Significance
Genome-scale models of metabolism are important tools for metabolic engineering and production strain development. We present an experimentally validated and manually curated model of metabolism in Synechococcus elongatus PCC 7942 that (i) leads to discovery of unique metabolic characteristics, such as the importance of a truncated, linear TCA pathway, (ii) highlights poorly understood areas of metabolism as exemplified by knowledge gaps in nucleotide salvage, and (iii) accurately quantifies light input and self-shading. We now have a metabolic model that can be used as a basis for metabolic design in S. elongatus.
Keywords: cyanobacteria, constraint-based modeling, TCA cycle, photosynthesis, Synechococcus elongatus
Abstract
The model cyanobacterium, Synechococcus elongatus PCC 7942, is a genetically tractable obligate phototroph that is being developed for the bioproduction of high-value chemicals. Genome-scale models (GEMs) have been successfully used to assess and engineer cellular metabolism; however, GEMs of phototrophic metabolism have been limited by the lack of experimental datasets for model validation and the challenges of incorporating photon uptake. Here, we develop a GEM of metabolism in S. elongatus using random barcode transposon site sequencing (RB-TnSeq) essential gene and physiological data specific to photoautotrophic metabolism. The model explicitly describes photon absorption and accounts for shading, resulting in the characteristic linear growth curve of photoautotrophs. GEM predictions of gene essentiality were compared with data obtained from recent dense-transposon mutagenesis experiments. This dataset allowed major improvements to the accuracy of the model. Furthermore, discrepancies between GEM predictions and the in vivo dataset revealed biological characteristics, such as the importance of a truncated, linear TCA pathway, low flux toward amino acid synthesis from photorespiration, and knowledge gaps within nucleotide metabolism. Coupling of strong experimental support and photoautotrophic modeling methods thus resulted in a highly accurate model of S. elongatus metabolism that highlights previously unknown areas of S. elongatus biology.
The unicellular cyanobacterium Synechococcus elongatus PCC 7942 is being developed as a photosynthetic bioproduction platform for an array of industrial products (1–3). This model strain is attractive for this purpose because of its genetic tractability (4) and its reliance on mainly CO2, H2O, and light for metabolism, reducing the environmental and economic costs of cultivation. For low-cost, high-volume products, such as biofuels, however, one of the biggest challenges is attaining profitable product yields (5, 6). Genome-scale models (GEMs) of metabolism provide a valuable tool for increasing product titers by optimizing yield in silico and then, reproducing the changes in vivo (7). For instance, GEMs were used to select the optimal synthetic pathway for 3-hydroxypropanoate biosynthesis in Saccharomyces cerevisiae (8). In Escherichia coli, GEM optimization was used to realize heterologous production of 1,4-butanediol synthesis and increase titers three orders of magnitude (9). Although there have been numerous modeling efforts in Synechocystis sp. PCC 6803 (here in referred to as PCC 6803), this organism is highly divergent from S. elongatus, where limited modeling has been done (10).
This deficit can partially be explained by the lack of in vivo validation datasets, such as 13C metabolic flux analysis (MFA), for obligate phototrophs (11). Development of metabolic models of S. elongatus with strong experimental support is necessary to exploit the organism as a bioproduction platform and advance models of obligate phototrophic metabolism.
A metabolic network reconstruction is a representation of all metabolic reactions, the enzymes responsible for their catalysis, and the genes that encode them. Genome-scale reconstructions have a proven record of contextualizing organism-specific information and facilitating the characterization and engineering of cellular metabolism (12, 13). When complete, the reconstruction enables quantitative prediction of metabolic phenotypes represented as reaction fluxes. The overall predictive power of a GEM is naturally dependent on its quality (14). Essentiality datasets have been successfully used to increase the accuracy of GEMs (15). We recently determined genome-wide gene essentiality by screening ∼250,000 pooled mutants for their survival under standard laboratory conditions with continuous light via random barcode transposon site sequencing (RB-TnSeq) (16). This dataset facilitated the generation and testing of a high-quality genome-scale reconstruction through comparison of the model outputs and in vivo phenotypes at the genome scale. Inconsistencies between model predictions and in vivo data can highlight parts of S. elongatus metabolism where current understanding is inadequate (17).
Another key characteristic of an accurate GEM is the application of constraints that place physical, chemical, and biological limitations on a culture and generate biologically relevant phenotypic predictions. Incorporating light, a dominant constraint on phototrophic growth, into a GEM remains a challenge (18). Photon uptake is typically fixed based on experimental results, an approach that allows retrospective analysis but not predictive modeling (19). Therefore, no current model inputs light quantity, quality, and shading, resulting in a linear growth curve characteristic of photoautotrophic batch culture.
Here, we present a comprehensive GEM of obligate phototrophic metabolism. We performed a complete reannotation and reconstruction of metabolic genes in the S. elongatus genome and developed an approach to incorporate light absorption that factors in the effects of cell shading. In addition, GEM predictions have been compared with and improved by essentiality data (16). These comparisons are also used to reveal unique attributes of the organism’s metabolism. The result is a comprehensive metabolic model of S. elongatus metabolism.
Results
Genome-Scale Reconstruction of Phototrophic Metabolism.
A metabolic reconstruction is a knowledge base that places biochemical, genetic, and genomic information into a structured framework. The reconstruction contains the functional annotation of the genome and defines the organism’s metabolic capability: the substrates that it can use and the reactions that it can perform. To properly define the metabolic capability of S. elongatus, we curated the genome annotation, leveraged state of the art in silico methods, incorporated comprehensive in vivo essentiality data, and included a detailed reconstruction of light harvesting. The resulting model is an organized collection of the extensive data available for S. elongatus in a format that enables accurate predictions of phototrophic metabolism (Fig. S1).
Fig. S1.
Model development flowchart. This diagram outlines the steps that went into genome reconstruction and model simulation. CDD, Conserved Domains Database; NCBI, National Center for Biotechnology Information.
Manual curation of S. elongatus genome annotation.
Because the metabolic capability reflected in a reconstructed network is dependent on the functional annotation of the organism, we reannotated the metabolic genes using amino acid as well as protein functional domain homology-based methods. This functional reannotation enabled the conversion of an initial draft reconstruction to a completed GEM (20) (SI Materials and Methods). Of 2,723 genes in the S. elongatus genome, 785 (29%) were included in the final version of the model, and 118 of these genes (15%) had updated functional annotations. This GEM is named iJB785 (model files are in Dataset S1, and the Excel file is in Dataset S2) following convention (21).
Protein structure-guided reconstruction.
Amino acid and protein domain-based annotations often do not provide sufficient detail to assign enzyme function. Therefore, enzymes are often incorrectly annotated as functionally equivalent (isozymes). In S. elongatus, this challenge resulted in instances where multiple genes that are essential in vivo were assigned to the same reaction; however, if the enzymes were truly compensatory, none should be essential. The importance of enzyme structure in catalytic activity suggests that structural homology modeling, which uses in silico-derived 3D analysis of a target protein based on the crystal structure of a similar enzyme, may provide additional insight into protein function.
Protein structure data have recently been applied in the global analysis of GEMs (22). We set out to apply structural modeling to protein annotation by determining the functional difference between four annotated phosphoglycerate mutases (PGMs; Synpcc7942_2078, Synpcc7942_1516, Synpcc7942_0485, EC 5.4.2.11; and Synpcc7942_0469, EC 5.4.1.12) in the S. elongatus genome, three of which are essential in vivo. Previous work in S. elongatus suggested that multiple PGMs work in concert to regulate metabolic flux during shifts in CO2 availability (23). However, it seemed unlikely that three of four PGMs would be essential for regulation in a stable CO2 environment. To test the hypothesis of divergent functions between the S. elongatus PGMs, structural homology models were generated and compared with published control crystal structures (SI Materials and Methods, Table S1).
Table S1.
Structural homology analysis of S. elongatus PGMs
| Gene | Organism | Annotation | C-terminal chain | His85 | Gln22 | Conclusion | Ref. |
| HTH_0103 | H. thermophilus | PSP (control) | + | + | + | Control | 24 |
| b0755 | E. coli | dPGM (control) | − | Tyr | Thr | Control | 86 |
| Synpcc7942_0485 | S. elongatus | PGM | + | + | + | PSP | |
| Synpcc7942_2078 | S. elongatus | PGM | − | Phe | Ser | dPGM | |
| Synpcc7942_1516 | S. elongatus | PGM | + | Leu | Leu | Histidine phosphatase | |
| Synpcc7942_0469 | S. elongatus | iPGM | iPGM |
Based on the structural comparison, it was possible to ascribe a more detailed function to each of the annotated PGMs. The Synpcc7942_0469 protein is structurally distinct from the three other PGMs and was annotated as the primary glycolytic PGM in S. elongatus based on its canonical PGM structure and the fact that it is essential. Previous work indicated that two PGMs are required to regulate central carbon flux during a transition from high to low CO2 (23). The Synpcc7942_2078 protein shares structural features with an E. coli PGM control but is nonessential; thus, it was annotated as a PGM performing this regulatory function. Synpcc7942_0485 shares strong structural similarity to a phosphoserine phosphatase (PSP) in Hydrogenobacter thermophiles (24) and has sequence homology to the recently characterized PSP in PCC 6803 (25). Thus this gene was confidently annotated as a PSP in S. elongatus. Synpcc7942_1516, however, has structural features that could not be classified as a traditional PGM or PSP and is essential in vivo. Based on genomic neighborhood analysis and transcriptome mapping data (26), we hypothesized that Synpcc7942_1516 plays a regulatory role in an uncharacterized signaling network. As a regulatory enzyme, it fell outside the scope of the metabolic model. These results indicate that structural homology modeling is a promising annotation tool to increase the quality of genome-scale reconstructions and hypothesize enzyme function.
Improved reconstruction through incorporation of essential gene data.
The essential gene calls for S. elongatus determined by RB-TnSeq-enabled refinement of the gene reaction annotations during development of the reconstruction (16). This in vivo dataset provides a gauge of gene importance by identifying genes that cannot sustain insertion mutants, which are interpreted to be essential, and the growth rate of those that can, which are interpreted to be beneficial or nonessential. Alternatively, in silico essentiality calls are made by quantifying the impact on growth when the flux through each enzyme in the model is independently set to zero (SI Materials and Methods). Discrepancies between essential gene calls in the in silico draft model and the RB-TnSeq results were investigated, and with sufficient evidence, the gene assignment for the model reaction was updated accordingly. For example, the S. elongatus genome encodes two annotated uroporphyrinogen methyltransferases (Synpcc7942_0271 and Synpcc7942_2610, EC 2.1.1.107), catalyzing an early step of both vitamin B12 and siroheme biosynthesis. However, both genes are essential in vivo, suggesting that they are not compensatory. Genomic neighborhood analysis indicated that synpcc7942_2610 is adjacent to an iron chelatase gene; thus, we proposed that it is dedicated to the biosynthesis of siroheme, not vitamin B12. However, discrepancies between the in silico and in vivo essential gene data were not forced into agreement without additional evidence, and these remaining discrepancies are contained in Dataset S3. For example, S. elongatus has two genes annotated for type II NADH oxidoreductases (synpcc7942_0101 and synpcc7942_0198, EC 1.6.5.9); however, the in vivo data indicated that one is essential (synpcc7942_0101) and thus, the enzymes are not redundant. Nevertheless, the manual curation process did not reveal any significant difference between the two genes, and they were annotated as isozymes, although the RB-TnSeq data suggest divergent functions.
Discrepancies also led to a more complete representation of cellular biomass. For example, alkanes were not initially included in the biomass, because their function is unknown. However, their synthesis is essential. Recently, it was discovered that alkanes can play a role in cyclic electron flow in PCC 6803 (27). Hypothesizing a homologous function in S. elongatus, they were added to the biomass equation. Thus, applying the essential gene data to the curation process increased the quality of the reconstruction.
Explicit modeling of light absorption.
The dominant constraints on photoautotrophic growth are light and CO2 availability. A fundamental barrier to predictive modeling of photoautotrophic growth has been the inability to translate light irradiance into a metabolite. Traditionally, light uptake has been inferred and not explicitly determined. A two-step optimization method is commonly used, where the CO2 uptake rate is fixed based on experimental values at the observed growth rate followed by minimization of the photon flux (19, 28). However, the resulting values reflect only the photons that perform metabolic work, not the totality of absorbed light. Excess photon absorption has a significant impact on growth rate and metabolism. Reactive oxygen species, such as singlet oxygen generated in the pigment antenna and photosystem II (29) as well as superoxide at photosystem I (30), damage the photosynthetic apparatus and consume metabolic resources. Thus, the two-step optimization method does not accurately account for total light absorption and is incapable of predictive modeling of phototrophic metabolism.
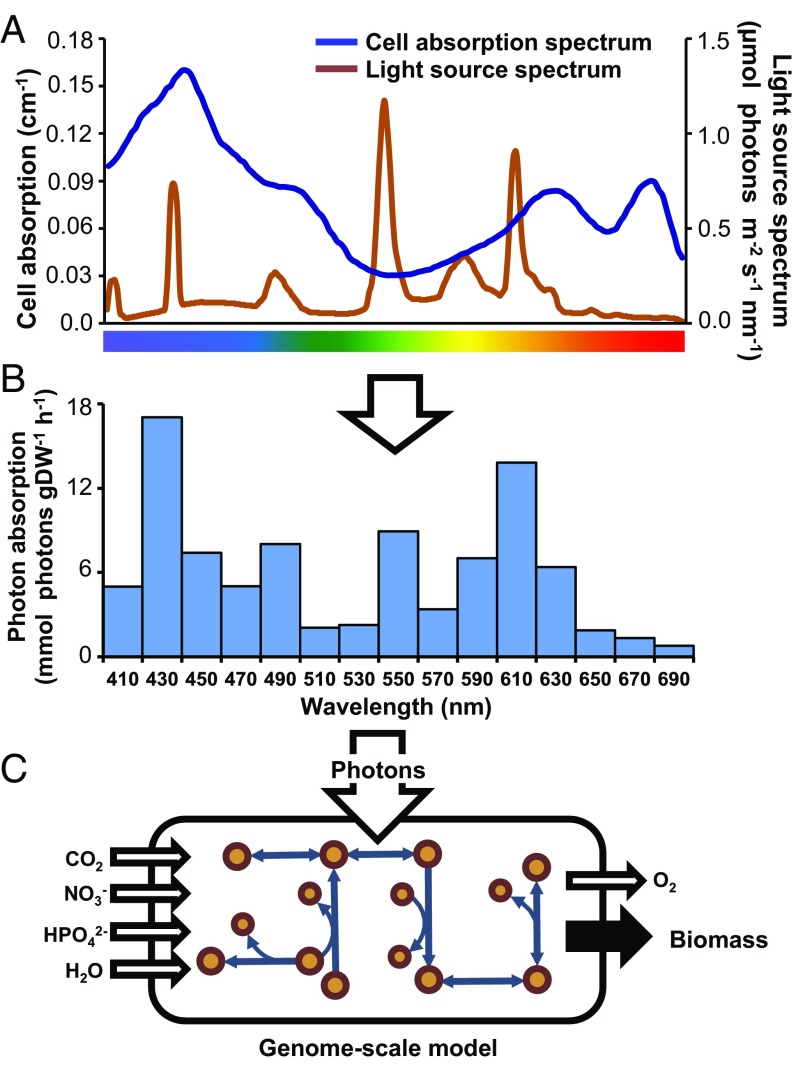
In a more extensive description of photon capture, a metabolic reconstruction of Chlamydomonas reinhardtii accounted for light source quality (31). This approach was more mechanistic than the two-step optimization but still did not consider cellular pigmentation as a factor in photon absorption, and therefore, light uptake could not be quantified into a typical modeling input flux. In another paper, a light distribution function was combined with flux-balance analysis to model cyanobacterial growth in photobioreactors; however, a mechanistic model of light harvesting was not included (32). Our approach goes further by combining light source irradiance with in vivo absorption to define photon use from measurements of incident light. We incorporated the chlorophyll-normalized optical absorption cross-section (33), derived from the in vivo absorption spectrum, to link photon uptake to cellular composition. Because the model’s biomass explicitly defines the cellular composition, the biomass-normalized photon absorption rate was calculated from the combination of irradiance, optical absorption cross-section, and the chlorophyll component of the biomass. This approach enabled comparison between the photon absorption capacity of the cell and the photon delivery rate of the light source at a given irradiance (Fig. 1).
Fig. 1.
Deriving light uptake rates for the GEM. (A) The chlorophyll a-normalized optical absorption cross-section was calculated from the cellular absorbance in the photosynthetically active range (400–700 nm) and compared with the spectral distribution of the incident light. (B) The photon absorption rate as a function of cell biomass [millimoles photons gram dry weight (DW)−1 hour−1] was determined by combining the optical absorption cross-section and incident light. (C) The GEM was constrained by setting the photon uptake flux to the calculated absorption rate, which was split into 15 20-nm bins across the photosynthetic range.
Accurate modeling of obligate phototrophic metabolism also required a new level of detail in the reconstruction of the photosystem. We used recent proteomics (34) and fluorescence microscopy data (35) to reconcile membrane localization of electron transport complexes that previous cyanobacterial models had included inaccurately in the cytoplasmic membrane. Additionally, the ferredoxin:plastoquinone oxidoreductase complex gene associations were updated to include additional subunits and the use of ferredoxin as the electron donor (36, 37). We also included the photoinactivation of the D1 subunit of photosystem II. Using the photodamage rates in PCC 6803 (38), we were able to calculate a D1 repair metabolic cost as a proportion of flux through photosystem II. Finally, we generated stoichiometric reactions accounting for the energy transfer efficiencies of each of the photosynthetic pigments. Targeted excitation of S. elongatus permaplasts provided relative efficiency metrics for the transfer of energy from a given pigment to the photosystems (39). Reactions including these efficiencies enabled the model to account for light spectrum-specific photosynthetic efficiency. This comprehensive reconstruction of light gathering set the framework for accurate constraint-based modeling of phototrophic metabolism.
The completed reconstruction, iJB785, consists of 785 genes, 850 metabolic and transport reactions, and 768 nonunique metabolites distributed over seven cellular compartments (Dataset S2). The reconstruction was completed in the BiGG Models format (40), enabling standardization and cross-referencing to external databases (bigg.ucsd.edu). Combining an updated annotation, whole-genome RB-TnSeq data, and an advanced representation of light harvesting resulted in a comprehensive phototrophic model.
Modeling Phototrophic Growth.
Conversion of a reconstruction into a mathematical model and the subsequent application of biologically relevant constraints enable the simulation of cellular phenotypes. Modeling cellular growth is typically computed as either yield or specific growth rate, which assumes exponential growth (41). In both cases the inputs are normalized, such that the simulation reports a single value representative of cellular growth as long as there is a constant ratio between the biomass and the input flux. However, as the culture becomes denser, the photon absorption capacity can exceed the amount of light delivered by the light source and in the process, alter the ratio of uptake to biomass. When this phenomenon occurs, it results in a linear growth curve caused by self-shading–induced light limitation. Cells closest to the light source absorb excess photons, preventing cells in the inner culture from achieving their maximum growth rate. We accounted for shading with an unsteady-state growth modeling methodology (42), which resulted in an accurate model of linear photoautotrophic growth over the duration of a batch culture.
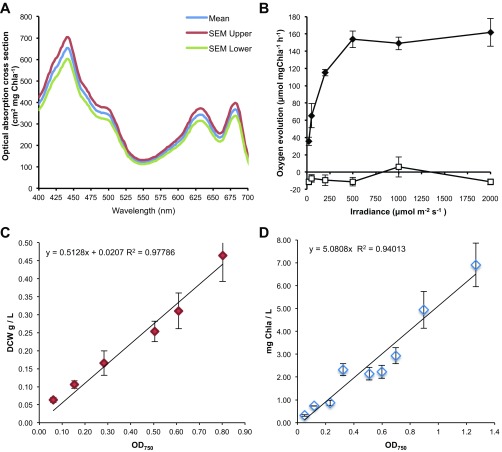
An additional constraint was required to capture the point when light absorption became excessive. As the other dominant constraint on growth, the maximum carbon uptake rate marks the transition between a light- and a carbon-limited culture. It is dependent on the availability of inorganic carbon in the media (43) and the acclimated state of the cell (44). However, an approximation of maximum photosynthetic output, a proxy for carbon uptake, can be captured in a single parameter: the oxygen evolution rate (45). We used experimentally determined oxygen evolution rates to constrain the maximum photosynthetic output at a given irradiance (Fig. S2). Excess photon absorption was allowed to leave the system in a manner that did not incur a metabolic cost, simulating loss as heat or fluorescence. Reactive oxygen species production in the light-harvesting antenna caused by excess light is currently not modeled.
Fig. S2.
In vivo physiological data incorporated as constraints in in silico flux balance analysis. (A) Whole-cell absorption spectra. (B) Oxygen evolution under illumination at various light intensities shown as ♦. Dark condition oxygen consumption after each light period shown as □. (C) Dry cell weight (DCW) vs. OD750. (D) Chlorophyll a concentration vs. OD750. The data shown for each graph are averages of three independent measurements, and error bars indicate SDs.
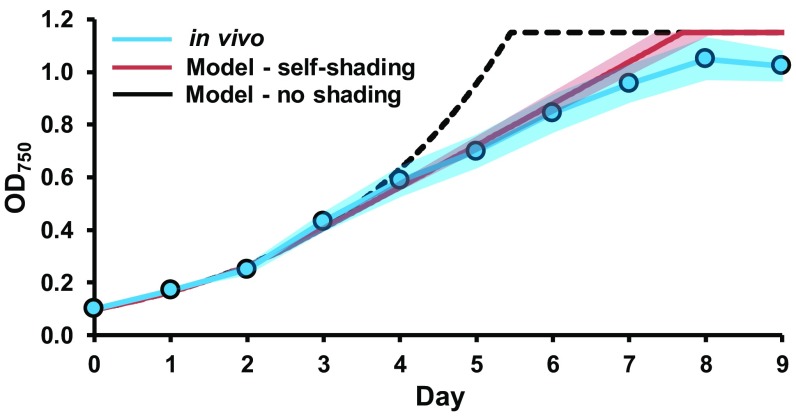
A growth curve for a typical S. elongatus culture was simulated using the oxygen evolution constraint and the calculated photon uptake rate. To account for self-shading, at 1-h intervals, the flask was sectioned into 50 concentric rings, with each ring modeling the biomass production of a 2% fraction of the culture. Light was modeled from the side of the flask, and the photon absorption of an outer ring was made unavailable to the remainder of the inner rings (Fig. S3). For the nonshading simulation, the growth rate matched the in vivo culture until light limitation, at which point the in silico growth remained exponential, whereas the in vivo curve became linear (Fig. 2). Growth simulations factoring in self-shading transitioned into linear phase on light limitation, characteristic of in vivo growth. This more accurate prediction of photoautotrophic growth was made possible by the combination of modeling photon uptake and shading as a function of culture density.
Fig. S3.
Diagram of cell-shading calculation. Increased light limitation caused by cell–cell shading was factored into the model simulations for biomass prediction in a typical 250-mL Erlenmeyer flask with 100 mL culture medium.
Fig. 2.
Modeling the self-shading of cultures increases the accuracy of growth rate prediction. OD750 values for the in vivo culture were the mean of three individual cultures. SE is represented by the shaded areas. SE for the in silico growth rate was determined from the SE of the in vivo inputs to the model.
Model parameters are specific to the cellular phenotype: in particular, the chlorophyll-normalized optical absorption cross-section, which depends on the photoacclimation state of the cell. The primary photon-harvesting complex in S. elongatus, the phycobilisome, which can efficiently deliver light energy to both photosystems (39), is highly adaptive (46) but devoid of chlorophyll. Photosystem I contains 80–95% of the chlorophyll a in S. elongatus (39). Because photoacclimation causes fluctuation in the photosystem I to photosystem II ratio and changes to the phycobilisome (47), a given chlorophyll-normalized optical absorption cross-section may no longer be representative. The model can be easily reparameterized to account for this adaptation. Additionally, the oxygen evolution rate can be adjusted to account for phenotypic changes in different carbon environments. The mechanistic nature of this approach is generally applicable to any phototrophic reconstruction and expands genome-scale modeling into new phenotypes, such as photoacclimation and with additional constraints, photoinhibition.
Validation and Refinement of the GEM Through Essential Gene Comparison.
The RB-TnSeq data served as not only a powerful guide during reconstruction but also a validation metric of the resulting model. The accuracy of the model was determined through a comparison of the in silico essential gene calls with the in vivo dataset. The gap between model predictions and in vivo realities highlights the limitations of the model as well as additional constraints on cellular metabolism. Evaluation of the disagreements enabled development of additional constraints beyond simple network connectivity (nonnetwork constraints), increasing the accuracy of the model predictions.
Essential gene-based model validation.
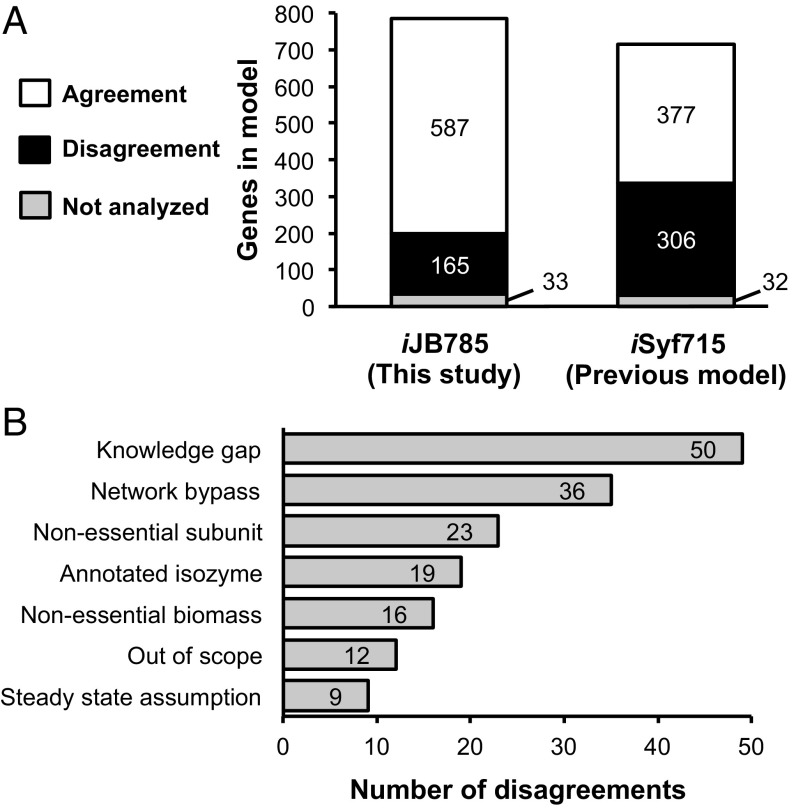
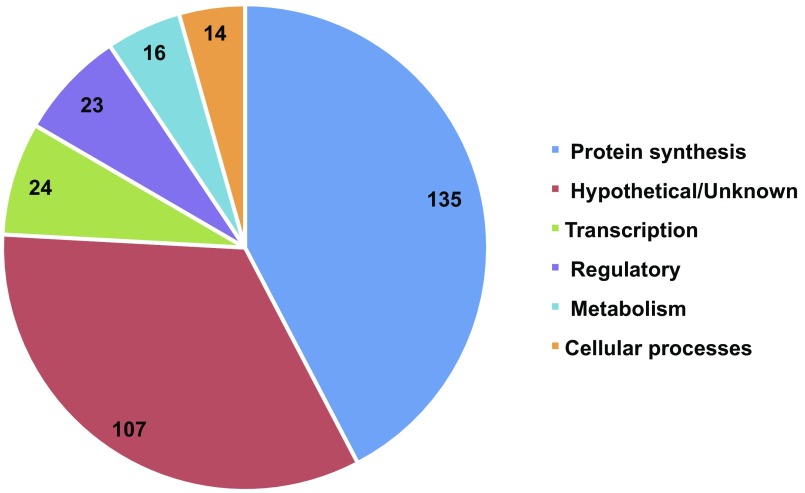
We compared the in silico essential gene calls between our reconstructed model, iJB785, and a previous model of S. elongatus, iSyf715 (48) (Table 1). Minimal standardized constraints were applied to both models, allowing a direct comparison of the metabolic flexibility of the two networks. Of 752 genes in iJB785 with in vivo data, 587 (78%) were correctly assigned as either essential or nonessential (Fig. 3A and Dataset S3). The 165 disagreements were separated into explanatory categories (Fig. 3B). iSyf715 contained 683 genes with in vivo data, of which 377 (55%) were correctly assigned. Neither model was able to accurately predict the reduced growth rate phenotype indicative of genes categorized as beneficial (growth defect when mutated). The 319 genes essential in vivo that were not included in iJB785 participate in cellular processes, such as protein synthesis and transcription, that are out of scope for this GEM (Fig. S4). Incorporating the RB-TnSeq data during manual curation prevented the addition of excess metabolic flexibility (i.e., metabolic capabilities that are implied from the genome annotation but not observed in vivo) as evident by the increased accuracy of iJB785 compared with iSyf715.
Table 1.
Comparison of essentiality results between iJB785 and iSyf715
| Gene category | ORFs | S. elongatus PCC 7942 | |||||
| iJB785 (this study) | iSyf715 (previous model) | ||||||
| Included in GEM | Essentiality prediction | Correct* (%) | Included in GEM | Essentiality prediction | Correct* (%) | ||
| Essential | 718 | 399 | 457 | 350 (88) | 360 | 134 | 118 (33) |
| Beneficial | 157 | 72 | 5 | 0 (0) | 56 | 1 | 0 (0) |
| Nonessential | 1,748 | 281 | 323 | 237 (85) | 266 | 579 | 258 (97) |
| No in vivo data | 100 | 33 | 0 | N/A | 32 | 0 | N/A |
| Total genes | 2,723 | 785 | 785 | 587 (78†) | 714 | 714 | 376 (55†) |
N/A, not applicable due to a lack of in vivo data.
Equal to the number of genes correctly predicted to be essential in silico.
Total correct genes/(total genes included in GEM − model genes with no in vivo data) × 100%.
Fig. 3.
Comparison of in vivo vs. in silico gene essentiality. (A) Comparison of in silico gene essentiality results for the model iJB785 and the previous model of S. elongatus iSyf715. The agreements/disagreements are based on the comparison with in vivo RB-TnSeq results. (B) Sources of disagreements between in silico iJB785 and in vivo gene essentiality.
Fig. S4.
Essential genes in S. elongatus not present in iJB785. The number of essential genes in each category is indicated.
Although both models incorporate only 25–30% of the ORFs identified in the S. elongatus genome, ∼50% of the experimentally shown essential genes and 44% of functionally annotated ORFs are represented in iJB785. Additionally, of 157 genes labeled beneficial in vivo, 46% are present in iJB785. This enrichment of genes that impact cellular fitness underscores the value of GEMs for contextualizing meaningful in vivo genetic perturbations.
Increased model accuracy through nonnetwork constraints.
GEMs offer a tool for visualizing the metabolic network use for a given KO genotype. The resulting flux map identifies alternate routes available to the network to respond to genetic perturbations. However, there are instances where the network connectivity indicates a metabolic pathway, but an additional constraint prevents its use in vivo. Such nonnetwork constraints resulted in a disagreement in the essentiality call for the pyruvate dehydrogenase (PDH) complex. Single-gene deletion of PDH in silico indicated that phosphoketolase (Synpcc7942_2080, EC 4.1.2.9) enables bypass of lower glycolysis by generating acetyl phosphate from the Calvin cycle intermediate fructose-6-phosphate or xylose-5-phosphate. Acetyl phosphate is converted to acetyl-CoA by the combined action of reversible acetate kinase and acetyl-CoA synthase, enabling bypass of PDH. This bypass has been investigated in PCC 6803 (49), and flux balance analysis in that organism also bypassed lower glycolysis with this pathway (50). The essential nature of PDH in vivo indicated an additional constraint that prevents this bypass from carrying sufficient flux to satisfy the acetyl-CoA needs of the cell. However, proteomics (51) and transcriptomics (26) datasets for S. elongatus indicated phosphoketolase abundance on the same order of magnitude as PDH subunits.
Because enzyme abundance could not explain the essentiality of PDH, we investigated metabolite channeling as a factor. Channeling is the result of spatial aggregation of pathway enzymes that prevents the intermediates from being acted on by enzymes outside of the pathway. MFA in PCC 6803 suggested metabolite channeling of Calvin cycle intermediates (11). This phenomenon can be modeled by coupling the flux between two reactions, forcing a ratio, and analyzing the result on the metabolic network. The analysis in silico of metabolite channeling indicated that, if more than 1% of Calvin cycle intermediates were allowed to enter the phosphoketolase bypass, PDH would be nonessential. These results indicate that either substantial metabolite channeling occurs in the Calvin cycle of S. elongatus or the phosphoketolase pathway is functioning in a yet uncharacterized way.
An additional nonnetwork constraint suggested by discrepancies between the model and the in vivo data is the phototrophic reaction catalyzed by ferredoxin-NADP oxidoreductase (FNOR; Synpcc7942_0978, EC 1.18.1.2), an essential reaction in vivo. The model indicated that FNOR was bypassed by NADPH:NAD+ transhydrogenase (Synpcc7942_1610, Synpcc7942_1611, and Synpcc7942_1612, EC 1.6.1.2), resulting in a discrepancy between the model and in vivo data. The canonical function of transhydrogenase is to provide metabolic flexibility by interconverting the two primary redox carriers. However, previous work in PCC 6803 had called into question the presence of an active transhydrogenase in cyanobacteria (52, 53). Previous modeling of PCC 6803 also observed dramatic changes in flux predictions depending on the activity of the transhydrogenase reaction (50), with the authors retaining the canonical transhydrogenase function. However, when we repeated the in silico essential gene assessment setting the transhydrogenase reaction bounds to zero; along with additional nonnetwork constraints, such as routing flux through PDH, 13 genes that had previously been discrepancies fell into alignment with in vivo data. These additional constraints, suggested for modeling in constant light, are provided in SI Results.
Central carbon metabolism flux predictions.
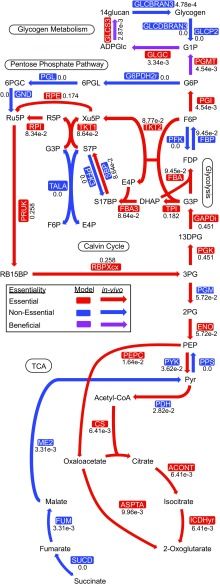
The intracellular flux distribution maps the metabolic reaction use in a given condition. This visualization provides insight into highly used pathways that can be drawn on for product synthesis. The photoautotrophic flux distribution for central carbon metabolism in S. elongatus predicted by our GEM is shown in Fig. 4. Our flux values were in close alignment with PCC 6803 13C MFA data (11), the in vivo equivalent of these in silico data. In PCC 6803, the flux ratio between carbon uptake and fixation was 1:1.27, whereas our model prediction for S. elongatus was 1:1.14. The flux ratio between the Calvin cycle and lower glycolysis, indicative of the biosynthetic carbon requirements of the cell, was 7.9:1 in our model prediction, in good agreement with the ratio of 9.8:1 observed in PCC 6803. The differences may be attributed to variations in the biomass composition for the two species or the fact that the model predicts optimality, whereas the in vivo data reflect inefficiencies naturally present in a living organism. Overall, the high accuracy of the essential gene assessments and consistency with published photoautotrophic flux data underscore the quality of the GEM.
Fig. 4.
Central carbon metabolism. In silico flux values are given numerically next to each reaction. The model essentiality calls are given by the color of the enzyme name, whereas arrow color indicates in vivo gene essentiality. Beneficial indicates a growth defect phenotype when the gene is mutated. Reaction and metabolite abbreviations are given in BiGG format (bigg.ucsd.edu) and found in Dataset S2.
Unusual Attributes of S. elongatus Metabolism.
Taking into account the essential gene dataset (16) during the modeling process greatly improved the consistency of the model and the in vivo data. Still, reactions for which no evidence existed to bring the in silico prediction into agreement with the in vivo data deserved additional attention. The network reconstruction represents a repository of current knowledge; thus, discrepancies between in silico and in vivo results highlighted potential gaps in understanding of S. elongatus metabolism. Disagreements between iJB785 and the RB-TnSeq essential gene calls were separated into categories reflecting the hypothesized source of the discrepancies (Fig. 3B and Dataset S3). Some categories, such as the Network Bypass and Annotated Isozymes, have already been discussed. Only 7% of discrepancies were categorized as Out of Scope, and these reactions were included in iJB785 for completeness but operate in cellular processes not necessary for in silico growth. For example, the tRNA modification queuosine (54) is beneficial according to the RB-TnSeq data but was not explicitly modeled in silico, resulting in a discrepancy for all genes in the pathway.
The Nonessential Biomass discrepancies include enzymes that synthesize biomass components of the WT cellular composition but do not result in a significant in vivo growth defect when missing. Because every component of the defined biomass is required for in silico growth, this category represents in vivo flexibility not present in iJB785. Two such examples of known nonessential biomass components are sulfoquinovosyl diacylglycerol, a component of the photosynthetic membranes (55), and the O-antigen polysaccharide, which even confers a fitness advantage against predators when mutated (56).
We also identified Nonessential Subunits of multiprotein complexes. If one gene in the complex is essential in silico, every subunit associated with that reaction is considered essential, even if the loss is tolerated in vivo. These disagreements included known nonessential subunits of the PCC 6803 photosynthetic electron transport chain (57) and ferredoxin:plastoquinone oxidoreductase complex (36).
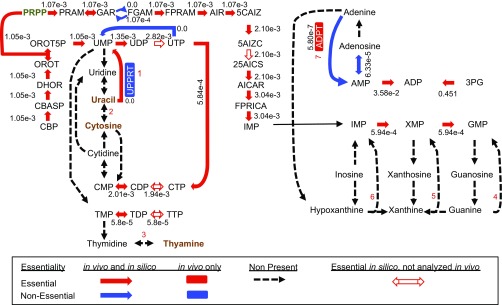
Nucleotide salvage metabolism.
Other discrepancies were placed into the categories Steady-State Assumption and Knowledge Gaps. Examples of both can be found in nucleotide salvage metabolism. Although S. elongatus encodes a complete set of enzymes for de novo biosynthesis of both purine and pyrimidine nucleotides, salvage reactions are mostly absent (Fig. S5). One exception is adenine phosphoribosyltransferase (Synpcc7942_2454, EC 2.4.2.7), which recycles adenine into AMP and is nonessential in vivo but essential in silico. Without this reaction, adenine produced during biosynthesis of polyamines would accumulate and violate the Steady-State Assumption in the model. However, the in vivo data suggest that adenine accumulation is not lethal to the organism, likely because of the small predicted flux through this pathway. Conversely, the catabolism of uracil into UMP by the enzyme uracil phosphoribosyltransferase (Synpcc7942_1715, EC 2.4.2.9) is essential in vivo but not in silico. The in vivo source of uracil and the metabolic requirement to salvage it to UMP represent a Knowledge Gap in our understanding of S. elongatus. Therefore, overlaying in vivo essentiality information over the model’s predictions reveals multiple classes of unknowns in nucleotide salvage alone.
Fig. S5.
Nucleotide salvage pathway. Reactions not present in S. elongatus are indicated by dashed black arrows, essential reactions are indicated by red arrows, nonessential reactions are indicated by blue arrows, and those essential in the model but not analyzed in vivo are indicated by red-outlined arrows. Reactions important for nucleotide salvage are numbered as follows: 1, uricil phosphoribosyltransferase (UPPRT); 2, cytosine deaminase; 3, thymidine phosphorylase; 4, guanine phosphoribosyltransferase; 5, xanthine phosphoribosyltransferase; 6, hypoxanthine phosphoribosyltransferase; 7, and adenine phosphoribosyltransferase (ADPT). Reaction and metabolite abbreviations are given in BiGG format (bigg.ucsd.edu) and found in Dataset S2.
Photorespiration.
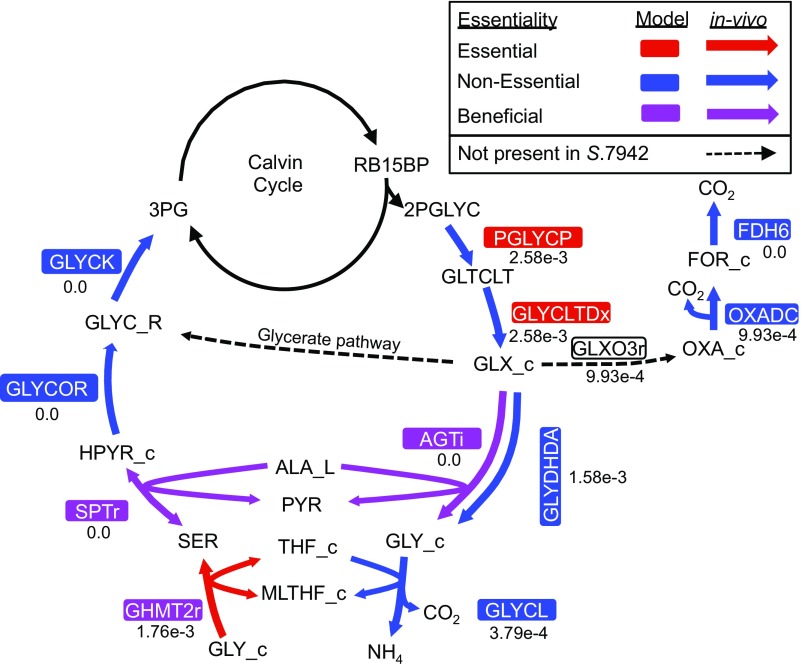
The key carbon-fixing enzyme of the Calvin cycle in photosynthetic metabolism is ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO; Synpcc7942_1426 and Synpcc7942_1427, EC 4.1.1.39). This enzyme can fix not only CO2 but O2 as well, generating 2-phosphoglycolate. Buildup of this molecule is toxic and needs to be recycled through the process of photorespiration (58). To represent photorespiration in silico, a basal level of oxygenase activity needed to be added to the model’s RuBisCO reaction. Based on extrapolation from 13C flux analysis in PCC 6803 (11) and metabolite concentrations in low- vs. high-carbon experiments in S. elongatus (59), we set the model RuBisCO oxygenase flux at 1% of total RuBisCO activity. Interestingly, the first step of the photorespiratory pathway is nonessential in vivo, although the model predicts it to be essential. In this step, phosphoglycolate phosphatase catalyzes the conversion of 2-phosphoglycolate to glycolate (Fig. S6). This enzyme, present upstream of the branching of photorespiration, has no high-confidence isozymes in S. elongatus. The nonessentiality of phosphoglycolate phosphatase suggests either an unknown enzyme for this function or dispensability of the pathway.
Fig. S6.
Photorespiration reactions. Beneficial arrows (purple) indicate a growth defect phenotype when a gene is mutated. Reaction and metabolite abbreviations are given in BiGG format (bigg.ucsd.edu) and found in Dataset S2.
It is improbable that the photorespiratory pathway is nonessential. In PCC 6803, three pathways of photorespiration exist and have been included in previous GEMs of this species (50): the plant-like C2 cycle, full decarboxylation, and the glycerate pathway—which when disrupted in concert, cause a high-CO2 dependency (60). S. elongatus may be missing the last of these pathways (Fig. S6). The glycerate pathway begins with glyoxylate carboligase (GCL), an enzyme that combines two molecules of glyoxylate and ends with the central carbon metabolite 3-phosphoglycerate after the investment of ATP, NAD(P)H, and the release of CO2. The gene found in PCC 6803 (sll1981) that is most similar to GCL in E. coli does not have a homolog in S. elongatus. Therefore, it is possible that only the plant-like C2 cycle and full decarboxylation via formate occur in S. elongatus.
The potential to generate glycine through photorespiration raised the possibility that the process could compensate for de novo glycine/serine biosynthesis. When RuBisCO oxygenase activity was set at 1%, the model predicted that sufficient glycine would be created to support growth, similar to predictions in previous cyanobacterial GEMs (50, 61). Therefore, even when the de novo synthesis pathway of glycine from 3-phosphoglycerate via serine was broken in the model, it still predicted that cells grow at 72% of their normal rate. This finding runs counter to the in vivo data, which show that the entire biosynthesis pathway of serine from 3-phosphoglycerate is essential. The experimental data suggest low flux to amino acid biosynthesis through photorespiration, which could be explained by lower than expected photorespiration activity. Along these lines, in silico essential gene results became consistent with the in vivo data only when RuBisCO’s oxygenase activity was lowered to 0.15% of its carbon-fixing activity. Another possibility is decreased flux specifically toward the amino acid biosynthesis pathway of photorespiration. Glycine hydroxymethyltransferase (Synpcc7942_0282, EC 2.1.2.1) in PCC 6803 is a choke point for the conversion of glycine from photorespiration into serine (62); it is possible that the same limitation exists in S. elongatus. There is also evidence for essentiality of the de novo serine biosynthetic pathway beyond simple metabolic requirements in PCC 6803 (25). Therefore, because of lower than expected photorespiration, limited flux toward glycine synthesis, or undiscovered requirements of de novo serine synthesis, photorespiration is not able to replace de novo amino acid biosynthesis from 3-phosphoglycerate.
A truncated TCA cycle.
The completeness of the cyanobacterial TCA cycle has been an oft-debated subject (63). Since the discovery that 2-oxogluterate dehydrogenase is missing in cyanobacteria, it was accepted for many years that the TCA cycle is incomplete (64). However, a complete TCA cycle is responsible for the majority of energy intermediates created by oxygenic metabolism and nearly ubiquitous throughout nature (65). For this reason, extensive effort has been applied to uncover routes that complete cyanobacterial TCA cycles. A number of bypasses of the missing 2-oxoglutarate dehydrogenase have been discovered, such as the 2-oxoglutarate decarboxylase pathway (66), the GABA shunt (67), and the glyoxylate shunt (68). These bypasses revealed that cyanobacteria harbor complete, albeit noncanonical TCA cycles (63). More recently, however, the necessity of the newly circularized TCA cycles of cyanobacteria has been called into question by experimental (16) and modeling studies (50). Therefore, the structure and biological relevance of a TCA cycle remains an open question in cyanobacteria.
For these reasons, we were particularly interested in discrepancies between the draft model simulations and the in vivo essentiality for two enzymes of the TCA cycle: fumarase (Synpcc7942_1007, EC 4.2.1.2) and malic enzyme (Synpcc7942_1297, EC 1.1.1.40). These enzymes are required by the model’s steady-state assumption for the recycling of fumarate, a by-product of both purine and arginine synthesis. However, the overlay of the in vivo data on the model not only suggests that this recycling is not an essential function but more broadly, led us to evaluate the importance of a complete TCA cycle in S. elongatus.
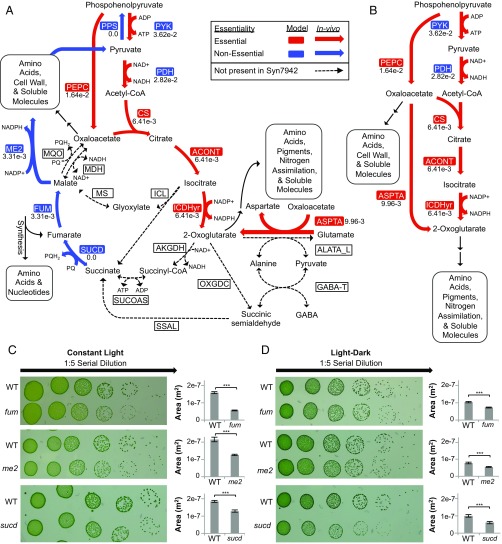
We began by examining the bypasses that complete TCA cycles in other model cyanobacteria (63) for their potential presence in S. elongatus. Our reconstruction, however, revealed none of the known cyanobacterial bypasses (Fig. 5A). Additionally, we were unable to find evidence of the core TCA-cycle enzymes malate dehydrogenase (EC 1.1.1.37), malate:quinone oxidoreductase (EC 1.1.5.4), and succinyl-CoA synthetase (EC 6.2.1.5). Furthermore, the succinate dehydrogenase genes (synpcc7942_0314, synpcc7942_0641, and synpcc7942_1533, EC 1.3.5.1) are nonessential in vivo in addition to genes for fumarase and malic enzyme. Together, these data provide evidence that the metabolically important portion of the TCA cycle in S. elongatus is highly abridged. We call this oxidative, noncyclic portion of the TCA cycle that is essential in S. elongatus the TCA pathway (Fig. 5B). To explain the feasibility of this TCA pathway, we examined whether it would be sufficient to accomplish the central functions of the TCA cycle: precursor metabolite production, by-product recycling, and energy generation.
Fig. 5.
The TCA pathway. (A) The canonical TCA cycle is shown, with bypasses and alternative reactions present in other cyanobacterial species indicated by dotted lines. (B) The TCA pathway, the proposed biologically relevant TCA process. Insertion loss of function mutants in fum (8S42-O6; synpcc7942_1007), me2 (8S29-J6; synpcc7942_1297), and sucd (8S1-JJ4; synpcc7942_1533) were made (SI Materials and Methods, Table S2), and growth was compared with the WT in both (C) continuous light and (D) cycles of alternating light–dark (12–12 h). Colony area was measured using ImageJ (80). Reaction and metabolite abbreviations are given in BiGG format (bigg.ucsd.edu) and found in Dataset S2. ***Significance level of 0.001 (t test).
The TCA pathway preserves the enzymes necessary for the synthesis of oxaloacetate and 2-oxoglutarate, which are precursors for many required biomass components; additionally, 2-oxoglutarate serves as the gateway to nitrogen assimilation. Therefore, functionality in producing precursor metabolites and nitrogen assimilation can be provided by the TCA pathway.
The TCA pathway does not include functionality for the recycling of fumarate. Fumarate is created as a by-product of nucleotide and arginine synthesis, the salvage of which is posited by previous models to be essential (69). However, loss of function mutants for fumarase and malic enzyme show that this recycling function is not required for viability on solid media or in liquid culture (Fig. 5C and Fig. S7 A and B). The dispensability of fumarate salvage could be explained by the ability of S. elongatus to excrete fumarate into the media (70), and when this possibility was added into the model, the in silico predictions for fumarase and malic enzyme become nonessential in agreement with the in vivo data. In addition, iJB785 shows a minor cost of excreting useable carbon backbones (Fig. S7C). This cost is commensurate with the in vivo data that show a significant decrease in colony size in the fumarase and malic enzyme mutants (Fig. 5C). Therefore, fumarate recycling is a dispensable function of the TCA cycle, despite a slight fitness cost.
Fig. S7.
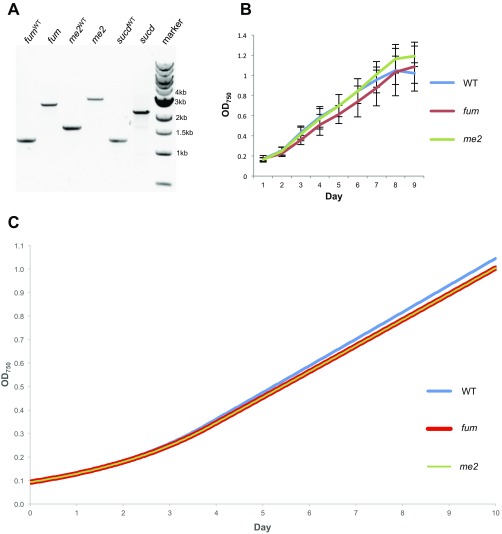
Genotypic characterization and growth curve of the TCA cycle mutants. (A) Lane 1 (fumWT), amplification of WT DNA with primers surrounding the fum gene (synpcc7942_1007); lane 2 (fum), amplification with the same primers from fum transposon insertion KO mutant (8S42-O6), in which a 1.3-kb insertion is present; lane 3 (me2WT), amplification of WT DNA with primers surrounding the gene encoding malic enzyme (synpcc7942_1297); lane 4 (me2), amplification with the same primers from the transposon insertion KO mutant for the gene encoding malic enzyme (8S29-J6); lane 5 (sucdWT), amplification of WT DNA with primers surrounding the sucd subunit B gene (synpcc7942_1533); lane 6 (sucd), amplification with the same primers from the sucd transposon insertion KO mutant (8S1-JJ4); and lane 7 (marker), standard 1-kb ladder (New England BioLabs). Each band is representative of three colonies tested. (B) Growth of WT, fumarase mutant, and malic enzyme mutant strains in liquid culture. Error bars show SDs of three independent replicates for each mutant. (C) Single-gene deletions of each of the TCA cycle genes fum (synpcc7942_1007) and me2 (synpcc7942_1297) were performed in silico, and the resulting impact to growth rate was analyzed.
The final core function of the TCA cycle is energy production. During photosynthetic metabolism in S. elongatus, however, full oxidation of pyruvate by the TCA cycle would amount to “metabolic suicide,” in which the cell is fixing and degrading the same carbon compounds concurrently (71). The wastefulness of a complete TCA cycle for energy generation in S. elongatus is supported by viability of mutants defective for succinate dehydrogenase subunit B (Synpcc7942_1533, EC 1.3.5.1) (Fig. 5C), which in addition to its importance for cycle flux, is an electron donor to the electron transport chain in PCC 6803 (53). At nighttime, however, when the cell switches from photosynthesis to glycogen as its energy source (72), we might expect that a cyclic TCA process would become essential, because it would enable further energy generation from glycogen. In fact, expression of TCA cycle enzymes has been shown to occur in the dark period during light–dark cultivation (73), and modeling has shown cyclic flux through the cycle in PCC6803 under these conditions (50). Therefore, we repeated the viability assay under day–night conditions but found that fumarase, malic enzyme, and succinate dehydrogenase remain nonessential (Fig. 5D). Furthermore, iJB785 simulations of dark metabolism indicated that full oxidation of glycogen through the oxidative pentose phosphate (OPP) pathway could generate equivalent ATP compared with a complete TCA cycle in S. elongatus (24.4 mol ATP/mol glucose in OPP vs. 24.7 mol ATP/mol glucose via TCA bypass). This prediction is supported by previous experimental evidence that the OPP pathway is important for diurnal survival in S. elongatus (74–76). Taken together, the model, the essential gene dataset, and our loss of function mutants support the hypothesis that an abridged TCA pathway, focused on generation of precursor metabolites, and not the traditional TCA cycle is the physiologically relevant TCA process for S. elongatus. This finding diverges from the current paradigm of complete TCA cycles in cyanobacteria (63).
TCA cycles focused on biosynthesis instead of energy generation have precedents. Green sulfur bacteria run their TCA cycle in reverse to fix CO2 in a process called the reductive TCA cycle (77). It also is common for obligate autotrophs to lack the enzyme 2-oxoglutarate dehydrogenase (71). The hypothesized TCA mechanism in the absence of 2-oxoglutarate is a bifurcated process, in which a reductive branch leads to succinyl-CoA and an oxidative branch leads to 2-oxoglutarate (71). According to iJB785, however, S. elongatus does not require succinyl-CoA or any other metabolites of the reductive branch of this TCA process. Thus, it is likely that just the oxidative wing of the TCA cycle, represented as the TCA pathway, is important largely as a biosynthetic pathway in S. elongatus.
The truncated TCA pathway of S. elongatus probably generalizes to other members of the phylum. Even if we artificially model the complete TCA cycle in S. elongatus by adding malate dehydrogenase and the 2-oxoglutarate dehydrogenase bypass present in Synechococcus sp. PCC 7002 (66), the model still predicts no cyclic flux through the completed TCA cycle. In PCC 6803, which contains a complete TCA cycle, both flux balance analysis (50) and 13C MFA (78) show negligible flux from 2-oxoglutarate to the rest of the TCA cycle. Furthermore, when the complete TCA cycle of PCC 6803 is blocked, only minor decreases in growth rate are observed (67). Finally, a bifurcated TCA cycle with a reductive branch for succinyl-CoA synthesis is likely to be nonessential in many cyanobacteria because of the presence of a heme biosynthesis pathway that begins with 2-oxoglutarate instead of succinyl-CoA (79). Based on these data, the abridged TCA pathway as opposed to a complete or bifurcated TCA process is likely relevant in other cyanobacteria, even those with genes making a complete TCA cycle.
SI Materials and Methods
Genome-Scale Reconstruction for Synechococcus elongatus.
The genome annotation of Synechococcus elongatus PCC 7942 was obtained from the Cyanobase database (genome.microbedb.jp/cyanobase/SYNPCC7942; date accessed: 9/2015). Functional annotation of the predicted ORFs was performed using the BLAST command line tool (81). Initially, the amino acid sequences of the S. elongatus proteins were compared with all reviewed cyanobacterial proteins in the Uniprot/Swissprot database (82) (BLASTp; e-value cutoff 1e−20). The S. elongatus genes without a best hit in the cyanobacterial subset of the database were then searched against all reviewed Uniprot/Swissprot sequences that had evidence at the protein or transcript level (BLASTp; e-value cutoff 1e−20). Additionally, the S. elongatus protein sequences were queried for conserved domains using the National Center for Biotechnology Information Batch CD-Search Tool (83) with the default settings. A draft reconstruction for S. elongatus based on a previous model for sp. PCC 6803 (50), provided by Henning Knoop and Ralf Steuer, Humboldt-Universität zu Berlin Institute for Theoretical Biology, Berlin, served as the starting point for the reconstruction. The genome functional annotation was curated based on the BLASTp and CD-Search outputs, and the network was reconstructed as described previously (20) using the COBRApy Python package (84) in iPython Notebook (85).
Structural Homology Modeling of Annotated PGMs.
All protein structures were downloaded from the Protein Data Bank (PDB). PDB ID codes 4IJ5 [Hydrogenobacter thermophilus TK-6 metal-independent phosphoserine phosphatase 1 (iPSP1)] (24), 1E59 [Escherichia coli cofactor-dependent phosphoglycerate mutase (dPGM)] (86), and 1H2F (Bacillus stearothermophilus PhoE) (87) were selected for this analysis. Full-length homology modeling of amino acid sequences was conducted using a locally downloaded version of the I-TASSER v4.4 (iterative threading assembly refinement) package (88). The COACH package (89), which is contained within I-TASSER, was used for binding site and substrate binding predictions, which were cross-referenced with known dPGM and iPSP binding residues annotated within UniProt (82) and various literature sources. Additionally, the full-length homology model of the E. coli dPGM (UniProt ID code P62707) was obtained from a database of E. coli homology models previously generated with I-TASSER (zhanglab.ccmb.med.umich.edu/Ecoli/). To compare the positions of binding and active residues, structure files were simultaneously loaded into VMD (90), and sequences were aligned using the MultiSeq tool (91). MultiSeq also contains the structural alignment tool STAMP (92), which was used to align the regions of interest. The positions of known histidine phosphatase residues and residues that are known to contribute to either iPSP or dPGM activity were then compared between all homology models and the selected experimental PDB files.
Strains and Culture Conditions.
All WT assays were done in S. elongatus PCC 7942, which is stored in our laboratory’s culture collection as AMC06. Mutants were also constructed in this WT background. All culturing occurred at 30 °C. Liquid cultures were grown in 100 mL BG-11 medium in 250-mL flasks (PYREX) and shaken at 150 rpm (Thermo Fischer MaxQ 2000 Orbital Shaker).
Chlorophyll Determination.
Working under reduced irradiance to prevent degradation of extracted pigments, 1–2 mL cultures were harvested in triplicate in 2-mL tubes by centrifugation at 15,000 × g at laboratory temperature for 7 min. The supernatant fraction was removed, and cells were resuspended in 1 mL cold 100% (vol/vol) methanol. Samples were placed in a light-free container incubated at 4.0 °C for 1 h to extract the pigments from the cells. After incubation, cellular material was centrifuged at 15,000 × g for 10 min at 4.0 °C, and the supernatant was used for spectrophotometry. A Beckman Coulter DU 640B Spectrophotometer was calibrated using methanol as a blank, and absorbance at 665 and 720 nm was measured. Chlorophyll a concentrations were determined using the following equation: chlorophyll a (micrograms/milliliter) = 12.9447 (A665 − A720) (93).
Whole-Spectrum Light Absorption.
Whole-cell absorption spectra were measured every 2 nm using an Infinite 200 PRO Multiplate Reader (Tecan) from 400 to 800 nm. Before measurements, culture densities were adjusted to an OD750 of 0.05. Measurements using BG-11 medium as a blank were subtracted from the sample results, and then, results were normalized to OD750 for comparison.
Modeling of Photon Uptake.
Absorption spectra were collected as described above, corrected for light scattering by subtracting the OD750 value from each absorbance value, and then, normalized to a 1-cm path length. The chlorophyll a-normalized optical absorption cross-section was calculated from the absorbance by the following equation (modified from ref. 33):
The photosynthetically active radiation (PAR) range (400–700 nm) was divided into 15 bins of 20 nm each, and the values across each bin were averaged to give a spectrally averaged, chlorophyll a-normalized absorbance cross-section for each bin. The SE for each bin was determined from the SD of the in vivo absorption spectrum. The spectral distribution of the Osram Sylvania Octron(R) 741 Fluorescent Lamp was obtained from the manufacturer (www.sylvania.com/en-us/Pages/default.aspx). The spectrum was divided into 15 bins of 20 nm each, and the area of each bin was calculated using the trapz function in the NumPy Python package (www.numpy.org/).
After dilution to minimize light scattering, chlorophyll a measurements were used to normalize the absorption spectrum. The ratio of chlorophyll a to OD remained constant during the time course, indicating a lack of significant photoacclimation (Fig. S1). Because the model’s biomass equation explicitly defines the cellular composition, the biomass-normalized photon absorption rate, [micromoles photons gram dry weight (DW)−1 hour−1], was calculated from the combination of irradiance (micromoles photons meter−2 second−1), optical absorption cross-section [centimeters2 milligram chlorophyll a (Chl a)−1], and the chlorophyll a component of the biomass equation [modified from P259 (94)]:
The resulting photon absorption flux was modeled as 15 metabolites, each representing a 20-nm segment of the 400- to 700-nm photosynthetic range.
Physiological Assays.
S. elongatus was grown under 100 μmol photons m−2 s−1 from cool white fluorescent bulbs. Growth was monitored by taking OD750 measurements and dry cell weight measurements at six time points over the course of 10 d. Cells were diluted appropriately to achieve OD750 measurements between 0.05 and 1.0. Dry cell weight was determined by vacuum filtration of 50 mL culture material using preweighed 0.45-μm hydrophilic polypropylene filters (47-mm GH Polypro; p/n 66548; Life Sciences). Filter disks containing cellular material were then placed in a large glass petri plate, dried at 90 °C for 1 h, allowed to cool to room temperature, and weighed. The initial weight of the filter disk was subtracted from the final to get the weight of the dry cells. Filters were placed back in for an additional 30 min and weighed again to ensure that drying was complete.
Oxygen Evolution.
Activities of photosynthetic oxygen evolution were determined using a Clark-type oxygen electrode (ALGinstruments). The cells grown under 100 μmol photons m−2 s−1 light at 30 °C were placed in 15-mL sterile conical tubes and quickly transported to another room for oxygen evolution analysis. Cells with chlorophyll a concentrations of 1–2 μg/mL were placed in a water-heated apparatus and kept at 30 °C. Light was provided at various intensities following a regimen of initial dark for 5 min followed by 2 min of constant illumination and 2 min of complete darkness. Light intensities included 0, 50, 100, 500, 1,000, and 2,000 μmol photons m−2 s−1. BG-11 medium was used as a blank, and rates were calculated by dividing by chlorophyll a concentration.
Simulations of in Silico Growth Rates.
Growth rates were simulated for a 100-mL culture of S. elongatus in a 250-mL shaken flask. Irradiance (micromoles photons meter−2 second−1) was measured outside the flask using a QSL-100 PAR Irradiance Sensor (Biospherical Instruments Inc.). A correction factor for light loss caused by the apparatus was calculated by taking light measurements inside an empty flask at different irradiances and plotting the outside irradiance vs. the inside irradiance. A correction factor was derived from a linear regression of the data. The photon delivery rate was determined from the light source irradiance and the surface area of the flask. The flask was modeled as a frustum of a cone, with the lower radius equal to the bottom of a 250-mL flask and the upper radius equal to the flask radius at the culture height (culture volume = 100 mL). The lateral surface area was calculated and used to derive the photon delivery rate (micromoles photons second−1) by multiplying the photon flux density (micromoles photons meter−2 second−1) by the calculated surface area (square meters). Self-shading was determined by dividing the 100 mL culture volume into 50 sections totaling 2% of the culture biomass in each section. For each section, the photon absorption rate and the photon delivery rate of each 20-nm bin were compared, and the lesser of two values was set as the upper and lower bounds of the photon exchange reaction. For each section (n), the photon delivery rate [] was equal to the initial photon delivery rate [] minus the photon absorption of all previous slices:
Growth curves were simulated by dividing the culture duration into 240 1-h segments. To avoid numerical precision issues, the flux units were converted to micromoles milligram DW−1 hour−1. At each time point, the biomass-normalized constraints were converted to the total metabolite flow across the reaction for the 1-h time period. For example, at a time t where the biomass equals 100 mg dry cell weight, the nongrowth-associated maintenance constraint, which is 0.071 μmol mg DW−1 h−1, becomes 7.1 μmol. After accounting for light attenuation caused by self-shading, biomass was maximized for each of 50 culture sections as follows:
Maximize .
Subject to
.
The following constraints were set before simulation:
,
,
,
,
,
, where I = irradiance [oxygen evolution set to the experimental values with Platt fitting (ref. 95, pp. 687–701)],
(maximum flux through photosystem II), and
(metabolite channeling constraint of phosphoketolase pathway),
where SmgDW is equal to 2% of the biomass of a given time point. Nongrowth-associated maintenance was set to 0.071 μmol mg DW−1 h−1 for all simulations. The biomass yield for each time point was the summation of the biomass output of each of the 50 sections. Inorganic phosphate use was tracked to determine when the culture entered phosphate limitation-induced stationary phase. Biomass yield was converted to OD using a standard curve (Fig. S2). Simulations were performed using the COBRAtoolbox (96) optimizeCbModel function in Matlab (Mathworks) with Gurobi 6.5.1 (Gurobi Optimization).
Intracellular Flux Distribution During Linear Growth.
Reaction fluxes were taken from a time point in the linear portion of the growth curve. Simulations were constrained as above, with the exception that a secondary objective of minimizing the taxicab norm of the flux vector was applied (97). The metabolite flow in micromoles was normalized to the biomass of the given time point to convert the units to millimoles gram DW−1 hour−1.
In Silico Essential Gene Comparison.
The previous S. elongatus model, iSyf715, was downloaded in SBML format, and the gene reaction rules were changed to a Boolean format and added to the model based on the supplemental files in the publication (48). The growth rate was fixed to 0.02 h−1, and the minimum photon uptake to achieve the set growth rate was determined using the optimize_minimal_flux function in the COBRApy Python package (84) in iPython Notebook (85) using the Gurobi 6.5.1 solver (Gurobi Optimization). Photon flux to the photosystem II reaction (_lightII_r) was set to 7.382 mmol g DW−1 h−1, and photon flux was set to 7.18 mmol g DW−1 h−1 for the photosystem I reaction (_lightI_r). All other constraints were left at their default values; the CO2 and bicarbonate default uptake rates were both 1.99 mmol g DW−1 h−1. The photon uptake rate for iJB785 was set by varying the light irradiance until a growth rate of 0.02 h−1 was achieved. All other constraints were set to the same value as in iSYF715, with the exception of the nongrowth-associated maintenance, which was set to the calculated value of 0.071 mmol g DW−1 h−1. The in silico essential gene assessment for both models was made using the single_gene_deletion function in COBRApy (84). Gene deletions where greater than 80% of the target growth rate was still achieved were considered to reflect nonessential genes, 80–10% were considered as beneficial genes, and below 10% were labeled essential genes. Comparison between RB-TnSeq in vivo essentiality calls and in silico assessments were performed using in-house Python scripts in iPython Notebook (76).
TCA in Silico Simulations.
The impact of TCA gene KOs on growth rate was simulated by setting the reaction flux to zero for the TCA enzymes fumarase and malic enzyme. Growth curves were simulated as outlined above at a model irradiance of 68 μmol m−2 s−1 (∼90–100 μmol m−2 s−1 in experimental conditions). The TCA cycle was completed in silico by adding the alpha-ketoglutate dehydrogenase and succinyl-CoA synthase genes from the E. coli model iJO1366 (98). Growth curves and reaction fluxes were determined as outlined above. The TCA bypass reaction alpha-ketoglutarate decarboxylase was manually added to the model, and the succinyl-CoA dehydrogenase reaction from the E. coli model iJO1366 was added to complete the bypass. Growth curves and reaction fluxes were determined as outlined above. The malate dehydrogenase (BiGG ID code MDH) reaction from the E. coli model iJO1366 was added to the model. Growth curves and reaction fluxes were determined as outlined above. For simulations in the dark, the light uptake was set to zero, and the glucose released by glycogen degradation was set to an arbitrary rate of 6 mmol g DW−1 h−1. The objective function was set to maximize ATP production.
Generating and Assaying TCA Cycle Mutants.
Mutants were generated from plasmids taken from the unigene set, an arrayed mutant library for S. elongatus (99, 100). Standard transformation protocols were used for mutant generation (4), and genotyping was done using colony PCR (Table S2) with Taq DNA Polymerase (NEB). Liquid culture assays were conducted in BG-11 medium containing kanamycin (5 μg/mL) under light levels of ∼100 μmol photons·m−2·s−1, with OD750 taken every 24 h.
Table S2.
Primers for validation of the TCA pathway mutants
| Purpose | Primer name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
| Segregation check of fumarate hydratase mutant | Synpcc 1007 F/R | AACATCAAATCCAGTCGGCG | TCATTTGCCCATTACTCGCG |
| Segregation check of malic enzyme | Synpcc 1297 F/R | CATTAAGACTCTTCGCAC | GAAACAATATCTGCCCTAC |
| Segregation check of succinate dehydrogenase subunit B | Synpcc 1533 F/R | TTTTGGGTACGGCCTATT | CTCTAGAACAACTGAATCC |
For spot plates, 4 μL culture was plated onto solid BG-11 kanamycin medium in a 1:5 dilution series. Constant light-incubated spot plates were put under light levels of ∼100 μmol photons·m−2·s−1 for 5 d. Light–dark-incubated plates were grown in a 12:12 -h cycle with square transitions at the same light intensity for 8–9 d. Colony area was measured using ImageJ analysis software (80).
SI Results
Structural Homology Modeling.
PGMs fall into two categories that are structurally distinct: dPGM (Synpcc7942_2078, Synpcc7942_1516, and Synpcc7942_0485, EC 5.4.2.11) and cofactor-independent phosphoglycerate mutase (iPGM; Synpcc7942_0469, EC 5.4.2.12). The iPGM family performs the mutase reaction exclusively, whereas the dPGM family has been assigned various catalytic functions (86). Because the enzymatic activity of a given PGM is structure-dependent, we attempted to categorize the S. elongatus PGM reaction specificity through structural analysis.
Because the iPGM family has only been shown to perform the mutase reaction, structural homology modeling focused on the dPGM family. We generated structural homology models for the S. elongatus dPGMs using an automated in silico platform for protein structural prediction (88). The resulting homologous enzyme scaffolds included a dPGM from E. coli (86) and a PSP from H. thermophiles. Interestingly, the S. elongatus network reconstruction indicated a gap in the gene assignment for PSP (EC 3.1.3.3). The H. thermophilus PSP is a member of the dPGM family, and the crystal structure, including features necessary for catalytic function, has been elucidated (24). Structural features of the resulting homology models were compared with the E. coli dPGM and the H. thermophilus PSP controls to refine the functional annotation of the enzymes (SI Materials and Methods).
S. elongatus synpcc7942_0469 is the only gene encoding an iPGM, and it is essential in vivo. Thus, synpcc7942_0469 was annotated as the primary glycolytic PGM in S. elongatus. The protein encoded by synpcc7942_2078 shares structural features with the E. coli dPGM control and lacks features that are important for PSP activity in H. thermophilus. Because it is nonessential in vivo, synpcc7942_2078 was annotated as a dPGM: possibly performing the “reverse regulatory” function with Synpcc7942_0469 as proposed previously (23). These researchers also suggested that Synpcc7942_0485 functions as a PSP, and the recently characterized PSP in PCC 6803 has amino acid homology to Synpcc7942_0485 (25) (Table S1).
Synpcc7942_0485, its homolog in PCC 6803 (slr1124), and the H. thermophilus PSP shared strong structural similarity; synpcc7942_1516 was essential in vivo, and the protein carried structural features that could not be classified into a dPGM or PSP. Its genomic neighbor, synpcc7942_1517, encodes an essential cyanobacterial-conserved histidine kinase, and transcriptome mapping data indicated that synpcc7942_1516 and synpcc7942_1517 are coexpressed on the same transcript (26). Cyanobacteria have a variety of two-component systems comprising a histidine kinase and at times, a phosphatase to regulate signal transduction activity (101). We hypothesized that synpcc7942_1516 encodes a histidine phosphatase regulator of an uncharacterized cyanobacterial two-component system. As a regulatory enzyme, Synpcc7942_1516 fell outside the scope of the metabolic model and was not included in the model gene list.
Suggested Constraints for Modeling in Constant Light.
Upper and lower flux bounds through NADP:NADH transhydrogenase reaction set to zero (). Justification is given in the text.
The lower bound of the transaldolase reaction set to zero (). Corrects a central carbon essentiality discrepancy for sedoheptulose-1,7-bisphosphatase (BiGG ID code SBP), resulting in a more accurate intracellular flux map.
Upper and lower flux bounds through ornithine transaminase set to zero (). Ornithine transaminase connects proline and arginine biosynthesis. If active, neither of those pathways would be essential. However, both proline and arginine biosynthesis pathways are essential in vivo.
Upper and lower flux bounds through cytochrome oxidase set to zero (). Presence allows for a biologically infeasible pseudocyclic electron flow. Fluorescence microscopy experiments in S. elongatus indicated that the concentration of this complex is very low based on the inability to visualize GFP constructs (35). Transcriptomics data also indicated very little expression of this complex (26).
The upper bound of the transaldolase reaction set to zero, making it irreversible away from pyruvate (). Avoids biologically irrelevant bypass of lower glycolysis through cell wall degradation into lactate and conversion back into pyruvate.
Conclusions
The iJB785 GEM of metabolism in S. elongatus presented here is a comprehensive representation of obligate phototrophic metabolism. Our mechanistic modeling of photon absorption and self-shading addresses the persistent challenge of accurately modeling light as a metabolite. This approach can be applied to any phototrophic GEM, enabling modeling of core aspects of light-driven metabolism. The predictive nature of the method also enables tailored light regimes for bioprocess optimization of photosynthetic platforms.
In addition to iJB785’s value for metabolic engineering and its technical improvement to phototrophic modeling, it serves as a platform for biological discovery. In synthesizing much of the physiological understanding available for S. elongatus, iJB785 reveals the holes in this knowledge. Some of these holes include missing elements of the nucleotide salvage system, the reason that phosphoketolase is unable to bypass lower glycolysis, and the apparent noncanonical activity of the transhydrogenase. Furthermore, the incorporation of essential gene data both improved the model’s accuracy and highlighted disagreements, which could not be explained by published data for S. elongatus. Many of these inconsistencies represent new biology for S. elongatus, such as the importance of a linear, noncyclic TCA pathway. Finally, as a representation of our current best understanding of S. elongatus, iJB785 is an ideal surface on which to overlay whole-genome datasets. The future addition of omic datasets to the model will both greatly improve in silico representation of S. elongatus metabolism and identify additional biological unknowns.
Materials and Methods
Methods used to generate the genome-scale reconstruction, derive constraints, and generate in silico results are presented in SI Materials and Methods. Briefly, the metabolic reconstruction was assembled using an established protocol (20), and biomass-normalized photon absorption rate for a given wavelength range was calculated from the combination of irradiance, optical absorption cross-section, and the chlorophyll a component of the biomass equation. Growth rates and reaction fluxes were simulated by maximizing the biomass objective function. All additional experimental protocols can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
A draft reconstruction of S. elongatus metabolism was provided by Henning Knoop and Ralf Steuer. This research was supported by NIH Cell and Molecular Genetics Training Grant T32GM00724 (to B.E.R.), University of California, San Diego Frontiers of Innovation Scholars Program 2-P3071 (to D.G.W), National Science Foundation Grant MCB1244108 (to S.S.G.), and US Department of Energy, Office of Science, Office of Biological and Environmental Research Award DE-SC0008593 (to B.O.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613446113/-/DCSupplemental.
References
- 1.Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 2015;33(6):352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27(12):1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 3.Oliver JW, Machado IM, Yoneda H, Atsumi S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc Natl Acad Sci USA. 2013;110(4):1249–1254. doi: 10.1073/pnas.1213024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clerico EM, Ditty JL, Golden SS. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol. 2007;362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsson S, Nogales J. Cyanobacteria as photosynthetic biocatalysts: A systems biology perspective. Mol Biosyst. 2015;11(1):60–70. doi: 10.1039/c4mb00335g. [DOI] [PubMed] [Google Scholar]
- 6.Savakis P, Hellingwerf KJ. Engineering cyanobacteria for direct biofuel production from CO2. Curr Opin Biotechnol. 2015;33:8–14. doi: 10.1016/j.copbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 8.Borodina I, et al. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab Eng. 2015;27:57–64. doi: 10.1016/j.ymben.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Yim H, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7(7):445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
- 10.Shih PM, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110(3):1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metab Eng. 2011;13(6):656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim TY, Sohn SB, Kim YB, Kim WJ, Lee SY. Recent advances in reconstruction and applications of genome-scale metabolic models. Curr Opin Biotechnol. 2012;23(4):617–623. doi: 10.1016/j.copbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10(4):291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat Biotechnol. 2014;32(5):447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury R, Chowdhury A, Maranas CD. Using gene essentiality and synthetic lethality information to correct yeast and CHO cell genome-scale models. Metabolites. 2015;5(4):536–570. doi: 10.3390/metabo5040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin BE, et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA. 2015;112(48):E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed JL, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci USA. 2006;103(46):17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baroukh C, Muñoz-Tamayo R, Steyer JP, Bernard O. A state of the art of metabolic networks of unicellular microalgae and cyanobacteria for biofuel production. Metab Eng. 2015;30:49–60. doi: 10.1016/j.ymben.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Shastri AA, Morgan JA. Flux balance analysis of photoautotrophic metabolism. Biotechnol Prog. 2005;21(6):1617–1626. doi: 10.1021/bp050246d. [DOI] [PubMed] [Google Scholar]
- 20.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4(9):R54. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunk E, et al. Systems biology of the structural proteome. BMC Syst Biol. 2016;10:26. doi: 10.1186/s12918-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jablonsky J, Hagemann M, Schwarz D, Wolkenhauer O. Phosphoglycerate mutases function as reverse regulated isoenzymes in Synechococcus elongatus PCC 7942. PLoS One. 2013;8(3):e58281. doi: 10.1371/journal.pone.0058281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba Y, et al. Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis. J Biol Chem. 2013;288(16):11448–11458. doi: 10.1074/jbc.M112.449561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klemke F, et al. Identification of the light-independent phosphoserine pathway as an additional source of serine in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2015;161(Pt 5):1050–1060. doi: 10.1099/mic.0.000055. [DOI] [PubMed] [Google Scholar]
- 26.Vijayan V, Jain IH, O’Shea EK. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12(5):R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berla BM, Saha R, Maranas CD, Pakrasi HB. Cyanobacterial alkanes modulate photosynthetic cyclic electron flow to assist growth under cold stress. Sci Rep. 2015;5:14894. doi: 10.1038/srep14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendry JI, Prasannan CB, Joshi A, Dasgupta S, Wangikar PP. Metabolic model of Synechococcus sp. PCC 7002: Prediction of flux distribution and network modification for enhanced biofuel production. Bioresour Technol. 2016;213:190–197. doi: 10.1016/j.biortech.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 29.Krieger-Liszkay A. Singlet oxygen production in photosynthesis. J Exp Bot. 2005;56(411):337–346. doi: 10.1093/jxb/erh237. [DOI] [PubMed] [Google Scholar]
- 30.Patterson CO, Myers J. Photosynthetic production of hydrogen peroxide by Anacystis nidulans. Plant Physiol. 1973;51(1):104–109. doi: 10.1104/pp.51.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang RL, et al. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol Syst Biol. 2011;7:518. doi: 10.1038/msb.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He L, Wu SG, Wan N, Reding AC, Tang YJ. Simulating cyanobacterial phenotypes by integrating flux balance analysis, kinetics, and a light distribution function. Microb Cell Fact. 2015;14:206. doi: 10.1186/s12934-015-0396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubinsky Z, Berman T, Schanz F. Field experiments for in situ measurement of photosynthetic efficiency and quantum yield. J Plankton Res. 1984;6(2):339–349. [Google Scholar]
- 34.Liberton M, et al. Global proteomic analysis reveals an exclusive role of thylakoid membranes in bioenergetics of a model cyanobacterium. Mol Cell Proteomics. 2016;15(6):2021–2032. doi: 10.1074/mcp.M115.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu LN, et al. Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc Natl Acad Sci USA. 2012;109(28):11431–11436. doi: 10.1073/pnas.1120960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao F, et al. NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. strain PCC 6803. Plant Physiol. 2016;170(2):752–760. doi: 10.1104/pp.15.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battchikova N, et al. Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J Biol Chem. 2011;286(42):36992–37001. doi: 10.1074/jbc.M111.263780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allakhverdiev SI, Murata N. Environmental stress inhibits the synthesis de novo of proteins involved in the photodamage-repair cycle of Photosystem II in Synechocystis sp. PCC 6803. Biochim Biophys Acta. 2004;1657(1):23–32. doi: 10.1016/j.bbabio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Stamatakis K, Tsimilli-Michael M, Papageorgiou GC. On the question of the light-harvesting role of β-carotene in photosystem II and photosystem I core complexes. Plant Physiol Biochem. 2014;81:121–127. doi: 10.1016/j.plaphy.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 40.King ZA, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44(D1):D515–D522. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feist AM, Palsson BO. The biomass objective function. Curr Opin Microbiol. 2010;13(3):344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol. 1994;60(10):3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO₂. Sci Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata M, et al. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: Genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA. 2001;98(20):11789–11794. doi: 10.1073/pnas.191258298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodger FJ, Badger MR, Price GD. Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol. 2005;139(4):1959–1969. doi: 10.1104/pp.105.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConnell MD, Koop R, Vasil’ev S, Bruce D. Regulation of the distribution of chlorophyll and phycobilin-absorbed excitation energy in cyanobacteria. A structure-based model for the light state transition. Plant Physiol. 2002;130(3):1201–1212. doi: 10.1104/pp.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muramatsu M, Hihara Y. Acclimation to high-light conditions in cyanobacteria: From gene expression to physiological responses. J Plant Res. 2012;125(1):11–39. doi: 10.1007/s10265-011-0454-6. [DOI] [PubMed] [Google Scholar]
- 48.Triana J, et al. Generation and evaluation of a genome-scale metabolic network model of Synechococcus elongatus PCC7942. Metabolites. 2014;4(3):680–698. doi: 10.3390/metabo4030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong W, et al. Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Nat Plants. 2015;2:15187. doi: 10.1038/nplants.2015.187. [DOI] [PubMed] [Google Scholar]
- 50.Knoop H, et al. Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLOS Comput Biol. 2013;9(6):e1003081. doi: 10.1371/journal.pcbi.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerreiro AC, et al. Daily rhythms in the cyanobacterium synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol Cell Proteomics. 2014;13(8):2042–2055. doi: 10.1074/mcp.M113.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angermayr SA, Paszota M, Hellingwerf KJ. Engineering a cyanobacterial cell factory for production of lactic acid. Appl Environ Microbiol. 2012;78(19):7098–7106. doi: 10.1128/AEM.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooley JW, Vermaas WF. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: Capacity comparisons and physiological function. J Bacteriol. 2001;183(14):4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J Biol Chem. 2004;279(8):6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 55.Aoki M, Sato N, Meguro A, Tsuzuki M. Differing involvement of sulfoquinovosyl diacylglycerol in photosystem II in two species of unicellular cyanobacteria. Eur J Biochem. 2004;271(4):685–693. doi: 10.1111/j.1432-1033.2003.03970.x. [DOI] [PubMed] [Google Scholar]
- 56.Simkovsky R, et al. Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc Natl Acad Sci USA. 2012;109(41):16678–16683. doi: 10.1073/pnas.1214904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider D, Volkmer T, Rögner M. PetG and PetN, but not PetL, are essential subunits of the cytochrome b6f complex from Synechocystis PCC 6803. Res Microbiol. 2007;158(1):45–50. doi: 10.1016/j.resmic.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Husic DW, Tolbert NE. Inhibition of glycolate and D-lactate metabolism in a Chlamydomonas reinhardtii mutant deficient in mitochondrial respiration. Proc Natl Acad Sci USA. 1987;84(6):1555–1559. doi: 10.1073/pnas.84.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz D, et al. Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 2011;155(4):1640–1655. doi: 10.1104/pp.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eisenhut M, et al. Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2008;148(4):2109–2120. doi: 10.1104/pp.108.129403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA. 2012;109(7):2678–2683. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenhut M, et al. The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 2006;142(1):333–342. doi: 10.1104/pp.106.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinhauser D, Fernie AR, Araújo WL. Unusual cyanobacterial TCA cycles: Not broken just different. Trends Plant Sci. 2012;17(9):503–509. doi: 10.1016/j.tplants.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Pearce J, Leach CK, Carr NG. The incomplete tricarboxylic acid cycle in the blue-green alga Anabaena variabilis. J Gen Microbiol. 1969;55(3):371–378. doi: 10.1099/00221287-55-3-371. [DOI] [PubMed] [Google Scholar]
- 65.Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7(3):254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334(6062):1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 67.Xiong W, Brune D, Vermaas WF. The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol Microbiol. 2014;93(4):786–796. doi: 10.1111/mmi.12699. [DOI] [PubMed] [Google Scholar]
- 68.Zhang S, Bryant DA. Biochemical validation of the glyoxylate cycle in the cyanobacterium Chlorogloeopsis fritschii strain PCC 9212. J Biol Chem. 2015;290(22):14019–14030. doi: 10.1074/jbc.M115.648170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beck C, Knoop H, Axmann IM, Steuer R. The diversity of cyanobacterial metabolism: Genome analysis of multiple phototrophic microorganisms. BMC Genomics. 2012;13:56. doi: 10.1186/1471-2164-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hickman JW, et al. Glycogen synthesis is a required component of the nitrogen stress response in Synechococcus elongatus PCC 7942. Algal Res. 2013;2(2):98–106. [Google Scholar]
- 71.Wood AP, Aurikko JP, Kelly DP. A challenge for 21st century molecular biology and biochemistry: What are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol Rev. 2004;28(3):335–352. doi: 10.1016/j.femsre.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA. 2015;112(15):E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Welkie D, et al. Transcriptomic and proteomic dynamics in the metabolism of a diazotrophic cyanobacterium, Cyanothece sp. PCC 7822 during a diurnal light-dark cycle. BMC Genomics. 2014;15:1185. doi: 10.1186/1471-2164-15-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doolittle WF, Singer RA. Mutational analysis of dark endogenous metabolism in the blue-green bacterium Anacystis nidulans. J Bacteriol. 1974;119(3):677–683. doi: 10.1128/jb.119.3.677-683.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Broedel SE, Jr, Wolf RE., Jr Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. J Bacteriol. 1990;172(7):4023–4031. doi: 10.1128/jb.172.7.4023-4031.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scanlan DJ, Sundaram S, Newman J, Mann NH, Carr NG. Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177(9):2550–2553. doi: 10.1128/jb.177.9.2550-2553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atomi H. Microbial enzymes involved in carbon dioxide fixation. J Biosci Bioeng. 2002;94(6):497–505. doi: 10.1016/s1389-1723(02)80186-4. [DOI] [PubMed] [Google Scholar]
- 78.You L, Berla B, He L, Pakrasi HB, Tang YJ. 13C-MFA delineates the photomixotrophic metabolism of Synechocystis sp. PCC 6803 under light- and carbon-sufficient conditions. Biotechnol J. 2014;9(5):684–692. doi: 10.1002/biot.201300477. [DOI] [PubMed] [Google Scholar]
- 79.Oborník M, Green BR. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol Biol Evol. 2005;22(12):2343–2353. doi: 10.1093/molbev/msi230. [DOI] [PubMed] [Google Scholar]
- 80.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7-8):518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perez F, Granger BE. IPython: A system for interactive scientific computing. Comput Sci Eng. 2007;9(3):21–29. [Google Scholar]
- 86.Bond CS, White MF, Hunter WN. High resolution structure of the phosphohistidine-activated form of Escherichia coli cofactor-dependent phosphoglycerate mutase. J Biol Chem. 2001;276(5):3247–3253. doi: 10.1074/jbc.M007318200. [DOI] [PubMed] [Google Scholar]
- 87.Rigden DJ, Littlejohn JE, Henderson K, Jedrzejas MJ. Structures of phosphate and trivanadate complexes of Bacillus stearothermophilus phosphatase PhoE: Structural and functional analysis in the cofactor-dependent phosphoglycerate mutase superfamily. J Mol Biol. 2003;325(3):411–420. doi: 10.1016/s0022-2836(02)01229-9. [DOI] [PubMed] [Google Scholar]
- 88.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29(20):2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 91.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins. 1992;14(2):309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 93.Ritchie RJ. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth Res. 2006;89(1):27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
- 94.Falkowski PG, Raven JA. Aquatic Photosynthesis. 2nd Ed Princeton Univ Press; Princeton: 2007. [Google Scholar]
- 95.Platt T, Gallegos CL, Harrison WG. Photoinhibition of photosynthesis in natural assemblages of marine-phytoplankton. J Mar Res. 1980;38(4):687–701. [Google Scholar]
- 96.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat Protoc. 2011;6(9):1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lewis NE, et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orth JD, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Y, Holtman CK, Taton A, Golden SS. Functional analysis of the Synechococcus elongatus PCC 7942 genome. In: Burnap R, Vermaas W, editors. Functional Genomics and Evolution of Photosynthetic Systems. Springer; Dordrecht, The Netherlands: 2012. pp. 119–137. [Google Scholar]
- 100.Holtman CK, et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12(2):103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 101.Ashby MK, Houmard J. Cyanobacterial two-component proteins: Structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev. 2006;70(2):472–509. doi: 10.1128/MMBR.00046-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.