at 8 min, (iii) 61–27% B at 8.2 min, and 27–95% B at 9 min. The column was reequilibrated with 95% B at the end of each run. The flow rate for all gradients was 0.5 mL/min, and the total run was 11 min. The optimized transitions for each metabolite, including the precursor and product ions, were set up after tuning with the standard. The chromatograms were analyzed by MassLynx (Waters).
Transfections.
IDH1 and IDH2 gene expressions were knocked down by transfecting fetal RPE using siGENOME Human IDH1 and IDH2 siRNA (Dharmacon) and transfection reagent (DharmaFECT 4; Dharmacon). Cells transfected with nontarget siRNA were used as a control. The efficiency of silencing was determined by Western blot analysis of IDH1 and IDH2 expression up to 8 d after seeding. In all experiments, cells were transfected on the initial day and the third day after plating with 60 nM siRNA and 0.2% transfection reagent in antibiotic-free RPE medium with 1% serum.
IDH Immunohistochemistry.
Tissue sections of a human globe (81-y-old female) were obtained from the University of Washington Tissue Bank for Ophthalmology Research (HSD 41470). Tissue sections (5 µm in thickness) were deparaffinized and rehydrated in an ethanol series. Sections were blocked for nonspecific binding with 10% (vol/vol) horse serum in PBS and incubated overnight with the following primary antibodies: IDH1 (1:1,000; Santa Cruz Biotechnology), IDH2 (1:1,000; Abcam), and cellular retinaldehyde binding protein (1:100; University of Washington Vision Research Core). Sections were incubated with AlexaFluor secondary antibodies (1:500) for 45 min at room temperature in the dark, costained with DAPI, and mounted with Fluoromount-G.
MitoSOX Assay.
hfRPE cells were plated at a density of 100,000–120,000 per well in an eight-chambered slide (Nunc Lab-Tek Permanox Slide; Sigma-Aldrich) with RPE medium containing 5% (vol/vol) FBS and 10 μM Y-27632. After 4–7 d in culture, the media were washed and replaced with DMEM containing 5.5 mM glucose, 2 mM glutamine, and 1% FBS (dye-free and no pyruvate) with or without 1 mM OMA overnight. An aliquot of MitoSOX (Molecular Probes; ThermoFisher Scientific) was prepared by reconstituting 50 μg with 13 μL DMSO for 5 mM stock MitoSOX, which then was diluted 1:1,000 in Krebs-Ringer bicarbonate (KRB) buffer. The cells were changed into KRB buffer containing 5 μM MitoSOX and incubated for 10 min at 37 °C. Pictures were captured under fluorescence microscopy, and red fluorescent cells were counted using the software program Fiji.
Ethidium Homodimer Staining.
hfRPE cells were grown for 4–5 wk in 24-well plates. They were treated (∼16 h) with control (media only), 250 μM H2O2, or 250 μM H2O2 and 1 mM OMA in DMEM clear media without glucose, glutamine, or pyruvate (A1443001; Gibco). The next day, media were removed and replaced with 500 μL per well KRB media and ethidium homodimer dye. Red fluorescent cells were counted under a fluorescence microscope.
LDH Activity Assay.
Twenty microliters media from hfRPE cell cultures were used in the assay, which was conducted per the manufacturer’s protocol (Genzyme). Absorbance was measured at 340 nm, and the change in absorbance over time was determined by subtracting the background control.
Statistics.
Data are expressed as mean ± SD. Significance of differences between means was determined by unpaired two-tailed t tests or ANOVA with an appropriate post hoc test. A P value < 0.05 was considered to be significant.
Acknowledgments
We thank the University of Washington Vision Research Core for use of the cellular retinaldehyde binding protein antibody, the Lynn and Mike Garvey Cell Imaging Laboratory for confocal imaging, and the University of Washington Departments of Ophthalmology and Neuropathology for human eye bank tissue sections. This work was supported by NIH Grants EY026030 (to J.D., J.B.H., and J.R.C.), DK082783 (to A.R.), HL099993 (to A.R.), EY06641 (to J.B.H.), EY017863 (to J.B.H.), EY019714 (to J.R.C.), and EY001730 (to National Eye Institute Vision Research Core); the Bill & Melinda Gates Foundation (J.R.C.); and an unrestricted grant from Research to Prevent Blindness (to J.D. and J.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.F. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604572113/-/DCSupplemental.
References
- 1.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85(3):845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 2.Sparrow JR, Hicks D, Hamel CP. The retinal pigment epithelium in health and disease. Curr Mol Med. 2010;10(9):802–823. doi: 10.2174/156652410793937813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75(1):26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein R, et al. Prevalence of age-related macular degeneration in the US population. Arch Ophthalmol. 2011;129(1):75–80. doi: 10.1001/archophthalmol.2010.318. [DOI] [PubMed] [Google Scholar]
- 5.Mettu PS, Wielgus AR, Ong SS, Cousins SW. Retinal pigment epithelium response to oxidant injury in the pathogenesis of early age-related macular degeneration. Mol Aspects Med. 2012;33(4):376–398. doi: 10.1016/j.mam.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Young RW. Solar radiation and age-related macular degeneration. Surv Ophthalmol. 1988;32(4):252–269. doi: 10.1016/0039-6257(88)90174-9. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Tombran-Tink J. Mitochondrial decay and impairment of antioxidant defenses in aging RPE cells. Adv Exp Med Biol. 2010;664:165–183. doi: 10.1007/978-1-4419-1399-9_20. [DOI] [PubMed] [Google Scholar]
- 8.Terluk MR, et al. Investigating mitochondria as a target for treating age-related macular degeneration. J Neurosci. 2015;35(18):7304–7311. doi: 10.1523/JNEUROSCI.0190-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao H, et al. Mitochondrial oxidative stress in the retinal pigment epithelium leads to localized retinal degeneration. Invest Ophthalmol Vis Sci. 2014;55(7):4613–4627. doi: 10.1167/iovs.14-14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao C, et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121(1):369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohrer B, Bandyopadhyay M, Beeson C. Reduced metabolic capacity in aged primary retinal pigment epithelium (RPE) is correlated with increased susceptibility to oxidative stress. Adv Exp Med Biol. 2016;854:793–798. doi: 10.1007/978-3-319-17121-0_106. [DOI] [PubMed] [Google Scholar]
- 12.Adijanto J, Philp NJ. Cultured primary human fetal retinal pigment epithelium (hfRPE) as a model for evaluating RPE metabolism. Exp Eye Res. 2014;126:77–84. doi: 10.1016/j.exer.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunihara T, et al. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife. 2016;5:e14319. doi: 10.7554/eLife.14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iacovelli J, et al. PGC-1α induces human RPE oxidative metabolism and antioxidant capacity. Invest Ophthalmol Vis Sci. 2016;57(3):1038–1051. doi: 10.1167/iovs.15-17758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang L, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemons JM, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8(10):e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comte B, Vincent G, Bouchard B, Benderdour M, Des Rosiers C. Reverse flux through cardiac NADP(+)-isocitrate dehydrogenase under normoxia and ischemia. Am J Physiol Heart Circ Physiol. 2002;283(4):H1505–H1514. doi: 10.1152/ajpheart.00287.2002. [DOI] [PubMed] [Google Scholar]
- 20.Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283(30):20621–20627. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo SH, et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2001;276(19):16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 22.Lee SM, et al. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32(11):1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 23.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287(17):14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itsumi M, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ. 2015;22(11):1837–1845. doi: 10.1038/cdd.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu B, Wu T, Miao L, Mei Y, Wu M. MiR-181a regulates lipid metabolism via IDH1. Sci Rep. 2015;5:8801. doi: 10.1038/srep08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ku HJ, Ahn Y, Lee JH, Park KM, Park JW. IDH2 deficiency promotes mitochondrial dysfunction and cardiac hypertrophy in mice. Free Radic Biol Med. 2015;80:84–92. doi: 10.1016/j.freeradbiomed.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Oka S, Hsu CP, Sadoshima J. Regulation of cell survival and death by pyridine nucleotides. Circ Res. 2012;111(5):611–627. doi: 10.1161/CIRCRESAHA.111.247932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol. 2009;144(3):167–174. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J Biol Chem. 2013;288(18):12967–12977. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adijanto J, et al. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287(24):20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballinger SW, Van Houten B, Jin GF, Conklin CA, Godley BF. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68(6):765–772. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- 32.Jarrett SG, Boulton ME. Poly(ADP-ribose) polymerase offers protection against oxidative and alkylation damage to the nuclear and mitochondrial genomes of the retinal pigment epithelium. Ophthalmic Res. 2007;39(4):213–223. doi: 10.1159/000104683. [DOI] [PubMed] [Google Scholar]
- 33.Bakondi E, et al. Detection of poly(ADP-ribose) polymerase activation in oxidatively stressed cells and tissues using biotinylated NAD substrate. J Histochem Cytochem. 2002;50(1):91–98. doi: 10.1177/002215540205000110. [DOI] [PubMed] [Google Scholar]
- 34.Robaszkiewicz A, et al. Hydrogen peroxide-induced poly(ADP-ribosyl)ation regulates osteogenic differentiation-associated cell death. Free Radic Biol Med. 2012;53(8):1552–1564. doi: 10.1016/j.freeradbiomed.2012.08.567. [DOI] [PubMed] [Google Scholar]
- 35.Fendt SM, et al. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopalan KN, et al. Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer Metab. 2015;3:7. doi: 10.1186/s40170-015-0134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai S, Sheline CT. NAD(+) maintenance attenuates light induced photoreceptor degeneration. Exp Eye Res. 2013;108:76–83. doi: 10.1016/j.exer.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zabka TS, et al. Retinal toxicity, in vivo and in vitro, associated with inhibition of nicotinamide phosphoribosyltransferase. Toxicol Sci. 2015;144(1):163–172. doi: 10.1093/toxsci/kfu268. [DOI] [PubMed] [Google Scholar]
- 39.Koenekoop RK, et al. Finding of Rare Disease Genes (FORGE) Canada Consortium Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012;44(9):1035–1039. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrault I, et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat Genet. 2012;44(9):975–977. doi: 10.1038/ng.2357. [DOI] [PubMed] [Google Scholar]
- 41.Chiang PW, et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat Genet. 2012;44(9):972–974. doi: 10.1038/ng.2370. [DOI] [PubMed] [Google Scholar]
- 42.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E and beta carotene for age-related cataract and vision loss: AREDS report no. 9. Arch Ophthalmol. 2001;119(10):1439–1452. doi: 10.1001/archopht.119.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 44.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299(5606):560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 46.Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: From bench to bedside. Eur Heart J. 2015;36(48):3404–3412. doi: 10.1093/eurheartj/ehv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braidy N, et al. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6(4):e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16(10):593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurley JB, Lindsay KJ, Du J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J Neurosci Res. 2015;93(7):1079–1092. doi: 10.1002/jnr.23583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rueda EM, et al. The cellular and compartmental profile of mouse retinal glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and ∼P transferring kinases. Mol Vis. 2016;22:847–885. [PMC free article] [PubMed] [Google Scholar]
- 51.Massudi H, et al. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One. 2012;7(7):e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gariani K, et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology. 2016;63(4):1190–1204. doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24(8):464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Picciotto NE, et al. Nicotinamide mononucleotide supplementation reverses vascular dysfunction and oxidative stress with aging in mice. Aging Cell. 2016;15(3):522–530. doi: 10.1111/acel.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ChromaDex I. 2015 ChromaDex's NiagenTM Nicotinamide Riboside Meets Primary Endpoint in First Human Clinical Study. Available at investors.chromadex.com/phoenix.zhtml?c=212121&p=irol-newsArticle&ID=2015560. Accessed November 15, 2016.
- 56.Conze DB, Crespo-Barreto J, Kruger CL. Safety assessment of nicotinamide riboside, a form of vitamin B3. Hum Exp Toxicol. January 20, 2016 doi: 10.1177/0960327115626254. [DOI] [PubMed] [Google Scholar]
- 57.Sonoda S, et al. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat Protoc. 2009;4(5):662–673. doi: 10.1038/nprot.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindsay KJ, et al. Pyruvate kinase and aspartate-glutamate carrier distributions reveal key metabolic links between neurons and glia in retina. Proc Natl Acad Sci USA. 2014;111(43):15579–15584. doi: 10.1073/pnas.1412441111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du J, et al. Cytosolic reducing power preserves glutamate in retina. Proc Natl Acad Sci USA. 2013;110(46):18501–18506. doi: 10.1073/pnas.1311193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du J, et al. Inhibition of mitochondrial pyruvate transport by zaprinast causes massive accumulation of aspartate at the expense of glutamate in the retina. J Biol Chem. 2013;288(50):36129–36140. doi: 10.1074/jbc.M113.507285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adijanto J, et al. The retinal pigment epithelium utilizes fatty acids for ketonegenesis. J Biol Chem. 2014;289(30):20570–20582. doi: 10.1074/jbc.M114.565457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du J, Linton JD, Hurley JB. Probing metabolism in the intact retina using stable isotope tracers. Methods Enzymol. 2015;561:149–170. doi: 10.1016/bs.mie.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchholz DE, et al. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med. 2013;2(5):384–393. doi: 10.5966/sctm.2012-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]