Significance
Methylation of cytosine bases in DNA is an epigenetic modification that influences gene expression. TET (ten-eleven translocation)-family dioxygenases catalyze conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and additional oxidized methylcytosines in DNA. Here, we show that both Tet3- and Tet1/2/3-deficient mouse ES cells showed impaired neural conversion, with skewing toward cardiac mesoderm. Genome-wide analyses showed that Tet3 mediates cell-fate decisions by inhibiting Wnt signaling. Consistent with these findings, Wnt signaling was hyperactivated in Tet1/2/3-deficient embryos, leading to aberrant differentiation of bipotent neuromesodermal progenitors into mesoderm at the expense of neuroectoderm. Our data demonstrate a key role for TET proteins in modulating Wnt signaling and establishing the proper balance between neural and mesoderm cell fate determination.
Keywords: TET methylcytosine oxidases, mouse embryonic stem cells, Wnt signaling, DNA demethylation, neuromesodermal progenitors
Abstract
TET-family dioxygenases catalyze conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and oxidized methylcytosines in DNA. Here, we show that mouse embryonic stem cells (mESCs), either lacking Tet3 alone or with triple deficiency of Tet1/2/3, displayed impaired adoption of neural cell fate and concomitantly skewed toward cardiac mesodermal fate. Conversely, ectopic expression of Tet3 enhanced neural differentiation and limited cardiac mesoderm specification. Genome-wide analyses showed that Tet3 mediates cell-fate decisions by inhibiting Wnt signaling, partly through promoter demethylation and transcriptional activation of the Wnt inhibitor secreted frizzled-related protein 4 (Sfrp4). Tet1/2/3-deficient embryos (embryonic day 8.0–8.5) showed hyperactivated Wnt signaling, as well as aberrant differentiation of bipotent neuromesodermal progenitors (NMPs) into mesoderm at the expense of neuroectoderm. Our data demonstrate a key role for TET proteins in modulating Wnt signaling and establishing the proper balance between neural and mesodermal cell fate determination in mouse embryos and ESCs.
TET (ten-eleven translocation) enzymes are a family of Fe(II) and 2-oxoglutarate–dependent dioxygenases, which successively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in DNA (1–3). The three mammalian Tet proteins, Tet1, Tet2, and Tet3, possess homologous C-terminal catalytic domains as well as CXXC domains that bind unmodified CpGs (4), except that the CXXC domain of TET2 became separated from the catalytic domain during evolution and is now a separate protein known as IDAX/CXXC4 (5). The oxidized methylcytosine (oxi-mC) species generated by Tet enzymes facilitate DNA demethylation through both passive (replication-dependent) and active (replication-independent) mechanisms (4); they also function as epigenetic marks that bind transcription factors and chromatin-associated proteins, thereby influencing chromatin structure and gene expression (6–8).
Mice lacking individual TET proteins develop relatively normally until birth and beyond (9). Specifically, Tet1-deficient mice on a mixed 129/Sv × C57BL/6 background produce normal-sized litters (10), but display minor behavioral abnormalities and defects in learning, memory, and expression of neuronal activation-related genes (11, 12), as well as a tendency to develop B-cell lymphomas relatively late in life (13). Tet2-deficient mice on a pure C57BL/6 background are also viable and fertile; they exhibit mild hematopoietic phenotypes and occasionally develop myeloid malignancies late in life (14, 15). Tet3-deficient mice die perinatally for unknown reasons (16). Deletion of two of the three TET family members has more serious consequences. A significant fraction of mice lacking both Tet1 and Tet2 survive to adulthood, whereas the remaining pups succumb late in embryogenesis or shortly after birth (17). Embryos lacking Tet1 and Tet3 only survive to embryonic day 10.5 (E10.5) and display poor forebrain formation and abnormal facial structures (18). The 5hmC generated by Tet family dioxygenases is abundant in neurons (19) and may be critical for neural development (20). In Xenopus, Tet3 plays a vital function in early eye and neural development by directly targeting several key developmental genes (21). All of these studies indicate that TET-family proteins are involved in neural development, which prompted us to further study—through triple deletion of all three TET proteins—the in vivo functions and redundancy of TET proteins during neural development.
In vivo neurulation is a fundamental event of embryogenesis that culminates in the formation of the anterior neural plate (ANP) and the posterior neural plate (PNP), which are the precursors of the brain and spinal cord, respectively (22). The ANP is derived directly from the epiblast (23), whereas the development of the PNP from the epiblast involves an intermediate state of bipotent neuromesodermal progenitors (NMPs), which can give rise to both the neurectoderm and mesoderm (24). Population fate maps at early somite stages have identified two regions containing NMPs: the dorsal layer of the node-streak border (NSB) and the caudal lateral epiblast (CLE) on either side of the primitive streak (PS) (24–27). In vitro, the induction of the neuroectoderm in ES cell (ESC) cultures is often referred to as the “default” pathway, because neuroectoderm readily develops in cultures that contain no serum or lack signaling through the Wnt, bone morphogenetic protein 4 (Bmp4), or activin signaling pathways (28–30).
Here we show that, in Tet3- and Tet1/2/3-deficient mouse ESCs (mESCs), differentiation of mESCs was skewed away from the default pathway of neuroectoderm lineage specification and toward the cardiac mesoderm fate. This phenotype appeared to be driven primarily by the absence of Tet3, because it was also observed in Tet3-deficient mESCs. Conversely, ectopic expression of Tet3 in mESCs promoted ectoderm differentiation and inhibited cardiac mesoderm differentiation. Through genome-wide chromatin immunoprecipitation-sequencing (ChIP-seq) and transcriptional profiling (RNA-sequencing; RNA-seq), we demonstrate that Tet3 mediates cell-fate decisions through inhibition of Wnt signaling, which was further corroborated by the fact that the skewed lineage specification in Tet3-deficient mESCs was rescued by treatment with Dickkopf1 (Dkk1), an extracellular inhibitor of Wnt signaling. Tet3-null mESCs showed decreased expression of genes encoding the Wnt signaling inhibitor secreted frizzled-related protein 4 (Sfrp4), correlating with increased DNA cytosine modification of the Sfrp4 promoter. Consistent with mESC findings, Wnt signaling was hyperactivated in Tet1/2/3-deficient embryos, as evidenced by increased levels of activated β-catenin, leading to aberrant differentiation of bipotent NMPs into mesodermal lineages at the expense of neural lineages. In summary, our data demonstrate a key role for TET proteins in modulating Wnt signaling and establishing the proper balance between neural and mesodermal cell fate specification in mouse embryos and ESCs.
Results
Tet3 Regulates the Balance Between Neuroectoderm and Cardiac Mesoderm Differentiation in mESCs.
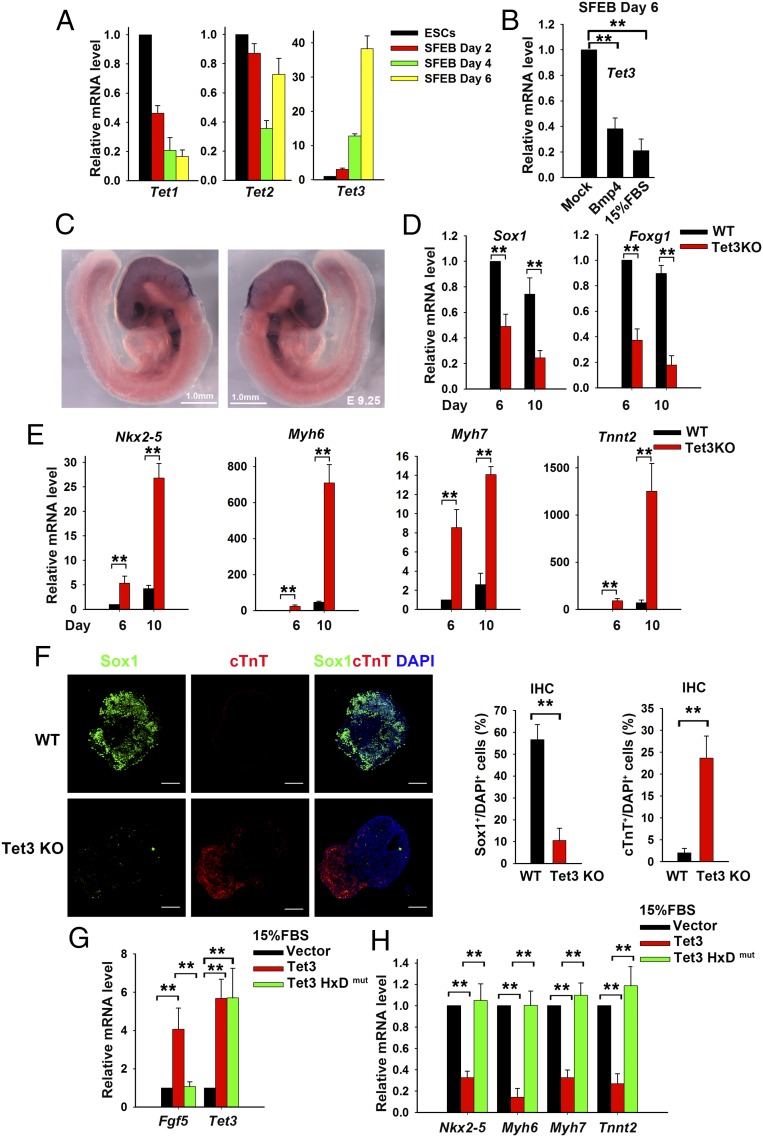
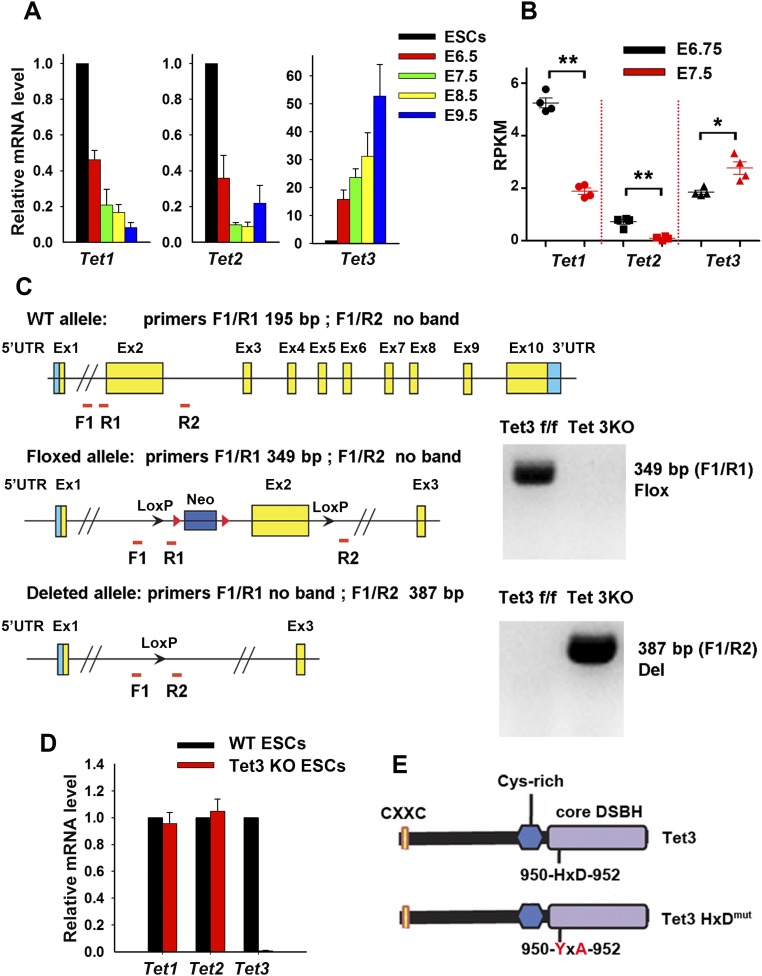
To investigate the role of Tet3 during neuroectoderm differentiation, we used an efficient system for in vitro neural differentiation of mESCs: serum-free floating cultures of embryoid-body like aggregates, referred to as “SFEB” (29, 31). In SFEB cultures, mESCs undergo neuroectoderm differentiation by default, without the induction of mesodermal and endodermal differentiation (29, 31). Consistent with our previous observation during retinoic acid-induced mESC differentiation (32), the expression levels of Tet1–3 displayed distinct patterns upon neural differentiation: Tet1 mRNA declined rapidly; Tet2 mRNA was initially maintained at a relatively steady level, then decreased on day 4, but recovered on day 6; and Tet3 mRNA showed a progressive increase, with >40-fold up-regulation on day 6 of SFEB culture compared with the starting mESCs (Fig. 1A). This increase in Tet3 transcripts was markedly reduced when the cultures were supplemented with inhibitory signals such as Bmp4 or FBS, both known to counteract neuroectoderm differentiation (28) (Fig. 1B). A similar pattern of Tet1–3 expression was observed in whole embryos during embryogenesis in vivo (Fig. S1A). RNA-seq analysis of early embryos at E6.75 and E7.5 further corroborated the quantitative PCR (qPCR) results, showing that Tet1 and Tet2 expression decreased, whereas Tet3 expression increased, upon development in vivo (Fig. S1B). Whole-mount in situ hybridization analysis of early embryos at E9.25 showed substantial Tet3 expression in the brain and optic vesicle (Fig. 1C). These expression patterns of Tet3 in vitro and in vivo suggested that Tet3 might have a role in neuroectoderm differentiation.
Fig. 1.
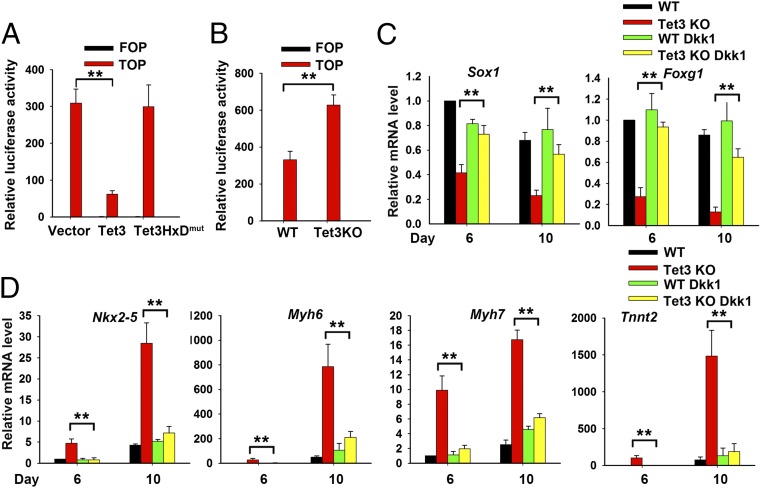
Tet3 mediates neuroectoderm and cardiac mesoderm cell fate determination in mESCs. (A) Quantitative real-time PCR (qRT-PCR) analysis of Tet1, Tet2, and Tet3 transcript levels in mESCs and neural cells differentiated by the SFEB method and normalized to the expression levels in mESCs (set at 1). Data are shown as mean ± SD (n = 3). (B) Tet3 expression level in mESCs cultured in differentiation medium alone or in the presence of 10 ng/mL BMP4 or 15% (vol/vol) FBS. Data are shown as mean ± SD (n = 3). (C) Whole-mount in situ hybridization for Tet3 mRNA at E9.25. (D and E) qRT-PCR analysis of transcripts of neural marker genes Sox1 and Foxg1 (D) and cardiomyocyte marker genes Nkx2-5, Myh6, Myh7, and Tnnt2 (E). WT or Tet3 KO mESCs were differentiated under SFEB culture conditions for 6 or 10 d. Data are shown as mean ± SD (n = 3). (F, Left) Immunocytochemistry of WT or Tet3 KO mESCs differentiated under SFEB culture conditions for 10 d. Cells were stained with anti-Sox1 (green, neural cell marker) and anti-cTnT (red, cardiac cell marker) antibodies. Nucleus staining: DAPI (blue). (Scale bars: 100 μm.) (F, Right) Percentage of Sox1+ or cTnT+ cells in the total cell population. IHC, immunohistochemistry. (G and H) qRT-PCR analysis of transcripts of Tet3 and ectoderm (Fgf5; G) or cardiomyocyte (Nkx2-5, Myh6, Myh7, and Tnnt2; H) marker genes in mESCs transfected with empty vector or vectors encoding Tet3 or Tet3HxDmut. The cells were cultured in differentiation medium containing 15% (vol/vol) FBS for 7 d. Data are shown as mean ± SD (n = 3). **P < 0.01.
Fig. S1.
Tet3 mediates neuroectoderm and cardiac mesoderm cell fate determination in mESCs. (A) qRT-PCR analysis of Tet1, Tet2, and Tet3 transcript levels on E6.5, E7.5, E8.5, and E9.5. For E6.5 and E7.5, three embryos are pooled. Data are shown as mean ± SD (n = 3). (B) RNA-seq data for Tet1, Tet2, and Tet3 mRNA expression in E6.75 and E7.5 embryos. RPKM, reads per kilobase of transcript per million mapped reads. (C, Left) Schematic view of targeting scheme for deletion of exon 2 in the endogenous Tet3 locus. (C, Right) Genotyping of Tet3 fl/fl and Tet3 KO mESCs. Bands for floxed (C, Upper) and Tet3 KO (C, Lower) alleles are shown. Genotyping primer sequences are provided in Dataset S4. (D) qRT-PCR analysis of Tet1, Tet2, and Tet3 transcript levels in WT and Tet3 KO mESCs. Data are shown as mean ± SD (n = 3). (E) Schematic representation of Tet3 and catalytically inactive TET3 HxD mutant. Cys-rich, cysteine-rich; DSBH, double-stranded β-helix. *P < 0.05; **P < 0.01.
To examine whether Tet3 is required for neuroectoderm differentiation in mESCs, we established a Tet3-deficient mESC line by Cre-mediated excision of exon 2 (Fig. S1C). mRNA levels of Tet1 and Tet2 were not altered, whereas Tet3 mRNA was almost undetectable, in Tet3 KO (Tet3−/−) mESCs (Fig. S1D). When WT and Tet3 KO mESCs were differentiated into embryoid bodies under serum-free conditions [SFEB (31)], neural marker genes Sox1 and Foxg1 showed a significant decrease at day 6, and the decrease was further enhanced at day 10 (Fig. 1D); moreover, mRNA expression levels of the cardiac precursor marker gene Nkx2–5 and the mature cardiomyocyte marker genes Myh6 and Myh7 (encoding cardiac myosin heavy chains) and Tnnt2 (encoding cardiac troponin T2; cTnT) increased dramatically in Tet3 KO mESCs (Fig. 1E). The skewed gene expression pattern of Tet3 KO cells was further corroborated at the protein level by immunostaining: A large portion of Tet3 KO cells were cTnT+ under conditions where the majority of WT mESCs had undergone neural differentiation, as judged by Sox1 expression (Fig. 1F). Collectively, in the absence of Tet3, the default tendency of mESCs to differentiate toward neuroectoderm was skewed toward cardiac mesoderm differentiation.
Conversely, we carried out gain-of-function experiments by stably overexpressing Tet3 in mESCs. To test the effects of Tet3 overexpression on mESC differentiation, we used culture medium containing FBS, which promotes differentiation toward mesoderm rather than neuroectoderm (30). Ectopic expression of Tet3 induced substantial expression of the ectoderm marker gene Fgf5 (Fig. 1G) and repressed expression of the cardiac lineage marker genes Nkx2–5, Myh6, Myh7, and Tnnt2 (Fig. 1H), consistent with our observations of increased cardiac skewing in Tet3 KO mESCs under serum-free (SFEB) culture conditions (Fig. 1E). This effect required the catalytic activity of Tet3, because expression of a catalytically inactive (HxD) mutant of Tet3, containing the amino acid substitutions H950D and Y952A (5) (Fig. S1E), did not alter Fgf5 or cardiac mesoderm marker gene expression (Fig. 1 G and H). Collectively, these findings demonstrate that Tet3 functions as a key determinant of the selective differentiation of mESCs toward the neuroectoderm vs. the cardiac mesoderm cell fate, in a manner dependent on Tet3 catalytic activity.
Tet3-Dependent Transcriptional Programs During mESC Differentiation.
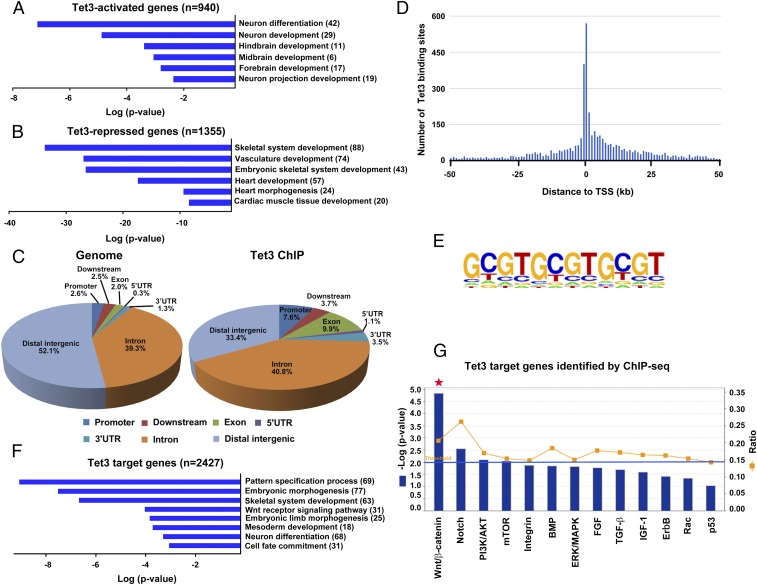
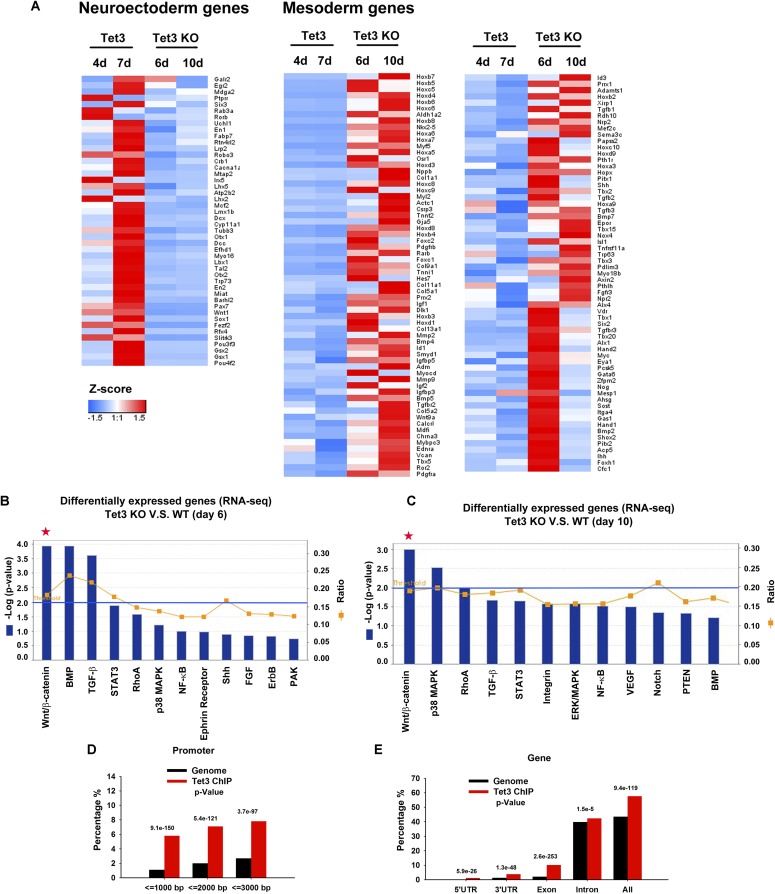
To determine the effects of Tet3 gain and loss on global gene-expression profiles during mESC differentiation, we performed whole-transcriptome RNA-seq analysis of (i) WT and Tet3 KO mESCs differentiating into neuroectoderm under SFEB culture conditions (days 6 and 10); and (ii) control and Tet3-expressing mESCs in nonpermissive neural differentiation conditions containing FBS (days 4 and 7). We defined 940 “Tet3-activated genes” that showed twofold or greater increase in expression in Tet3-expressing cells on day 4 or 7 and also twofold or greater decrease in expression in Tet3 KO cells on day 6 or 10; similarly, we defined 1,355 “Tet3-repressed genes” that showed twofold or greater decrease in expression in Tet3-expressing cells on day 4 or 7, and also a twofold or greater increase in expression in Tet3 KO cells on day 6 or 10 (Dataset S1). Gene Ontology (GO) analysis of Tet3-activated genes revealed a strong enrichment in functional categories relevant to neural developmental processes, including neuronal differentiation and brain development (Fig. 2A); conversely, Tet3-repressed genes were significantly implicated in mesoderm differentiation, including the skeletal system, vasculature, and heart development (Fig. 2B). Based on GO terms, we identified 46 Tet3-activated neural genes and 127 Tet3-repressed mesoderm genes (Fig. S2A and Dataset S1). To determine whether Tet3 directs neural differentiation by affecting extrinsic signaling pathways, we performed Ingenuity pathway analysis on genes differentially expressed in WT or Tet3 KO mESCs on days 6 and 10 of SFEB culture. For all time points analyzed, differentially expressed genes were highly enriched in components of Wnt/β-catenin pathways (Fig. S2 B and C).
Fig. 2.
Tet3-dependent transcriptional programs during mESC differentiation. (A) GO biological process analysis of Tet3-activated genes (defined as genes with twofold or greater increase in Tet3-overexpressing cells at day 4 or 7, concomitantly with twofold or greater decrease in Tet3 KO cells at day 6 or 10, relative to WT control cells). (B) GO biological process analysis of Tet3-repressed genes (defined as genes with twofold or greater decrease in Tet3-overexpressing cells at day 4 or 7, concomitantly with twofold or greater increase in Tet3 KO cells at day 6 or 10, relative to WT control cells). (C) Genomic distribution of Tet3-binding sites relative to their nearest RefSeq genes using the cis-regulatory element annotation system. “Promoter” was defined as 3 kb upstream from the TSS. “Downstream” was defined as 3 kb downstream from the 3′ end of the gene. “Distal intergenic region” refers to all locations outside the boundaries of a gene and the 3-kb region flanking the gene on either end. (D) Histogram showing the distribution of Tet3 ChIP-seq peaks relative to the nearest TSS. The majority of sites occupied by Tet3 in the genome are near the TSS (within ∼10 kb 5′ of the TSS and ∼25 kb 3′ of the TSS). (E) The highest-ranked DNA motif conserved in Tet3-bound loci revealed by de novo motif discovery analysis. (F) GO biological process analysis of Tet3 target genes (with at least one Tet3 ChIP-seq peak within 10 kb flanking the TSS). (G) Signaling pathway analysis (using Ingenuity Pathway Analysis software) of Tet3 target genes, defined as possessing a Tet3 ChIP-seq peak within the 10 kb flanking the TSS. The statistically significant canonical pathways are listed according to their P values (−Log10) (blue bars) and the ratio of Tet3 target genes found in each pathway over the total number of genes in that pathway (ratio) is shown by orange squares. The threshold line corresponds to a P value of 0.01. Note the predominance of genes in the Wnt/β-catenin signaling pathway among Tet3 target genes.
Fig. S2.
Tet3-dependent transcriptional programs during mESC differentiation. (A) Heat map of genes related to neuroectoderm differentiation, which were significantly activated by Tet3, and genes related to mesoderm differentiation, which were significantly repressed by Tet3. Red, high expression; blue, low expression using Z-score values normalized. (B and C) Signaling pathway analysis (using Ingenuity Pathway Analysis software) of genes differentially expressed (at least twofold change) in Tet3 KO relative to WT mESCs on days 6 (B) and 10 (C) of SFEB culture. The statistically significant canonical pathways are listed according to their P value (−Log10) (blue bars), and the ratio of differentially expressed genes found in each pathway over the total number of genes in that pathway (ratio) is shown by orange squares. The threshold line corresponds to a P value of 0.01. Note the predominance of the Wnt/β-catenin signaling pathway (red asterisks) at both 6 and 10 d. (D and E) Tet3 ChIP-seq peaks in NPCs were enriched in gene promoters (D) and gene bodies (E) above the genomic background by using randomly generated peaks.
To determine the global distribution of Tet3 DNA occupancy, we performed ChIP coupled to high-throughput sequencing (ChIP-seq) in neural precursor cells (NPCs). We chose to use NPCs for the ChIP-seq experiments because these cells display high expression of endogenous Tet3 (Fig. 1A) and constitute the endpoint of mESC differentiation under SFEB conditions (31). We identified 6,121 Tet3-bound regions (ChIP-seq peaks) in NPCs (Dataset S2), most of which were enriched in gene bodies and their immediate vicinity: By comparison with the mm9 reference genome, Tet3-binding sites were enriched in promoters and exons and to some extent also in 5′ and 3′ untranslated regions (UTRs), whereas peaks mapping to distal intergenic regions were substantially underrepresented (Fig. 2C and Fig. S2 D and E). Tet3-binding sites clustered close to transcription start sites (TSSs), with a low frequency of binding at distal regions relative to the TSSs (Fig. 2D), consistent with a previous report in which Tet3 binding regions were defined by pulldown with a GST fusion of the Tet3 CXXC domain (21). DNA motif analysis identified a CpG-rich sequence as the highest-ranked motif among Tet3-binding sites (Fig. 2E), also consistent with the previous report (21).
Validating the results of our RNA-seq data, GO analysis of Tet3 target genes (based on the presence of a Tet3 ChIP-seq peak within 10 kb of the TSS) showed enrichment for genes related to mesoderm development and neural differentiation (Fig. 2F). Ingenuity pathway analysis showed that Tet3 target genes were also strongly enriched for components of the Wnt signaling pathway (Fig. 2G), with the Notch signaling pathway being the second most highly enriched.
Tet3 Regulates mESC Differentiation via Modulation of the Wnt Signaling Pathway.
To further investigate the relation between Tet3 and the Wnt signaling pathway, we assessed the effects of Tet3 on β-catenin/TCF-mediated transcription using the TOP-Flash luciferase reporter, which contains a minimal promoter adjacent to seven tandem TCF-binding sites; the FOP-Flash luciferase reporter, which contains mutated TCF binding sites, was used as a negative control (33). The activity of the β-catenin/TCF reporter decreased to 20% of control levels in Tet3-expressing cells (Fig. 3A), whereas it increased by almost twofold in Tet3 KO cells relative to WT cells (Fig. 3B); again, expression of the catalytically inactive form of Tet3 (Tet3 HxD mutant) had no effect on reporter activity (Fig. 3A). These data suggest that Tet3 normally inhibits the Wnt/β-catenin signaling pathway and that the skewed differentiation observed in Tet3 KO mESCs could be due to defective repression of the Wnt/β-catenin signaling pathway in the absence of Tet3.
Fig. 3.
Tet3 regulates mESC differentiation by modulating Wnt/β-catenin signaling. (A and B) TOP/FOP-Flash luciferase reporter assay in vector, Tet3, or Tet3-HxDmut transduced mESCs (A) and in WT and Tet3 KO mESCs (B) induced to differentiate by withdrawal of LIF plus addition of 0.1 μM all-trans retinoic acid (RA) for 4 d. Data are shown as mean ± SD (n = 3). (C and D) qRT-PCR analysis of transcripts of neural marker genes Sox1 and Foxg1 (C) and cardiomyocyte marker genes Nkx2-5, Myh6, Myh7, and Tnnt2 (D). WT and Tet3 KO mESCs were differentiated under SFEB culture conditions for 6 or 10 d in the absence or presence of the Wnt inhibitor Dkk1 (100 ng/mL). Data are shown as mean ± SD (n = 3). **P < 0.01.
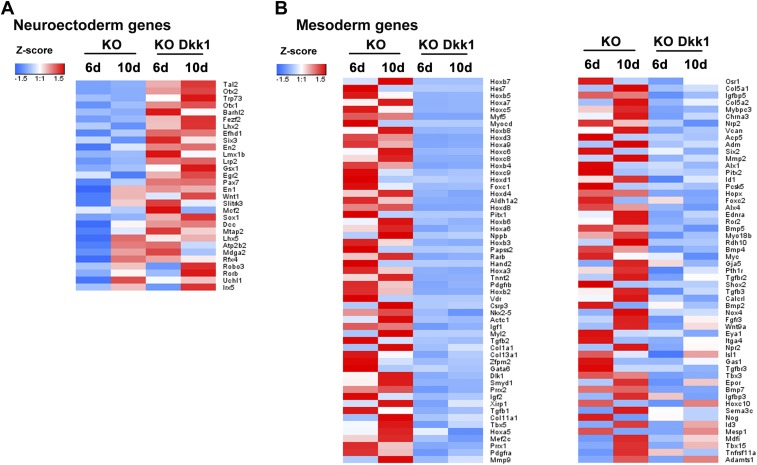
To test this hypothesis, we used a well-known negative regulator of Wnt signaling—Dkk1, a secreted protein that acts extracellularly to inhibit Wnt-receptor interactions (34). Addition of Dkk1 to SFEB cultures reversed the skewed differentiation of Tet3 KO mESCs away from neuroectoderm and toward cardiac mesoderm, as indicated by restored expression of the neural marker genes Sox1 and Foxg1 (Fig. 3C), and inhibited up-regulation of the cardiac mesoderm marker genes Nkx2-5, Myh6, Myh7, and Tnnt2 (Fig. 3D). Whole-transcriptome RNA-seq analysis revealed that the expression of 30 of 46 Tet3-related neural genes was restored, and the expression of 110 of 127 Tet3-related mesoderm genes was repressed, in Tet3 KO mESCs of SFEB culture in the presence of Dkk1 (Fig. S3 and Dataset S1). Collectively, these functional data combined with our genome-wide transcriptional and ChIP-seq data suggest that Tet3 regulates mESC differentiation by repressing Wnt signaling to regulate neuroectoderm vs. cardiac mesoderm cell fate.
Fig. S3.
Tet3 regulates mESC differentiation by modulating Wnt/β-catenin signaling. (A) Heat map of genes related to neuroectoderm differentiation, which were repressed in the absence of Tet3 and whose expression was restored by the addition of Dkk1 to the culture. Red, high expression; blue, low expression using Z-score values normalized. (B) Heat map of genes involved in mesoderm differentiation, which were activated in the absence of Tet3 and whose expression was inhibited by addition of Dkk1 to the culture. Red, high expression; blue, low expression using Z-score values normalized.
Tet3 Influences the Expression and DNA Modification Status of the Wnt Inhibitor Sfrp4.
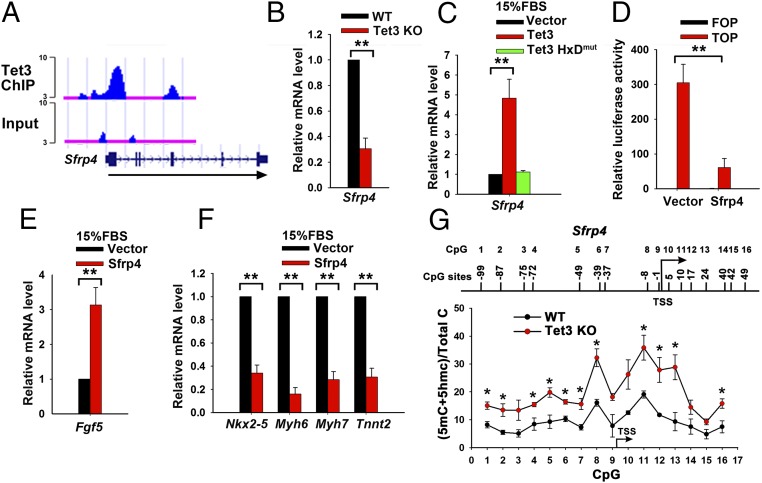
Combining our RNA- and ChIP-seq data, we identified 19 Tet3 target genes related to the Wnt/β-catenin signaling pathway (Dataset S1). Of these, the gene encoding Sfrp4 drew our attention: Sfrp4 possesses a domain similar to one in the Wnt receptor Frizzled protein and therefore can act as an inhibitor of Wnt signaling by preventing Wnt receptor binding: Epigenetic inactivation of the Sfrp4 gene results in constitutive Wnt/β-catenin signaling (35, 36). In ChIP-seq assays, Tet3 directly bound the promoter region of Sfrp4 in NPCs (Fig. 4A); moreover, Sfrp4 mRNA expression was dramatically down-regulated in Tet3 KO mESCs on day 6 of SFEB culture relative to WT control cells (Fig. 4B), whereas ectopic Tet3 expression induced substantial expression of Sfrp4, an effect that depended on the catalytic activity of Tet3 based on the fact that catalytically inactive (HxD mutant) Tet3 had no effect (Fig. 4C). Finally, stable expression of Sfrp4 in differentiating mESCs mimicked the effect of ectopic Tet3 expression: In luciferase assays using the β-catenin/TCF reporter, Wnt activity was inhibited (Fig. 4D), and there was substantial induction of the ectoderm marker gene Fgf5 (Fig. 4E) and repression of the cardiac lineage marker genes Nkx2-5, Myh6, Myh7, and Tnnt2 (Fig. 4F).
Fig. 4.
Tet3 regulates mESC differentiation through Sfrp4, an inhibitor of the Wnt signaling pathway. (A) University of California Santa Cruz Genome Browser snapshots showing Tet3 ChIP-seq peaks at the TSS and in the gene body of Sfrp4. The exon–intron structure of the Sfrp4 gene is shown below. The arrow shows the direction of transcription. The y axis of binding profiles denotes numbers of sequence tag reads. (B) qRT-PCR analysis of Sfrp4 transcripts in WT and Tet3 KO mESCs on day 6 of SFEB culture. Data are shown as mean ± SD (n = 3). (C) qRT-PCR analysis of Sfrp4 transcripts. Vector-, Tet3-, or Tet3HxDmut-transduced mESCs were cultured in differentiation medium containing 15% (vol/vol) FBS for 7 d. Data are shown as mean ± SD (n = 3). (D) TOP/FOP-Flash luciferase reporter assay in vector and Sfrp4-transduced mESCs induced to differentiate by withdrawal of LIF plus addition of 0.1 μM all-trans retinoic acid for 4 d. Data are shown as mean ± SD (n = 3). (E and F) qRT-PCR analysis of transcripts of ectoderm marker gene Fgf5 (E) and cardiomyocyte maker genes Nkx2-5, Myh6, Myh7, and Tnnt2 (F). Vector or Sfrp4-transduced mESCs were cultured in differentiation medium containing 15% (vol/vol) FBS for 7 d. Data are shown as mean ± SD (n = 3). (G) Bisulfite sequencing showing the percentage of 5mC+5hmC at each CpG site in the promoter region of Sfrp4 in WT and Tet3 KO mESCs on day 6 of SFEB culture. The positions of CpG sites are indicated relative to the TSS. Data are shown as mean ± SD (n = 2). *P < 0.05; **P < 0.01.
It is known that Sfrp4 expression is highly correlated to the methylation status of its promoter region (36). To investigate the relation of Sfrp4 expression to Tet3 function and changes in DNA methylation, we performed bisulfite-sequencing (BS-seq) to examine whether the down-regulation of Sfrp4 transcripts in differentiating Tet3 KO cells correlated with alterations of DNA cytosine modification. Because BS-seq cannot distinguish 5mC and 5hmC (37), cytosines not deaminated by bisulfite treatment are designated “5mC+5hmC.” In Tet3 KO mESCs on day 6 of SFEB culture, the percentage of “5mC+5hmC” in the promoter region of Sfrp4 increased compared with WT control cells across all of the CpGs analyzed (Fig. 4G). Together, these data suggest that Tet3 directly binds to and alters the DNA modification status of the Sfrp4 locus and regulates Sfrp4 transcription; in turn, Sfrp4 expression results in decreased Wnt signaling.
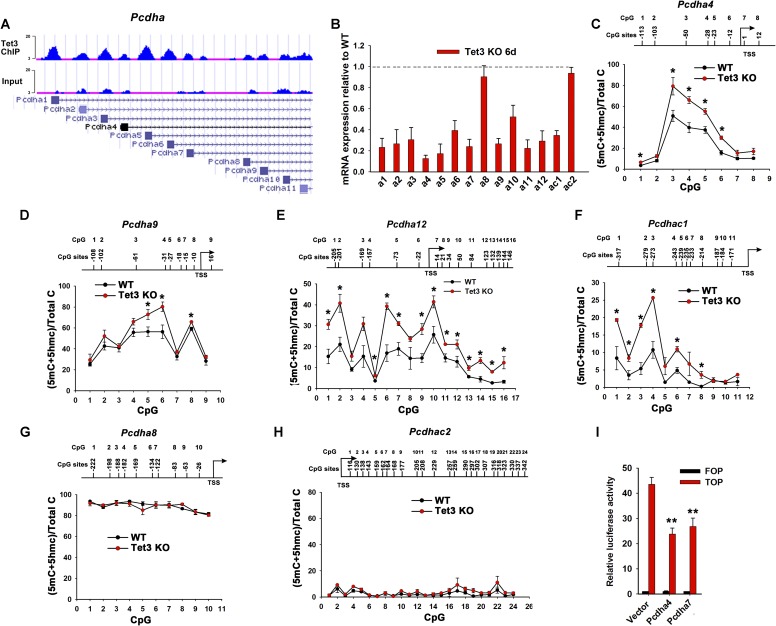
In addition to Sfrp4, our ChIP-seq data also showed that Tet3 directly bound each promoter region of a gene cluster encoding the protocadherin-alpha cluster (Pcdha) in NPCs (Fig. S4A). The Pcdha cluster contains 14 variable exons, each regulated by its own promoter, followed by three constant exons, and is highly expressed in neurons (38). Except for Pcdha8 and Pcdhac2, expression of all genes in the Pcdha cluster was dramatically down-regulated in Tet3 KO mESCs on day 6 of SFEB culture relative to WT control cells (Fig. S4B). The transcription of specific Pcdha isoforms has been shown to correlate significantly with the methylation status of the promoter and 5′ region of the first exon (39). In Tet3 KO mESCs on day 6 of SFEB culture, the percentage of 5mC+5hmC in the promoter regions of Pcdha4, Pcdha9, Pcdha12, and Pcdhac1 increased compared with WT control cells across the majority of CpGs analyzed (Fig. S4 C–F), whereas the methylation status of the promoter regions of Pcdha8 and Pcdhac2—the two isoforms whose mRNA expression was unaltered—remained unchanged (Fig. S4 G and H).
Fig. S4.
Tet3 regulates mESC differentiation through Pcdha cluster. (A) University of California Santa Cruz Genome Browser snapshots showing Tet3 binding sites in Pcdha. Gene tracks are shown at the bottom. The y axis of binding profiles denotes the numbers of sequence tag reads. (B) qRT-PCR analysis of transcripts of Pcdha in Tet3 KO mESCs on day 6 of SFEB culture. The transcript level in WT cells was set as 1. Data are shown as mean ± SD (n = 3). All bars are <1 with P < 0.05. (C–H) BS-seq showing the percentage of 5mC+5hmC at each CpG site in the promoter regions of Pcdha4 (C), Pcdha9 (D), Pcdha12 (E), Pcdhac1 (F), Pcdha8 (G), and Pcdhac2 (H) in WT and Tet3 KO mESCs on day 6 of SFEB culture. The positions of CpG sites are indicated relative to the TSS. Data are shown as mean ± SD (n = 2). *P < 0.05. (I) TOP/FOP-Flash luciferase reporter assay after transient expression of vector, Pcdha4 or Pcdha7 in mESCs induced to differentiate by withdrawal of LIF plus addition of 0.1 μM all-trans retinoic acid (RA) for 4 d. Data are shown as mean ± SD (n = 3). **P < 0.01.
It has been reported that the Pcdh superfamily functions to inhibit Wnt signaling (40–42). However, in our hands, transient expression of Pcdha4 and Pcdha7 during mESC differentiation inhibited Wnt signaling activity only moderately, based on a β-catenin/TCF reporter assay (less than twofold; Fig. S4I). Thus, Tet3 regulates the expression levels and the DNA modification status of the promoter regions of Sfrp4 and certain genes of the Pcdha cluster, and at least part of the ability of Tet3 to diminish Wnt signaling may be mediated through increased Tet3-mediated expression of Sfrp4. Given the lack of an embryonic phenotype in Sfrp4-deficient mice (43), additional factors likely contribute to the increased Wnt signaling activity in Tet3 KO cells.
Phenotype of Tet3-Deficient mESCs Is Exacerbated by Concurrent Deficiency of Tet1 and Tet2.
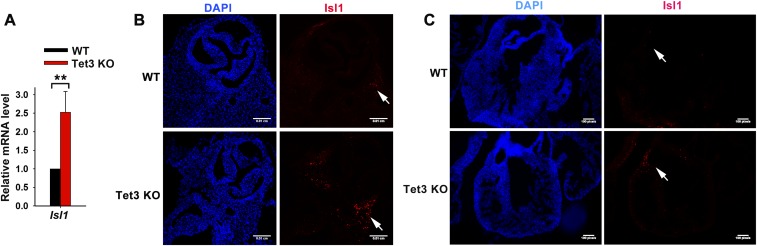
The data from Tet3 KO mESCs prompted us to look for developmental abnormalities in Tet3-deficient mice. As reported for 129Sv-background Tet3 KO mice (16), the C57BL/6-background Tet3-deficient pups generated in our laboratory survived until birth, but all pups showed perinatal lethality. Phase-contrast images of E18.5 embryos and hearts from WT and Tet3 KO mice showed no obvious developmental abnormalities of cardiac or other tissues. Nevertheless, qPCR and immunostaining analysis of the whole hearts from WT and Tet3 KO embryos (E18.5) showed an increase of the cardiac progenitor marker Isl-1, a target gene of Wnt/β-catenin signaling (44) in the Tet3 KO heart, especially in the right ventricle (Fig. S5). Thus, deficiency of Tet3 alone does not severely block embryonic development in vivo.
Fig. S5.
Cardiac progenitor marker Isl-1 expression is increased in the Tet3 KO heart. (A) qRT-PCR analysis of the transcript level of Isl1 of whole hearts from E18.5 WT and Tet3 KO embryos. Data are shown as mean ± SD (n = 3). (B and C) Immunostaining for Isl1 in transverse (B) and sagittal section (C) of heart from E18.5 WT and Tet3 KO embryos. Nucleus staining: DAPI (blue). **P < 0.01. (Scale bars: B, 100 μM; C, 62.5 μM.)
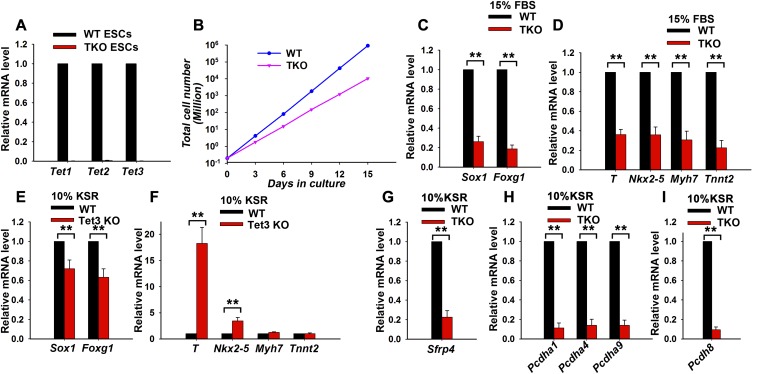
We hypothesized that the lack of overt cardiac and neuronal phenotypes in Tet3-deficient mice was most likely due to functional redundancy from the other two Tet family members, Tet1 and Tet2. To investigate this potential redundancy between Tet proteins, we generated Tet1/2/3 fl/fl mice from mice bearing the individual floxed alleles (14, 18) and used them to generate Tet1/2/3 fl/fl mESCs. These were then transiently transfected with a Cre expression plasmid to establish a Tet1/2/3 triple-deficient mESC line (Tet1/2/3 TKO mESCs) in which Tet1, Tet2, and Tet3 mRNAs were completely undetectable (Fig. S6A). The proliferation of these Tet1/2/3 TKO mESCs was significantly slower compared with the WT controls (Fig. S6B). Consistent with a previous report (45), neuroectoderm and cardiac mesoderm markers were all decreased during differentiation of Tet1/2/3 TKO mESCs in serum-containing cultures compared with the WT controls (Fig. S6 C and D).
Fig. S6.
The phenotype of Tet3-deficient mESCs is exacerbated by concurrent deficiency of Tet1 and Tet2. (A) qRT-PCR analysis of Tet1, Tet2, and Tet3 transcript levels in WT and Tet1/2/3 TKO mESCs. Data are shown as mean ± SD (n = 3). (B) Growth curves of WT and Tet1/2/3 TKO mESCs. Cells were split every 3 d, and cells were counted. (C and D) qRT-PCR analysis of transcripts of neural maker genes Sox1 and Foxg1 (C), mesoderm marker T (Brachyury), and cardiomyocyte marker genes Nkx2-5, Myh7, and Tnnt2 (D). WT or Tet1/2/3 TKO mESCs were cultured in differentiation medium containing 15% (vol/vol) FBS for 7 d. Data are shown as the mean ± SD (n = 3). (E and F) qRT-PCR analysis of transcripts of neural marker genes Sox1 and Foxg1 (E) and cardiac mesoderm marker T (Brachyury), Nkx2-5, Myh7, and Tnnt2 (F). WT or Tet3 KO mESCs were differentiated under SFEB culture conditions for 7 d in 10% (vol/vol) KSR. Data are shown as mean ± SD (n = 3). (G–I) qRT-PCR analysis of transcripts of Sfrp4 (G), Pcdha1, Pcdha4, Pcdha9 (H), and Pcdh8 (I). WT or Tet1/2/3 TKO mESCs were differentiated under SFEB culture conditions for 7 d in 10% (vol/vol) KSR. Data are shown as mean ± SD (n = 3). **P < 0.01.
We used medium supplemented with 10% (vol/vol) knockout serum replacement (KSR) medium to compare WT, Tet3 KO, and Tet1/2/3 TKO mESC differentiation in serum-free conditions; this culture condition was chosen because it supported Tet1/2/3 TKO mESC survival and differentiation better than the conditions used for Fig. 1D (N2B27 media; SI Materials and Methods). At day 7 of SFEB culture in 10% (vol/vol) KSR, Tet3 KO mESC showed a milder decrease of the neuroectodermal markers Sox1 and Foxg1 relative to WT cells than in N2B27 medium (compare Fig. S6E with Fig.1D); they also showed a significant increase of the mesoderm marker T (Brachyury) and a mild increase of the cardiac precursor marker Nkx2–5, but no change in the expression of the mature cardiomyocyte markers Myh7 and Tnnt2 (Fig. S6F). In contrast, compared with Tet3 KO cells, Tet1/2/3 TKO cells showed a more significant decrease of the neuroectoderm marker genes Sox1 and Foxg1 (compare Fig. 5A with Fig. S6E) and a more dramatic increase of the cardiac precursor marker Nkx2-5 and the mature cardiomyocyte markers Myh7 and Tnnt2 (compare Fig. 5B with Fig. S6F). Therefore, in 10% (vol/vol) KSR serum-free conditions, cell differentiation was skewed only toward early mesoderm in Tet3 KO mESCs; however, it could go further toward the cardiac mesoderm stage in Tet1/2/3 TKO ESCs. The previously mentioned Tet3 target gene, Sfrp4, which encodes a Wnt inhibitor, was also decreased in Tet1/2/3 TKO mESCs after differentiation under SFEB conditions (Fig. S6 G and H). In addition, we found that the expression level of another Pcdh superfamily member, Pcdh8, was decreased significantly in Tet1/2/3 TKO mESCs after differentiation under SFEB conditions (Fig. S6I). In light of a previous report showing that Pcdh8 can inhibit Wnt signaling activity (42), it is possible that down-regulation of both Sfrp4 and Pcdh8 contribute to increasing Wnt signaling in Tet1/2/3 TKO cells.
Fig. 5.
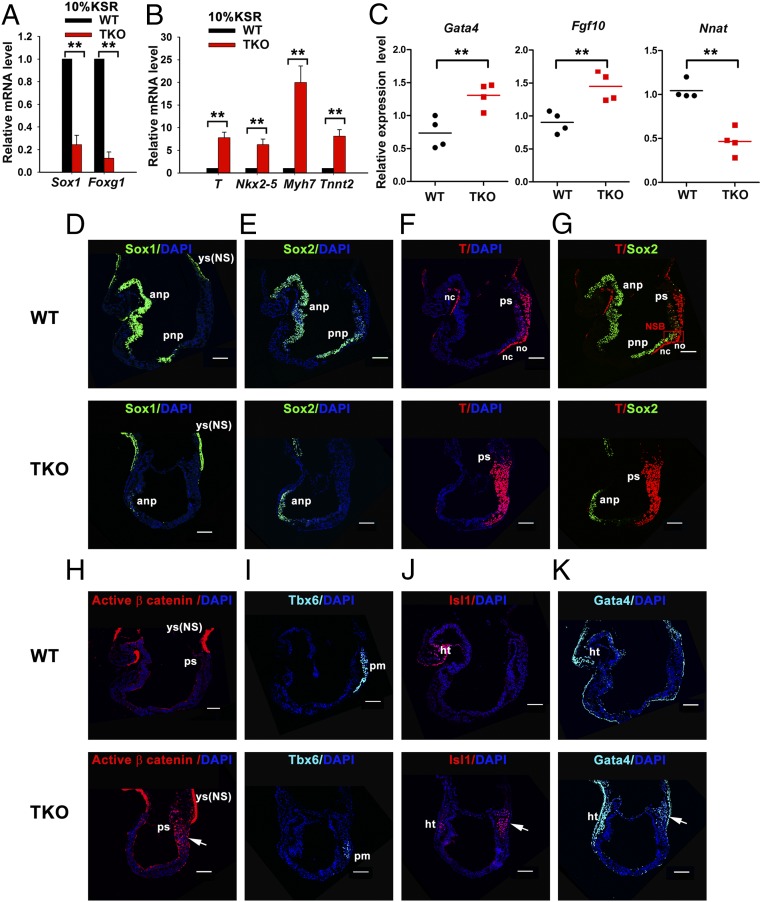
TET proteins control the balanced differentiation of NMPs during early embryogenesis in vivo. (A and B) qRT-PCR analysis of transcripts of neural marker genes Sox1 and Foxg1 (A) and cardiac mesoderm marker T (Brachyury), Nkx2-5, Myh7, and Tnnt2 (B). WT or Tet1/2/3 KO (TKO) mESCs were differentiated under SFEB culture conditions for 7 d in 10% (vol/vol) KSR. Data are shown as mean ± SD (n = 3). (C) RNA-seq data for cardiac progenitor marker genes (Gata4 and Fgf10) and neural initiator gene Nnat at E6.75 of WT and Tet1/2/3 TKO embryos. (D–K) Immunocytochemistry of WT or Tet1/2/3 TKO at E8.0–E8.25 for Sox1 (D), Sox2 (E), T (Brachyury) (F), Sox2 and T (G), active β-catenin (H), Tbx6 (I), Isl1 (J), and Gata4 (K). ht, heart; no, node; NS, nonspecific; pm, paraxial mesoderm; so, somites; yc, yolk sac. Nucleus staining: DAPI (blue). (Scale bars: 100 μm.) WT (n = 3); TKO (n = 4). **P < 0.01.
TET Proteins Control the Balanced Differentiation of Neuromesodermal Progenitors During Early Embryogenesis in Vivo.
To examine phenotypes of embryos lacking all three TET proteins, we generated Tet1/2/3 triple-deficient progeny by crossing Zp3- and Stra8-Cre mice with Tet1/2/3 fl/fl mice to generate mice in which expression of all three Tet proteins was abrogated in oocytes and sperm respectively. The progeny of Tet1/2/3 fl/fl Zp3-Cre female and Tet1/2/3 fl/fl Stra8-Cre male (hereafter termed Tet1/2/3 TKO or TKO mice/embryos) lack all three TET proteins beginning at the zygotic stage.
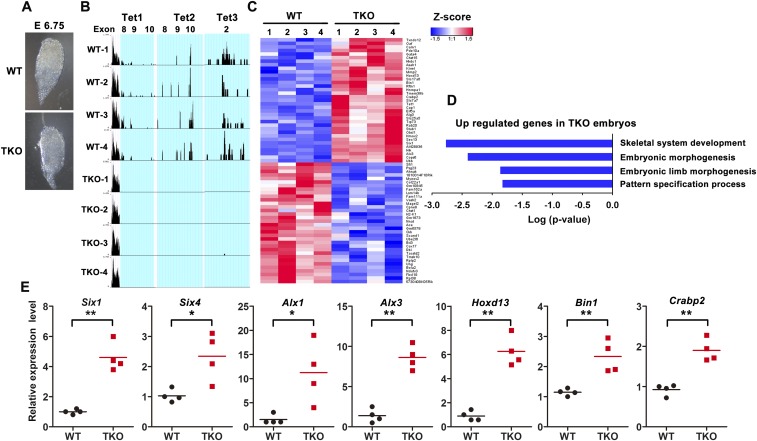
In Tet1/2/3 TKO embryos at early streak stages (E6.75), most decidua (31 of 42) were empty, and no degenerating embryos were observed, which may be due to the defects in oocytes in the absence of Tet proteins. However, surviving embryos (11 of 42) were morphologically indistinguishable from controls (Fig. S7A). Single-embryo RNA-seq of control and Tet1/2/3 TKO embryos (E6.75; four embryos from each group) showed efficient deletion of the targeted “floxed” exons (flanked by LoxP sites) of each Tet gene (Fig. S7B). GO analysis of the 69 genes whose expression levels were most significantly altered in Tet1/2/3 TKO embryos relative to controls (Fig. S7C and Dataset S3) revealed that up-regulated genes were implicated in mesoderm formation, including skeletal system development and limb morphogenesis (Fig. S7D): Cardiac progenitor marker genes (Gata4 and Fgf10) and genes related to embryonic limb and skeletal development (Six1, Six4, Alx1, Alx3, Bin1, Hoxd13, and Crabp2) were all significantly increased in Tet1/2/3 TKO embryos compared with control embryos (Fig. 5C and Fig. S7E). In contrast, the neural initiator Neuronatin (Nnat), which acts as an intrinsic factor to promote neural fate in mammals and Xenopus (46), was decreased in Tet1/2/3 TKO embryos (Fig. 5C). Thus, triple TET deficiency promotes expression of mesoderm-related genes during early embryogenesis in vivo.
Fig. S7.
Triple Tet deficiency promotes the expression of mesoderm-related genes during early embryogenesis. (A) Phase contrast images of WT (Tet1/2/3 fl/fl) and Tet1/2/3 TKO embryos at E6.75. (B) Genome browser snapshots showing RNA-seq reads for targeted exons of Tet1, Tet2, and Tet3. Reads corresponding to deleted Tet1 exons 8–10, Tet2 exons 8–10, and Tet3 exon 2 are specifically absent in Tet1/2/3 TKO embryos. (C) Hierarchical clustering of gene expression data for the 69 most significant differentially expressed genes between WT and Tet1/2/3 TKO E6.75 embryos. P < 0.0005; adjusted P value < 0.1. Red, high expression; blue, low expression using Z-score values normalized. (D) GO biological process analysis of up-regulated genes in Tet1/2/3 TKO embryos at E6.75. (E) RNA-seq data for seven mesoderm genes (Six1, Six4, Alx1, Alx3, Bin1, Hoxd13, and Crabp2) involved in regulation of embryonic limb and skeletal development. *P < 0.05; **P < 0.01.
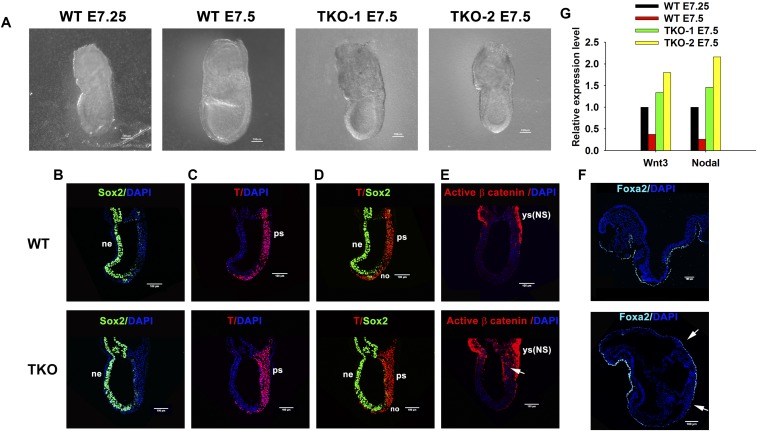
At mid-/late-streak stages (E7.25–7.5), we observed a developmental delay in Tet1/2/3 TKO embryos: When the control embryos were at late-streak stage, the Tet1/2/3 TKO embryos appeared to be at the midstreak stage (Fig. S8A). However, Tet1/2/3 TKO embryos at E7.5 were morphologically indistinguishable from control embryos at E7.25. Immunostaining results showed that in Tet1/2/3 TKO embryos at E7.5, the neural marker Sox2 was expressed in neuroectoderm (Fig. S8 B, Lower), the T-box transcription factor T (Brachyury) was expressed in the PS (Fig. S8 C, Lower), whereas Sox2+T+ NMPs, which give rise to both neuroectoderm and mesoderm, were detected in the PS close to the node (Fig. S8 D, Lower). These staining patterns were similar to those in WT control embryos at E7.25 (Fig. S8 B–D, Upper).
Fig. S8.
TET proteins control the balanced differentiation of NMPs during early embryogenesis in vivo. (A) Phase contrast image of WT and Tet1/2/3 TKO embryo at E7.25–E7.5. (Scale bars: 100 μM.) (B–E) Immunocytochemistry of WT (E7.25) or Tet1/2/3 TKO (E7.5) for Sox2 (B), T (C), Sox2 and T (D), active β-catenin (E). ne, neuroectoderm; no, node; NS, nonspecific; ys, yolk sac. Nucleus staining: DAPI (blue). (Scale bars: 100 μm.) WT (n = 3); TKO (n = 3). (F) Immunocytochemistry of WT or Tet1/2/3 TKO at E8.25-E8.5 for Foxa2. Due to the failure of neural plate closure, both left and right sides of the embryo are shown in Tet1/2/3 TKO embryo sections. Nucleus staining: DAPI (blue). (Scale bars: 100 μm.) WT (n = 3); TKO (n = 3). (G) RNA-seq data for Wnt3 and Nodal expression in WT and Tet1/2/3 TKO embryos at E7.25–7.5 shown in A.
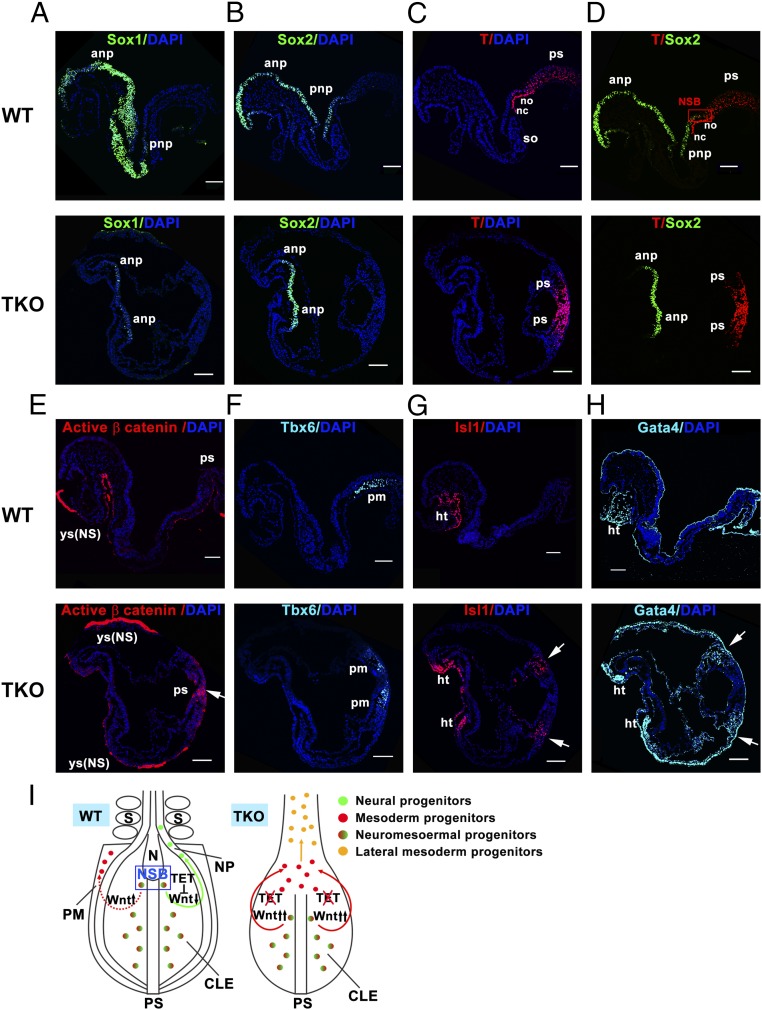
At head-fold stages (E8.0–8.25), clear morphological differences between WT and Tet1/2/3 TKO embryos were observed (Fig. 5 D–K). To investigate the nature of these patterning defects, we analyzed expression of characteristic lineage markers by immunocytochemistry. In WT embryos, the primordial neural gene Sox1 was maintained throughout the ANP, which later differentiates into the brain, and the PNP; in contrast, in Tet1/2/3 TKO embryos, Sox1 expression was significantly reduced (Fig. 5D). Note that in these figure panels, we observed nonspecific staining of the yolk sac [indicated by ys(NS)].
In WT embryos, strong expression of Sox2 was observed in ANP and PNP (Fig. 5E, Upper), whereas T (Brachyury) was expressed in the PS, ventral node (no), and notochord (NC) (Fig. 5F, Upper). Sox2 and T were coexpressed in the NSB, where the dual-fated NMPs are located. NMPs have the capacity to develop into body midline tissues—PNP, somites (so), and NC (Fig. 5G, Upper). However, in Tet1/2/3 TKO embryos, Sox2 expression was only observed in ANP, and was completely absent in posterior regions of the embryos (Fig. 5E, Lower), whereas T (Brachyury) was readily detected in PS (Fig. 5F, Lower). Moreover, colocalization of Sox2 and T was not detected in the NSB region in Tet1/2/3 TKO embryos (Fig. 5G, Lower).
Activation of Wnt signaling can prevent NMPs from adopting a neural cell fate, because high Wnt3a levels in PS down-regulate Sox2 expression (47). To examine whether Wnt activity was altered in Tet1/2/3 TKO embryos, we analyzed expression of the active, dephosphorylated form of β-catenin. Immunostaining results showed that, in comparison with WT control embryos, there were strong signals for the active form of β-catenin in Tet1/2/3 TKO embryos overlapping the T (Brachyury)-positive PS region at both midstreak (E7.25–7.5) and head-fold (E8.0–8.25) stages (Fig. 5H and Fig. S8E, arrow).
In WT embryos, NMPs give rise to both neuroectoderm and paraxial mesoderm, as indicated by immunostaining for the neural marker Sox2 and the paraxial mesoderm marker Tbx6 (Fig. 5 E and I, Upper). However, in Tet1/2/3 TKO embryos, in addition to loss of Sox2 expression, expression of Tbx6 was significantly reduced (Fig. 5 I, Lower), suggesting an inability of NMPs to develop into paraxial mesoderm in Tet1/2/3 TKO embryos. Next, we examined expression of lateral mesodermal markers Isl1 and Gata4. Anterior lateral mesoderm can give rise to cardiac mesoderm and further form the heart (48). We found that both Isl1 and Gata4 were ectopically coexpressed in the caudal region of Tet1/2/3 TKO embryos, in addition to their normal expression in the heart region (Fig. 5 J and K, Lower, arrow). In contrast, in WT control embryos at this stage, Isl1 and Gata4 were coexpressed only in the cardiogenic region (Fig. 5 J and K, Upper). The observation in Tet1/2/3 TKO embryos of ectopic expression of cardiac mesodermal markers in the absence of neural plate is consistent with prior studies, which demonstrated that factors within neural plate signal to adjacent paraxial mesoderm to prevent its adopting a cardiac mesodermal fate (49). Thus, in Tet1/2/3 TKO embryos, the absence of PNP would lead to a lack of repressive signals from neural plate, and therefore mutant NMPs, unable to form neuroectoderm, would adopt cardiac mesodermal rather than paraxial mesodermal cell fate.
At slightly later stages (E8.25–8.5), a striking feature of Tet1/2/3 TKO embryos was the complete absence of neural plate, somite, and NC (Fig. 6 A–H). In Tet1/2/3 TKO embryos, neural markers Sox1 and Sox2 were expressed in the ANP, but the territory of expression was reduced to varying extents (Fig. 6 A and B, Lower); T (Brachyury) was expressed in the PS at the caudal end of the embryo, where Wnt signaling was hyperactivated, as indicated by ectopic expression of active β-catenin (Fig. 6 C and E, Lower, arrow). We also observed that in Tet1/2/3 TKO embryos, the paraxial mesoderm marker Tbx6 was again reduced in anterior PS (Fig. 6F, Lower), and the lateral mesoderm markers Gata4 and Isl1 continued to be ectopically expressed in the anterior PS region (Fig. 6 G and H, Lower, arrow). Because Gata4 and Isl1 can also mark endodermal tissues in the cardiogenic region, we examined expression of the endodermal marker Foxa2. Notably, cells ectopically expressing Gata4 and Isl1 were negative for the endoderm marker Foxa2 (Fig. S8F, Lower, arrow), affirming their mesodermal identity. Together, these observations demonstrated that the fate of NMPs residing in the PS was skewed toward lateral mesoderm instead of neuroectoderm and paraxial mesoderm.
Fig. 6.
Molecular analysis of Tet1/2/3 TKO embryos at E8.25–8.5. (A–H) Immunocytochemistry of Tet1/2/3 TKO at E8.25–8.5 for Sox1 (A), Sox2 (B), T (Brachyury) (C), Sox2 and T (D), active β-catenin (E), Tbx6 (F), Isl1 (G), and Gata4 (H). Due to the failure of neural plate closure, both left and right sides of the embryo are shown in Tet1/2/3 TKO embryo sections. ht, heart; no, node; NS, nonspecific; pm, paraxial mesoderm; so, somites; yc, yolk sac. Nucleus staining: DAPI (blue). (Scale bars: 100 μm.) WT (n = 3); TKO (n = 3). (I) A model for TET function in developmental specification of NMPs in the E8.5 mouse embryo. In WT embryos, NMPs (red/green) are located in the NSB and CLE. When Wnt signaling is inhibited by Tet proteins, NMPs give rise to neural progenitors (green) which will contribute to the neural plate (NP), whereas activated Wnt signaling promotes the development of NMPs to mesoderm progenitors (red), which will contribute to paraxial mesoderm (PM). In Tet1/2/3 TKO embryos, Wnt signaling is abnormally activated in the absence of TET proteins. Overactivation of Wnt signaling in NMPs residing in the PS leads to their differentiation into mesoderm progenitors (red) and then further into lateral mesoderm (orange), resulting in abrogated development of body midline structures.
Together, these results indicated that, in Tet1/2/3 TKO embryos, anterior patterning was severely disrupted, with deficiencies in head development. In posterior development, NMPs located in the NSB/CLE, which are normally committed to develop into both neural plate and paraxial mesoderm all along the anterior–posterior (AP) body axis, were skewed to a lateral mesodermal cell fate, likely owing to overactivation of Wnt signaling (model in Fig. 6I). These in vivo observations are completely consistent with the aberrant differentiation of Tet3 KO and Tet1/2/3 TKO ESCs in serum-free conditions in vitro, in which we observed a decrease of neuroectodermal markers and an increase of cardiac mesodermal markers.
SI Materials and Methods
Plasmids.
Mutant Tet3 (H950D and Y952A) was generated by PCR using Pfu polymerase, Dpn I treatment, and transformation (Stratagene). The ORF of mouse Tet3, mutant Tet3 (H950D and Y952A), Sfpr4, Pcdha4, and Pcdha7 were cloned into pPyCAGIP vector (60).
Generation of Tet3 KO and Tet1/2/3 TKO mESCs.
Tet3 KO and Tet1/2/3 TKO mouse ESCs were generated from mice (C57BL/6 background) bearing the individual floxed alleles (Fig. S1C) (14, 18), followed by excision of the floxed exon by transient expression of Cre recombinase.
mESC Culture and Differentiation.
mESCs were maintained on mitomycin C-treated mouse embryonic fibroblasts (MEFs; feeders) in standard medium (61). Neural differentiation in SFEB culture was performed as described (31) with a minor modification. Briefly, mESCs were dissociated to single cells in 0.25% trypsin–EDTA (Invitrogen). Dissociated ESCs (1 × 105 cells per mL) were seeded onto a nonadhesive bacterial-grade dish and cultured in Knockout DMEM supplemented with 10% (vol/vol) KSR, 2 mM glutamine, 0.1 mM nonessential amino acids, and 0.1 mM 2-mercaptoethanol (2-ME). On day 2, the medium was changed to N2B27 medium (62). The medium was changed every other day. The day on which ESCs were seeded to differentiate is defined as differentiation day 0. Because Tet1/2/3 TKO mESCs did not differentiate well under SFEB culture in N2B27 medium, they were differentiated by using the SFEB method in 10% (vol/vol) KSR medium continuously. For nonpermissive neural differentiation, dissociated ESCs (1 × 105 cells per mL) were seeded onto an ultralow attachment culture dish (Corning) and cultured in Knockout DMEM supplemented with 15% (vol/vol) FBS, 2 mM glutamine, 0.1 mM nonessential amino acids, and 0.1 mM 2-ME for 4 or 7 d.
Generation of Tet1/2/3-Deficient Mice.
The Tet1 gene was inactivated by targeting its exons 8–10 encoding part of the catalytic domain (18). Tet2 targeting was described (14). The Tet3 gene was inactivated by targeting exon 2 (18). The deleted allele bears a frame-shift mutation that results in a truncated protein (Fig. S1B). The strains of Zp3-Cre and Stra8-Cre mice used in this study are FVB/N-TgN(Zp3-Cre)3Mrt and Tg(Stra8-Cre)1Reb/J (Jackson Laboratories) respectively. Zp3-Cre and Stra8-Cre are exclusively expressed in growing oocytes and spermatogonia, respectively (63, 64). We generated Tet1/2/3 triple-deficient mice by crossing Zp3-Cre and Stra8-Cre mice with Tet1/2/3fl/fl mice to generate mice in which expression of all three Tet proteins was abrogated in oocytes and sperm, respectively. The progeny of Tet1/2/3fl/fl Zp3-Cre female and Tet1/2/3fl/fl Stra8-Cre male lack all three Tet proteins beginning at the zygotic stage. Tet1/2/3 triple-deficient mice were on C57BL/6 background.
Whole-Mount in Situ Hybridization.
Embryos from E9.25 were dissected, fixed at 4 °C overnight in 4% (wt/vol) paraformaldehyde/PBS, dehydrated through a series of increasing methanol concentrations, and stored at −20 °C. Whole-mount in situ hybridization was performed as described (65). Tet3 probe was amplified from mouse cDNA (GenBank accession no. NM_183138.2) and corresponds to the nucleotide 569–1,443 region.
Luciferase Reporter Assay.
β-catenin/TCF reporter assay was performed as described (61). Briefly, cells were seeded at a density of 1 × 105 per well into a 24-well tissue-culture plate 24 h before transfection. Cells were transfected with reporter constructs TOP- or FOP-Flash using Lipofectamine 2000 (Invitrogen). Cell extracts were prepared 48 h after transfection. The luciferase activity was evaluated by using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s recommendations.
qRT-PCR.
qRT-PCR was performed by using Universal SYBR Green Master Mix (Roche) and analyzed by a StepOne Plus real-time PCR system (Applied Biosystems), according to the manufacturer’s instructions, and the data were normalized for Gapdh expression. The primers used for qRT-PCR are listed in Dataset S4.
Immunohistochemistry.
Immunohistochemistry was performed as described (61) with the primary antibodies described below. For statistical analysis, ∼300 cells were examined for each experiment, which was repeated four times. Mouse embryos were fixed overnight in 4% (wt/vol) paraformaldehyde, saturated with 20% (wt/vol) sucrose, and frozen in O.C.T. embedding medium, and 8-μm sections were prepared on a cryotome. The following antibodies were used: anti-Sox1 (no. 4194; Cell Signaling), anti-Sox2 (NL20181V; R&D), anti-cTnT (CT3; Hybridoma Bank), anti-T (Brachyury) (AF2085; R&D), anti-Gata4 (sc-1237; Santa Cruz), anti-Tbx6 (AF4744; R&D), anti-Isl1 (NL1837R; R&D), anti-Foxa2 (IC2400G; R&D), and anti–Active-β-Catenin (05-665; Millipore) generated against the peptide 36-44, HSGATTTAP, dephosphorylated at Ser-37 and Thr-41.
Whole-Transcriptome RNA-Seq.
Total RNA was extracted by using an RNeasy Plus Mini Kit (Qiagen). Multiplexed libraries were constructed by using a SOLiD Total RNA-seq kit. Libraries were sequenced on the SOLiD 5500 platforms according to the manufacturer’s instructions. Approximately 10–15 million uniquely mapped reads per sample were generated. Reads were aligned to the mouse genome (mm9; NCBI37) by using Tophat. DE-Seq was performed to identify differentially expressed genes. GO biological process analysis was performed by using DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/). Pathway analyses were performed by using Ingenuity Pathway Analysis software.
ChIP-Seq.
ChIP using anti-Tet3 antibody (gift of Guoliang Xu, Institute of Biochemistry and Cell Biology, Shanghai, China) (16) was performed in NPCs. To generate NPCs, mESCs were differentiated in SFEB culture conditions [10% (vol/vol) KSR] for 5 d, followed by adherent culture for 6 d in N2B27 with 10 ng/mL EGF and basic FGF. ChIP assays were carried out as described (5). Briefly, chromatin was sheared by using truChIP High Cell Chromatin Shearing Kit with Nonionic Shearing Buffer (Covaris). Chromatin fragments from two biologically independent NPCs were immunoprecipitated by using anti-Tet3 antibody. ChIP-seq libraries were constructed by using a 5500 SOLiD Fragment 48 Library Core Kit. Libraries were sequenced on the SOLiD 5500 platforms, according to the manufacturer’s instructions. Sequencing reads were mapped against mm9 by using Bowtie. Tet3-enriched regions were identified by MACS peak-calling software (Version 1.4.2) (66). We had performed pulldowns by using control IgG antibody, but no chromatin was recovered. Therefore, sequencing reads from input were used as negative controls in MACS. The statistical cutoff used for identifying Tet3-binding sites was P < 0.0005. Genomic distribution of Tet3-bound sites was performed by using the cis-regulatory element annotation system (67). The distribution of Tet3-binding sites to TSS was undertaken by using ChIPseek (68). De novo motif discovery was performed by using the findMotifsGenome.pl function of the Homer software package (69) using default parameter settings. Tet3-specific peaks within 100 kb flanking the TSS were used for motif analysis.
BS-Seq.
BS-seq was performed as described (70). Briefly, genomic DNA was extracted by using PureLink Genomic DNA mini Kit (Invitrogen) and treated with sodium bisulfite (MethylCode Bisulfite Conversion Kit; Invitrogen). The PCR amplicons were generated by using a PyroMark PCR kit (Qiagen) and quantified by using a Quant-iT PicoGreen dsDNA reagent (Invitrogen). The PCR amplicon was generated by using a PyroMark PCR kit (Qiagen). PCR amplicons were then mixed together for 1 μg final quantity and used for the library preparation using NEBNext DNA Library Modules for illumine platform (NEB). The final libraries were then combined together and quantified by using a KAPA library quantification kit for Illumina (KAPA Biosystems), then sequenced on Miseq (300 bp, paired end; Illumina). The data are based on thousands of sequence reads per amplicon. To monitor bisulfite conversion efficiency, a 210-bp spike-in control was generated by using unmethylated lambda DNA (Promega) as a template and added to the genomic DNA to a final ratio of 0.5%. Based on C-to-T conversion efficiencies of spike-in controls, the average bisulfite conversion efficiencies were >99.5%. Reads were aligned to the genome and used to measure methylation using BS-Seeker2 (71). The primer sequences are shown in Dataset S4.
Single-Embryo RNA Sequencing.
CTL and Tet1/2/3 TKO embryos were collected at E6.5. Single embryos were lysed and converted into double-stranded cDNA by using the SMARTer ultralow RNA kit for Illumina sequencing (Clontech). A total of 1 ng of cDNA for each sample was used for preparing libraries by using the Nextera XT DNA sample preparation kit (IIlumina). The good quality of the prepared libraries was validated by using the Bioanalyzer high-sensitivity DNA kit (Agilent). Libraries were sequenced on Illumina HiSeq 2500 platforms according to the manufacturer’s instructions. Approximately 10–15 million uniquely mapped reads per sample were generated. Reads were aligned to the mouse genome (mm9; NCBI37) by using Tophat. DE-Seq was performed to identify differentially expressed genes. GO biological process analysis was performed by using DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/).
Statistical Analysis.
All values are shown as means ± SD. To determine the significance between groups, comparison was made by using Student’s t test. For all statistical tests, the 0.05 confidence level was considered statistically significant. In all figures, * denotes P < 0.05 and ** denotes P < 0.01 in a two-tailed Student’s t test.
Discussion
Here, we show that Tet proteins, especially Tet3, are key transcriptional regulators that maintain the balance between neuroectodermal and mesodermal cell fate determination by repressing Wnt signaling. Both Tet3- and Tet1/2/3-deficient mESCs showed impaired neural conversion, with skewing toward cardiac mesoderm in serum free conditions. Conversely, ectopic Tet3 expression enhanced neuroectoderm differentiation and limited cardiac mesoderm specification. Impaired neuroectodermal differentiation and increased cardiac mesodermal specification of Tet3-deficient mESCs was rescued by addition of the extracellular Wnt inhibitor Dkk1. Our results demonstrate a key role for Tet3 in establishing the correct balance between neural and mesodermal cell fate through modulation of Wnt signaling in mESCs and emphasize a clear functional redundancy between TET-family enzymes in vivo.
Although Tet3 KO embryos developed until birth and showed no overt cardiac and neuronal phenotypes other than an increase in Isl-1 expression in the right ventricle, Tet1/2/3-deficient embryos showed major defects in neural development, manifesting a complete absence of midline structures, including the posterior neural tube, somites, and NC. Differences in single Tet3 vs. triple Tet KOs is most likely due to functional redundancy from the other two Tet family members, Tet1 and Tet2. In WT embryos, NMPs residing in the NSB and CLE contribute to both neural tube and paraxial mesoderm all along the AP body axis (24–27) (Fig. 6 I, Left). Timing and duration of Wnt activity are important parameters for the induction and maintenance of NMPs, which maintain self-renewal and bipotency at moderate levels of Wnt signaling: Cells exposed to a low-Wnt environment become neural, whereas cells exposed to high levels of Wnt transition to a mesodermal fate (47, 50, 51). Our studies of Tet1/2/3-deficient embryos and Tet3-deficient mESCs indicated that Wnt signaling was abnormally activated in the absence of TET proteins, particularly Tet3. Overactivation of Wnt signaling in NMPs residing in the PS region led to their differentiation into lateral mesoderm at the expense of neuroectoderm and paraxial mesoderm fate, resulting in abrogated development of body midline structures (Fig. 6 I, Right).
A precedent for this phenotype occurring downstream of hyperactivated Wnt signaling is provided by a transgenic mouse line in which ectopic Wnt3a is driven by a Cdx2 enhancer in the posterior epiblast. In Cdx2P–Wnt3a embryos, neuroectodermal specification is adversely affected, as evidenced by strong down-regulation of Sox2. However, activation of Wnt3a does not result in overproduction of paraxial mesoderm, but, rather, resulted in ectopic lateral mesoderm expressing Wnt2 and Tbx4 (47). Previous studies have demonstrated that specification of lateral mesoderm at the expense of paraxial mesoderm occurs in the absence of signaling from the neural tube, offering an explanation as to why lateral mesodermal fate, rather than paraxial mesodermal fate, is observed with loss of neural specification (49).
Tet1/2/3-deficient embryos also displayed defects in development of ANP. Indeed, formation of anterior neural structures requires suppression of Wnt signals, because mouse embryos deficient for the Wnt antagonist Dkk1 are headless (52). However, defects in ANP development were not fully penetrant and therefore could be secondary: Recent studies point to a role of node derivatives such as the NC and somites in maintaining and stabilizing anterior neural specification (53). In Hnf3b/Foxa2 conditional mutants, which lack paraxial mesoderm, specification of the ANP occurs, but is labile (54).
Besides their functionally redundant roles in the generation of oxi-mC, TET-family members also display distinct roles, in part because they are expressed in different cellular locations or at different developmental stages (18, 32), and regulate oxi-mC levels at different genomic locations (55, 56). Although there is some overlap, Tet1 primarily regulates 5hmC levels at gene promoters and TSSs, whereas Tet2 mainly regulates 5hmC levels in gene bodies (55). Our Tet3 ChIP-seq data in NPCs show that Tet3-binding sites also cluster close to TSSs, with a low frequency of binding at distal regions relative to the TSS, suggesting that Tet1 and Tet3 may have similar functions, despite their distinct temporal expression patterns during mESC differentiation (Fig.1A) (32). To gain further insights into the redundant and specific functions of TET proteins during embryogenesis, we will need to develop new techniques that enable genome-wide profiling of distinct cellular lineages, ideally at a single-cell level, in TET-deficient embryos.
A recent publication (57) showed that Tet1/2/3 TKO embryos display patterning defects in association with impaired specification of paraxial mesoderm. The authors suggested that hyperactive Nodal signaling contributed to the patterning defects in Tet1/2/3 TKO embryos. Our data extend their findings. We show that overactivation of Wnt signaling in NMPs leads to their differentiation into lateral mesoderm at the expense of neuroectoderm and paraxial mesoderm fate. We also show a strong correlation between the expression levels of Wnt3 and Nodal in both control and Tet1/2/3 TKO embryos at the gastrulation stage: Expression of both Wnt3 and Nodal RNAs decreased sharply from E7.25 to E7.5 in WT control embryos, but was high in Tet1/2/3 TKO embryos at day 7.5 (Fig. S8G). Notably, Wnt3 was shown to activate Nodal expression directly through its proximal epiblast enhancer at the gastrulation stage (58, 59). Xu and colleagues (57) observed that Wnt3 is strongly expressed in posterior of PS in Tet1/2/3 TKO embryos at E7.5, where we also found strong signals for the active form of β-catenin (Fig. S8E). Therefore, it is likely that Wnt and Nodal signaling pathways are both involved in Tet1/2/3-regulated embryonic patterning processes.
Materials and Methods
Detailed materials and methods are described in SI Materials and Methods. Dataset S4 lists the primers used for qRT-PCR, bisulfite sequencing and genotyping.
Supplementary Material
Acknowledgments
We thank Dr. Guoliang Xu for generously providing Tet3 antibody; Susan Togher and Ryan Hastie for animal genotyping; the La Jolla Institute sequencing facility for next-generation sequencing; and the La Jolla Institute microscopy and histology facility for help with histological and microscopic analysis. This work was supported by NIH Grants R01 CA151535 and R01 HD065812 (to A.R.). X.L. was supported by CIRM UCSD Interdisciplinary Stem Cell Research & Training Grant II Postdoctoral Fellowship TG2-01154.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE89313, GSE89314, GSE89315, and GSE89316).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617802113/-/DCSupplemental.
References
- 1.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ko M, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497(7447):122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto H, et al. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014;28(20):2304–2313. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79(6):1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13(2):237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16(6):653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko M, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 17.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang J, et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci USA. 2015;112(31):E4236–E4245. doi: 10.1073/pnas.1510510112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151(6):1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4(10):784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- 23.Iwafuchi-Doi M, et al. The Pou5f1/Pou3f-dependent but SoxB-independent regulation of conserved enhancer N2 initiates Sox2 expression during epiblast to neural plate stages in vertebrates. Dev Biol. 2011;352(2):354–366. doi: 10.1016/j.ydbio.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Henrique D, Abranches E, Verrier L, Storey KG. Neuromesodermal progenitors and the making of the spinal cord. Development. 2015;142(17):2864–2875. doi: 10.1242/dev.119768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cambray N, Wilson V. Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development. 2002;129(20):4855–4866. doi: 10.1242/dev.129.20.4855. [DOI] [PubMed] [Google Scholar]
- 26.Cambray N, Wilson V. Two distinct sources for a population of maturing axial progenitors. Development. 2007;134(15):2829–2840. doi: 10.1242/dev.02877. [DOI] [PubMed] [Google Scholar]
- 27.Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas JF. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev Cell. 2009;17(3):365–376. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz-Sanjuán I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3(4):271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 29.Kamiya D, et al. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470(7335):503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- 30.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132(4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe K, et al. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8(3):288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 32.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 34.Semënov MV, et al. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11(12):951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 35.Linhart HG, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21(23):3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyoda S, et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82(1):94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Kawaguchi M, et al. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. 2008;283(18):12064–12075. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- 40.Dallosso AR, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5(11):e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallosso AR, et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31(40):4409–4419. doi: 10.1038/onc.2011.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kietzmann A, Wang Y, Weber D, Steinbeisser H. Xenopus paraxial protocadherin inhibits Wnt/β-catenin signalling via casein kinase 2β. EMBO Rep. 2012;13(2):129–134. doi: 10.1038/embor.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christov M, Koren S, Yuan Q, Baron R, Lanske B. Genetic ablation of sfrp4 in mice does not affect serum phosphate homeostasis. Endocrinology. 2011;152(5):2031–2036. doi: 10.1210/en.2010-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qyang Y, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1(2):165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 45.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29(1):102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin HH, et al. Neuronatin promotes neural lineage in ESCs via Ca(2+) signaling. Stem Cells. 2010;28(11):1950–1960. doi: 10.1002/stem.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurberg AD, Aires R, Nóvoa A, Rowland JE, Mallo M. Compartment-dependent activities of Wnt3a/β-catenin signaling during vertebrate axial extension. Dev Biol. 2014;394(2):253–263. doi: 10.1016/j.ydbio.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91(6):457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 49.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11(4):451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 50.Martin BL, Kimelman D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev Cell. 2012;22(1):223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouldin CM, et al. Wnt signaling and tbx16 form a bistable switch to commit bipotential progenitors to mesoderm. Development. 2015;142(14):2499–2507. doi: 10.1242/dev.124024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukhopadhyay M, et al. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1(3):423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- 53.Camus A, et al. The morphogenetic role of midline mesendoderm and ectoderm in the development of the forebrain and the midbrain of the mouse embryo. Development. 2000;127(9):1799–1813. doi: 10.1242/dev.127.9.1799. [DOI] [PubMed] [Google Scholar]
- 54.Hallonet M, et al. Maintenance of the specification of the anterior definitive endoderm and forebrain depends on the axial mesendoderm: A study using HNF3beta/Foxa2 conditional mutants. Dev Biol. 2002;243(1):20–33. doi: 10.1006/dbio.2001.0536. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(4):1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56(2):286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai HQ, et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538(7626):528–532. doi: 10.1038/nature20095. [DOI] [PubMed] [Google Scholar]
- 58.Ben-Haim N, et al. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11(3):313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Arnold SJ, Robertson EJ. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10(2):91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 60.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 61.Li X, et al. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell. 2011;8(1):46–58. doi: 10.1016/j.stem.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 62.Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 63.Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7(2):148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 64.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46(12):738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin L, et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci USA. 2007;104(22):9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: Cis-regulatory element annotation system. Nucleic Acids Res. 2006;34(Web Server issue):W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen TW, et al. ChIPseek, a web-based analysis tool for ChIP data. BMC Genomics. 2014;15:539. doi: 10.1186/1471-2164-15-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue X, et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med. 2016;213(3):377–397. doi: 10.1084/jem.20151438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo W, et al. BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.