Significance
Light, a prevailing environmental factor, plays important roles in various processes during plant development and stress response. Whether light could also regulate stress-induced transcriptional memory, however, is not clear. Herein we reported that light signal is positively involved in salt-induced transcriptional memory of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) and subsequent proline accumulation. Furthermore, HY5-dependent light signaling is required for the maintenance of salt-induced H3K4me3 in P5CS1 during the recovery stage. This mechanism is likely operating during other stress as well, and could shed light on future research into the concerted effects of different environmental factors on plant response to stresses.
Keywords: transcriptional memory, light, H3K4me3, HY5/HYH, P5CS1
Abstract
To cope with environmental stresses, plants often adopt a memory response upon primary stress exposure to facilitate a quicker and stronger reaction to recurring stresses. However, it remains unknown whether light is involved in the manifestation of stress memory. Proline accumulation is a striking metabolic adaptation of higher plants during various environmental stresses. Here we show that salinity-induced proline accumulation is memorable and HY5-dependent light signaling is required for such a memory response. Primary salt stress induced the expression of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1), encoding a proline biosynthetic enzyme and proline accumulation, which were reduced to basal level during the recovery stage. Reoccurring salt stress-induced stronger P5CS1 expression and proline accumulation were dependent upon light exposure during the recovery stage. Further studies demonstrated that salt-induced transcriptional memory of P5CS1 is associated with the retention of increased H3K4me3 level at P5CS1 during the recovery stage. HY5 binds directly to light-responsive element, C/A-box, in the P5CS1 promoter. Deletion of the C/A-box or hy5 hyh mutations caused rapid reduction of H3K4me3 level at P5CS1 during the recovery stage, resulting in impairment of the stress memory response. These results unveil a previously unrecognized mechanism whereby light regulates salt-induced transcriptional memory via the function of HY5 in maintaining H3K4me3 level at the memory gene.
Proline accumulation is an adaptive mechanism of higher plants to cope with various environmental stresses (1). It is well-documented that the enhanced proline biosynthesis from glutamate is the main contributor for proline accumulation under stress conditions, which is essentially regulated at the transcription level of Δ1-pyrroline-5-carboxylate synthetase 1 (P5CS1) encoding the rate-limiting enzyme of the pathway. As a result, P5CS1 has been overexpressed in many different plant species to achieve enhanced proline content and stress tolerance (1). On the other hand, P5CS1 knock-out mutants displayed impaired proline accumulation and increased sensitivity to osmotic stresses, providing genetic evidences that P5CS1 is necessary and sufficient for proline accumulation (2). However, the studies on long-term stress memory within generation, as well as transgenerational stress memory of proline accumulation and the underlying regulatory mechanisms, are just starting to emerge in the context of rapid progress in plant epigenetic research. It has recently been reported that pretreatment of Arabidopsis with menadione sodium bisulphite could induce more rapid and higher proline accumulation upon subsequent salt stress, which was associated with a hypomethylation state at the promoter region of P5CS1 and proline dehydrogenase 1 (PDH1) (3). In rice, the progenies of osmotically stressed plants were found to acquire altered DNA demethylation in P5CS1 and ornithine-δ-animotransferase (δ-OAT), but not in pyrroline-5-carboxylate reductase (P5CR), correlated with the inheritable up-regulation of P5CS1 and δ-OAT and elevated proline accumulation (3, 4). However, the effect of histone modification on the memory response of proline accumulation upon stress has not been investigated.
Recently, histone modification has been implicated in transcriptional memory of stress responsive genes (5–7). The retention of active transcription marks, H3K4me3 (trimethylation of histone H3 at lysine 4) and Ser5P polymerase II (phosphorylated serine 5 at consensus repeat sequences of the C-terminal domain of RNA polymerase II), on RESPONSIVE TO DESICCATION 29B (RD29B) and RESPONSIVE TO ABA 18 genes in the recovery phase was found to be correlated with more robust induction of their transcription during repeated dehydration (8). For salt stress, on the other hand, the dynamic of a transcriptional repressive mark, H3K27me3, on salt stress-responsive genes was proposed to contribute to their long-term stress memory in Arabidopsis (9). So far, there is no evidence that the enzyme responsible for epigenetic modification, like DNA or histone methyltransferase and histone acetyltransferase, could interact directly with the target genomic DNA region (10). Thus, other proteins, such as transcription factors (TFs) and associated proteins, might function as a mediator for those enzymes to achieve a coordinated and specific gene regulation. Transcription factors WRKY38 and WRKY62 could interact with HDA19 (histone deacetylase 19) to fine-tune the basal resistance to biotic stress in Arabidopsis (11). MYC2 was reported to mediate the memory behavior of a subset of MYC2-regulated genes (12). In addition, a memory recruitment sequence in promoter and nuclear pore proteins were found to be required for the deposition of Ser5P polymerase II and H3K4me2, and hence account for the transcriptional memory of inositol-3-phosphate synthase gene INO1 in yeast responding to inositol starvation and that of HLA-DRA in HeLa cells responding to IFN-γ (13, 14). In the study of RD29B, the abscisic acid (ABA) response elements in the promoter are necessary and sufficient for transcriptional memory responding to dehydration (15). However, environmental factors that might regulate maintenance of stress memory are poorly understood. Light is one of the most important environmental factors for plants, not only in providing the source of energy, but also in its function as a signal to shape plant development and stress responses (16), including proline accumulation during salt stress (17). It has also been reported that light could cooperatively and extensively influence histone modification, which might be involved in light-regulated gene expression (18). However, little is known about the effect of light on transcriptional memory responses during environmental stresses.
In this report, memory response of proline accumulation to repeated salt stresses was found to be caused by transcriptional memory of P5CS1: namely, subsequent stress induces significantly higher transcript levels than initial stress. We have shown that salt stress-induced transcriptional memory of P5CS1 is light-dependent and restricted only to the aerial part. Further study identified a promoter region, which could recruit light-responsive transcriptional factor HY5 to facilitate the maintenance of H3K4me3 at P5CS1 during the recovery stage, accounting for the transcriptional memory of P5CS1.
Results
Transcriptional Memory of P5CS1 Upon Repeated Salt Stress.
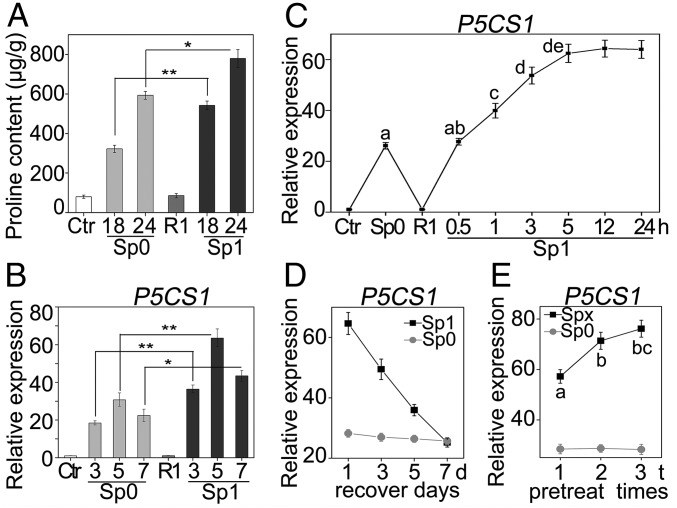
Proline has long been considered as a compatible osmolyte in many plant species, which accumulates in response to a variety of environmental stresses (1). In our experimental condition, the free proline content of 10-d-old Arabidopsis seedlings was less than 100-μg/g fresh weight, while it increased significantly after 8 h of 200-mM NaCl treatment, and reached up to 600-μg/g fresh weight after 24 h (Fig. S1). To understand whether proline accumulation is memorable during repeated salt stress, the free proline content in 10-d-old Arabidopsis seedlings was examined with or without prestress treatment. The pretreatment was performed by incubating the seedlings in liquid Murashige and Skoog medium (MS) supplemented with 100 mM NaCl for 24 h. Pretreated seedlings were allowed to recover (R1) for 48 h to reset the proline content to the control (Ctr) basal level before a second treatment (Sp1, one-time pretreatment) with 200 mM NaCl. Pretreated seedlings showed more rapid and higher accumulation of proline during the second salt treatment compared with the seedlings without pretreatment (Sp0) (Fig. 1A). The result indicated that Arabidopsis memorized the information obtained from previous stress experience to manipulate proline biosynthesis upon upcoming stress.
Fig. S1.
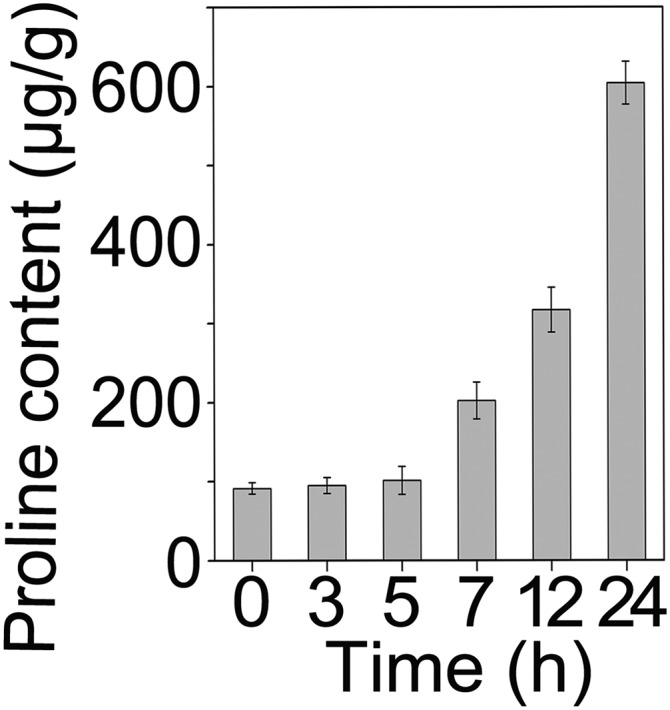
Time course of proline accumulation under 200 mM NaCl. Data are shown as mean ± SEM (n = 3). More than three biological replicates were performed and one representative is shown.
Fig. 1.
Memory responses of proline accumulation and P5CS1 transcription under salt stress. (A) Seedlings were pretreated with 100 mM NaCl for 24 h and then recovered for 48 h before being stressed with 200 mM NaCl for 18 or 24 h. Proline content was measured at different stages. Ctr, nonstressed control; R1, recovered plant after pretreatment; Sp0, stressed plant without pretreatment; Sp1, stressed plant with pretreatment. (B) Seedlings were treated similarly to A, except that the recovery was for 24 h and the second stress was for 3, 5, or 7 h. Transcript levels of P5CS1 were measured at different stages. (C) Seedlings were pretreated for 0.5, 1, 3, 5, 12, or 24 h or not; P5CS1 transcript levels were measured after 5 h of second stress to show the pretreatment time needed to establish memory. (D) Seedlings were pretreated for 24 h or not; P5CS1 transcript levels were measured after 1, 3, 5, or 7 d of recovery and 5 h of the second stress to investigate the maintenance of transcriptional memory. (E) After one, two, or three pretreatments (each for 5 h), 19 h of recovery, and 5 h of the second stress, P5CS1 transcript levels were measured. The transcript levels were measured by real-time qPCR with ACT 7 as an internal control. A and B were repeated at least five times, and C–E were repeated three times, each with three replicates. A representative result was shown as mean ± SEM (n = 3). Asterisks in A, B, and different letters on the value in C and E indicate statistical significance based on Student’s t test. **P < 0.01; *P < 0.05.
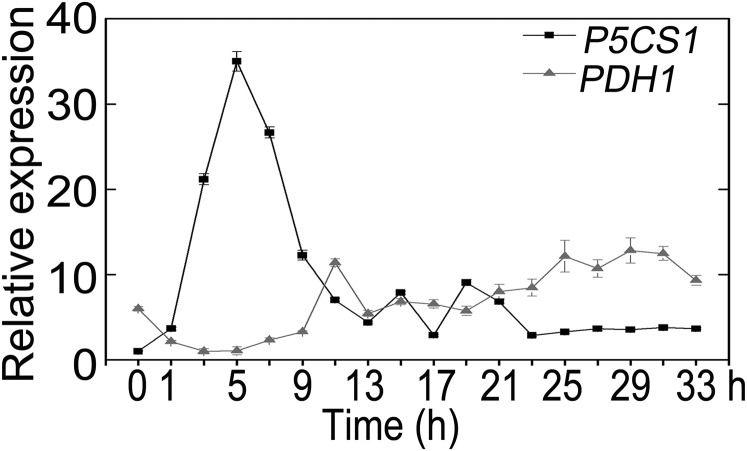
Given the fact that proline accumulation results mainly from enhanced biosynthesis from glutamate and decreased degradation, and P5CS1 and PDH1 catalyzes the rate-limiting step in two pathways, respectively (19, 20), we investigated the time course expression of these two genes under salt stress. The transcript level of P5CS1 was transiently induced, after 5 h of salt stress, to about 30-fold and then progressively declined to a stable level about 5-fold after 24 h. As expected, PDH1 expression was down-regulated initially to about 20% of the original level and fluctuated around the basal level afterward (Fig. S2). This result was consistent with previous reports (20, 21) and P5CS1 was likely to be the major player for proline accumulation during salt stress. Therefore, we next examined whether transcription of P5CS1 can also be memorized during repeated salt stress. For this purpose, 10-d-old seedlings were pretreated with 100 mM NaCl for 24 h, recovered for 24 h, and subsequently subjected to 200 mM NaCl for 3, 5, and 7 h, because P5CS1 expression peaked at 5 h after salt stress. The P5CS1 transcript level was measured at the end of recovering period (R1) and after 200 mM NaCl treatment with (Sp1) or without (Sp0) pretreatment. Similar to proline accumulation, subsequent exposure to 200 mM NaCl caused quicker and more potent induction of P5CS1 (Fig. 1B). Two additional reference genes were used as internal controls to calibrate the P5CS1 transcript level and similar results were obtained (Fig. S3). Hereafter, we named the quicker and more potent induction of P5CS1 as “transcriptional memory of P5CS1” (TMP).
Fig. S2.
Time course of P5CS1 and PDH1 transcript levels under 200 mM NaCl. Data are shown as mean ± SEM (n = 3).
Fig. S3.
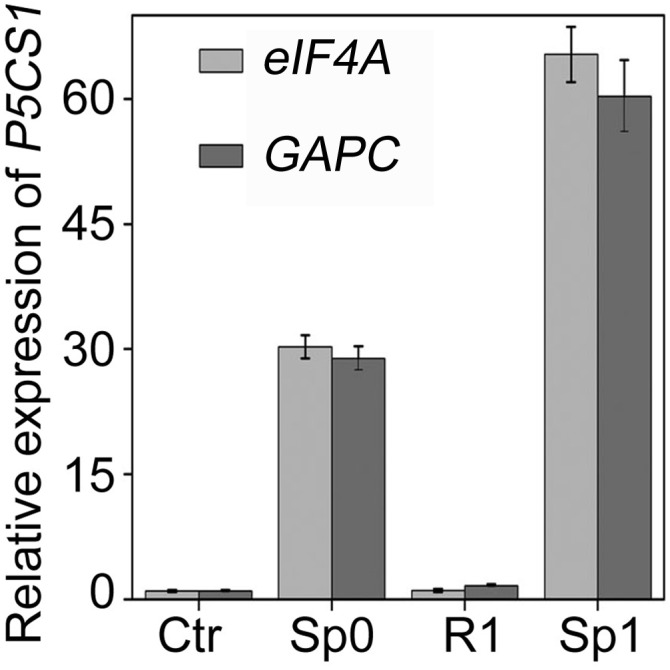
Different reference genes were used as internal control to test TMP. The transcript levels of P5CS1 in Ctr, Sp0, R1, and Sp1 states were measured. Data are shown as mean ± SEM (n = 3).
To better understand TMP, we then determined the minimum length of the pretreatment that is sufficient for evoking TMP during subsequent stress, and how long the TMP could persist before the subsequent stress. To this end, 10-d-old seedlings were pretreated for 0, 0.5, 1, 3, 5, 12, or 24 h in saline MS liquid. After 24 h of recovery and 5 h of subsequent salt stress the transcript levels of P5CS1 in Ctr, Sp0, R1, and Sp1 plants were measured. The result demonstrated that 1 h of pretreatment was sufficient for inducing TMP (Fig. 1C). In addition, the longer the pretreatment, the stronger the TMP would be in the subsequent stress. Consistent with the transcription level of P5CS1, proline accumulation also exhibited a similar tendency (Fig. S4). To establish how long the TMP could last, 24-h pretreated seedlings were recovered for 1, 3, 5, or 7 d before the next stress was applied. After 1, 3, or 5 d of recovery, TMP could still be detected during subsequent salt stress, but could not after 7 d of recovery (Fig. 1D). This result suggested that there is a time limit beyond which Arabidopsis will not remember the salt stress it has experienced in terms of P5CS1 expression.
Fig. S4.
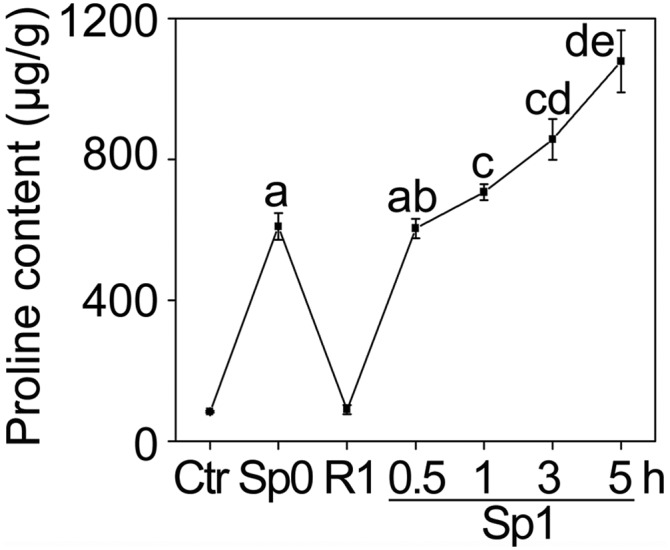
Memory response of proline accumulation after salt pretreatment for different times. Plants were pretreated for different hours and proline content in Ctr, Sp0, R1, and Sp1 were measured. Different letters on the value indicate statistical significance based on Student’s t test. The experiment was repeated at least three times. The representative experiment shown indicates the mean ± SEM (n = 3).
Next, we investigated if repeated pretreatment could enhance TMP. Seedlings were pretreated in saline solution one to three times (Sp1–Sp3) separated by a 19-h interval of recovery (R1–R3). The repetitious exposure could strengthen the TMP, as the transcript level of P5CS1 in Sp3 was higher than in Sp2 and Sp1 (Fig. 1E). Thereafter, we pretreated seedlings twice for 5 h each to analyze transcriptional memory.
TMP Is Light-Dependent and Restricted to the Aerial Part.
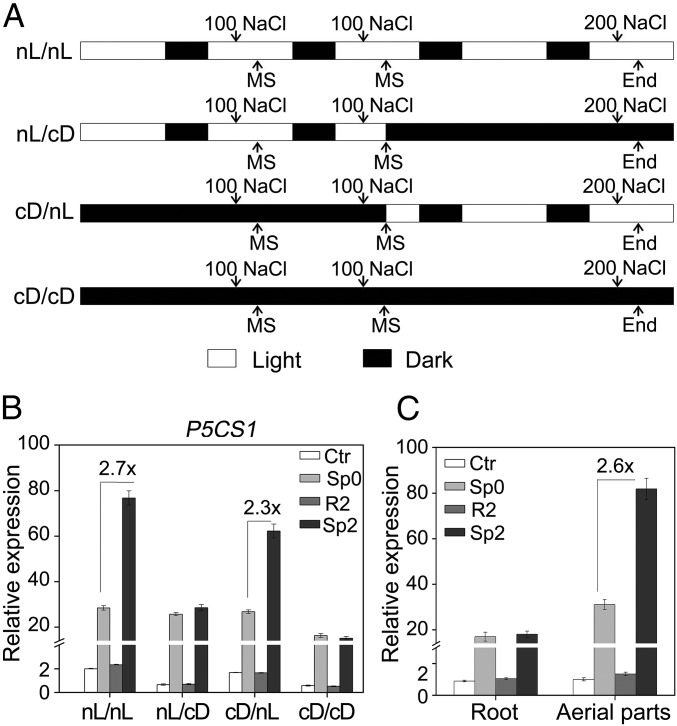
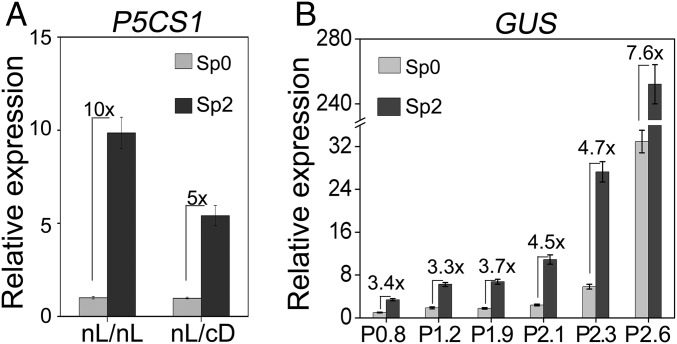
It has been previously reported that the induction of P5CS1 by salt or ABA was attenuated in dark (17); thus, we asked whether darkness could also impede the salt stress-induced TMP. To answer this question, pretreatment (two times) and the last stress treatment were performed under either normal light/dark period (nL) or continuous dark (cD), with four different kinds of pretreatment/stress combinations: nL/nL, nL/cD, cD/nL, and cD/cD (Fig. 2A). A period of dark adaptation was allowed for pretreatment (cD/nL, cD/cD) or the last stress (nL/cD) in the dark. The P5CS1 transcript level was measured in the control (Ctr) plants, the plants stressed with (Sp2) or without (Sp0) pretreatment (twice), and the plants recovered from the second pretreatment (R2). Pretreatment in the nL or cD could both potentiate TMP, as long as the second recovery and the last stress treatment were performed in the nL (nL/nL and cD/nL). However, TMP could not be observed if the second recovery and the last stress were carried out under cD, regardless the light condition during pretreatment (Fig. 2B). These results indicated that light during the final recovery and stress treatment was required for manifestation of TMP, but light was not essential for establishing the potential of TMP during the pretreatment period.
Fig. 2.
TMP is affected by light conditions and restricted to the aerial part. (A) The diagram of experimental design with four different light conditions during two-time pretreatment and the last stress (nL/nL, nL/cD, cD/nL, and cD/cD). Open box, light period; solid box, dark period. Transcript levels of Ctr, Sp0, R2, and Sp2 under different conditions (B) or in different organs (C) were measured from intact seedlings (B) or the root and aerial part (C), respectively. Values between bars in B and C denote fold-change. All experiments were repeated three times, each with three replicates. A representative result was shown as mean ± SEM (n = 3).
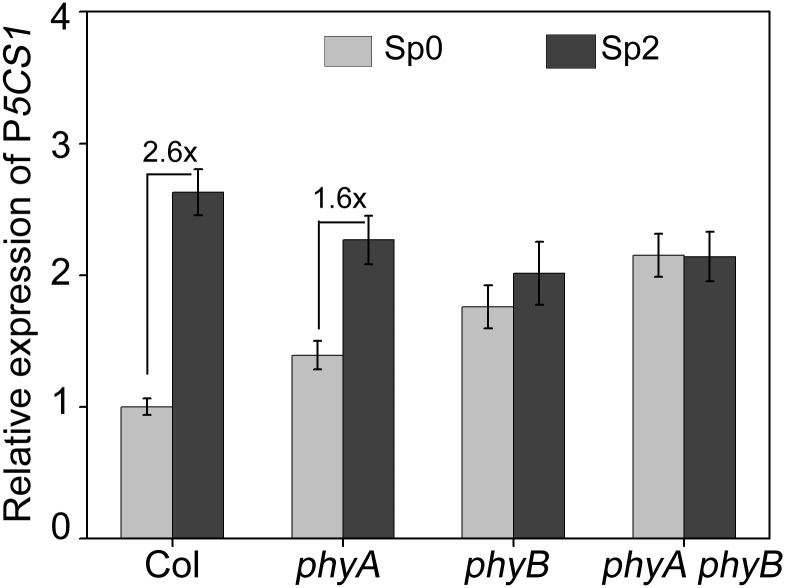
Given that phytochrome is the upstream component in the light-signaling pathway, we tested the memory response of P5CS1 in two photoreceptor mutants, phyA, phyB, and phyA phyB growing under the nL. Results showed that transcriptional memory was reduced in phyA and lost in phyB and phyA phyB (Fig. S5), suggesting that phyB may play a more vital role in TMP.
Fig. S5.
The transcript levels of P5CS1 for Ctr, Sp0, R2, and Sp2 in Col, phyA, phyB, and phyA phyB mutant seedlings. The experiment was repeated at least three times. The representative experiment shown indicates the mean ± SEM (n = 3). Values between bars indicate fold-change.
In nature, roots grow underground and seldom experience the alternation of light and darkness. It has been recently reported that light differentially regulated a group of Small Auxin Up RNA genes through auxin and phytochrome interacting factor, which further mediate the differential growth of different organs (22). Thus, transcriptional memory was further analyzed separately in roots and in aerial parts under nL. Interestingly, TMP was only detected in aerial parts, but not in roots, and the magnitude of P5CS1 salt-induction was also higher in aerial parts than that in roots (Fig. 2C).
The Dynamic of H3K4me3 Is Correlated with Salt-Induced TMP.
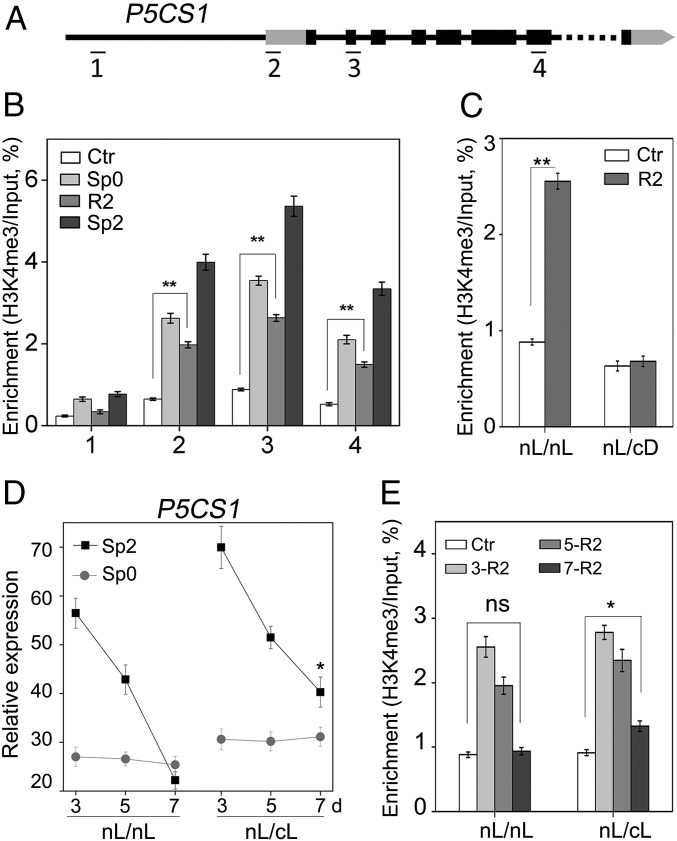
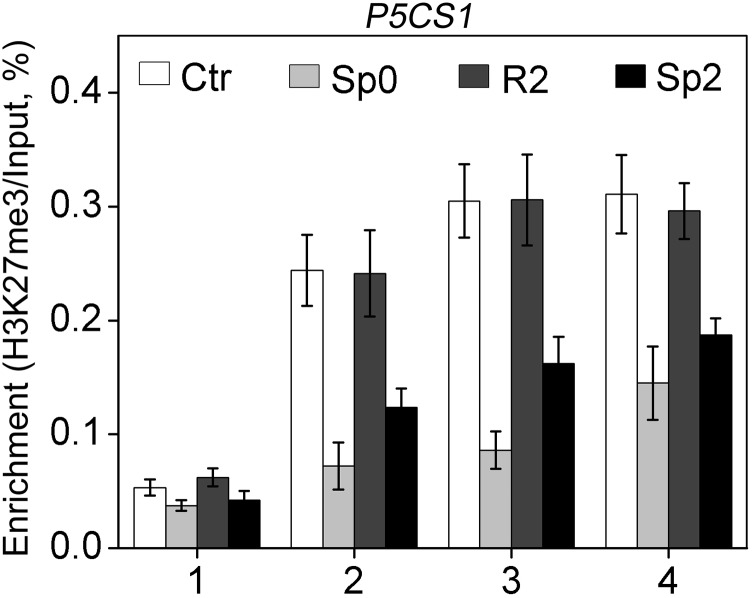
It has been shown that the retention of H3K4me3 at P5CS1 during the recovery stage after dehydration pretreatment was associated with dehydration-induced transcriptional memory of P5CS1 (8). In addition, the fluctuation of H3K27me3 level was also found to contribute to salt-induced transcriptional memory response (9). Therefore, we tested the deposition of H3K4me3 and H3K27me3 on P5CS1 in different stages (Ctr, Sp0, R2, and Sp2). Four regions in the P5CS1 genomic sequence, namely, ∼1,600 bp upstream of the translation start site (TSS) (region 1), the 5′-UTR (region 2), the second exon (region 3), and a midcoding region (region 4), were examined (Fig. 3A). H3K4me3 modification was enriched around the TSS (5′UTR and second exon) of P5CS1, consistent with previous reports (23, 24). H3K4me3 levels induced by salt pretreatment at the 5′UTR, the second exon, and the midcoding region, but not the promoter region, of P5CS1 were maintained at a level significantly higher than that in Ctr at the end of the recovery stage (R2) (Fig. 3B) when the amount of P5CS1 transcript has returned to the Ctr level (Fig. 2B). Consistent with the quicker and more potent induction of P5CS1 upon subsequent exposure, H3K4me3 levels at the three regions of P5CS1 were higher in Sp2 compared with that of Sp0 (Fig. 3B). In contrast, we failed to detect a correlation between the fluctuation of H3K27me3 modification and P5CS1 transcriptional memory (Fig. S6). Thus, it is possible that the retention of H3K4me3 at P5CS1 during the recovery stage accounted for salt stress-induced TMP; however, other chromatin modifications could not be ruled out either.
Fig. 3.
TMP during salt stress was correlated to H3K4me3 retention. (A) Schematic diagram of P5CS1, including the promoter region (thin line to the left), 5′ and 3′UTR (gray box), exons (dark box), introns (thin line between exons), and omitted region (dotted line). The DNA regions analyzed by ChIP-qPCR are indicated with numbered bars underneath. Plants were grown under nL/nL (B) or nL/cD (C) conditions, as shown in Fig. 2A. H3K4me3 levels in regions 1–4 (B) or region 3 (C) of P5CS1 at Ctr, Sp0, R2, and Sp2 (B) or Ctr and R2 stages (C) were measured. (D) Transcript levels were measured in Sp0 and Sp2 plants, which were recovered for 3, 5, or 7 d under nL or cL before the last stress. (E) H3K4me3 levels were measured in Ctr and R2 plants recovered for 3, 5, or 7 d under nL or cL conditions. Experiments were repeated twice, each with three replicates. A representative result was shown as mean ± SEM (n = 3) with ACT7 as an internal control. Asterisks indicate statistical significance of transcript levels (D) between Sp2 and Sp0 or H3K4me3 levels (B, C, and E) between R2 and Ctr based on Student’s t test. **P < 0.01; *P < 0.05.
Fig. S6.
H3K27me3 levels on P5CS1 in Ctr, Sp0, R2, and Sp2 stage. One to four regions (shown in Fig. 2A) of P5CS1 were tested. ACT7 was used as an internal control. The experiment was repeated two times, each with three replicates. The representative experiment shown indicates the mean ± SEM (n = 3).
Because TMP was light-dependent and the retention of H3K4me3 during the recovery stage may be crucial for TMP, we went on to study how the H3K4me3 level at the R2 stage is affected by the light regime. To this end, the H3K4me3 level after recovery for 48 h was compared between nL/nL and nL/cD, where the two pretreatments were identically performed under nL, but the recovery was either under nL or cD. In R2 seedlings, pretreatment-induced H3K4me3 remained at a higher level under nL recovery, but returned to Ctr level under cD recovery (Fig. 3C), reinforcing the correlation between H3K4me3 retention and TMP.
To further verify the effect of light on TMP and H3K4me3 level, the seedlings were recovered for different days under continuous light (cL) or nL. TMP can still be observed after 7 d of recovery under cL, but not for recovery under nL (Fig. 3D). Accordingly, pretreatment-induced H3K4me3 modification was maintained longer when recovered under cL (Fig. 3E). Thus, we conclude that light is a positive regulator for TMP and probably affects TMP by helping the retention of H3K4me3 modification during the recovery stage.
A Fragment in the P5CS1 Promoter Is Essential for TMP.
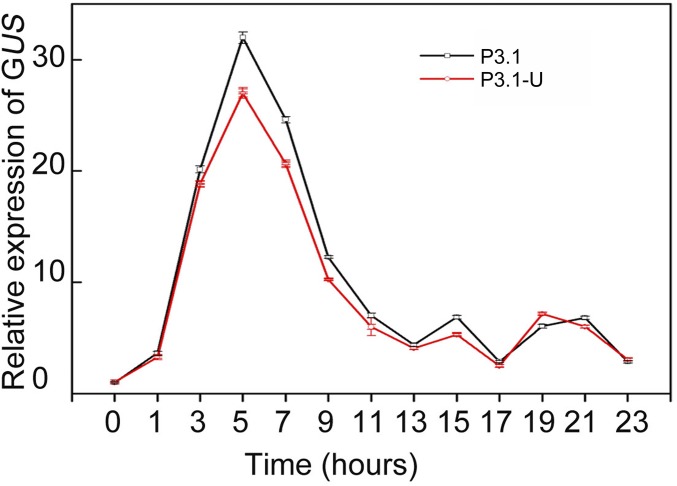
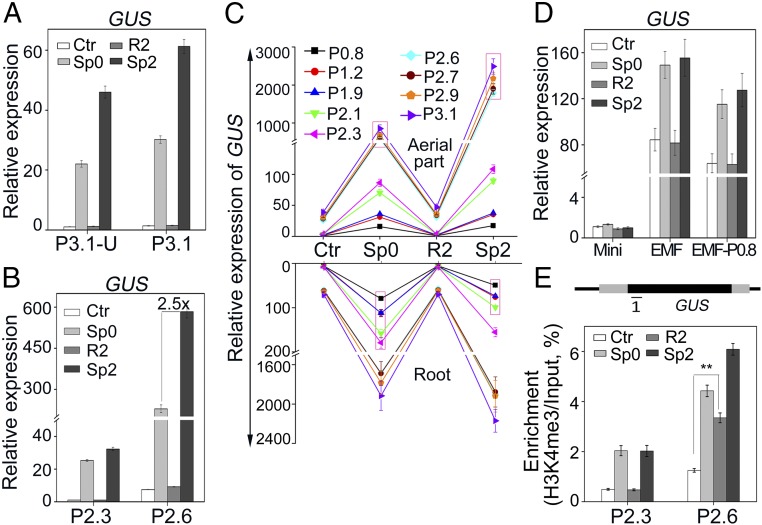
To identify whether the promoter or the 3′UTR of P5CS1 play any roles in TMP, we constructed the β-glucuronidase (GUS) fusion with the 3.1-kb P5CS1 promoter, with (p3.1-U) or without (p3.1) 3′UTR of P5CS1. Transgenic plants harboring two constructs (P3.1-U and P3.1) were exposed to salt stress and GUS transcript levels were measured every 2 h. The result indicated that the GUS reporter gene could mimic the induction pattern of P5CS1 in both P3.1-U and P3.1 transgenic plants under salt stress (Fig. S7). In addition, the transcriptional memory of GUS could be observed in both P3.1-U and P3.1 transgenic lines at a similar level (Fig. 4A), indicating that the promoter region is sufficient and the 3′UTR of P5CS1 is not required for transcriptional memory.
Fig. S7.
Time course transcript levels of GUS of P3.1 and P3.1-U transgenic plants under 200-mM NaCl stress. More than five independent transgenic lines were measured. One representative result is shown as mean ± SEM (n = 3).
Fig. 4.
The levels of transcript and H3K4me3 of GUS in different transgenic plants under salt stress. Transcript levels of GUS were measured in P3.1/P3.1-U (A), P2.3/P2.6 (B), and Mini/EMF/EMF-P0.8 (D) intact transgenic plants at Ctr, Sp0, R2, and Sp2 stage. (C) The transcript levels of GUS in roots and aerial part of different transgenic plants (from P0.8 to P3.1). Equal portions of three representative transgenic lines with the same construct were mixed and data are from three biological sample replicates. (E) H3K4me3 levels at region 1 of GUS in P2.3 and P2.6 transgenic plants are shown. Experiments were performed with three independent transgenic lines in A, B, D, and E with similar results, and one representative result is shown as mean ± SEM (n = 3). Asterisks in E indicate statistical significance based on Student’s t test. **P < 0.01; *P < 0.05.
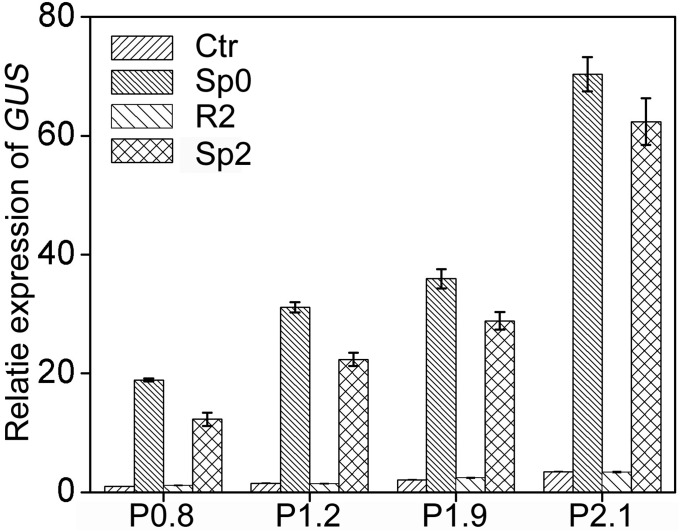
To define the promoter region that affects TMP, a series of 5′ truncated promoters were fused to GUS and used to generate transgenic lines of P2.9, P2.7, P2.6, P2.3, P2.1, P1.9, P1.2, and P0.8, with promoters of 2.9, 2.7, 2.6, 2.3, 2.1, 1.9, 1.2, and 0.8 kb upstream the TSS, respectively. Transcriptional memory of GUS was not significantly altered when the promoter was longer than 2.6 kb, but disappeared when the promoter was truncated to 2.3 kb or shorter (Fig. 4B and Fig. S8), suggesting that the fragment between −2.6 kb and −2.3 kb was essential for the TMP. For simplicity, we refer to such a fragment (between −2.6 and −2.3 kb) as an “essential for memory fragment” (EMF). Given that the TMP was restricted to aerial parts in wild-type seedlings, the stress memory of GUS expression was further investigated in roots and aerial parts of different transgenic seedlings (Fig. 4C). Similar to endogenous P5CS1, the transcriptional memory of GUS was also restricted to aerial parts and disappeared when the promoter was truncated to 2.3 kb or shorter.
Fig. S8.
Short P5CS1 promoters driven GUS genes did not show transcriptional memory. Transcript levels of GUS at the Ctr, Sp0, R2, and Sp2 stage were measured in P0.8, P1.2, P1.9, and P2.1 transgenic plants. Three independent transgenic lines were measured. One representative result is shown as mean ± SEM (n = 3).
To investigate whether the EMF is sufficient for the transcriptional memory, it was fused upstream of a 54-bp minimal CaMV 35S promoter or a 0.8-kb P5CS1 promoter, which, by themselves, do not show any stress memory when driving GUS. In transgenic Arabidopsis seedlings, these constructs did not confer transcriptional memory response and the GUS transcript was only weakly induced by salt stress (about twofold) (Fig. 4D), indicating that the EMF was not sufficient for the TMP.
Deposition of H3K4me3 on the GUS Gene.
Given that the EMF is essential for salt-induced TMP, we measured the H3K4me3 level of the GUS gene in P2.3 (does not contain the EMF) and P2.6 (contains the EMF) transgenic plants. H3K4me3 modification of GUS was higher in P2.6 than in P2.3. In addition, pretreatment-induced H3K4me3 was maintained at a higher level in P2.6 but dropped to the Ctr level in P2.3 after 24 h of recovery (Fig. 4E). Together with the results above, the findings reinforce that the EMF is essential for the salt-induced TMP. Thus, we proposed that some cis-elements in the EMF may control the dynamics of H3K4me3 at P5CS1.
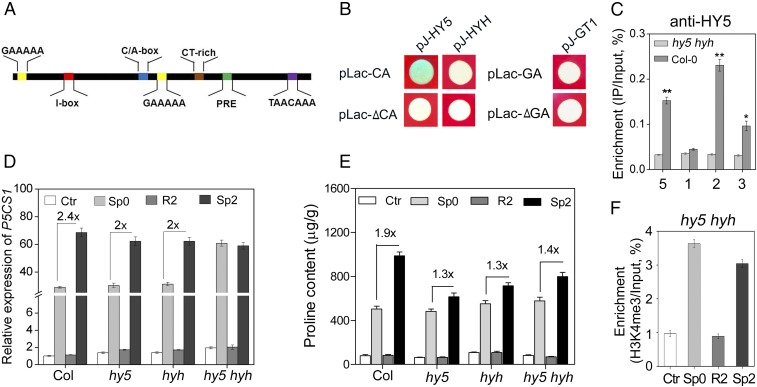
HY5 Binds Directly with C/A-box Element in EMF.
To advance further our understanding of light-dependent TMP, we performed in silico analysis for the cis-acting elements in the EMF (25, 26). Five kinds of light- or abiotic stress-responsive cis-acting elements were identified (Fig. 5A). It has been reported that GAAAAA elements, the binding site of GT-1, a C/A-box (ACGT-containing element), and the binding site of HY5 were responsible for plant response to both light and abiotic stress (27, 28). In addition, two putative HY5 binding sites (C/A box) were also found in the 5′UTR of P5CS1. Based on above results that the EMF was required for light-dependent TMP, the binding of HY5, HYH to C/A-box, and GT-1 to the GAAAAA element within the EMF were then tested using a yeast one-hybrid assay. As shown in Fig. 5B, HY5 could bind to the C/A-box (pLac-CA) but not to a mutated C/A-box (pLac-ΔCA, CCACGT changed to ACAACT). On the other hand, HYH and GT-1 did not bind to the C/A-box and GAAAAA element, respectively (Fig. 5B). To confirm the binding of HY5 to the C/A-box in vivo, we performed a chromatin immunoprecipitation with quantitative PCR (ChIP-qPCR) analysis using anti-HY5 antibody. Indeed, the EMF, 5′UTR, and second exon were significantly enriched in DNA precipitated from Col, but not in DNA pulled down from the hy5 hyh mutant (Fig. 5C).
Fig. 5.
HY5 binds to EMF and influences TMP. (A) Six kinds of element were predicted in the EMF. (B) Yeast one-hybrid assay tested the ability of HY5, HYH, and GT-1 to bind to C/A-box (pLac-CA), GAAAA(pLac-GA), mutational C/A-box (pLac-▵CA), or GAAAA(pLac-▵GA). The blue plaque indicates positive interaction between the tested proteins and corresponding elements. (C) The enrichment of regions 1, 2, 3 (as shown in Fig. 3A), and 5 (EMF) in P5CS1 precipitated by HY5 antibody in Col-0 and hy5 hyh. (D) The transcript levels of P5CS1 and (E) proline content at the Ctr, Sp0, R2, and Sp2 stage in wild-type and mutant plants. Values between bars denote fold-change. (F) H3K4me3 levels of P5CS1 at region 3 (Fig. 3A) at the Ctr, Sp0, R2, and Sp2 stage in hy5 hyh double mutants. C–F were repeated three times and a representative result is shown as mean ± SEM (n = 3). Asterisks in C indicate statistical significance between hy5 hyh and Col-0 based on Student’s t test. **P < 0.01; *P < 0.05.
HY5/HYH May Positively Regulate Salt-Induced TMP.
To further understand whether HY5 is necessary for the TMP, the transcript levels of P5CS1 at different stages (Ctr, Sp0, R2, and Sp2) were measured in hy5, hyh, and hy5 hyh under nL. Interestingly, compared with the wild-type, the intensity of transcriptional memory was slightly affected in hy5 and hyh, and abolished in hy5 hyh (Fig. 5D). The salinity-induced proline accumulation was also determined in these three mutants. The memory intensity of proline accumulation was impaired remarkably in hy5, hyh, and hy5 hyh (Fig. 5E). Subsequently, the H3K4me3 dynamic on P5CS1 was also measured in hy5 hyh under nL. The result showed that the pretreatment-induced H3K4me3 deposition, which could be retained for about 5 d in the wild-type, was erased after merely 19 h of recovery in hy5 hyh (Fig. 5F), suggesting that HY5/HYH were required for maintenance of pretreatment-induced H3K4me3 on P5CS1. Taken together, these results indicate that HY5 and HYH are required for salinity-induced stress memory of P5CS1 expression and proline accumulation.
Light Also Affects Dehydration-Induced TMP.
It has been reported that P5CS1 displayed transcriptional memory in repeated dehydration stress (8). We are curious whether light is also necessary for TMP induced by dehydration. Seedlings were treated with dehydration stress under nL/nL and nL/cD (see details in Materials and Methods). As shown in Fig. 6A, transcriptional memory of P5CS1 could be detected when seedlings were recovered in darkness, but the intensity was significantly reduced, suggesting that full activation of dehydration-induced TMP is light-dependent. To test whether the EMF was also essential for dehydration-induced TMP, the GUS transcript levels at the Sp0 and Sp2 stage in various transgenic plants (P2.6–P0.8) were measured. Under dehydration stress, a higher transcript level was detected in the Sp2 than in Sp0 stage in all transgenic plants, but the intensity of the memory gradually dropped as the P5CS1 promoter was shortened (Fig. 6B), suggesting that the EMF is not the only promoter region required for the dehydration-induced TMP.
Fig. 6.
Light also affects dehydration stress-induced TMP. (A) Transcript levels of P5CS1 at Sp0 and Sp2 stage under nL/nL or nL/cD conditions (similar to Fig. 2A, with modification stated in Materials and Methods). (B) Transcript levels of GUS at Sp0 and Sp2 stage in P0.8–P2.6 transgenic plants. Equal portions of three independent representative transgenic lines were mixed and data are from three biological sample replicates and shown as mean ± SEM (n = 3). Experiments were repeated twice, each with three replicates and a representative result was shown as mean ± SEM (n = 3). Values between bars denote fold-change.
Discussion
Transcriptional memory response to adverse stimuli could help plants to better cope with recurring stresses and improve the acclimation to a temperamental environment. In this report, we have presented evidence that salt stress-induced P5CS1 expression, as well as proline accumulation, can be memorized during repeated salt stress, which is associated with retention of H3K4me3 at P5CS1 during the recovery stage. Interestingly, we also revealed that light, a distal promoter fragment EMF, and HY5/HYH are required for the manifestation of salt-induced TMP, by helping in the maintenance of salt-induced H3K4me3 state.
The involvement of the distal promoter element and binding protein in transcriptional memory has been reported in human and yeast, where the binding of nuclear pore proteins to a promoter element of the memory genes was found to be essential for their transcriptional memory by affecting the histone modification and their targeting to particular nuclear loci (13, 14). In Arabidopsis, ABA response elements and their flanking sequence in the promoter of RD29B, a memory gene during dehydration, were required for promoting transcriptional memory of RD29B. The ABA response elements binding factors, ABF2/3/4, induce RD29B expression during primary stress, but triple mutations in ABF2/3/4 (abf2 abf3 abf4) did not affect the transcriptional memory of RD29B, suggesting that activation of the memory gene and stress-induced transcriptional memory are regulated by different sets of transcriptional cofactors (15). In our study, we found that the EMF, required for TMP, could interact with an HY5 transcriptional factor. However, HY5 and HYH are not responsible for the salt-induced P5CS1 expression (and actually showed a negative effect on basal expression of P5CS1, as shown in Fig. 5D), but rather involved in maintenance of the TMP. This result suggests that in our case transcriptional memory was not attributable to a poised TF regulating the target gene during pretreatment.
The dependence of transcriptional memory on light has not been reported previously. In our study, several lines of evidences supported this conclusion. First, the quicker and stronger response to subsequent salt stress was suppressed by dark during the recovery stage. Second, continuous light in the recovery state would prolong the memory time. Third, the loss-of-function of several key light-signaling components, HY5/HYH, phyA, and phyB, would impede the transcriptional memory response. Finally, HY5 could interact, both in vivo and in vitro, with the EMF in the P5CS1 promoter.
HY5 is a versatile transcriptional factor, involved in the regulation of multiple developmental processes and stress responses (29–31). Our results indicated that the level of H3K4me3 during the recovery stage, but not during the pretreatment stage, is significantly affected in hy5 hyh (Fig. 5F). Similarly, dark treatment during the recovery stage also impaired the retention of H3K4me3 (Fig. 3C). These findings could be explained by the fact that cD accelerates the degradation of HY5/HYH (16). In contrast, cL, which would further stabilize HY5/HYH (16), also favored the retention of H3K4me3 for a longer time (Fig. 3E) and prolonged transcriptional memory (Fig. 3D). It is interesting that HY5 was found to bind not only to the EMF, but also to the regions flanking the TSS, where the H3K4me3 levels are high. This finding raises the possibility that HY5 proteins bound to the EMF and downstream region, collaborating as a homodimer to maintain H3K4me3 level, because HY5/HYH was reported to form homodimers to regulate reactive oxygen species-responsive gene expression in the cross-talk between light and reactive oxygen species signaling (28). It is somehow unexpected to detect that the memory response of proline accumulation was significantly impaired in hy5 and hyh single mutants (1.9× in Col-0 vs. 1.3× in hy5 and hyh), whereas the TMP was only marginally affected (2.4× in Col-0 vs. 2.0× in hy5 and hyh) (Fig. 5 D and E). This result is probably an indication that HY5 and HYH mutations might affect other aspects of proline accumulation, such as translational regulation of P5CS1 or the supply of glutamate, the precursor for proline biosynthesis.
Currently, it is not clear how HY5/HYH is functioning in maintaining the H3K4me3 state during the recovery stage. Although there is no evidence that HY5 could directly interact with H3K4 histone methyltransferase or demethylase, HY5 was proposed to recruit GCN5 to acetylate histones and to activate light-responsive gene expression (32), and histone acetylation has been reported to associate with H3K4me3 to activate gene transcription (23, 33). The result in Fig. 5D demonstrated that HYH is functioning redundantly or synergistically with HY5 in mediating the TMP. However, we failed to detect the interaction of HYH with the C/A-box in a yeast one-hybrid assay (Fig. 5B). It is possible that HYH and HY5 are components of a multiprotein complex and HYH could bind to the C/A-box indirectly via other proteins in the complex. This possibility was supported by previous reports that HY5 and HYH could form heterodimer with other TFs (34), such as G-box binding factor 1 (GBF1), to regulate its genome-wide DNA binding (35). Because GBFs are also predicted to bind to C/A-box cis-element in the EMF of the P5CS1 promoter, it is possible that HY5/HYH-GBF1 heterodimers may be required for the maintenance of the TMP. In addition, the possibility that the binding to the C/A-box of HYH itself is too weak to be detected by yeast one-hybrid assay cannot be ruled out.
It is not surprising that salt-induced TMP was not detected in roots (Fig. 4C), because roots are natively grown in dark. Given the fact that the stability and activity of HY5/HYH are subjected to different levels of light-mediated regulation, such as that by proteasome and kinase (36), it is possible that HY5/HYH is differentially fine-tuned for specific tissue. Actually, many TFs have been reported to function in organ-specific manners (10, 37, 38).
The memory response of salt-induced P5CS1 expression and proline accumulation under the light condition is biologically relevant, because proline accumulation has been proposed to ameliorate the damage caused by photoinhibition during most of the stresses by consuming NADPH as a cofactor for proline biosynthesis (39). Photoinhibition, resulting from overreduction of photosystem II (40, 41), is likely more severe under light than in dark. Therefore, being able to activate proline biosynthesis more rapidly upon repeated stress would certainly be beneficial for the adaptation of the plant to such stresses.
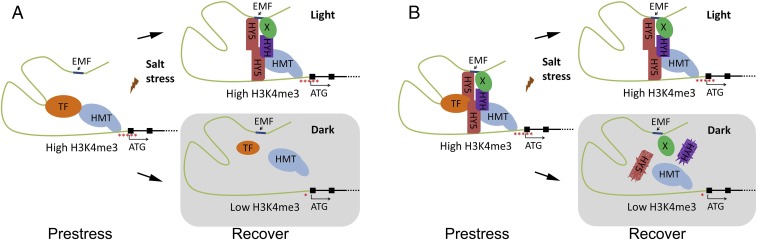
Based on these observations, we propose a working model (Fig. 7). During prestress, the transcriptional regulatory complex, containing salt stress-responsive TF and H3K4 methyltransferase, establishes a high H3K4me3 level at P5CS1 and activates its transcription. However, whether HY5 and HYH are included in the complex during the prestress condition is unknown. During recovery, the TF may dissociate from the complex or be inactivated, because the transcription level of P5CS1 was reduced to the control level at this stage (Figs. 1 and 2). If recovered in the light, COP1 activity is low, and subsequently HY5 and HYH are accumulated (42), which is beneficial for the maintenance of salinity-induced TMP. Because ChIP assays showed that HY5 associates with both the EMF and 5′UTR of P5CS1 (Fig. 5C), it is possible that HY5 and HYH proteins could form homodimer or heterodimer with itself or another factor (X) to maintain the high H3K4me3 level in P5CS1 in the absence of the TF, which would increase the probability of more robust transcription during subsequent stress. If recovered in darkness, COP1 mediates ubiquitination and degradation of HY5 and HYH rapidly (42), resulting in the dissociation of the complex and reduction of H3K4me3 levels at the P5CS1 locus.
Fig. 7.
The working model of how light regulates the salt stress-induced TMP. We propose that (A) during prestress conditions, a salt stress-responsive TF binds to the promoter of P5CS1 and recruits histone H3K4 methyltransferase (HMT), which results in a high H3K4me3 level (shown as red asterisk) and transcriptional activation of P5CS1. During recovery in the light, HY5 and HYH are recruited to the transcriptional regulator complex as a homodimer or heterodimer. HY5 directly associates with the EMF and 5′UTR of P5CS1, and possibly form a supercomplex to enhance the maintenance of H3K4me3 at the P5CS1 locus. On the other hand, HYH may indirectly (through unknown factor X) associate with the EMF of P5CS1 promoter to maintain H3K4me3 levels at P5CS1 locus. In darkness, extreme low levels of HY5 and HYH cannot maintain prestress-induced high levels of H3K4me3 at P5CS1 locus. (B) Alternatively, at prestress conditions, the transcriptional regulatory complex may already contain HY5 and HYH. During recovery in the light, the TF is possibly dissociated from the transcriptional regulatory complex but HY5 and HYH are escaped from COP1-mediated ubiquitination and degradation. Thus, the HY5/HYH-containing the transcriptional regulatory complex may maintain prestress-induced H3K4me3 levels at the P5CS1 locus. However, if recovered in darkness, HY5 and HYH are rapidly degraded, which results in the dissociation of complex and reduction of H3K4me3 levels at the P5CS1 locus.
In summary, our study provides mechanistic insight into the effect of light on salt stress-induced TMP, which is likely to operate during dehydration stress as well (Fig. 6). The present study provides a framework for future research into the concerted effects of different environmental factors on plant response to stresses.
Materials and Methods
Plant Materials and Growth Conditions.
Surface-sterilized seeds of Arabidopsis wild-type (Columbia-0), transgenic plants and mutants were sown on solid MS medium, containing 10 g/L and 2.5 g/L phytogel, in Petri dishes and stratified at 4 °C for 2 d in the dark, before being transferred to an incubator for germination at 21 °C with a 16-h light/8-h dark photoperiod (light intensity of 110 μmol⋅m−2⋅s−1) (nL). All subsequent treatments in this study were under nL unless specified otherwise. The salt pretreatment was applied by incubating 10-d-old seedlings in liquid MS supplemented with 100 mM NaCl for indicated period. The pretreated seedlings were then recovered in liquid MS for different hours (R), before the seedlings—together with nonpretreated seedlings (Sp0)—were subjected to the final stress with 200 mM NaCl for the indicated period (Sp1 or Sp2 depending on the number of pretreatment/recovery cycle). Control plants were treated in parallel using liquid MS. The dehydration stress was performed similarly to that described previously (8). Briefly, 10-d-old seedlings were gently blotted onto filter paper to remove liquid and subjected to a priming of air-drying stress for 2 h in a 60% humidity conditions and were placed subsequently in sugar-free MS solution for 22 h for recovery (R1). This priming/recovery cycle was repeated twice to obtain R2. Nonpretreated plants and R2 plants were subjected to air-drying stress for 5 h, giving rise to Sp0 and Sp2, respectively. Control plants were treated in parallel under well-watered conditions. After treatment, plant tissues were harvested for histone modification assay and stored immediately in liquid nitrogen for subsequent determination of proline content and gene transcript level.
Plasmid Construction and Plant Transformation.
P5CS1 promoter fragments (with 5′UTR), varying from 0.8 kb to 3.1 kb, were amplified from Col-0 genomic DNA using primers listed in Table S1, and were ligated with pMD19-T vector. These promoter fragments were further cloned into a binary vector pBI101released again by HindIII and BamHI digestion, resulting in GUS fusion constructs p3.1, p2.9, p2.7, p2.6, p2.3, p2.1, p1.9, p1.2, p0.8. The 3′UTR of P5CS1 was amplified from Col-0 genomic DNA using primer p3u-F and p3u-R, and the GUS coding sequence was amplified from pBI101 using primers gus-F and gus-R. GUS and the 3′UTR of P5CS1 were fused by overlap extension PCR. The resulting GUS-3′UTRP5CS1 fragment was ligated with a pMD19-T vector and further cloned into BamH I and SacI sites to replace the GUS coding region in p3.1, resulting in p3.1-U. For pC1391-mini, a 54-bp minimal 35S promoter was synthesized and inserted into EcoR I and NcoI sites of pCAMBIA1391. For pC1391-P0.8, the 0.8-kb P5CS1 promoter fragment, amplified from p0.8 by primer ep0.8-F and ep0.8-R, was digested with EcoR I and NcoI and then inserted into the EcoR I and NcoI sites of pCAMBIA1391. To test the function of the EMF, it was amplified from p3.1 using primer p2.6-F and emf-R and cloned first into the pEZ-blunt. The resulting plasmid was digested with HindIII and EcoRI to release the EMF, which was finally inserted into HindIII and EcoRI sites in pC1391-mini and pC1391-P0.8 upstream of the minimal 35S promoter and 0.8-kb P5CS1 promoter, respectively, to generate pEMF and pEMF-P0.8. For the yeast one-hybrid assay, the ORFs of HY5, HYH and GT-1 were amplified using primers listed in Table S1 and cloned into the pJG4-5 vector via recombination (Seamless Assembly Cloning Kit, Cat. No. C5891-50) to generate pJ-HY5, pJ-HYH, and pJ-GT1, respectively. The promoter fragments containing the C/A-box (−2,567 to −2,559), GAAAAA element (−2,535 to −2,527), and their mutated forms (▵CA and ▵GA; see Table S1 for details) were synthesized and inserted into the EcoR I–SalI site of pLacZi2u, generating pLac-CA, pLac-GA, pLac-▵CA, and pLac-▵GA, respectively. All constructs were validated by sequencing.
Table S1.
Primers and synthetic nucleotides used in this study
| Primer or nucleotide | Sequence | Annotation | |
| For transgenetic plants | |||
| Primers | p0.8-F | AAGCTTGGGGTGGACAATAAACCGC | With p-R, for the 800 bp P5CS1 promoter |
| P1.2-F | AAGCTTCTGGAGAATCTCGAGGTTGGC | With p-R, for the 1,200 bp P5CS1 promoter | |
| P1.9-F | AAGCTTGGCTTTGAACAAAACGCATG | With p-R, for the 1,900 bp P5CS1 promoter | |
| P2.1-F | AAGCTTCCCCAGAGACAATCAGACATC | With p-R, for the 2,100 bp P5CS1 promoter | |
| p2.3-F | AAGCTTACGCATGTGCAGCTGCTG | With p-R, for the 2,300 bp P5CS1 promoter | |
| p2.6-F | AAGCTTTCATGCGAGAAAGAAAAAGTCGTT | With p-R, for the 2,600 bp P5CS1 promoter | |
| p2.7-F | AAGCTTCGTATCGACGCCATCATTTCATC | With p-R, for the 2,700 bp P5CS1 promoter | |
| P2.9-F | AAGCTTCGAGGCGTATGCGTTCAC | With p-R, for the 2,900 bp P5CS1 promoter | |
| p3.1-F | AAGCTTAAGCAGGATCGTTATTAACCATC | With p-R, for the 3,100 bp P5CS1 promoter | |
| p-R | GGATCCTATCGTCGTCGTCGTCTACCAAAAC | ||
| p3u-F | ACAATGAATCAACAAGACTTCCGAGTGTGTG | Primers used, for amplification GUS-P5CS1 3′UTR fused fragment by overlap extension PCR | |
| p3u-R | GAGCTCCTAAGAATGAGATTGTGAGGTTC | ||
| gus-F | GGATCCCCGGGTGGTCAGTC | ||
| gus-R | GGAAGTCTTGTTGATTCATTGTTTGCCTCCCTG | ||
| emf-R | GAATTCGAAAAGAAAGATGCGATTTTGATTGGTACG | ||
| ep0.8-F | GAATTCAATAAACCGCTGCGGACCAAG | ||
| ep0.8-R | CCATGGTATCGTCGTCGTCGTCTACCAA | ||
| Synthetic nucleotides | Minimal 35S-F | AATTCCTCTATATAAGGAAGTTCATTTCATTTGGAGAGAACACGGGGGACTCTTGAC | For pC1391-mini construction |
| Minimal 35S-R | CATGGTCAAGAGTCCCCCGTGTTCTCTCCAAATGAAATGAACTTCCTTATATAGAGG | ||
| For yeast one-hybrid assay | |||
| AT5G11260 | HY5-F | CCAGATTATGCCTCTCCCGAATTCCAGGAACAAGCGACTAGCTC | |
| HY5-R | GAAGAAGTCCAAAGCTTCTCGAGAAGGCTTGCATCAGCATTAGAAC | ||
| AT3G17609 | HYH-F | CCAGATTATGCCTCTCCCGAATTCTCTCTCCAACGACCCAATGGG | |
| HYH-R | GAAGAAGTCCAAAGCTTCTCGAGGTGATTGTCATCATTTTAGGCCT | ||
| AT1G13450 | GT-F | CCAGATTATGCCTCTCCCGAATTCTTCATTTCCGACAAATCTCGTC | |
| GT-R | GAAGAAGTCCAAAGCTTCTCGAGTCTCACACCTCGATACACAG | ||
| Synthetic nucleotides | CA-F | AATTCATTTTAAAACCTATTTCTCCACGTAAACAAAAAAACTACG | Fragments used to construct pLac-CA, pLac-GA, pLac-▵CA, and pLac-▵GA |
| CA-R | TCGACGTAGTTTTTTTGTTTACGTGGAGAAATAGGTTTTAAAATG | ||
| ▵CA-F | AATTCATTTTAAAACCTATTTCTACAACTAAACAAAAAAACTACG | ||
| ▵CA-R | TCGACGTAGTTTTTTTGTTTAGTTGTAGAAATAGGTTTTAAAATG | ||
| GA-F | AATTCGCAAAAAATCGAAAAAGTAGTTTTTTCG | ||
| GA-R | TCGACGAAAAAACTACTTTTTCGATTTTTTGCG | ||
| ▵GA-F | AATTCGCAAAAAATCTACATAGTAGTTTTTTCG | ||
| ▵GA-R | TCGACGAAAAAACTACTATGTAGATTTTTTGCG | ||
| Primers for qPCR | |||
| AT2G39800 | P5CS1-F | TGTGTAAAGACCTTCAACATCGCTC | |
| P5CS1-R | AAGAGCCCCATATCAGGATTCTTCT | ||
| AT3G30775 | PDH1-F | CAACCCGTCTTCTCCGAACA | |
| PDH1-R | CGGTGCTTGTTGTCCAAAGG | ||
| GUS | GUS-F | CCGACATGTGGAGTGAAGAG | |
| GUS-R | GCAAAATCGGCGAAATTCCA | ||
| AT5G09810 | ACT7-F | CGCTTTCCTTAGTGTTAGCTGC | |
| ACT7-R | GAATATCCTCACCATCGGCCA | ||
| AT3G19760 | eIF4A-F | TCAGAAGGAGAGAGACGCCA | |
| eIF4A-R | CACGGTTGTTGGGGAGATCA | ||
| AT1G13440 | GAPC-F | TGAGGCTGGAGCTGACTTTG | |
| GAPC-R | TTGGGGCAGAGATGACAACC | ||
| Primers for ChIP-qPCR | |||
| AT2G39800 | Pair1-F | AGCATCATGAAGCGGTATCCA | Used for detect the enrichment of regions 1, 2, 3, 4, and 5 of the P5CS1 gene |
| Pair1-R | ATGAGGGGCTTCCACAATCG | ||
| Pair2-F | GTTCACTTCCACGGCGTTTC | ||
| Pair2-R | CAAATGAAGTGGCGCGTGTT | ||
| Pair3-F | GGTTGGGACAGCAGTTGTTAC | ||
| Pair3-R | ACAATTACCTGTTCACACAGTGC | ||
| Pair4-F | GGGTATTCAGCTGAGAACATAG | ||
| Pair4-R | TCTGGAACTTTCCCTCGCAG | ||
| Pair5-F | GTCGTTTAGACATACACATCACTTC | ||
| Pair5-R | AGAGAGAGAGAAAAAACTACTTTTTCG | ||
| GUS | Chip-GUS-F | TCAGCGTTGGTGGGAAAGCG | Used for detect the enrichment of region 1 of GUS gene |
| Chip-GUS-R | GCTGATACCAGACGTTGCCC | ||
| AT5G09810 | Chip-ACT7-R | CTAGCGAAATCGGCATAGAGAA | Primer used with ACT7-F to detect the enrichment of ACT7, as internal control |
The Arabidopsis plants were transformed by the floral dip method (43) and screened on kanamycin or hygromycin B medium, depending on the vector used. More than 30 independent transformants of each construct were obtained and, among them, five single-copied and representative transformants were selected from T2 generation via PCR and histochemical GUS staining. Three of five independent T3 homozygous lines were analyzed for measuring GUS transcript level, GUS activity, and H3K4me3 level.
Proline Assay.
Free proline content in fresh plant materials was measured as described by BATES using l-proline as the standard (44).
Reverse Transcription and Real-Time PCR.
Total RNAs were isolated and reverse-transcribed by SuperScript III reverse transcriptase (Invitrogen) with an oligo (dT) primer. To measure the relative transcript levels of P5CS1, PDH1, and GUS, the cDNA was used as templates in real-time PCR using SYBR Green mixture (KaPa Biosystem) with corresponding gene-specific primers. PCR was performed in 96-well optical reaction plates in a cycler apparatus (Agilent; Stratagene Mx3000P). Quantification was performed with the 2−∆∆Ct calculation (45), where ∆∆Ct is the difference in the threshold cycles between specific genes and the reference housekeeping genes, which were ACT7, eIF4A, and GAPC for expression analyses or input DNA for ChIP assays. The primers used are listed in Table S1.
ChIP Assay.
ChIP was performed using a previously described method (46). Briefly, seedlings were submerged in 50 mM PBS buffer containing 1% (vol/vol) formaldehyde with vacuum-infiltration for 12 min to cross-link DNA and binding proteins, followed by vacuum-infiltration for 5 min in PBS containing 0.125 M glycine to terminate the cross-linking. Chromatin complexes were isolated and sonicated to reduce the average DNA size to ∼500 bp. Antibodies against HY5 (28), H3K4me3 (Abcam, ab1012), or H3K27me3 (Abcam, ab6002) were mixed with protein A (Millipore, 16-157) beads and incubated at 4 °C under gentle rotation for 4 h, before being added to 450-μL chromatin suspensions and incubated overnight at 4 °C under rotation. An equal amount of chromatin suspension without antibody was used as a mock control. After several washes with low-salt, high-salt, LiCl washing buffer and TE buffer, the immune-complexes were eluted from the protein A beads in elution buffer, incubated at 65 °C overnight to reverse the cross-link, and treated with proteinase K at 45 °C for 1 h to digest proteins. DNA was purified by phenol-chloroform extraction and ethanol precipitation with sodium acetate and glycogen (Fermentas # R0561), and quantified by qPCR. Relative enrichment of a DNA fragment was normalized to the corresponding input DNA.
Yeast One-Hybrid Assay.
Plasmid pJ-HY5, pJ-HYH or pJ-GT1 with TF-activating domain fusion and plasmid pLac-CA, pLac-GA, pLac-▵CA, or pLac-▵GA with cis-elements upstream of a LacZ reporter were cotransformed into yeast strain EGY48 using a standard transformation protocol. Positive clones were selected from Trp/Ura drop-out plates and validated by PCR. Next, transformants with both plasmids were grown on Trp/Ura drop-out plates containing galactose, raffinose, and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development.
Acknowledgments
We thank Prof. Dr. Rong Cheng Lin (Institute of Botany, Chinese Academy of Sciences) for kindly providing hy5 hyh, phyA phyB seeds, and HY5 antibody. This work was supported by National Natural Science Foundation of China Grant 31271308 (to X.J.H.); Ministry of Science and Technology of the People’s Republic of China Grant 2012CB114302 (to J.B.J.); and Chinese Academy of Sciences Grant XDA08010105 (to J.B.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610670114/-/DCSupplemental.
References
- 1.Szabados L, Savouré A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010;15(2):89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Szekely G, et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53(1):11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CY, et al. DNA methylation involved in proline accumulation in response to osmotic stress in rice (Oryza sativa) Genet Mol Res. 2013;12(2):1269–1277. doi: 10.4238/2013.April.17.5. [DOI] [PubMed] [Google Scholar]
- 4.Jiménez-Arias D, et al. Priming effect of menadione sodium bisulphite against salinity stress in Arabidopsis involves epigenetic changes in genes controlling proline metabolism. Environ Exp Bot. 2015;120:23–30. [Google Scholar]
- 5.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Sailaja BS, Cohen-Carmon D, Zimmerman G, Soreq H, Meshorer E. Stress-induced epigenetic transcriptional memory of acetylcholinesterase by HDAC4. Proc Natl Acad Sci USA. 2012;109(52):E3687–E3695. doi: 10.1073/pnas.1209990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding Y, Fromm M, Avramova Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 9.Sani E, Herzyk P, Perrella G, Colot V, Amtmann A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013;14(6):R59. doi: 10.1186/gb-2013-14-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos AP, et al. Transcription regulation of abiotic stress responses in rice: A combined action of transcription factors and epigenetic mechanisms. OMICS. 2011;15(12):839–857. doi: 10.1089/omi.2011.0095. [DOI] [PubMed] [Google Scholar]
- 11.Kim K-C, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20(9):2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Ding Y, Fromm M, Avramova Z. Different gene-specific mechanisms determine the ‘revised-response’ memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res. 2014;42(9):5556–5566. doi: 10.1093/nar/gku220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40(1):112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Light WH, et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11(3):e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virlouvet L, Ding Y, Fujii H, Avramova Z, Fromm M. ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana. Plant J. 2014;79(1):150–161. doi: 10.1111/tpj.12548. [DOI] [PubMed] [Google Scholar]
- 16.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8(3):217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 17.Abrahám E, et al. Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol Biol. 2003;51(3):363–372. doi: 10.1023/a:1022043000516. [DOI] [PubMed] [Google Scholar]
- 18.Guo L, Zhou J, Elling AA, Charron J-BF, Deng XW. Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 2008;147(4):2070–2083. doi: 10.1104/pp.108.122929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hare PD, Cress WA, van Staden J. Proline synthesis and degradation: A model system for elucidating stress-related signal transduction. J Exp Bot. 1999;50(333):413–434. [Google Scholar]
- 20.Savouré A, et al. Isolation, characterization, and chromosomal location of a gene encoding the delta 1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995;372(1):13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- 21.Verbruggen N, Hua XJ, May M, Van Montagu M. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc Natl Acad Sci USA. 1996;93(16):8787–8791. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun N, et al. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA. 2016;113(21):6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roudier F, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30(10):1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solovyev VV, Shahmuradov IA, Salamov AA. Identification of promoter regions and regulatory sites. Methods Mol Biol. 2010;674:57–83. doi: 10.1007/978-1-60761-854-6_5. [DOI] [PubMed] [Google Scholar]
- 26.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou DX. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 1999;4(6):210–214. doi: 10.1016/s1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
- 28.Chen D, et al. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell. 2013;25(5):1657–1673. doi: 10.1105/tpc.112.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, et al. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA. 2008;105(11):4495–4500. doi: 10.1073/pnas.0710778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catalá R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA. 2011;108(39):16475–16480. doi: 10.1073/pnas.1107161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Servet C, Conde e Silva N, Zhou DX. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol Plant. 2010;3(4):670–677. doi: 10.1093/mp/ssq018. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Ram H, Abbas N, Chattopadhyay S. Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J Biol Chem. 2012;287(31):25995–26009. doi: 10.1074/jbc.M111.333906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ram H, Priya P, Jain M, Chattopadhyay S. Genome-wide DNA binding of GBF1 is modulated by its heterodimerizing protein partners, HY5 and HYH. Mol Plant. 2014;7(2):448–451. doi: 10.1093/mp/sst143. [DOI] [PubMed] [Google Scholar]
- 36.Hardtke CS, et al. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000;19(18):4997–5006. doi: 10.1093/emboj/19.18.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics. 2008;280(6):547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol. 2005;57(2):155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- 39.Hare PD, Cress WA. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997;21(2):79–102. [Google Scholar]
- 40.Aro E-M, McCaffery S, Anderson JM. Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol. 1993;103(3):835–843. doi: 10.1104/pp.103.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XP, Muller-Moule P, Gilmore AM, Niyogi KK. PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA. 2002;99(23):15222–15227. doi: 10.1073/pnas.232447699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X, Ouyang X, Deng XW. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol. 2014;21:96–103. doi: 10.1016/j.pbi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Bates LS, Walden RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39(1):205–207. [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Gendrel A-V, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2(3):213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]