Significance
Receptor-like kinase FERONIA (FER) is a versatile regulator of cell growth under both normal and stress environments. FER binds its peptide ligand, rapid alkalinization factor 1 (RALF1), and triggers downstream events to inhibit cell growth in primary roots. However, the mechanism of RALF1 reception by FER is still largely unknown. In this study, we identified a receptor-like cytoplasmic kinase (RPM1-induced protein kinase, RIPK) that directly interacts with and is phosphorylated by FER in a RALF1 peptide-dependent manner. The defects of fer-4 mutant in RALF1 response and root hair development are mimicked by ripk loss-of-function but partially compensated by RIPK overexpression. These and other data suggest that formation of the FER–RIPK complex serves as a crucial step in the RALF1 signaling pathway.
Keywords: plant hormone, feronia, phosphorylation
Abstract
A number of hormones work together to control plant cell growth. Rapid Alkalinization Factor 1 (RALF1), a plant-derived small regulatory peptide, inhibits cell elongation through suppression of rhizosphere acidification in plants. Although a receptor-like kinase, FERONIA (FER), has been shown to act as a receptor for RALF1, the signaling mechanism remains unknown. In this study, we identified a receptor-like cytoplasmic kinase (RPM1-induced protein kinase, RIPK), a plasma membrane-associated member of the RLCK-VII subfamily, that is recruited to the receptor complex through interacting with FER in response to RALF1. RALF1 triggers the phosphorylation of both FER and RIPK in a mutually dependent manner. Genetic analysis of the fer-4 and ripk mutants reveals RIPK, as well as FER, to be required for RALF1 response in roots. The RALF1–FER–RIPK interactions may thus represent a mechanism for peptide signaling in plants.
The mechanism by which cells perceive and respond to external signals to control cell growth remains an area of central interest in cell biology. Receptor-like kinase (RLK) superfamily, with more than 600 members encoded in Arabidopsis genome, is believed to play a key role in coupling external signals to the regulation of plant growth and development (1–6). However, the function of the large majority of these RLKs remains to be explored. Among the RLKs, there is a 17-member subfamily referred to as Catharanthus roseus RLK1 (CrRLK1)-like (CrRLK1L). CrRLK1L proteins feature a predicted intracellular Ser/Thr kinase domain highly conserved among all RLKs, a transmembrane domain, and a variable extracellular domain. Within the extracellular domain of CrRLK1L proteins reside two malectin-like modules that bear limited homology to the endoplasmic reticulum (ER)-localized, carbohydrate-binding malectin protein of Xenopus laevis, suggesting that the ligands of CrRLK1L subfamily may be glycosylated or carbohydrate-rich (7). Several CrRLK1L family members have been shown to play a role in cell growth regulation. For example, THESEUS1 (THE1), located to the plasma membrane, detects perturbation of cellulose synthesis and may function as a cell wall integrity sensor (8, 9). ANXUR1 (ANX1) and ANXUR2 (ANX2), two redundant members of the CrRLK1L subfamily, play a role in controlling cell wall integrity during pollen tube (PT) growth (10–12). Loss of function of the ANX1 and ANX2 leads to precocious PT rupture shortly after germination, resulting in male sterility (11, 12).
FERONIA (FER), also a CrRLK1L subfamily member, was originally identified as a regulator for the communication between the male and female gametophytes during fertilization (13). Arabidopsis fer mutants have severe defects in fertility because PTs continue to grow inside the mutant female gametophyte and fail to rupture and release the sperm (13–15). Recent studies have revealed function of FER in a variety of other processes in Arabidopsis, including cell growth control in leaves (16), hormonal and stress responses (17, 18), mechanical signaling (19), root-hair development (20), and seed size control (21). A recent study further showed that FER serves as a receptor for a peptide ligand (RALF1) that binds FER and triggers inhibition of rhizosphere acidification, thereby suppressing cell elongation in roots (22). In addition to FER, RALF1 peptide also interacts with LLG1, which forms a complex with FER by interacting with the extracellular juxtamembrane region of FER, and the assembly of RALF1–LLG1/FER complex triggers activation of the GEF–ROP/ARAC pathway that mediates auxin-dependent root hair development (23). Interestingly, FER also uses the GEF1/4/10–ROP11 pathway to activate ABI2 phosphatases to inhibit ABA response (17). Furthermore, ABA signals cross-talk with RALF1 signaling through ABI2, which interacts directly with FER to dephosphorylate and inactivate FER (24). To date, however, the precise biochemical mechanism of perception and transduction of the RALF1 signal downstream of FER receptor remains unclear.
Studies on other subfamilies of RLKs have identified receptor-like cytoplasmic kinases (RLCKs) as downstream kinases that further transduce RLK–ligand signals. For instance, BIK1, an RLCK-VII subfamily protein kinase, works together with a leucine-rich repeat RLK (LRR-RLK), FLS2, in plant innate immunity to control NADPH oxidase activity and production of reactive oxygen species (ROS) (25–28). RIPK (RPM1-induced protein kinase), like BIK1, also belongs to the RLCK-VII subfamily and has been originally identified as a crucial component in plant immunity (29, 30). In the presence of Pseudomonas syringae effector AvrB, RIPK interacts with and phosphorylates an RPM1-interacting protein (RIN4), leading to activation of the NLR immune receptor RPM1 to initiate host immunity (30). Plant immunity and plant cell growth are closely connected with each other. For example, BSK1, a member of the RLCK-XII subfamily, is involved in both brassinosteroid (BR) signaling and innate immunity in Arabidopsis. BSK1 can form a BRI1–BAK1–BSK1 complex in BR signaling pathway to regulate cell growth. Meanwhile, BSK1 also works as a downstream kinase with FLS2 in plant innate immunity (31, 32).
In search of downstream players of RALF1–FER signaling, we show here that RIPK directly interacts with FER and is rapidly phosphorylated after RALF1 peptide treatments. Compared with wild-type plants, ripk mutants, like fer-4 mutant, display defects in root hair development and RALF1 response. Together with cross-phosphorylation of FER and RIPK in response to RALF1, these results suggest that RIPK, originally identified for its role in immunity, works together with FER to transduce RALF1 signal in the control of cell growth in roots.
Results
RIPK Interacts with FER at the Plasma Membrane.
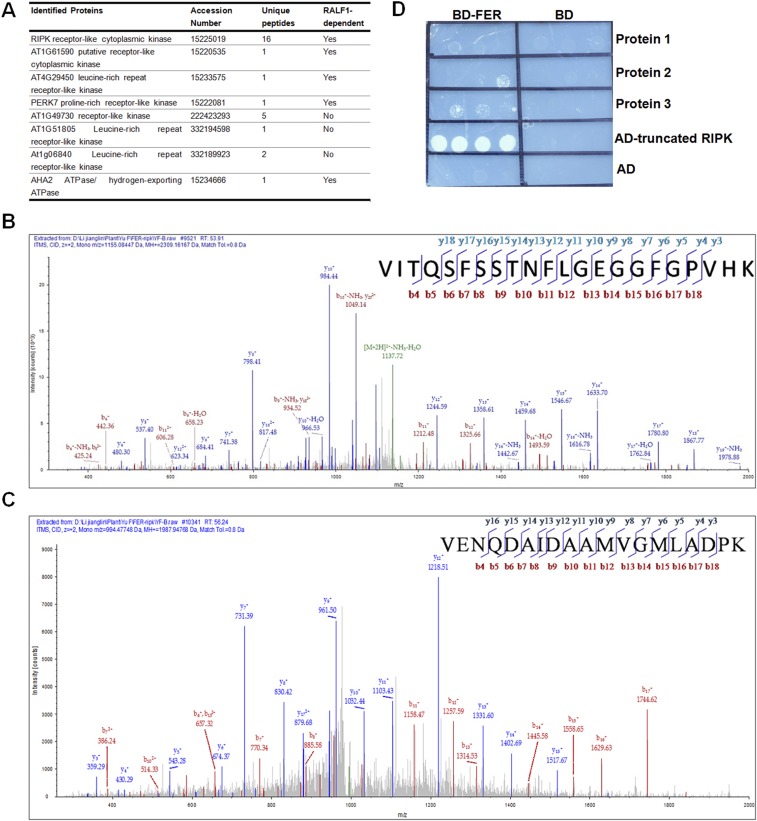
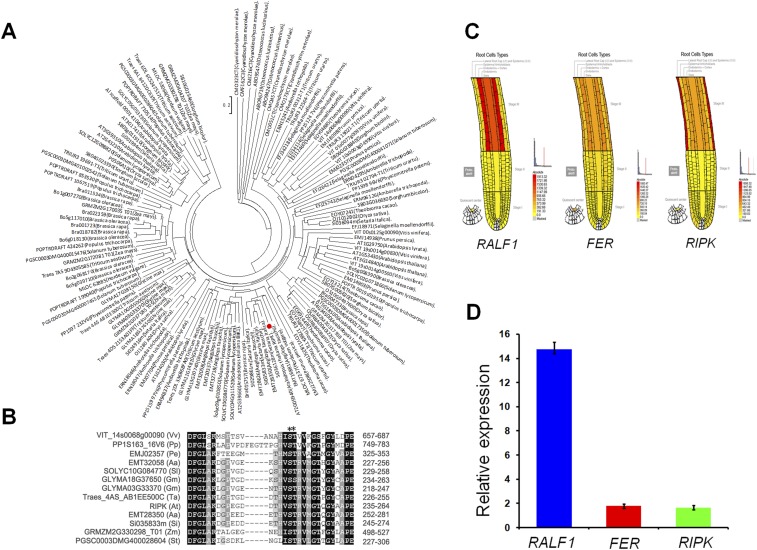
In the RALF1–FER signaling pathway, a critical question that remains to be answered is whether FER receptor kinase requires other partner proteins to control RALF1 response at the plasma membrane. To answer this question, we performed immunoprecipitation coupled with mass spectrometry (IP-MS/MS) to identify potential partner proteins for FER. FER-FLAG transgenic plants and Col.0 plants were treated with or without RALF1 peptide following the method described earlier (18). Protein extracts were subjected to IP procedure using anti-FLAG affinity gel, followed by 10% (mass/vol) SDS/PAGE and MS analysis of isolated gel slices. The proteins that were present only in the IP product from FER-FLAG transgenic plants but not in the IP product from Col.0 plants were considered as FER-associated proteins. Further, we especially focused our attention on proteins present in RALF1-treated samples. We identified several proteins in the IP product treated with RALF1 peptide (Fig. S1 A–C), including AHA2, the target H+-ATPase in the plasma membrane, which was also identified previously as a target that is regulated by RALF1 peptide (22). One of the RLCK-VII subfamily members, RIPK, attracted our interest because it was not only identified as FER interacting proteins through IP-MS/MS after RALF1 peptide treatments of the FER-FLAG transgenic plants, but a truncated RIPK protein (225–462 aa) was also identified as a positive clone in our yeast two-hybrid (Y2H) screen using FER kinase domain as a bait (Fig. S1D). It has been shown that FER is conserved among land plants (13). We performed phylogenetic analysis using Arabidopsis RIPK as a query and found that, like FER, RIPK homologs are also widely distributed among plants (Fig. S2 A and B). In addition, the expression pattern of RALF1, FER, and RIPK overlapped in a number of plant tissues, making it possible for them to function in similar processes (Fig. S2 C and D). We decided to test the possibility that FER and RIPK are functionally linked in the context of RALF1 response.
Fig. S1.
MS analysis of affinity-purified FER-interacting proteins and FER-interacting protein screening in Y2H. (A) Summary of LC–MS/MS analysis of affinity-purified FER-interacting proteins with or without RALF1 treatments. (B) Representative high-resolution fragmentation spectra of peptide [VITQSFSSTNFLGEGGFGPVHK] (+2 charged, m/z 1155.084) of RIPK protein. (C) Representative high-resolution fragmentation spectra of peptide [VENQDAIDAAMVGMLADPK] (+2 charged, m/z 994.477) present in AHA protein family. (D) FER-interacting protein screening in Y2H. The truncated RIPK was shown here.
Fig. S2.
Phylogenetic analysis of RIPK and expression profiles of RALF1, FER, and RIPK. (A) Phylogenetic tree of RIPK and related homologs from different plant species was constructed by neighbor-joining method using ClustalX (version 1.8) and MEGA 4.0. The bootstrap values (1,000 replicates) are indicated. The length of the branch is proportional to the degree of divergence. (B) Conserved sequence alignment of RIPK and related homologs from different plant species. An asterisk indicates conserved phosphorylation sites. (C) Expression profiles of RALF1 (Left), FER (Middle), and RIPK (Right) show that they are preferentially expressed in the root hair zone of roots. The data are from Botany Array Resource. Signal threshold was set to 2,000 for the data extraction. (D) Quantitative RT-PCR analysis for RALF1, FER, and RIPK expression in roots. Three independent experiments were done with similar results.
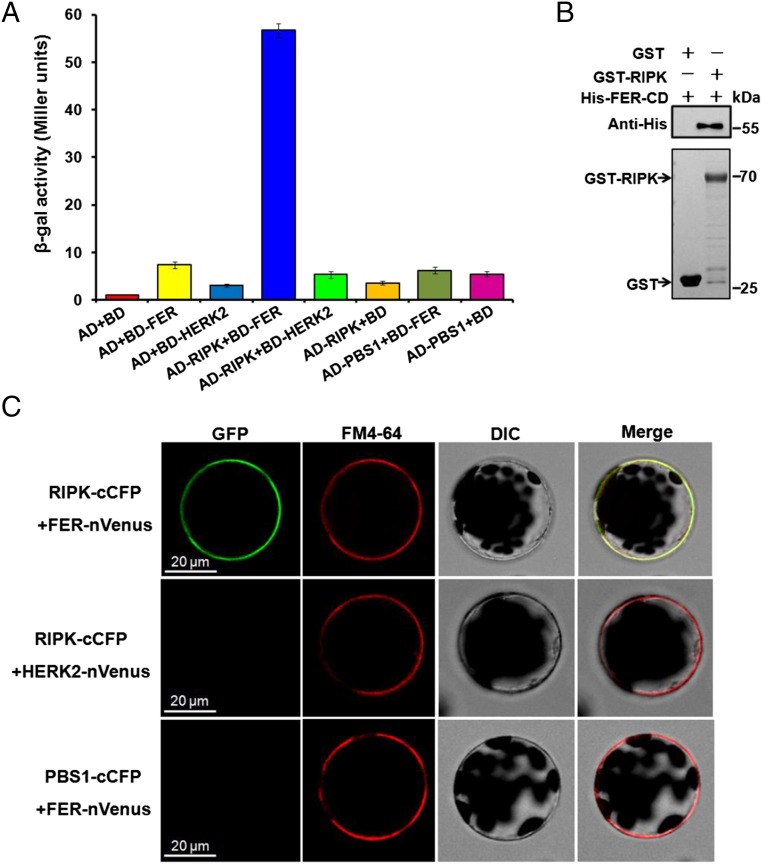
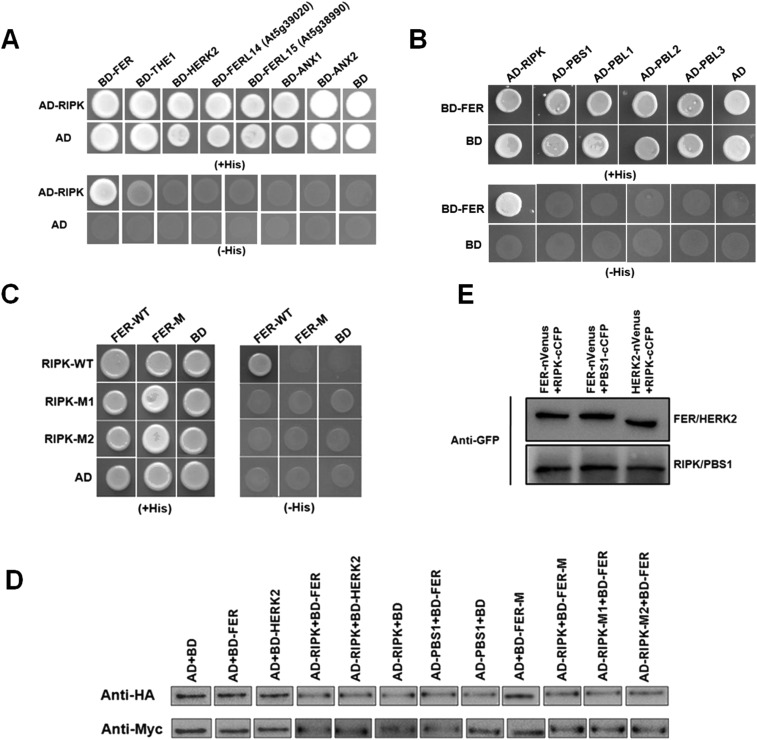
We further confirmed that RIPK interacts directly with FER by several independent assays. When full-length coding regions of RIPK in the pGADT7 prey vector and FER cytoplasmic kinase domain (FER-CD) sequence in the pGBKT7 bait vector were cotransformed into yeast cell AH109, they supported cell growth, indicating an interaction between FER and RIPK (Fig. 1A). We next explored the specificity of the FER–RIPK interaction by including some different members in the FER subfamily and RIPK subfamily using the Y2H system. These results showed that the FER–RIPK interaction is rather specific. Among the seven FER family members, there was a strong interaction between RIPK and FER, a weak interaction between RIPK and THE1, but no detectable interaction between RIPK with HERK2, FERL14 (AT5G39020), FERL15 (AT5G38990), ANX1, or ANX2 (Fig. S3A). Also, among the five RIPK family members (RIPK, PBS1, PBL1, PBL2, and PBL3) that we checked, only RIPK interacted with FER (Fig. S3B).
Fig. 1.
RIPK interacts with FER. (A) β-galactosidase assay in the Y2H system. AvrPphB susceptible protein 1 (PBS1) and HERKULES Receptor Kinase 2 (HERK2) were used as negative controls. RIPK and PBS1 were cloned into the pGADT7 vector (AD-RIPK, AD-PBS1) and FER-CD and HERK2-CD into the pGBKT7 vector (BD-FER, BD-HERK2). Experiments were repeated three times with similar results. (B) GST pull-down assay. Input GST (27 kDa, 2 μg) or GST-RIPK (70 kDa, 2 μg) protein was visualized by Coomassie Brilliant Blue staining (Lower). The eluted proteins were separated by a SDS/PAGE gel and probed with anti-His antibody (1:5,000) (Upper). Experiments were performed three times with similar results. (C) BiFC assay of FER–RIPK interaction in Arabidopsis protoplasts. Before imaging, the protoplasts were treated with FM4-64 (2 µM) for 5 min. GFP fluorescence was detected in protoplasts coexpressing RIPK-cCFP and FER-nVenus (Upper) but not in the controls (RIPK-cCFP + HERK2-nVenus or PBS1-cCFP + FER-nVenus; Middle and Lower). (Scale bar, 20 μm.)
Fig. S3.
RIPK interacts with FER in the Y2H and BiFC system. (A) Y2H analysis of the interaction between RIPK and multiple CrRLK1L subfamily members. RIPK was cloned into the AD vector; FER and other CrRLK1L subfamily members were cloned into the BD vector. Cells were grown on medium with (Upper) or without (Bottom) His. (B) Y2H analysis of interaction between FER and multiple RLCK-VII subfamily members. RIPK and other RLCK-VII subfamily members were cloned into the AD vector; FER was cloned into the BD vector. Cells were grown on medium with (Upper) or without (Bottom) His. (C) RIPK, RIPK-M1 (S251R), and RIPK-M2 (T252R) were cloned into the AD vector; FER and FER-M (K565R) were cloned into the BD vector. Cells were grown on medium with (Left) or without (Right) His. Four independent experiments were conducted, and a representative picture was shown. (D) Protein accumulation of the assay and controls in the BiFC construct as detected by Western blot using GFP antibody (1:5,000). (E) Protein accumulation of the BD baits (fused with the Myc-tag) and AD preys (fused with the HA-tag) in the Y2H system as detected by Western blot.
Studies showed that K565R mutation in the ATP-binding domain of FER-CD disrupts kinase activity (13). In addition, the S251 and T252 amino acid sites in both the BIK1 and RIPK kinase domain were highly conserved and probably required for kinase activity (33). We used this information and tested whether FER and RIPK mutants lacking kinase activity can interact with each other. In the Y2H assays, the RIPK and FER mutant versions containing kinase-dead mutations failed to interact (Fig. S3C). To make sure that all proteins were expressed in the yeast cells, we performed Western blots to monitor the accumulation of the BD baits (fused with the Myc-tag) and AD preys (fused with the HA-tag) in the Y2H system (Fig. S3D).
In the GST pull-down assay, GST-RIPK was shown to associate with the cytoplasmic kinase domain of FER (His-FER-CD) (Fig. 1B). We further tested the FER–RIPK interaction using bimolecular fluorescence complementation (BiFC) combined with FM4-64 staining to label the plasma membrane in Arabidopsis protoplast cells. When RIPK and FER coding regions were fused with C-terminal CFP (cCFP) and N-terminal Venus (nVenus), respectively, and cotransformed into Arabidopsis protoplasts, we observed cell surface-localized fluorescence signal that overlapped with the FM4-64 signal (Fig. 1C). No fluorescence signal was observed in the negative controls (RIPK-cCFP + HERK2-nVenus or PBS1-cCFP + FER-nVenus) performed with the same procedure. We also confirmed the expression of all proteins in the protoplasts using Western blot (Fig. S3E). These data suggest that the FER–RIPK interaction occurred at the plasma membrane.
RIPK-Null Mutant Mimics fer-4 Mutant Phenotypes.
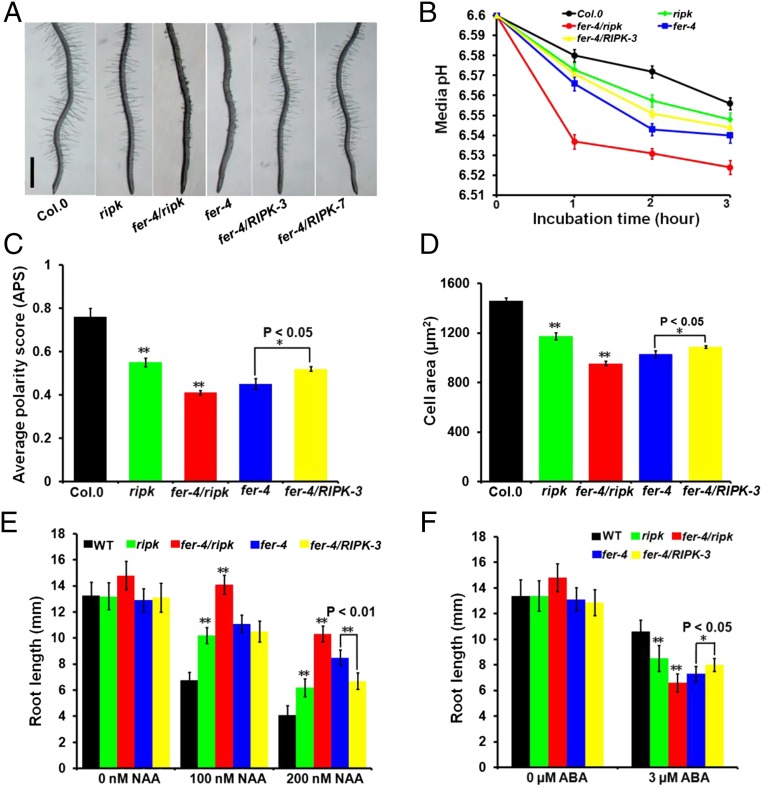
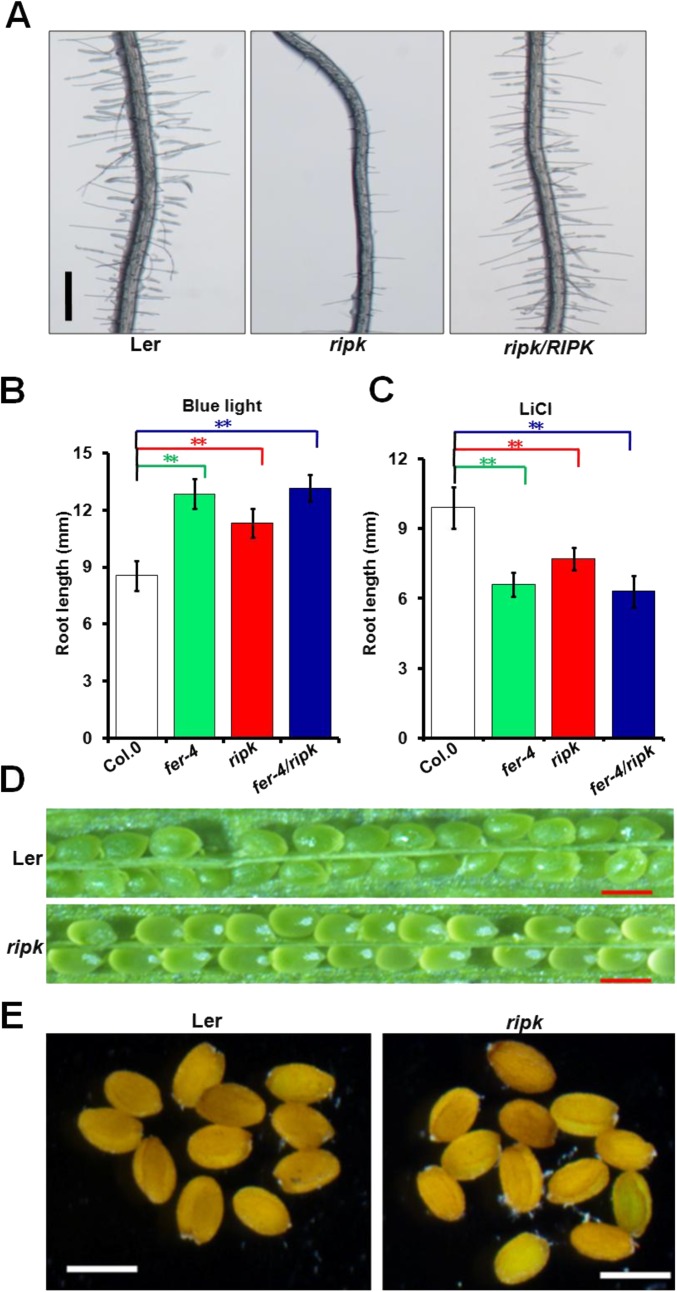
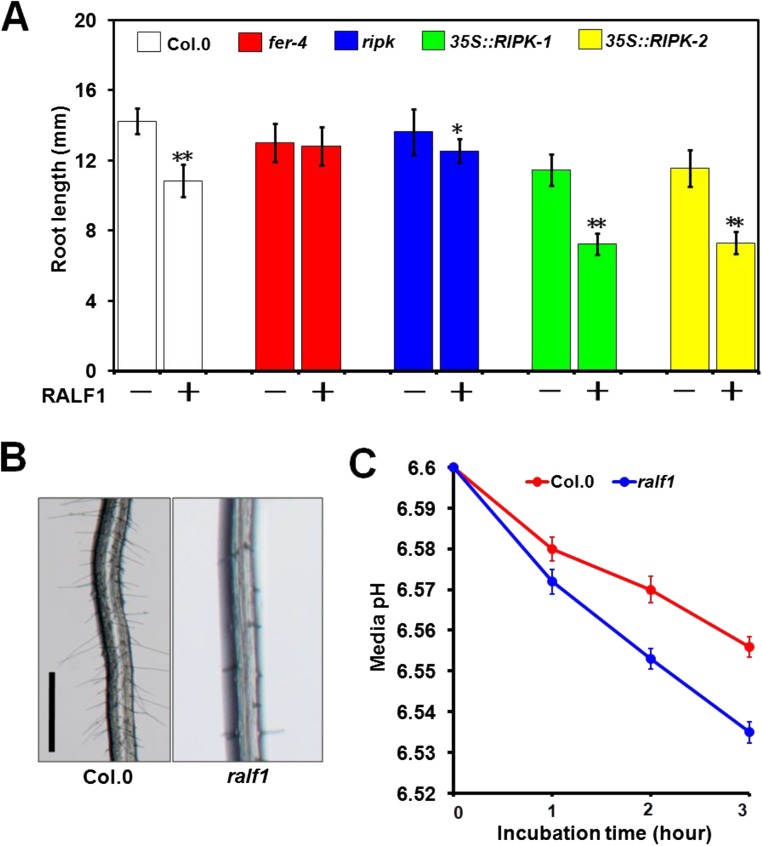
To test if FER–RIPK association is functionally relevant, we obtained a T-DNA insertional mutant of the RIPK gene in the Ler background (ripk-Ler) as reported previously (30) and also a ripk-Col.0 mutant derived from back-crossing ripk-Ler mutant with wild-type Col.0 for several generations to get the homozygous mutant in Col.0 background (30). As FER acts in a number of processes that are related to vegetative growth (16, 20) and hormonal responses (17, 18), we examined the ripk mutant together with the fer-4 mutant in several assays side by side. First, we analyzed the root hair phenotype that has been shown to be closely related to FER function in auxin response (20). The ripk-Col.0 mutant, like the fer-4 mutant, exhibited shorter root hairs compared with the wild-type control (Fig. 2A). The ripk-Ler mutant also showed the same phenotype (Fig. S4A). In addition, the ripk-Ler mutant phenotype was complemented by a genomic fragment fused in frame to a FLAG tag (pRIPK::RIPK-FLAG, abbreviated as ripk/RIPK; Fig. S4A), confirming that the short-root-hair phenotype in the ripk mutants resulted from loss of function of RIPK. The fer-4/ripk double mutant showed the same root hair phenotype as the fer-4 single mutant (Fig. 2A), suggesting that FER and RIPK may function in the same pathway. To further examine if RIPK functions downstream of FER, we produced transgenic lines overexpressing RIPK in the fer-4 background (fer-4/RIPK) by introducing the 35S::RIPK-FLAG construct into the fer-4 mutant. We chose two independent lines (fer-4/RIPK-3 and fer-4/RIPK-7) to examine root hair development and found that RIPK overexpression could partially rescue root hair defects in fer-4 mutant (Fig. 2A). We then used the fer-4/RIPK-3 line in more assays described below.
Fig. 2.
RIPK loss-of-function mutants mimic fer-4 mutants in genetic phenotypes. (A) Root-hair phenotype in Col.0, ripk-Col.0, fer-4/ripk, fer-4, and fer-4/RIPK plants. Photo shows 7-d-old Arabidopsis seedlings vertically grown on 1/2 MS plate. (Scale bar, 1 mm.) (B) Quantitative assay of the rhizosphere acidification rate in Col.0, ripk-Col.0, fer-4/ripk, fer-4, and fer-4/RIPK plants using a pH indicator, fluorescein-Dextran conjugate. Values represent the means ± SD obtained from three biological replicates (n = 7 seedlings). (C) Average polarity score (APS) of epidermal pavement cells from 7-d-old seedlings was determined. Values represent averages ± SD (n = 15 cells). (D) Area of epidermal pavement cells in Col.0, ripk-Col.0, fer-4/ripk, fer-4, and fer-4/RIPK leaves. Values represent averages ± SD (n = 15 cells). (E) Root length comparison of Col.0, ripk-Col.0, fer-4/ripk, fer-4, and fer-4/RIPK plants under different NAA concentrations (0, 100, and 200 nM) and treatments. Values represent the means ± SD obtained from three biological replicates. (F) Root length comparison of Col.0, ripk-Col.0, fer-4/ripk, fer-4, and fer-4/RIPK plants under different ABA concentrations (0, 3 µM) and treatments. Values represent the means ± SD obtained from three biological replicates. In C–F, asterisks indicate significant difference from the control as determined by one-way ANOVA (**P < 0.01 and *P < 0.05).
Fig. S4.
RIPK KO mutant phenotype assays. (A) Root hair phenotypic analysis of Ler, ripk-Ler, and ripk/RIPK. Root hairs of 7-d-old seedlings vertically grown on 1/2 MS were examined under a dissecting microscope. (Scale bar, 0.5 mm.) Three independent experiments were conducted, and a representative picture was shown. (B) Seeds of Col.0, fer-4, ripk-Col.0, and fer-4/ripk mutants were exposed to blue light (2.2 μmol·m2·s1) for 7 d before primary root length was measured. (C) Inhibition of root growth by LiCl. Three-day-old seedlings of Col.0, fer-4, ripk-Col.0, and fer-4/ripk mutants were transferred to 1/2 MS medium containing 12 mM LiCl. Root length was measured 4 d after transfer. In A and B, values represent averages ± SD (n = 15 seedlings). Asterisks indicate significant difference from the control as determined by one-way ANOVA (**P < 0.01). (D) A silique of self-pollinated Ler or ripk mutant. (Scale bar, 100 µm.) (E) Seed size of Ler or ripk mutant. (Scale bar, 200 µm.)
Previous studies showed that the fer-4 mutant promotes root elongation under blue light and increases uptake of cations into the cytoplasm, which will lead to hypersensitivity to LiCl (22). When grown under continuous blue light, the ripk-Col.0 mutant, like the fer-4 mutant, showed longer main roots than the wild-type (Fig. S4B). The ripk-Col.0 mutant seedlings were also hypersensitive to LiCl, similar to the fer-4 mutant than the wild-type (Fig. S4C). Because both blue light and LiCl assays reflect rhizosphere acidification ability (22), we further examined the rhizosphere acidification rate in the ripk mutants using a quantitative assay described by Haruta (22) and Gujas (34) and found that the ripk-Col.0 mutant, like the fer-4 mutant and the fer-4/ripk double mutant, had a higher rhizosphere acidification rate than the wild-type Col.0 plants (Fig. 2B), suggesting that disruption of the RALF1 signaling pathway leads to less inhibition of extracellular acidification. In contrast, overexpression of RIPK in the fer-4 mutant (fer-4/RIPK-3) had a slightly lower rhizosphere acidification rate than the fer-4 mutant (Fig. 2B).
Apart from a role in root-hair development, both fer-4 and ripk mutants have smaller leaves compared with their respective wild-type controls, as previously described (20, 30). In this study, we found that leaf epidermal (pavement) cells of both ripk-Col.0 and fer-4 were smaller than the wild-type Col.0. In addition, the complementary lobes and indents of neighboring pavement cells were less obvious than those in the wild-type Col.0, again indicating that fer-4 and ripk mutants showed similar phenotypes. In addition, the defects of the fer-4 mutant in pavement cell morphology were also partially suppressed by RIPK overexpression (Fig. 2 C and D).
In addition to the cell growth defects, the ripk mutant also displays similar hormone-regulated phenotypes as the fer-4 mutant. For example, the ripk-Col.0 mutant was less sensitive to auxin but more sensitive to ABA (Fig. 2 E and F), as reported for the fer-4 mutant (17, 20, 23). In addition, overexpressing RIPK in the fer-4 mutant background partially restored fer-4 mutant phenotypes. However, the ripk mutant phenotypes appeared weaker than those of fer-4 in most cases. Moreover, ripk, unlike fer-4, did not seem to show defects in PT reception (Fig. S4D) nor in seed size control (Fig. S4E). It is likely that FER works together with multiple RLCK partner proteins (such as MARIS) to control its multiple downstream responses (35). As a result, mutation of one of these partners (such as in the ripk mutant) disrupts only part of these responses, leading to weaker phenotypes compared with those in fer-4. Taken together, these results indicate that FER and RIPK may work together in several processes that control cell growth and hormone responses.
RALF1 Enhances Phosphorylation of FER and RIPK in a Mutually Dependent Manner.
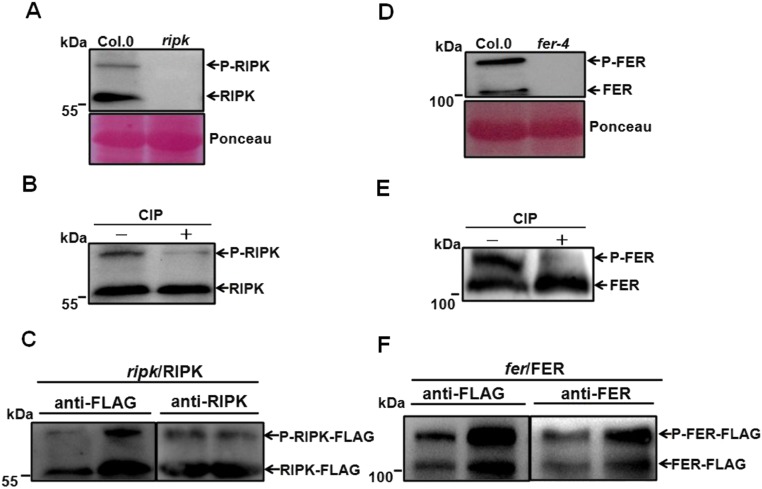
To investigate how RIPK and FER, both protein kinases, interact biochemically and function together, we examined their phosphorylation status in the context of their interaction. First, we raised an antibody that appeared to specifically react with the RIPK protein. The RIPK antibody detected two bands between 70 kDa and 55 kDa in the Col.0 but not in the ripk-Col.0 mutant (Fig. S5A), suggesting that RIPK is the major form of RLCKs recognized by the antibody in the samples prepared in our experiment. Adding a generic phosphatase calf intestinal alkaline phosphatase (CIP) to the protein extract reduced the intensity of the upper band, indicating that the upper band represents a phosphorylated form of RIPK: We named the phosphorylated form (the upper band) P-RIPK and the other nonphosphorylated form (the lower band) RIPK (Fig. S5B). To further confirm antibody specificity, we extracted proteins from the complementation lines that express RIPK-FLAG fusion in the ripk mutant background (ripk/RIPK-FLAG) and ran Western blots. The RIPK antibody detected two bands (P-RIPK-FLAG, RIPK-FLAG) that matched the bands detected by the FLAG antibody (Fig. S5C).
Fig. S5.
Specificity of FER and RIPK antibodies. (A) Anti-RIPK antibody recognized two bands in total protein extract from Col.0 but not ripk mutant. (B) RIPK has two forms: Without CIP treatment, the upper band is phosphorylated form (we named P-RIPK), and the lower band is nonphosphorylated form (we named RIPK), prominent after CIP treatment. (C) Specificity analysis of anti-RIPK antibody. Total protein was extracted from leaves of 7-d-old ripk/RIPK-FLAG complemented lines and probed with the anti-RIPK antibody (1:3,000) and anti-FLAG antibody (1:5,000), respectively. (D) Total protein was extracted from leaves of 45-d-old Col.0 and fer-4 mutant plants and was probed using the FER antibody (1:4,000) in a Western blot procedure. (E) FER was present in two forms: without CIP treatment, the upper band is phosphorylated form (we named P-FER), and the lower band is nonphosphorylated form (we named FER), prominent after CIP treatment. (F) Specificity analysis of anti-FER antibody using fer/FER complementation lines expressing FER-FLAG in the wild-type Col.0 background. Total protein was extracted from leaves of 35-d-old FER-FLAG plants and analyzed using the anti-FER antibody (1:4,000) and anti-FLAG antibody (1:5,000), respectively.
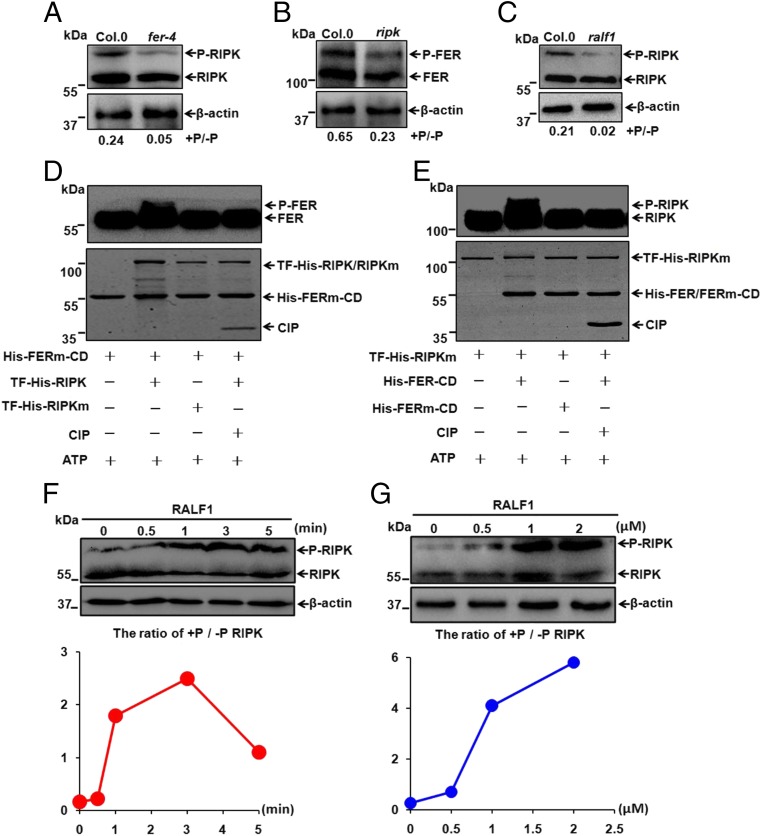
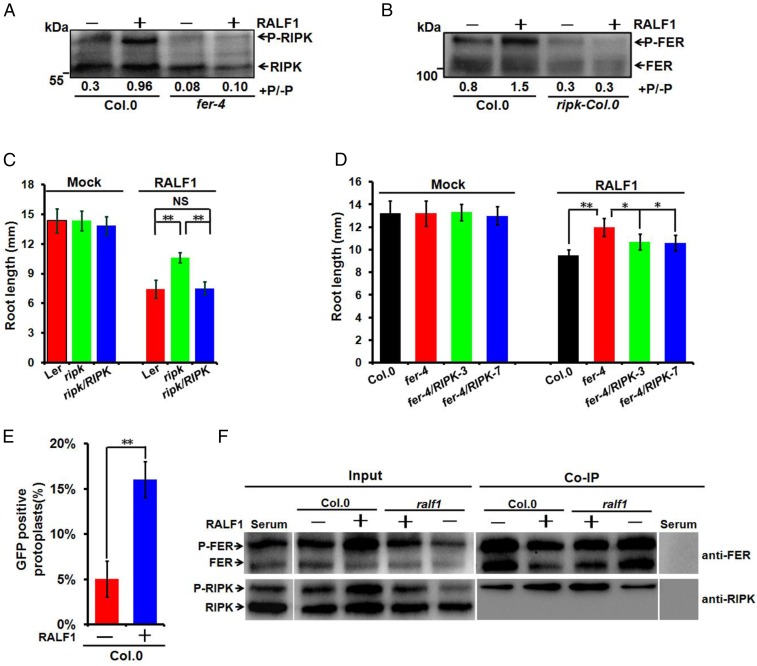
To test if RIPK phosphorylation depends on FER, we measured RIPK phosphorylation status in the Col.0 and fer-4 mutant using anti-RIPK antibody and found that the RIPK phosphorylation level was reduced in the fer-4 mutant compared with the wild-type Col.0 plants (Fig. 3A). Using a similar procedure, we raised another antibody that specifically reacted with FER protein. Two bands between 130 kDa and 110 kDa were detected in the wild-type Col.0 plants (but not in the fer-4 mutant): a phosphorylated FER (the upper band; P-FER) and the nonphosphorylated FER (the lower band; FER) (Fig. S5 D and E). To confirm FER antibody specificity, we extracted total protein from the complementation lines expressing FER-FLAG fusion protein in the fer-4 mutant background (fer/FER-FLAG) and performed Western blots. As in the case of RIPK, the FER antibody also detected two bands (P-FER-FLAG and FER-FLAG) that matched the bands detected by the FLAG antibody (Fig. S5F). We next examined FER phosphorylation status in the Col.0 and ripk mutant using anti-FER antibody. The phosphorylated band was reduced in the ripk mutant compared with that in the Col.0 (Fig. 3B). These results, together with those showing their physical interaction, suggested that RIPK and FER may form a protein kinase complex and phosphorylate each other in a mutually dependent manner in vivo. We further confirmed the FER-RIPK transphosphorylation in vitro using a kinase-dead version of FER (His-FERm-CD, about 55 kDa) or RIPK (TF-His-RIPKm, a fusion protein about 100 kDa) as a substrate. As shown in Fig. 3, RIPK phosphorylated kinase dead FER-CD (His-FERm-CD) (Fig. 3D), and the His-FER-CD also phosphorylated the kinase-dead version of RIPK (TF-His-RIPKm) (Fig. 3E).
Fig. 3.
Phosphorylations of RIPK and FER are interdependent in response to RALF1 peptide. (A) Phosphorylation status of RIPK in 7-d-old Col.0 and fer-4 Arabidopsis seedlings. The ratio of p-RIPK/RIPK was displayed below the gel, and β-actin was shown as a loading control in A–C, F, and G. (B) Phosphorylation status of FER in 45-d-old Col.0 and ripk-Col.0 plants. (C) Phosphorylation status of RIPK in 7-d-old Col.0 and RALF1-RNAi (ralf1) seedlings. (D and E) In vitro kinase activity assays showing cross-phosphorylation of FER and RIPK kinase domains. These assays were started by adding ATP and analyzed by Western blot using His antibody (1:5,000). Input proteins were visualized by Coomassie Brilliant Blue staining (Lower). Data shown are representative of at least three independent experiments with similar results. (F) Time courses (0, 0.5, 1, 3, and 5 min) of RIPK phosphorylation in response to RALF1 peptide (1 μM) using 7-d-old Col.0 Arabidopsis seedlings. (G) RIPK phosphorylation in 7-d-old Col.0 Arabidopsis seedlings after being exposed to different RALF1 concentrations (0, 0.5, 1, and 2 μM) for 3 min.
As RALF1 is a peptide ligand that activates FER phosphorylation (22), we examined if the phosphorylation of RIPK is also regulated by the RALF1 ligand. First, we analyzed the phosphorylation level of RIPK in the RALF1-RNAi (ralf1) plant in which RALF1 expression level was reduced (36) and found that the level of RIPK phosphorylation in ralf1 plants was lower than that in Col.0 (Fig. 3C). We further analyzed the phosphorylation status of RIPK in shoot or root separately using 1-wk-old Arabidopsis seedlings treated by RALF1 peptide for 3 min. Results showed that RALF1 treatment rapidly increased the phosphorylation of RIPK in roots and to a lesser extent in shoots (Fig. S6). We further analyzed the time course and dosage dependence of RIPK phosphorylation under RALF1 treatments and found that RIPK phosphorylation was elevated within 30 s after application of 1 µM RALF1 peptide (Fig. 3F), and the RIPK phosphorylation level was up-regulated in a RALF1 dosage-dependent fashion (Fig. 3G). These results strongly suggest that RIPK phosphorylation is an early event of RALF1 signaling, and FER–RIPK may form a protein kinase complex to perceive RALF1 signal through phosphorylating each other.
Fig. S6.
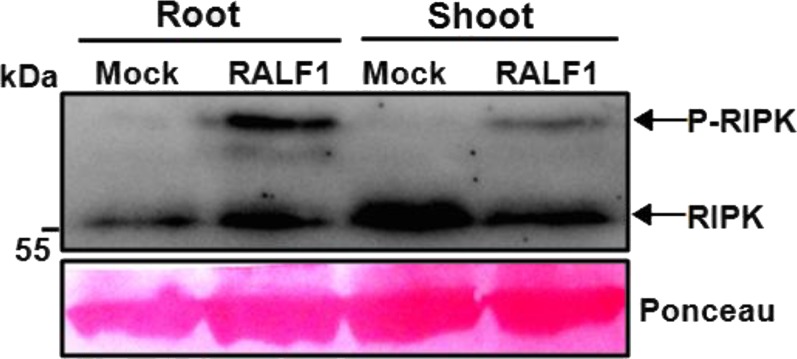
RIPK phosphorylation in response to RALF1 treatments in both roots and shoots. The level of RIPK phosphorylation was enhanced in both roots and shoots after being exposed to RALF1 peptide (1 μM) for 1 min.
If the RALF1 signal induces formation of the FER/RIPK kinase complex, it is possible that RALF1 addition would enhance the phosphorylation of both kinases. We thus examined RIPK phosphorylation status in Col.0 and fer-4 mutant plants with or without RALF1 treatment. We found that RIPK phosphorylation was reduced in fer-4 mutant regardless of the addition of RALF1 (Fig. 4A). Likewise, we also observed a decrease in FER phosphorylation level in the ripk-Col.0 mutant compared with that in Col.0 (Fig. 4B).
Fig. 4.
RALF1 enhances interaction and phosphorylation of FER and RIPK. (A) Phosphorylation status of RIPK in 7-d-old Col.0 and fer-4 Arabidopsis seedlings in the absence or presence of RALF1 peptide (1 µM) for 3 min. The ratio of p-RIPK/RIPK was shown below the gel. The data shown are representative of at least three independent experiments with similar results. (B) Phosphorylation status of FER in 45-d-old Col.0 and ripk-Col.0 Arabidopsis plants in the absence or presence of RALF1 peptide (1 µM) for 3 min. The ratio of p-FER/FER was shown below the gel. The data shown are representative of at least three independent experiments with similar results. (C) RALF1 inhibition of root growth in Ler, ripk-Ler, and the complementation line (ripk/RIPK). (D) RALF1 inhibition of root growth in Col.0, fer-4, fer-4/RIPK-3, and fer-4/RIPK-7 plants. (E) Interaction between FER and RIPK in Col.0 protoplasts was enhanced by RALF1 peptide (1 µM). In C–E, values represent the means ± SD obtained from three biological replicates. NS, no significant difference (P > 0.05). Asterisks indicate significant difference from the control as determined by one-way ANOVA (*P < 0.05; **P < 0.01). (F) Co-IP experiments showing RALF1-dependent FER–RIPK complex formation in Arabidopsis cells. Ten-day-old wild-type (Col.0) and RALF1-RNAi (ralf1) plants were treated with RALF1 (1 µM) for 10 min. Total protein extracts (Input) or proteins eluted from anti-FER IP were probed by FER antibody (1:4,000) or RIPK antibody (1:3,000). Preimmune serum (Preim) was used for negative controls.
RIPK, Like FER, Positively Regulates RALF1 Response in Roots.
We further directly tested whether RIPK is required for RALF1 response in plants using primary root growth inhibition assay and found that the ripk-Ler mutant exhibited reduced sensitivity to RALF1 compared with Ler, whereas the ripk/RIPK complementation lines showed the similar response as Ler (Fig. 4C). The ripk-Col.0 mutant also exhibited reduced sensitivity to RALF1 as in fer-4 mutant (Fig. S7A). We further analyzed RIPK overexpression lines in Col.0 background and found that they had shorter roots than the Col.0 (Fig. S7A). When treated with the RALF1 peptide, the RIPK overexpression lines were more sensitive to RALF1 than the Col.0 (Fig. S7A). In addition, overexpression of RIPK into the fer-4 background partially complemented the RALF1 response defect in the fer-4 mutant (Fig. 4D). We confirmed the idea that RALF1, FER, and RIPK act as a signaling module in the same pathway by examining ralf1 plants in their root hair development and rhizosphere acidification rate and found that the ralf1 plants, just like fer-4 and ripk mutant plants, had shorter root hairs and a faster rhizosphere acidification rate than those in Col.0 (Fig. S7 B and C).
Fig. S7.
RALF1 inhibition of root growth. (A) Root length analysis of Col.0, fer-4, ripk-Col.0, and 35S::RIPK-FLAG lines after RALF1 (1 μM) treatment. Values represent averages ± SD (n = 15 seedlings). Asterisks indicate significant difference from the control as determined by one-way ANOVA (*P < 0.05; **P < 0.01). (B) Root hair phenotype in Col.0 and the ralf1 mutant vertically grown on 1/2 MS for 7 d. (Scale bar, 0.5 mm.) (C) Quantitative analysis of rhizosphere acidification rate in Col.0 and the ralf1 mutant by using a pH indicator, fluorescein-Dextran conjugate. Values represent the means ± SD obtained from three biological replicates (n = 7 seedlings).
RALF1 Enhances Formation of the FER/RIPK Kinase Complex.
Studies have shown that the interaction between RLK and RLCK is regulated by the ligands (25–27). To explore whether the formation of the FER/RIPK kinase complex is regulated by the RALF1 peptide, we tested the FER–RIPK interaction in the BiFC system with or without addition of RALF1 peptide using Col.0 protoplasts. Interestingly, when RALF1 peptide was added, we observed a higher percentage of cells showing fluorescence signal than the control (Fig. 4E). This result indicated that RALF1 enhanced the interaction of the two kinases, thus assembling more functional fluorescence molecules. Furthermore, co-IP assay was performed using anti-FER antibody to test whether the association of endogenous RIPK and FER is affected in the RALF1-RNAi (ralf1) mutant and whether this defect can be complemented by treatment with the synthetic RALF1 peptide. We found that the association of RIPK and FER is reduced in the ralf1 mutant and this defect can be complemented by addition of the RALF1 peptide. Consistent with the results of BiFC assays, RALF1 enhanced the interaction between RIPK and FER in co-IP assays as well, and we found that FER preferably copurified with P-RIPK during co-IP (Fig. 4F). Taken together, the interaction between FER and RIPK is enhanced by RALF1 peptide, indicating that RALF1 ligand binding to FER recruits RIPK into the FER-based complex.
Discussion
Recent studies highlight the importance of RLCKs in controlling plant immunity. For example, RLCK-VII subfamily member BIK1 has been found to play an essential role in PAMP-triggered immunity (PTI) through its interaction with and phosphorylation by FLS2 receptor kinase (25–28). Another RLCK-VII subfamily member, PBS1, is targeted by the RPS5 NB-LRR immune receptor (37). RIPK, like BIK1 and PBS1, also belongs to the RLCK-VII subfamily and is shown to play a role in plant immune response (29, 30). In this study, we provide evidence that RIPK associates with the FER RLK and functions in RALF1-mediated cell growth regulation, establishing another RLK–RLCK partnership that regulates a number of cellular processes including RALF1-mediated cell growth inhibition in roots. However, the disruption of RIPK function did not phenocopy all of the defects in fer-4 mutants, such as notably PT reception and seed size control. This suggests that one specific RLK may partner with different downstream RLCKs to fulfill distinct functions. Likewise, each RLCK may partner with more than one RLK to regulate distinct responses. In support of this notion, a RLCK-VIII subfamily member, MARIS, was recently reported to participate in the ANX1 pathway in PT growth and in the FER pathway for root hair tip growth (35). It is yet to be determined whether MARIS, like RIPK, can directly interact with FER or ANX1. It will be interesting to find out whether one RLK can interact with multiple RLCKs to regulate the same or different downstream responses. In fact, recent work showed that BRI1 interacts with and phosphorylates multiple members of the BSK subfamily and even distantly related RLCK subfamily members (38). This could be a general mechanism that integrates distinct processes and mediates cross-talks among signaling pathways in plants.
Both FER and RIPK are active kinases and can transphosphorylate each other. As FER regulates growth in many different cell types and in response to various environment stress conditions, we speculate that FER may be phosphorylated at distinct sites in response to different factors. Such differential phosphorylation may be further diversified by interacting with different downstream partners such as RLCKs. Indeed, previous work and our study here showed that RALF1 triggers FER phosphorylation, and we further showed that both FER and RIPK phosphorylation levels are up-regulated quickly by RALF1. More importantly, phosphorylation of FER or RIPK is mutually dependent, indicating that not only the upstream kinase FER phosphorylates downstream RIPK but the reverse is true, supporting the emerging hypothesis that the phosphorylation pattern of the RLKs can depend on partner RLCKs. In this context, another finding is relevant: FERK565R mutation removes autophosphorylation activity, but it can still complement fer-4 PT reception defect (39), suggesting the possibility that there exists a RLCK that transphosphorylates FERK565R mutant and initiates downstream events during fertilization. The detailed functional analysis of different phosphorylation sites on FER and RIPK will shed light on the early events in the RALF1–FER/RIPK pathway.
Several RLCKs have been identified as interacting proteins of receptor kinases to transduce intracellular signaling. This RLK–RLCK complex may have become a general theme in RLK-facilitated signaling processes. For instance, BIK1, an RLCK-VII subfamily protein kinase, can work together with FLS2 RLK and coreceptor BAK1 to transduce flg22 signal in plant innate immunity (25–28). In parallel to the FLS2/BAK1/BIK1 receptor kinase complex, it will be crucial to identify the partner RLK that interacts with FER to function as the coreceptor of the RALF1 peptide. We noticed an interesting difference between the FLS2–BAK1–BIK1 and FER–RIPK pathway: BIK1 is associated with the FLS2 receptor kinase in the absence of ligand flg22 (25). Upon ligand binding, BIK1 is phosphorylated and then disassociated from the receptor complex to activate downstream signaling. In contrast, RIPK is largely disassociated from FER receptor in the absence of ligand RALF1. Upon ligand binding, RIPK is recruited into the receptor complex and then transphosphorylated by FER. Such a difference could implicate mechanistic diversity among RLK–RLCK interactions, although more work in various RLK–RLCK pathways is required to confirm such diversity.
Considering signaling events downstream of the receptor complex, it is interesting to note that RALF1 signaling also results in production of ROS (20) and calcium spikes (15, 19, 21, 22), reminiscent of the events triggered by the FLS2/BAK1/BIK1 pathway. Following the paradigm of BIK1 action (27, 28), further work should explore whether RIPK can regulate RBOHD or other enzymes leading to ROS production and whether RIPK controls calcium spikes in response to RALF1. It is also important to examine whether the FER/RIPK kinase complex can directly interact with and regulate the activity of H+-ATPase (such as AHA2) by phosphorylation. A thorough investigation on the RALF1–FER/RIPK pathway will help in understanding how plants respond to peptide signal and other hormones and how FER functions in the cross-talk of these signals that regulate plant cell growth.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana seeds were surface-sterilized and, unless stated otherwise, stratified at 4 °C for 2–3 d before growing on 1/2 MS with 0.8% sucrose and 1% Phytagel (Sigma-Aldrich) for subsequent analysis. Wild-type (Col.0, Ler), ripk mutants (30), fer-4 (20), fer-4/ripk, fer-4/RIPK, FER-FLAG, RIPK-FLAG, and pRIPK::RIPK-FLAG (ripk/RIPK) (30) Arabidopsis plants were all grown at 23 °C with a photoperiod of 16-h light/8-h dark. The fer-4/ripk double mutant was obtained by crossing fer-4 and ripk-Col.0 and confirmed by PCR analysis. T3 generation homozygous seeds were used for phenotypic analysis. For overexpression assays, full-length FER coding sequence fused with a C-terminal FLAG tag driven by Ubi promoter was cloned into pCAMBIA1301. Similarly, full-length RIPK coding sequence fused with a C-terminal FLAG tag driven by the 35S promoter was cloned into the pBI121. FER-FLAG, RIPK-FLAG, and fer-4/RIPK lines were generated using Agrobacterium-mediated floral dip method.
Y2H Assays.
The cytoplasmic kinase domain of FER and its kinase dead mutation (K565R) was amplified and constructed into pGBKT7 to make an in-frame fusion with GAL4-BD as “bait.” The kinase domain of RIPK and its kinase-dead mutations (S251R) and (T252R) were amplified and constructed into pGADT7 to make an in-frame fusion with GAL4-AD as “prey.” Some RLCK-VII subfamily members (e.g., PBS1, PBL1, PBL2, PBL3) were cloned into pGADT7 and some CrRLK1L subfamily members [e.g., THE1, HERK2, FERL14 (FER Like, At5g39020), FERL15 (At5g38990), ANX1, ANX2] were cloned into pGBKT7 as controls. Sequences of some primers are listed in Table S1.
Table S1.
Primers used for cloning purposes in this study
| Primers | Sequence, 5′–3′ |
| pGBKT7-FER-F | CGAATTCGCTTACCGCAGACGTAAGC |
| pGBKT7-FER-R | GCTGCAGCTAACGTCCCTTTGGATTCATG |
| pGBKT7-FER (K565R)-F | CAGGAGAGGCAACCCAATGTCC |
| pGBKT7-FER (K565R)-R | TTGGGTTGCCTCTCCTGATGGCTACCTTTG |
| pGADT7-RIPK-F | CCCGGGTATGGCGGTGAAGAAGAAAGTTTC |
| pGADT7-RIPK-R | GGATCCATGTACCGTTCCCCACCTGCCT |
| pGADT7-RIPK (S251R)-F | CTAACTCGAGTGATGGGCACAC |
| pGADT7-RIPK (S251R)-R | CCCATCACTCGAGTTAGCACATGAGTGTCG |
| pGADT7-RIPK (T252R)-F | CTACGAGTGATGGGCACACAAG |
| pGADT7-RIPK (T252R)-R | GTGCCCATCACTCGTAGTGACACATGAGTG |
| RIPK-kinase-F | GGATCCGAAACCCTAATGGCATCTTCAAAG |
| RIPK-kinase-R | GTCGACGTACCGTTCCCCACCTGCCT |
| FER-kinase-F | TCACGAATTCCGTGGTGATTACCAGCCTGC |
| FER-kinase-R | CTGAGTCGACGTCCCTTTGGATTCATG |
| BiFC-FER-F | CGAGCTCGAAGATCACAGAGGGACGATTCC |
| BiFC-FER-R | TGGATCCAACGTCCCTTTGGATTCATGATCTG |
| BiFC-RIPK-F | TAGTCGACTGGCGGTGAAGAAGAAAGTTTC |
| BiFC-RIPK-R | ATGGATCCAGTACCGTTCCCCACCTGCC |
For the FER–RIPK interaction analysis, different plasmid pairs [pGBKT7-FER vs. pGADT7-RIPK; pGBKT7-FER vs. RIPK (S251R); pGBKT7-FER vs. RIPK (T252R); pGBKT7-FER (K565R) vs. pGADT7-RIPK] were cotransformed into yeast cell AH109, respectively. A dilution series of transformants were plated onto synthetic dropout medium lacking tryptophan and leucine (SD/–Trp–Leu) or dropout medium lacking tryptophan, leucine, and histidine (SD/–Trp–Leu–His) supplemented with 10 mM 3-AT (3-Amino-1, 2, 4-triazole) for 3–4 d to monitor cell growth. The β-galactosidase assay was carried out according to the method described by Miller (40).
BiFC Assay.
For the BiFC assay, the FER-nVenus and RIPK-cCFP were cloned into the vectors pE3308 and pE3449 (17), respectively, using primers listed in Table S1. Meanwhile, HERK2-nVenus was cloned into the vector pE3308 and PBS1-cCFP was cloned into pE3449 as the negative controls. Protoplasts were isolated from well-expanded rosette leaves of 4-wk-old Arabidopsis plants, and then protoplasts were cotransfected with different construct pairs [FER-nVenus vs. RIPK-cCFP; HERK2-nVenus vs. RIPK-cCFP; FER-nVenus vs. PBS1-cCFP] using the polyethylene glycol (PEG) transformation method as described (17). The cotransfected protoplasts were incubated in the dark at 23 °C to allow expression of the BiFC proteins. Fluorescence was monitored 16–18 h after transformation with a confocal microscope at an excitation wavelength of 488 nm. Before imaging, the protoplasts were treated with FM4-64 (2 µM) for 5 min. To test the effect of RALF1 peptide on the FER–RIPK interaction, FER-nVenus and RIPK-cCFP constructs were transformed into wild-type Col.0 protoplasts and incubated following the same procedure described above. The cotransfected protoplasts were divided into two aliquots: One was treated with 1 μM RALF1 and the other treated with control buffer for 30 min before fluorescence was examined. The percentage of GFP-positive protoplasts in each aliquot was counted.
Expression and Purification of GST–RIPK Fusion Protein.
The coding sequence of RIPK (21–462 aa) was PCR-amplified with BamHI and SalI sites at the 5′- and 3′-ends. The amplified PCR products were then digested by BamHI and SalI and ligated into the pGEX-4T-1 vector. The resulting construct pGEX-4T-1-RIPK was transferred into the Escherichia coli BL21 (DE3), and expression of the fusion protein (GST-RIPK) was induced by isopropylthio-β-d-1-galactopyranoside (IPTG, 0.5 mM) after the culture reached an OD600 of 0.6 at 37 °C. The culture continued at 28 °C for 6 h with continuous shaking (230 rpm) before cells were collected by centrifugation for protein purification.
The protein purification follows these steps: First, six volumes of lysis buffer [25 mM Tris·HCI (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 10% (vol/vol) glycerol] were added to one volume of the collected cells and suspended for sonication. After sonication on ice and centrifugation at 10,621 × g for 15 min at 4 °C, the soluble fraction containing GST-RIPK was loaded to the GST agarose resin (Thermo Fisher Scientific, 20211) and incubated for 3 h at 4 °C. The agarose beads were washed with six volumes of buffer [25 mM Tris·HCI (pH 7.5), 300 mM NaCl, 10 mM MgCl2, 1 mM DTT, 10% (vol/vol) glycerol, and 1% Triton X-100] for 1 h at 4 °C, and the fusion protein was eluted with one volume of elution buffer [25 mM Tris·HCI (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, 10% (vol/vol) glycerol, and 15 mM glutathione] for 2 h at 4 °C. Recombinant protein purity was assessed by SDS/PAGE.
GST Pull-Down Assay.
GST agarose resin (Thermo Fisher Scientific, 20211) was prewashed three times using 1 mL binding buffer [25 mM Tris·HCl (pH 7.5), 100 mM NaCl, and 1 mM DTT]. GST-RIPK vs. His-FER-CD (2 μg each) and GST vs. His-FER-CD (negative control) were then incubated with prewashed GST beads (10 µL) in the same binding buffer with agitation for 4 h. The sample was then centrifuged at 10,621 × g for 5 min under 4 °C, and the resin was washed five times with 500 µL of washing buffer [25 mM Tris·HCl (pH 7.5), 300 mM NaCl, 10 mM MgCl2, 1 mM DTT, 10% (vol/vol) glycerol, and 0.1% Triton X-100] to remove nonspecific proteins. Proteins were eluted from the beads with 30 µL elute buffer [25 mM Tris·HCl (pH 7.5), 20 mM glutathione, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 10% (vol/vol) glycerol] and analyzed by SDS/PAGE and immunoblot with anti-His (1:5,000). The secondary goat anti-mouse IgG-HRP conjugates (1:8,000) were used for detection via enhanced chemiluminescence (Thermo Fisher Scientific, 34075).
Co-IP and Liquid Chromatography–MS/MS Assay.
To identify potential partner proteins for FER, co-IP assay was performed using protein extracts from FER-FLAG transgenic plants and wild-type Col.0 plants with or without RALF1 peptide as described (18) with some modifications. Anti-FLAG affinity gel (Sigma, A2220) was used to immunoprecipitate FER–RIPK protein complexes. Briefly, 1 g fresh samples were thoroughly ground in 0.3 mL NEB buffer [20 mM Hepes (pH 7.5), 40 mM KCl, 1 mM EDTA] + 0.3 mL NEB-T (NEB buffer containing 1% Triton X-100) buffer containing 1 mM PMSF and 1% protease inhibitor mixture (Thermo Fisher Scientific, 78420) at 4 °C. After incubation at 4 °C for 30 min with continuous shaking, the homogenized sample was centrifuged twice at 17,949 × g for 15 min each under 4 °C. We saved 50 μL of supernatant for SDS/PAGE loading control (Input). To the rest of protein extracts, 20 μL prewashed anti-FLAG affinity gel was added and the mixture was rotated at 4 °C for 3–4 h. Then the beads were centrifuged at 239 × g for 3 min and washed three times at 4 °C by washing buffer [20 mM Hepes (pH 7.5), 40 mM KCl, 0.1% Triton X-100] to remove nonspecific proteins. Finally, the bound proteins were eluted by adding 3× FLAG peptide (Sigma, F4799) to a final concentration of 0.5 mg/mL, rotated at 4 °C for 1 h. The eluate was boiled in 1× SDS loading buffer for 5 min, separated on a 10% (mass/vol) SDS/PAGE gel, and probed with anti-FLAG antibody (Abmart, M20008) and anti-RIPK antibody, respectively.
FER-FLAG IP samples were analyzed, with two technical repetitions each, using an EASY-nano liquid chromatography (LC) system (Proxeon Biosystems) coupled with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Peptides were loaded to a PepMap C18 trap column (75 μm, 15 cm; Dionex Corp) and eluted using a gradient from 100% solvent A (0.1% formic acid) to 35% (vol/vol) solvent B (0.1% formic acid, 100% acetonitrile) for 38 min, 35–90% (vol/vol) solvent B for 15 min, and 100% solvent B for 5 min (a total of 65 min at 200 nL/min). After each run, the column was washed with 90% (vol/vol) solvent B and re-equilibrated with solvent A. Mass spectra were acquired in positive ion mode applying data-dependent automatic survey MS scan and tandem mass spectra (MS/MS) acquisition modes. Each MS scan in the Orbitrap analyzer (mass range, m/z 350–1,800; resolution, 100,000) was followed by MS/MS of the seven most intense ions in the LTQ. Fragmentation in the LTQ was performed by high-energy collision-activated dissociation, and selected sequenced ions were dynamically excluded for 30 s. Raw data were viewed in Xcalibur v.2.1 (Thermo Fisher Scientific), and data processing was performed using Proteome Discoverer v.1.3 beta (Thermo Fisher Scientific). The raw files were submitted to a database search using Proteome Discoverer with an in-house sequence algorithm against the Arabidopsis database, downloaded (in early 2014.10.1) using the Database found in UniProt/Swiss-Prot and UniProt/TrEMBL. The searches were performed with the following parameters: MS accuracy, 10 ppm; MS/MS accuracy, 0.05 Da; two missed cleavage sites allowed; carbamidomethylation of cysteine as a fixed modification; and oxidation of methionine, as variable modifications. The numbers of proteins, protein groups, and peptides were filtered for false discovery rates less than 1% and only peptides with rank 1. The identification lists from technical repetitions were merged, and repeated protein groups were removed.
To investigate whether the formation of FER/RIPK kinase complex was enhanced by RALF1, co-IP assay was performed using RALF1-RNAi (ralf1) and Col.0 plants with or without RALF1 treatment as described above. Protein A/G agrose (Biotool, B23201) was used to immunoprecipitate FER–RIPK protein complexes. The IP samples were separated by SDS/PAGE and probed with anti-FER antibody (1:4,000) and anti-RIPK antibody (1:3,000), respectively.
Plant Phenotype Analysis.
Root hairs from the primary root tip of 7-d-old seedlings were examined.
Quantitative assays of rhizosphere acidification rate were performed following the method described by Haruta (22) and Gujas (34).
For LiCl hypersensitivity assay, 4-d-old vertically grown Arabidopsis seedlings were first placed under constant white light (25 µmol·m2·s1) for 3 d before transferring to medium supplemented with 12 mM LiCl for 4 d, followed by root length measurements for statistical analysis.
To measure blue light response, surface-sterilized seeds were placed onto 1/2 MS agar medium and stratified at 4 °C for 2 d. Seedlings were then vertically grown under blue light (2.0 µmol·m2·s1) for 7 d. After treatment, the root length was measured for statistical analysis.
For auxin treatments, 1-naphthaleneacetic acid (NAA) was added at concentrations indicated in the figures.
For ABA treatments, assays were performed as described (17).
Epidermal cell analysis was carried out as described (41, 42).
RALF1 Treatment Assay.
RALF1 treatment assays were performed following the method described by Haruta (22).
Polyclonal Antibody Preparation.
For RIPK polyclonal antibody preparation, GST-RIPK (21–462 aa) was expressed and purified as described above and used for the immunization of three 6-wk-old healthy mice. The mice were immunized once with 1 mg of purified GST-RIPK protein suspended in complete Freund’s adjuvant (CFA, Sigma, F5881). The mice were reimmunized twice, 14 and 21 d after the first immunization, with 0.5 mg protein each mixed with incomplete Freund’s adjuvant (iCFA, Sigma, F5506). At 28 d following the first immunization, mice were killed and serum was collected by centrifugation of total blood at 10,621 × g for 10 min under 4 °C and then stored at –80 °C.
For FER polyclonal antibody preparation, FER ectodomain (1–446 aa) was expressed as a GST fusion protein and purified as described above and used as an antigen for the immunization of healthy rabbits. The rabbits were immunized once by multipoint s.c. injection with 1 mg of purified GST-FER protein suspended in CFA (Sigma, F5881). The rabbits were boosted twice, 14 and 21 d after the first immunization, with 0.5 mg protein each mixed with iCFA (Sigma, F5506). At 28 d following the first immunization, blood was taken from the aortic, and serum was collected by centrifugation of total blood at 10,621 × g for 10 min under 4 °C and then aliquoted and stored at –80 °C.
Phosphorylation Assays.
Total protein extracts were prepared from Arabidopsis plants using NEB + T buffer [50 mM Tris·HCl (pH 8.0), 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 1% Triton X-100] containing 1 mM PMSF and 1% protease inhibitor mixture (Thermo Fisher Scientific, 78420). To distinguish the phosphorylated and dephosphorylated forms of FER or RIPK, we improved the protein gel quality by (i) adding 28% (vol/vol) glycerol into the separation gel and (ii) running longer time to allow the 35-kDa marker protein to run off the gel, maximizing the separation of high molecular weight proteins.
For FER phosphorylation assay in Arabidopsis plants, FER immunoblotting was performed with rabbit polyclonal anti-FER antibody at a 1:4,000 dilution. Secondary goat anti-rabbit IgG-HRP conjugate was used at a concentration of 1:5,000 for detection via enhanced chemiluminescence (Thermo Fisher Scientific, 34075).
For RIPK phosphorylation assay in Arabidopsis plants, RIPK immunoblotting was performed with mice polyclonal anti-RIPK antibody at a concentration of 1:3,000. Secondary goat anti-mouse IgG-HRP conjugate was used at a concentration of 1:8,000 for detection via enhanced chemilluminescence (Thermo Fisher Scientific, 34075).
CIP Assays.
Total protein extracts were prepared from 10-d-old Arabidopsis plants using CIP buffer [100 mM NaCl, 50 mM Tris·HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, and 0.5% Triton X-100] and divided into two aliquots and preheated at 65 °C for 15 min to inactivate endogenous enzymes. Alkaline phosphatase (New England Biolabs, M0290S) was added to one of the two aliquots, and both were incubated at 30 °C for 10 min. The reaction was terminated by the addition of 1× protein loading buffer and subsequent incubation at 95 °C for 6 min. The samples were then separated on SDS/PAGE gel and examined by immunoblot analysis as described earlier.
In Vitro Kinase Activity Assays.
RIPK (21–462 aa) and RIPKm (K122R) were cloned into pCold-TF-His expression vector for producing His-tagged recombinant protein TF-His-RIPK/RIPKm in E. coli. FER-CD (440–807 aa) and FERm-CD (K565R) were cloned into pET28a expression vector for producing His-tagged recombinant protein His-FER-CD/FERm-CD in E. coli. Protein purity was assessed by SDS/PAGE. Kinase activity assays were performed using the reaction buffer [25 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM CaCl2, 1 mM ATP, 1 mM DTT], 1 μg recombinant protein (TF-His-RIPK/RIPKm and/or His-FER-CD/FERm-CD) as substrates, in a total volume of 50 µL. The assay was initiated by adding 1 mM ATP and incubated for 30 min at 30 °C. The reaction was terminated by the addition of 1× protein loading buffer and subsequent incubation at 95 °C for 5 min. The proteins were then separated on a 10% (mass/vol) SDS/PAGE gel and analyzed by Western blot using anti-His antibody (1:5,000).
Acknowledgments
We thank Drs. Jia Li and Zhiyong Wang for reading the manuscript and helpful comments; Drs. Alice Cheung, Jun Liu, Gitta Coaker, and Daniel Moura for providing plant materials; and Drs. Legong Li and Lei Yang for assistance in laser confocal analysis. This work was supported by National Natural Science Foundation of China Grants NSFC-31571444 and -31400232, the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province Grant 20134486, and a grant from the National Science Foundation (to S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.Z. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609626113/-/DCSupplemental.
References
- 1.Walker JC, Zhang R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature. 1990;345(6277):743–746. doi: 10.1038/345743a0. [DOI] [PubMed] [Google Scholar]
- 2.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelsdorf T, Hamann T. An update on receptor-like kinase involvement in the maintenance of plant cell wall integrity. Ann Bot (Lond) 2014;114(6):1339–1347. doi: 10.1093/aob/mcu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smet I, Voss U, Jürgens G, Beeckman T. Receptor-like kinases shape the plant. Nat Cell Biol. 2009;11(10):1166–1173. doi: 10.1038/ncb1009-1166. [DOI] [PubMed] [Google Scholar]
- 5.Niederhuth CE, Cho SK, Seitz K, Walker JC. Letting go is never easy: Abscission and receptor-like protein kinases. J Integr Plant Biol. 2013;55(12):1251–1263. doi: 10.1111/jipb.12116. [DOI] [PubMed] [Google Scholar]
- 6.Antolín-Llovera M, Ried MK, Binder A, Parniske M. Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol. 2012;50:451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- 7.Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot. 2011;62(5):1581–1591. doi: 10.1093/jxb/erq445. [DOI] [PubMed] [Google Scholar]
- 8.Cheung AY, Wu HM. THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr Opin Plant Biol. 2011;14(6):632–641. doi: 10.1016/j.pbi.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Hématy K, et al. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007;17(11):922–931. doi: 10.1016/j.cub.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Boisson-Dernier A, et al. ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 2013;11(11):e1001719. doi: 10.1371/journal.pbio.1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boisson-Dernier A, et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009;136(19):3279–3288. doi: 10.1242/dev.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyazaki S, et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol. 2009;19(15):1327–1331. doi: 10.1016/j.cub.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 13.Escobar-Restrepo JM, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317(5838):656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 14.Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130(10):2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- 15.Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev Cell. 2014;29(4):491–500. doi: 10.1016/j.devcel.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, et al. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009;106(18):7648–7653. doi: 10.1073/pnas.0812346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu F, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA. 2012;109(36):14693–14698. doi: 10.1073/pnas.1212547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao D, et al. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ. 2015;38(12):2566–2574. doi: 10.1111/pce.12570. [DOI] [PubMed] [Google Scholar]
- 19.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol. 2014;24(16):1887–1892. doi: 10.1016/j.cub.2014.06.064. [DOI] [PubMed] [Google Scholar]
- 20.Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA. 2010;107(41):17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F, et al. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol Plant. 2014;7(5):920–922. doi: 10.1093/mp/ssu010. [DOI] [PubMed] [Google Scholar]
- 22.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014;343(6169):408–411. doi: 10.1126/science.1244454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, et al. 2015. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife doi: 10.7554/eLife.06587.
- 24.Chen J, et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci USA. 2016;113(37):E5519–E5527. doi: 10.1073/pnas.1608449113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Li L, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15(3):329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Kadota Y, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54(1):43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, et al. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7(6):e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9(2):137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, et al. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25(3):1143–1157. doi: 10.1105/tpc.112.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng F, et al. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485(7396):114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 34.Gujas B, Alonso-Blanco C, Hardtke CS. Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr Biol. 2012;22(20):1962–1968. doi: 10.1016/j.cub.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 35.Boisson-Dernier A, Franck CM, Lituiev DS, Grossniklaus U. Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc Natl Acad Sci USA. 2015;112(39):12211–12216. doi: 10.1073/pnas.1512375112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergonci T, et al. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J Exp Bot. 2014;65(8):2219–2230. doi: 10.1093/jxb/eru099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301(5637):1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 38.Xu P, et al. A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol Plant. 2014;7(2):441–444. doi: 10.1093/mp/sst105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler SA, Lindner H, Jones DS, Grossniklaus U. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 2015;16(1):107–115. doi: 10.15252/embr.201438801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. pp. 352–355. [Google Scholar]
- 41.Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB. Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol. 2006;16(9):895–901. doi: 10.1016/j.cub.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 42.Sorek N, et al. Differential effects of prenylation and s-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 2011;155(2):706–720. doi: 10.1104/pp.110.166850. [DOI] [PMC free article] [PubMed] [Google Scholar]