Significance
Inflammatory bowel disease (IBD) is a devastating illness that affects 1.6 million people in the United States and disproportionately affects males in early onset disease. However, IBD genes that contribute to sex-specific risk are unexplored. The gut microbiota interfaces the host with its environment and exhibits alterations in spatial organization in IBD with dysbiosis in the mucosa but a relatively unaffected lumen. Core 1 β3GalT-specific molecular chaperone (Cosmc) was recently identified as an IBD risk gene on the X chromosome by genome-wide association study. We functionally evaluated Cosmc in IBD and discovered that the loss of Cosmc leads to gut inflammation in males but not females and a spatial pattern of dysbiosis resembling IBD. Thus, Cosmc contributes to sex bias in IBD and spatially regulates the gut microbiota.
Keywords: Cosmc, IBD, inflammation, microbiota, sex
Abstract
Inflammatory bowel disease (IBD) results from aberrant immune stimulation against a dysbiotic mucosal but relatively preserved luminal microbiota and preferentially affects males in early onset disease. However, factors contributing to sex-specific risk and the pattern of dysbiosis are largely unexplored. Core 1 β3GalT-specific molecular chaperone (Cosmc), which encodes an X-linked chaperone important for glycocalyx formation, was recently identified as an IBD risk factor by genome-wide association study. We deleted Cosmc in mouse intestinal epithelial cells (IECs) and found marked reduction of microbiota diversity in progression from the proximal to the distal gut mucosa, but not in the overlying lumen, as seen in IBD. This loss of diversity coincided with local emergence of a proinflammatory pathobiont and distal gut restricted pathology. Mechanistically, we found that Cosmc regulates host genes, bacterial ligands, and nutrient availability to control microbiota biogeography. Loss of one Cosmc allele in males (IEC-Cosmc-/y) resulted in a compromised mucus layer, spontaneous microbe-dependent inflammation, and enhanced experimental colitis; however, females with loss of one allele and mosaic deletion of Cosmc in 50% of crypts (IEC-Cosmc+/−) were protected from spontaneous inflammation and partially protected from experimental colitis, likely due to lateral migration of normal mucin glycocalyx from WT cells over KO crypts. These studies functionally validate Cosmc as an IBD risk factor and implicate it in regulating the spatial pattern of dysbiosis and sex bias in IBD.
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is a devastating disease that affects 1.6 million people in the United States. Symptoms include abdominal pain, diarrhea, rectal bleeding, and growth failure. Severe cases require bowel resection, with current treatments aimed at management rather than cure. Mechanistically, immune hyperactivation to gut bacteria damages the intestine. Early onset or pediatric IBD (<15 y old) occurs in 10–20% of patients and is a model to understand genetic contributions to disease (1). CD and possibly UC in patients <15 or <8 y old, respectively, preferentially affects males (male:female is ∼1.5:1 for CD), indicating that genetic factors, such as risk genes on the X chromosome, may explain this sex bias and provide new insights into disease mechanisms and targets (1).
The gut microbiota can contribute to IBD with changes in community structure and functional capacity (2). Normally, the bacterial distribution and composition vary greatly from ∼1011 to 1012 cells per g within the ascending colon, to ∼107 to 108 in the distal ileum, and ∼102 to 103 in proximal ileum and jejunum (2). Specific members of the microbiota may occupy a planktonic niche, free-living in the luminal fecal stream, or adherent to the mucosa (3). The distinct geographical and ecological niches in the intestine (biogeographies) expand microbial metabolic potential and help maintain diversity in a competitive ecosystem (3). A recent study of pediatric IBD identified a dysbiotic microbiota in the intestinal mucosa but not in the lumen, suggesting that specific factors regulate the gut microbe biogeography in the healthy intestine and that these factors are altered in IBD (4). Nevertheless, no IBD risk genes are known to regulate gut microbe biogeography or the pattern of dysbiosis in patients.
Core 1 β3GalT-specific molecular chaperone (Cosmc) on the X chromosome encodes a chaperone for the T-synthase glycosyltransferase that extends O-glycans on >80% of secretory pathway proteins to form the glycocalyx and mature mucins (Fig. 1A) (5–7). Cosmc was implicated in sex-specific risk in Crohn’s and UC by genome-wide association study (8); however, its functional role in the intestine and IBD is unexplored. To address its role, we deleted Cosmc in mouse intestinal epithelial cells (IECs). Loss of one allele in male mice (IEC-Cosmc-/y) led to spontaneous inflammation, enhanced experimental colitis, and altered microbe biogeography, as seen in IBD. Significantly, female mice with loss of one allele (IEC-Cosmc−/+) were protected from spontaneous inflammation and partially protected from experimental colitis, despite complete KO of Cosmc in ∼50% of crypts (due to random X inactivation). Thus, Cosmc is an IBD risk gene that regulates microbe biogeography and disproportionately affects males.
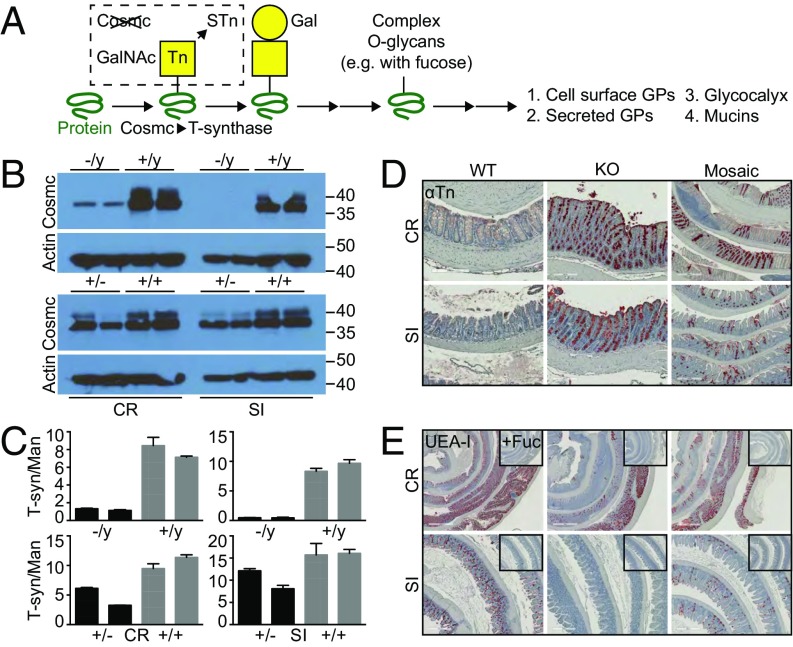
Fig. 1.
Characterization of IEC-Cosmc mice. (A) Schematic of O-glycan biosynthesis: Cosmc is the chaperone for the T-synthase and required to extend O-glycans beyond the Tn antigen. GPs, glycoproteins. Purified IECs from CR and SI from KO, mosaic, and gender-matched WT mice were probed with anti-Cosmc and anti-Actin antibodies by Western blot (B) and assayed for T-synthase and mannosidase activities (2 mice pooled per group; 2 groups evaluated/tissue per genotype; age 2–3 mo) (C). (D) Tn expression was analyzed by IHC on formalin fixed colorectum (CR) and ileum (SI) from KO, mosaic, and WT mice (n = 3–5 mice per group, 3 mo old). (Scale bars: Left and Center, 200 μM; Right, 500 μM.) (E) UEA-I lectin was used to analyze α1,2-fucose expression in the colorectum and ileum from KO, mosaic, and WT mice; Insets show UEA-I preincubated with 100 mM free fucose denoted as “+Fuc” (n = 3–5 mice per group, 3 mo old). Representative images shown. (Scale bars: Top, 1 mm; Bottom, 500 μM.)
Results
Spontaneous Inflammation in Male KO Mice.
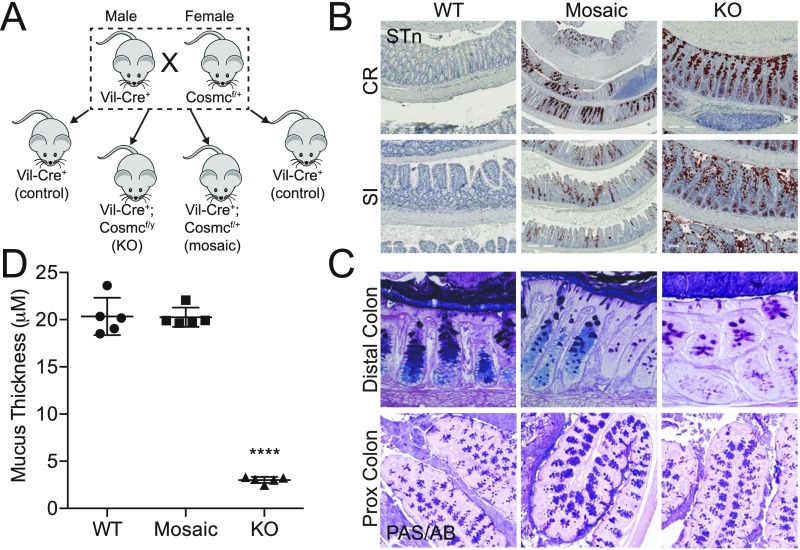
To delete Cosmc in the intestinal epithelia, we crossed Vil-Cre+ male mice with Cosmcf/+ female mice and obtained Vil-Cre+; Cosmcf/y (KO), Vil-Cre+; Cosmcf/+ (mosaic), and Vil-Cre+ (WT) controls (Fig. S1A) at Mendelian ratios (n = 500, Pχ2 = 0.17). We confirmed near-complete loss of Cosmc protein (Fig. 1B) and T-synthase activity (Fig. 1C) in purified IECs from male KOs and ∼50% loss in female mosaics. Cosmc loss led to >90% expression of the Tn (Fig. 1D) and STn (Fig. S1B) truncated O-glycans by IHC (Fig. 1A) in colorectum and small intestine in male KOs and ∼50% expression in female mosaics with patches of Tn/STn(+) crypts and Tn/STn(−) crypts, as predicted from random X inactivation of Cosmc (7).
Fig. S1.
Breeding strategy and characterization of IEC-Cosmc mice. (A) Breeding strategy: Vil-Cre+ males were bred to Cosmcf/+ females to generate intestinal epithelia-specific KO (Vil-Cre+;Cosmcf/y), mosaic (Vil-Cre+;Cosmcf/+), and WT control (Vil-Cre+;Cosmc+/y OR +/+) mice. (B) Sialyl-Tn expression was analyzed by monoclonal antibody on FFPE, swiss-rolled CR, and SI from KO, mosaic, and WT mice (distal segment in center of swiss-roll). (Scale bars: Left and Center, 200 μM; Right, 500 μM.) PAS/AB staining was performed on Carnoy’s fixed distal (magnification: 40×) and proximal (magnification: 10×) colon from WT, mosaic, and KO mice (C), and the inner mucus layer thickness was quantified in the distal colorectum (5 measurements per mouse per replicate, n = 5 mice per group) (D). n = 3–5 mice per group, age 3 mo (B and C). ****P ≤ 0.0001 from one-way ANOVA with Tukey’s post hoc test for multiple comparisons; mean ± SD.
To examine whether glycan truncation results in loss of terminal epitopes, including known bacterial nutrients and ligands (9), we assessed terminal α1,2-linked fucose and sulfates/sialic acids with UEA-I and alcian blue (AB), respectively. WT colon and ileum IECs strongly bound UEA-I (Fig. 1E) and AB (Fig. S1C), and UEA-I was specific because it was blocked by free fucose (Fig. 1E, Inset). In contrast, male KO crypts lost binding to UEA-I and AB, and female mosaic crypts partially lost binding in patches. Unexpectedly, both male KOs and female mosaics retained binding of UEA-I and AB in the proximal colon (Fig. 1E and Fig. S1C). Thus, Cosmc differentially regulates expression of terminal epitopes of the glycocalyx in different regions of the gut that are lost in the distal colon of male KOs.
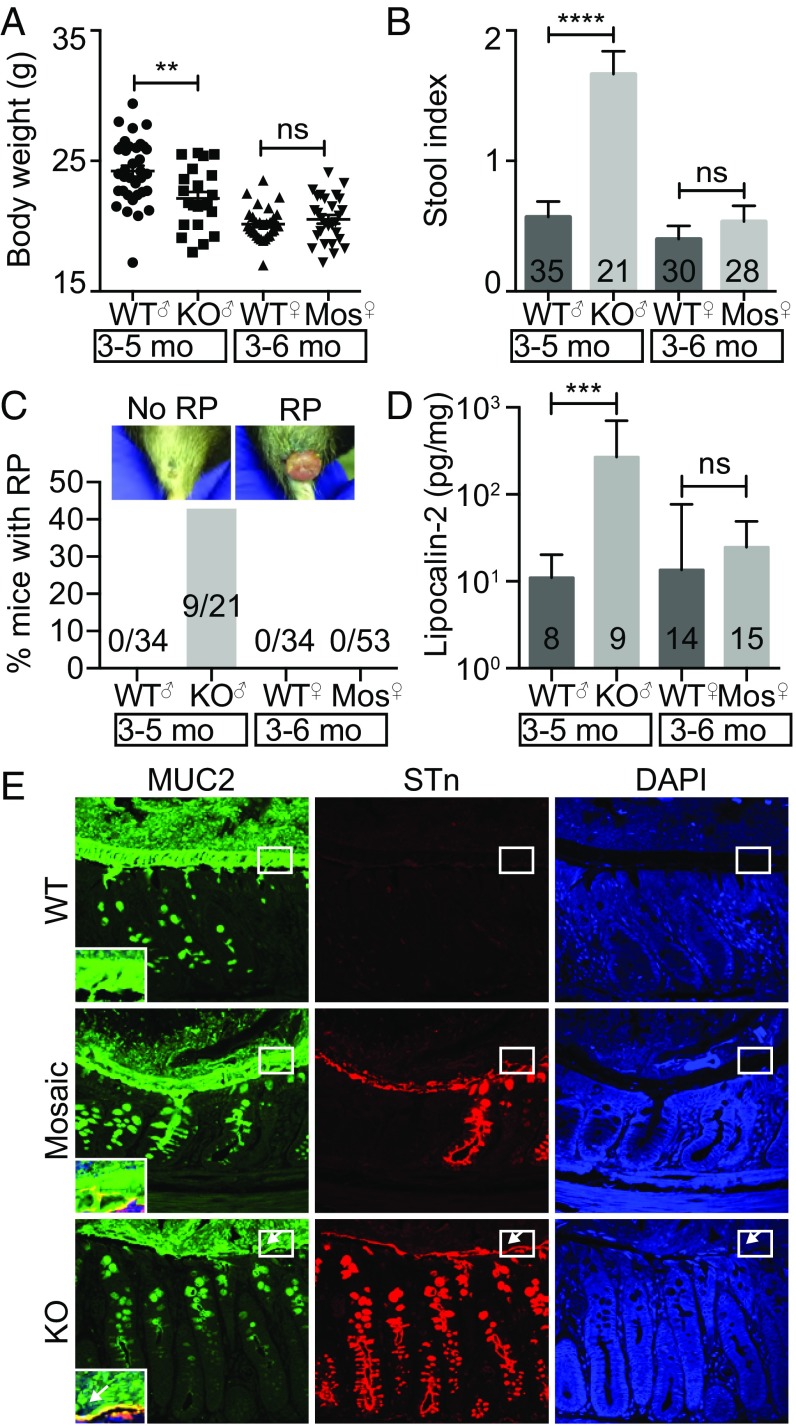
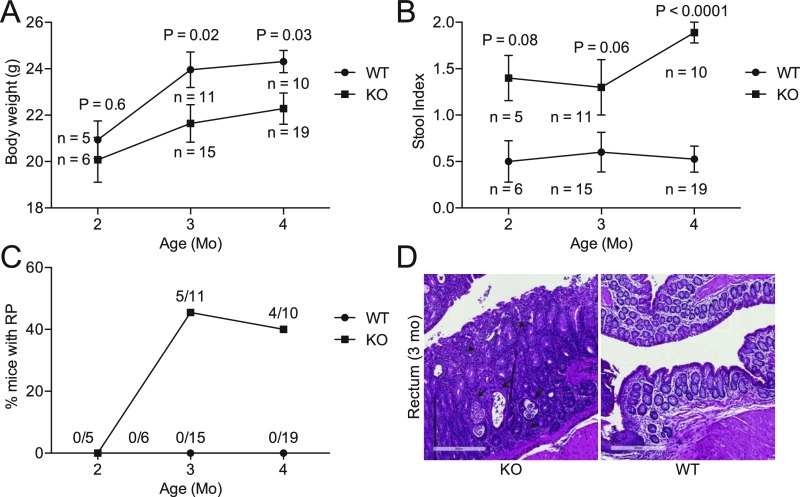
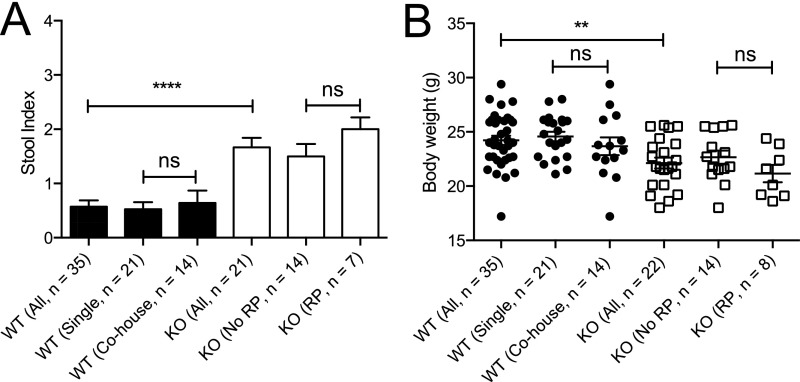
We next assessed the consequence of Cosmc deletion and glycocalyx loss in the intestine, which results in profound pathology when deleted in other tissues (7, 10). Beginning at 3 mo of age, male KOs exhibited multiple signs of inflammation, including weight loss (Fig. 2A and Fig. S2A), elevated stool index (stool softness, blood in the stool) (Fig. 2B and Fig. S2B), rectal prolapse (RP) in ∼40% of mice (Fig. 2C and Fig. S2C), inflammatory infiltrates limited to the distal colon (Fig. S2D), and elevated fecal lipocalin-2, an inflammatory biomarker (11) (Fig. 2D). Notably, RP alone did not drive inflammation, because male KOs with or without RP both had weight loss and an elevated stool index (Fig. S3).
Fig. 2.
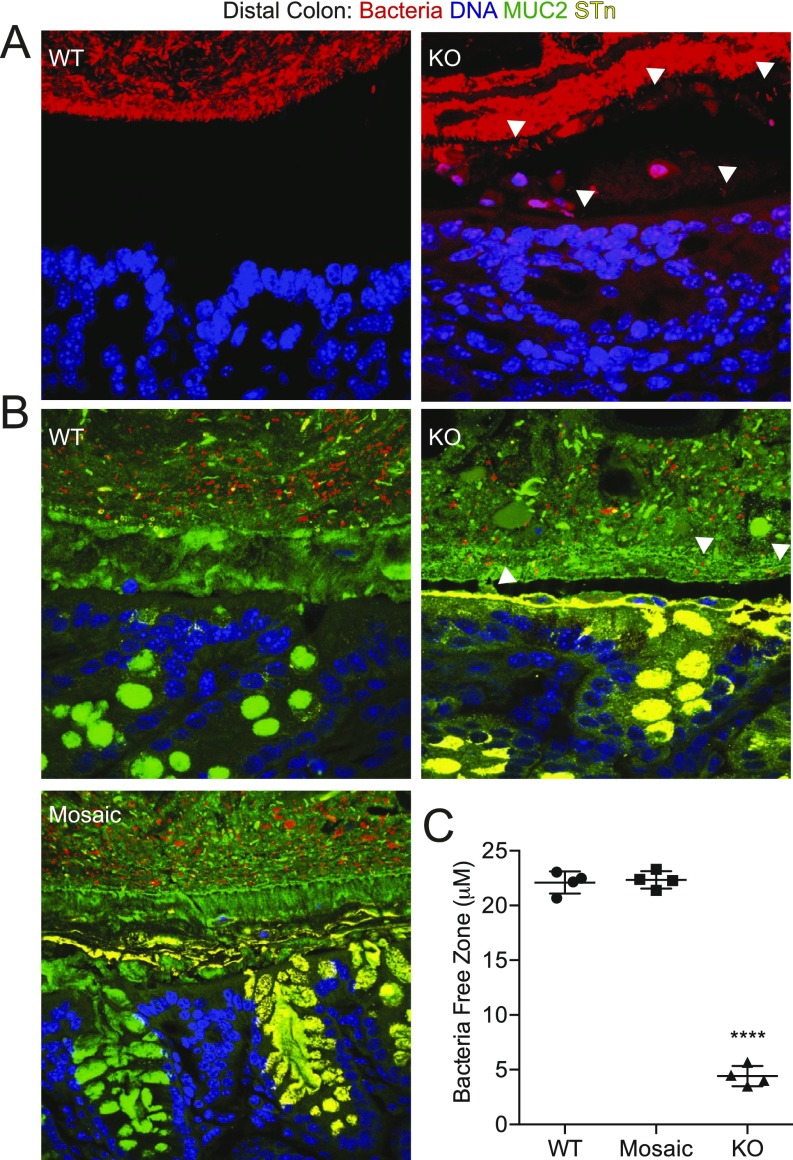
Spontaneous inflammation in IEC-Cosmc-KO but not mosaic mice. Body weight (n = 35 WT males, 21 KOs; 30 WT females, 28 mosaics) (A), stool index (average of softness and blood content) (B), and rectal prolapse for KO, mosaics, and gender matched WTs (C). (D) Fecal lipocalin-2 in KO, mosaics, and gender-matched WTs. (E) Distal colon from WT, mosaic, and KO were fixed with Carnoy’s reagent and stained with antibodies against MUC2 (green), STn (red), or with DAPI (blue) (3 mice per group, representative images shown); white boxes indicate the inner mucus layer, enlarged and merged in lower left corner; white arrows show where the inner mucus layer was lost. (Magnification: 40×.) **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns, not signficant from two-tailed Student’s t test (A) or Mann–Whitney (B and D); mean ± SE (A and B) or median ± interquartile range (D); number of mice are indicated on the graph (B–D).
Fig. S2.
Time course of clinical inflammation. Body weight (A), stool index (B), and percent of mice with RP (C) were measured in KO and WT mice at 2, 3, and 4 mo of age. Measurements are the same as in Fig. 2. (D) Histologic evidence for active inflammation in distal colon of KO but not WT: crypt abscesses (arrows) and inflammatory infiltrate consisting of neutrophils, lymphocytes, and plasma cells (arrowheads) (n = 4 mice, < 1 y, 3 mo old mice shown). (Scale bars: 300 μM.) Number of mice indicated on graphs (A–C); P values from two-tailed Student’s t test (A) or Mann–Whitney (B); mean ± SE (A and B).
Fig. S3.
Effects of cohousing and rectal prolapse on inflammation in KO mice. Stool index (average of softness and blood content) (A) and body weight (B) in 3- to 5-mo-old KO and WT mice. WT mice segregated by single housing (WT with WT) or cohousing (WT with KO); KO mice segregated by presence or absence of rectal prolapse. **P ≤ 0.01, ****P ≤ 0.0001 from Kruskal Wallis test with Dunn’s post hoc test for multiple comparisons (A) or one-way ANOVA with Sidak’s post hoc test for multiple comparisons (B); mean ± SE (A and B); number of mice indicated on graph (A and B). ns, not significant.
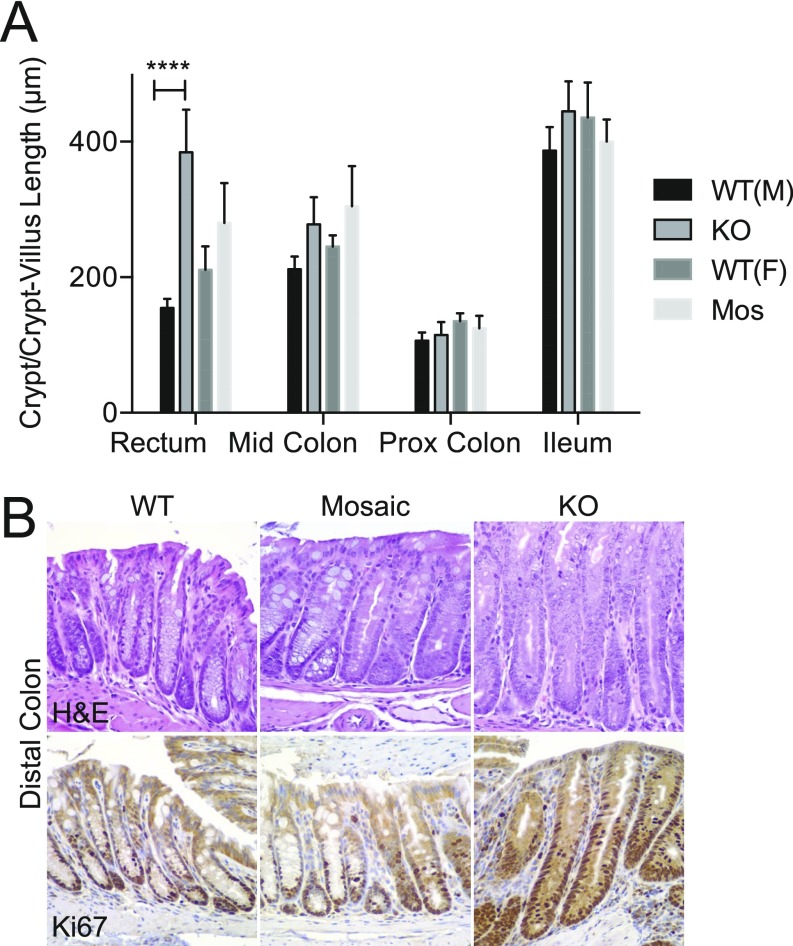
To further evaluate inflammation, we analyzed cell proliferation, which can increase by an uncontrolled injury-repair cycle or through immune drivers (12). Before the development of RP, male KOs showed histologic evidence of mucosal regeneration in the distal colon (Fig. S4) characterized by crypt elongation (154.7 ± 5.5 μm in WT versus 384.3 ± 28.1 μm in KO, mean ± SE) (Fig. S4A), epithelial hyperplasia (Fig. S4B), and increased mitoses and Ki67 proliferation indices (Fig. S4B). Other regions of the small and large bowel were unaffected, showing that loss of Cosmc in male KOs leads to spontaneous inflammation that is unexpectedly localized to the distal colon.
Fig. S4.
Hyperplasia in KO mice but not mosaic mice. Crypt length (A) and Ki67 (B) were analyzed in WT, mosaic, and KO mice. Summary data (n = 4–6 mice per group, 20 individual measurements/region per mouse for each replicate value) (A) and representative images (n = 3 mice per group) (B) are shown (magnification: 40×); ****P ≤ 0.0001 from two-way ANOVA with Sidak’s post hoc test for multiple comparisons (A and B).
O-glycans, dependent on Cosmc, account for 80% of MUC2’s mass and, thus, may be important for its function (13). To explore a possible mechanism for spontaneous inflammation (14), we preserved the mucus layer with Carnoy’s fixative and analyzed mucus integrity in the distal colon with immunofluorescence (IF) (Fig. 2E), AB staining (Fig. S1 C and D), and a 16S universal fluorescence in situ hybridization (FISH) probe against bacteria (Fig. S5). WT mice contained a thick, stratified AB(+) (Fig. S1 C and D) and MUC2(+) inner mucus layer that separated the DAPI(+) luminal contents (15) (Fig. 2E, Top, box) and FISH(+) bacteria (Fig. S5 A and B) from the epithelia. In contrast, male KOs showed thinning, fragmentation, and/or absence of this layer (Fig. 2E, Bottom and Fig. S1C and S5 A and B) with 85% loss by AB staining (Fig. S1D), resulting in direct bacterial-epithelial contact, including helical-shaped bacteria nearing or contacting superficial colonocytes (Fig. S5A, bottom two arrows). Thus, Cosmc is required to form an intact mucus layer in the distal colon.
Fig. S5.
Bacteria–epithelial interactions in the distal colon. Bacteria were visualized with EUB-16S rRNA Universal Bacteria FISH probe (red) and epithelia by DAPI (blue) (n = 2–3 mice per group, representative images shown) in A or additionally with antibodies against MUC2 (green) and STn (yellow) in Lower (n = 3 mice per group, representative images shown) (B) and the bacterial–epithelial distance was quantified (5 measurements per mouse per replicate from B, n = 4 mice per group) (C). White arrows indicate invading bacteria. ****P ≤ 0.0001 from one-way ANOVA with Tukey’s post hoc test for multiple comparisons; mean ± SD. (Magnification: A, 100×; B, 40×.)
No Spontaneous Inflammation in Female Mosaic Mice.
KO males with loss of one allele have near complete loss of Cosmc throughout the intestinal tract. To understand how the loss of one allele affects mosaic females, we assessed inflammation. We expected to see an intermediate level of inflammation, because ∼50% of crypts lack Cosmc and the mucin glycocalyx. Surprisingly, we found no difference between female mosaic and control mice in multiple metrics of inflammation [body weight (Fig. 2A), stool index (Fig. 2B), RP (Fig. 2C), fecal lipocalin-2 levels (Fig. 2D), and IEC proliferation (Fig. S4B)], indicating that female mosaic mice were protected from inflammation.
To determine the mechanism, we fixed distal colons from female mosaic mice with Carnoy’s and performed IF (Fig. 2E), AB staining (Fig. S1 C and D), and FISH (Fig. S5 B and C) to identify STn(+) KO cells, AB(+) WT cells, and their products in a female mosaic colon. In contrast to male KOs, female mosaic mice had a continuous, intact MUC2(+) mucus layer that covered both KO and WT cells, clearly separating luminal contents (Fig. 2E, Middle, box) and bacteria (Fig. S5 B and C) from the epithelium. Mechanistically, STn(+) KO MUC2 (Fig. 2E and Fig. S5) and AB(+) WT MUC2 (Fig. S1 C and D) formed a continuous layer over both WT and KO crypts, indicating that WT MUC2 intermixed with KO MUC2 to form an intact mucus layer. Thus, local disruptions in Cosmc do not result in spontaneous inflammation because adjacent crypts are able to produce WT MUC2 that diffuses laterally over the crypt surface to protect KO cells.
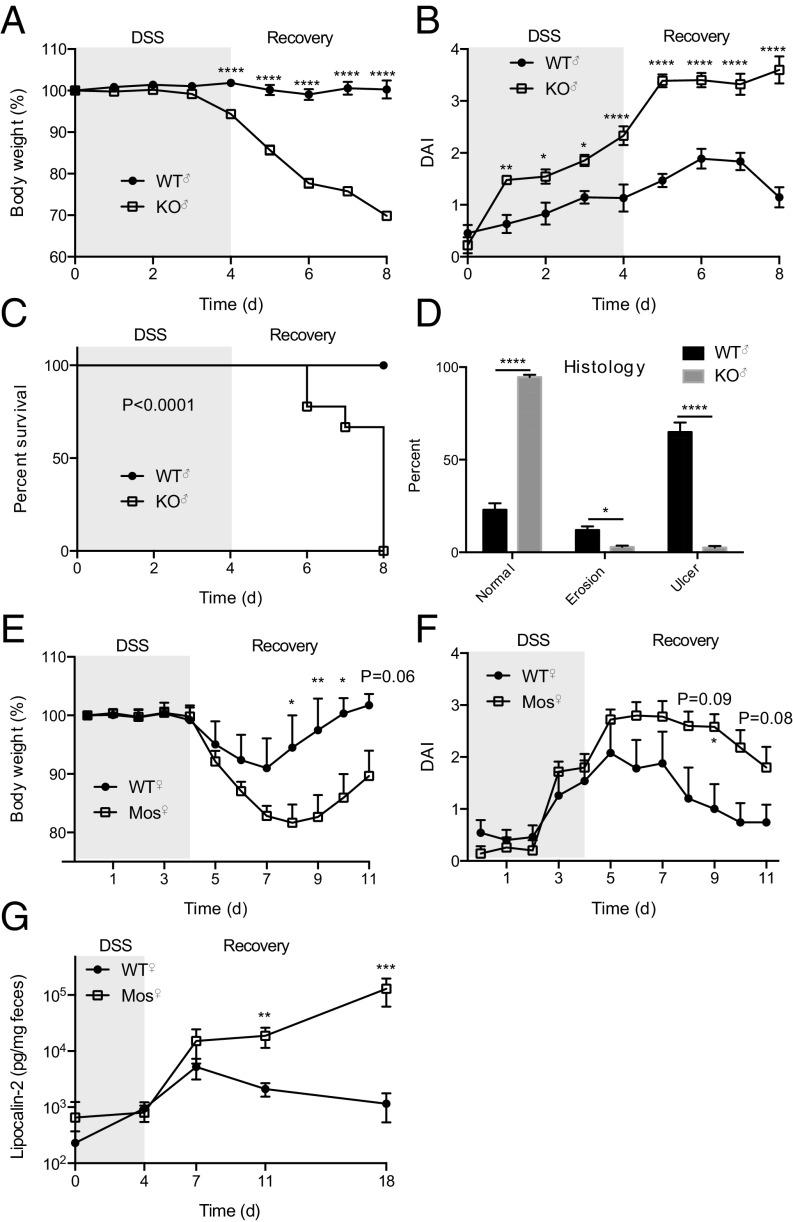
To further investigate the extent of WT Cosmc compensation in female mosaic mice, we induced colitis with a 4-d course of 2.5% (wt/vol) dextran sulfate sodium (DSS) in drinking water and assessed body weight and disease activity index (average of body weight change, blood in stool, and stool softness) in male KO, female mosaic, and control mice (Fig. 3). Male WTs were largely unaffected by DSS treatment, but male KOs exhibited profound weight loss (Fig. 3A) and rise in disease activity index (DAI) (Fig. 3B) throughout the acquisition and recovery phase, necessitating euthanasia in all of the male KOs but none of the controls (Fig. 3C). Histologically, ∼75% of the tissue was ulcerated or eroded in male KOs compared with less than 5% in male WTs (Fig. 3D). Female mosaics had less severe colitis than male KOs that did not differ from female WTs in the acquisition phase but had enhanced colitis in the recovery phase (Fig. 3 E and F). Notably, fecal lipocalin-2 from female mosaics remained elevated for at least 18 d in contrast to female WTs that recovered by day 11 (Fig. 3G). Although we could only generate a few KO females (Vil-Cre+;Cosmcf/f) precluding statistical analysis, we saw RP and histologic inflammation in one of these mice, and WT females had more severe DSS colitis than WT males, arguing that WT females are not inherently protected from gut inflammation (16). These data suggest that the apparently normal mucus layer in female mosaic mice is able to protect from spontaneous inflammation and enhanced acquisition of colitis seen in male KOs, but once DSS disrupts the mucus barrier that developed in the absence of bacteria, WT MUC2 is unable to protect KO cells that have now contacted bacteria of an adult gut for the first time, resulting in prolonged inflammation.
Fig. 3.
DSS colitis in KO and mosaic mice. KO (A–D), mosaic (E–G), and WT mice (A–G) were treated with 2.5% DSS for 4 d and then switched to drinking water. Body weight (A and E), disease activity index (% weight change, stool softness, blood in stool) (B and F), survival (Mantel–Cox test) (C), histology (D), and fecal lipocalin-2 (G) were measured. n = 9 mice per group, age 3 mo (A–C); n = 6 KO, 9 WT, killed on day 8 and the percent normal, eroded, and ulcerated epithelia on H&E-stained CR was quantified (D); n = 5–6 mice per group, age 3 mo (E–G): repeated two independent times, one representative experiment shown (E and F) or pooled data (G); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 from ANOVA with Sidak’s post hoc test for multiple comparisons (A, B, and D–F) or Mann–Whitney (G); 0.05 < P < 0.1 listed; mean ± SE (A, B, and D–G).
Microbes Drive Inflammation.
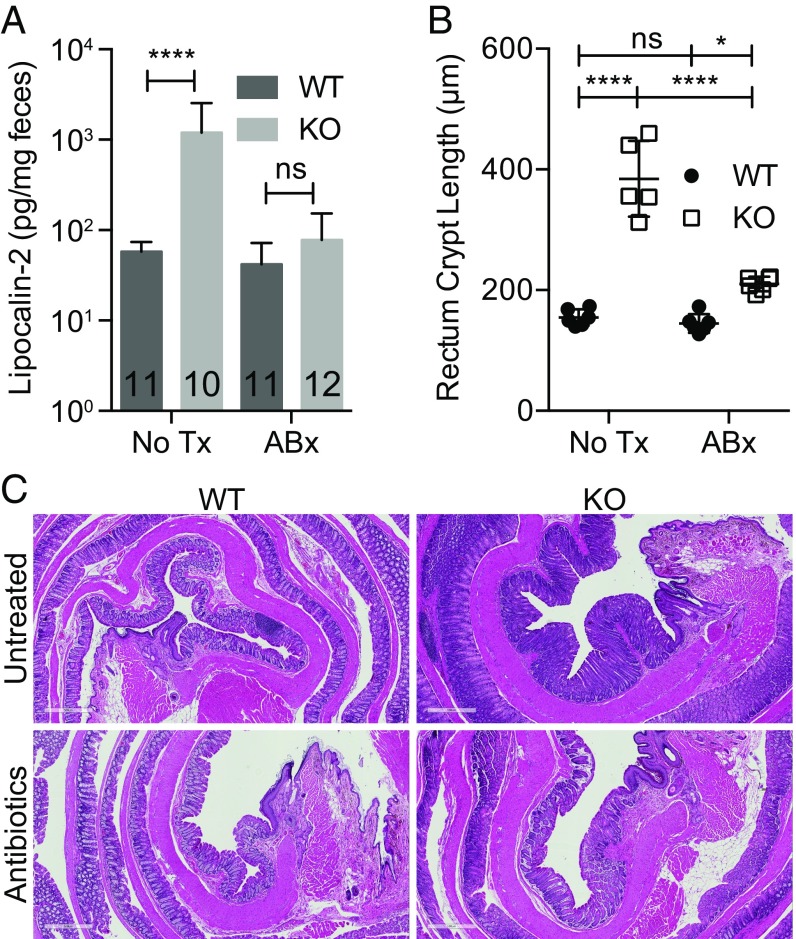
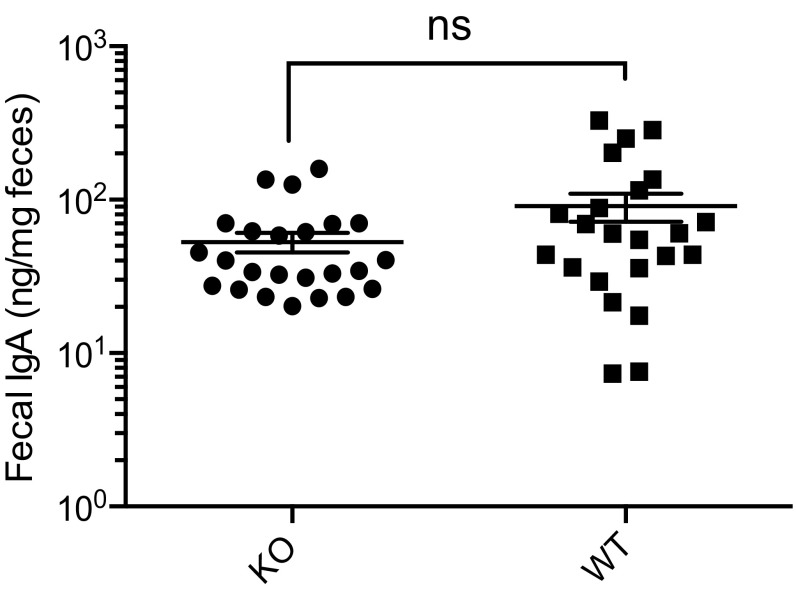
To assess whether microbes drive inflammation, we treated male KO and WT mice with neomycin, metronidazole, vancomycin, ampicillin (NMVA) antibiotics for 4 wk. Antibiotics depleted bacteria to undetectable levels (Fig. S6), resulting in normalization of fecal lipocalin-2 to WT levels (Fig. 4A) and partial normalization of IEC proliferation (Fig. 4 B and C). We then examined whether inflammation was transmissible by fecal-oral route (17). Clinical signs of inflammation (stool index, body weight) were similar between WTs housed with other WTs or WTs housed with KO males (Fig. S3). Interaction of the microbiota with small intestine epithelia induces fecal IgA (18). However, fecal IgA levels were equivalent in 3- to 5-mo-old male KO and WT mice (Fig. S7). Collectively, these data show that whereas microbes drive inflammation in male KOs, the inflammation is not transmissible by coprophagy nor associated with a change in fecal IgA.
Fig. S6.
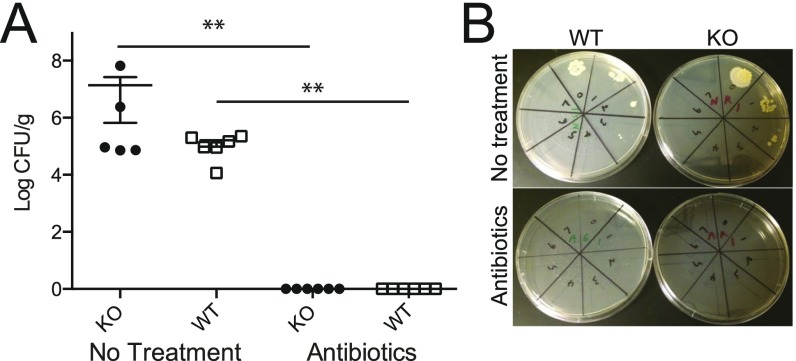
Bacterial counts of mice treated with antibiotics. (A) Two-mo-old KO and WT mice were treated with NMVA antibiotics for 4 wk, and CFUs were assessed from feces (n = 6 per group) with untreated KO (n = 5) and WT (n = 6) mice for comparison (two independent experiments, one representative shown; all antibiotic-treated groups had 0 CFUs/g, which were set at log 0 for display on the graph). (B) Representative plates are shown. **P ≤ 0.01 from Mann–Whitney test; mean ± SE.
Fig. 4.
Microbes drive inflammation in KO mice. Fecal lipocalin-2 (number of mice indicated on graph, data pooled from two independent experiments) (A), summary data (B), and representative images of rectum crypt length (C) in WT and KO mice treated with NMVA antibiotics or untreated for 4 wk beginning at 2 mo of age (n = 4–6 mice per group, 20 individual measurements/region per mouse for each replicate value); *P ≤ 0.05, ****P ≤ 0.0001 from two-way ANOVA with Sidak’s post hoc test for multiple comparisons (A and B). ns, not significant. (Scale bars: 500 μM.)
Fig. S7.
Analysis of fecal IgA. Fecal IgA was analyzed in KO (n = 24) and WT (n = 23) mice by ELISA (age-matched, 3–5 mo). ns, not significant from Mann–Whitney; mean ± SE.
Regional Changes in the Microbiota.
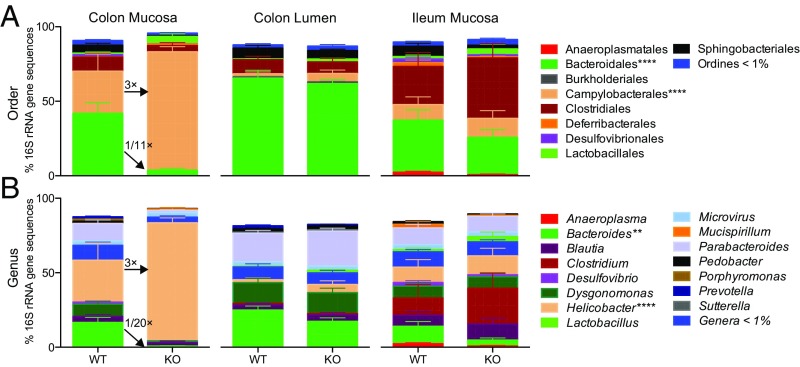
Distal gut localized pathology led us to ask whether we could identify proinflammatory microbes or regional changes in microbiota community structure. We sequenced the V4 region of 16S rRNA gene from multiple luminal and mucosal regions from ∼2-mo-old male KO and WT mice (Fig. 5) to identify changes that precede clinical signs of inflammation. Composition of the microbiota from control mice compared well with previous reports, including greater abundance of Bacteroidiales in the lumen versus mucosa of the colon (3).
Fig. 5.
Cosmc regionally regulates the gut microbiota community structure. (A and B) The microbiota was harvested from the gut lumen (distal colon) and mucosa (distal colon, ileum) from KO and WT mice for amplification of the V4 region and 16S rRNA gene sequencing (n = 7–8 mice per group, 2 mo old). Microbiota composition at the order (A) and genus (B) levels were plotted for all regions. Only colon mucosa contained significantly altered taxa (indicated with asterisks); ****P ≤ 0.0001 from two-way ANOVA with Sidak post hoc test for multiple comparisons.
Male KOs had a dramatic loss of diversity in the colonic mucosa, as indicated by a loss of comparative taxonomic representation at both the order (Fig. 5A) and genus (Fig. 5B) levels. Notably, male KOs had an 11-fold loss of the Bacteroidiales (42.0 ± 7.1% in WT to 3.8 ± 1.2% in KO, mean ± SE), including a 20-fold loss of Bacteroides (16.7 ± 9.5% in WT to 0.8 ± 0.2% in KO, mean ± SE), coinciding with markedly increased representation of Helicobacter (28.1 ± 11.6% in WT to 79.2 ± 3.3% in KO, mean ± SE), a proinflammatory pathobiont.
We next examined whether similar changes were observed along the mucosal-luminal and cephalo-caudal axis. Unexpectedly, no changes were found in the luminal contents directly overlying the colonic mucosa or in the mucosa of the ileum. Our functional data support the sequencing results: (i) Distal gut localized pathology corresponds to the site of dysbiosis; (ii) inflammation was not transferrable by coprophagy as predicted by an unaffected lumen; and (iii) IgA levels, regulated by the SI microbiota, were similar in male KO and WT mice along with a normal SI microbiota. Collectively, these results demonstrate that Cosmc differentially regulates the microbiota in different regions of the gut.
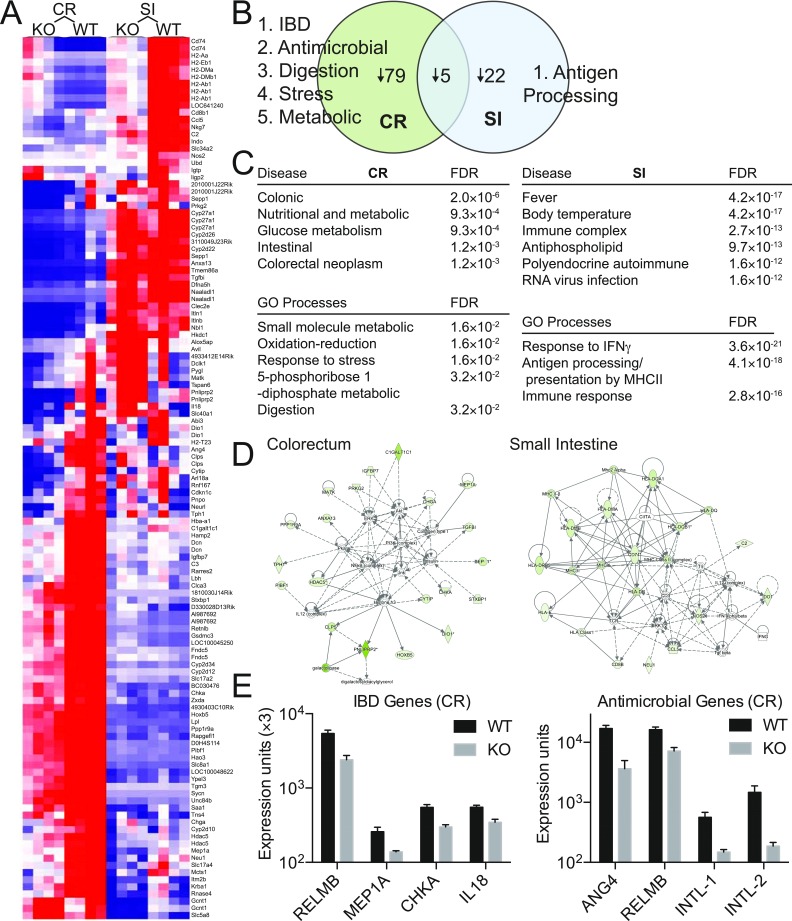
To identify host pathways that may contribute to regional changes in the microbiota, we isolated colorectal (CR) and small intestine (SI) epithelia from ∼2-mo-old male WT and KO mice, before clinical inflammation and at the same age as 16S sequencing, and analyzed transcripts with Illumina Mouse-Ref v2.0 gene chip array (n = 4 mice/group per region) (Fig. S8). Male KOs had a decrease in 79 genes in the CR, 22 genes in the SI, and only 5 genes in both (including Cosmc) [Fig. S8 A and B; false discovery rate (FDR) 5%, fold change ≥ 1.5]. Further, different gene sets (small molecule metabolism, redox, stress response, digestion) [Fig. S8C, Metacore Gene Set Enrichment Analysis (GSEA)], pathways (Fig. S8D, Ingenuity Pathway Analysis), and transcripts (RELMB, MEP1A, CHKA, IL18, ANG4, INTL-1, INTL-2) (19–26) (Fig. S8E) important for regulating the microbiota were altered in CR versus SI. These data suggest that Cosmc differentially regulates gene expression in different regions of the gut to help control microbial biogeography.
Fig. S8.
Cosmc controls diverse host pathways that regulate the microbiota and inflammation in the distal colon. Epithelial cells were purified from the CR or SI from KO and WT mice, and gene expression was performed on Illumina mouse ref 8 v2.0 expression chip. (A) Heat map indicating genes significantly altered by Cosmc KO (n = 4 mice per group, age 2 mo; fold change > 1.5, FDR < 0.05). (B) The number of genes altered in only CR, SI, or both are indicated with the most relevant gene sets from C or individual genes from E bulleted. No change in ST6GALNAC1 was seen. (C) Metacore GSEA was performed on CR and SI for gene sets enriched for diseases or GO processes. (D) Ingenuity pathway analysis used for gene expression from KO and WT epithelia isolated from colorectum. (E) Expression of individual IBD or antimicrobial genes from the CR as determined from the expression array.
Discussion
Here, in a report of IEC-specific deletion of Cosmc in mice, we have provided insight into whether the IBD risk gene Cosmc functionally contributes to IBD and sex bias in pathology. Deleting one allele of Cosmc in males resulted in the loss of T-synthase activity and O-glycan extension, compromising the mucus layer, epithelial glycoproteins, and downstream host pathways important for maintaining homeostasis of the gut and its microbiota. Ultimately these direct and indirect effects led to dysbiosis and bacterial-epithelial contact, resulting in spontaneous inflammation and enhanced experimental colitis.
Unexpectedly, the loss of one allele of Cosmc in females did not induce spontaneous inflammation, despite Cosmc KO and glycocalyx loss in 50% of crypts. Normally, the colonic mucus has an apparently attached sterile inner layer and an outer loose layer occupied by bacteria (27). Although prior work hypothesized that the inner layer covalently links to goblet cells (27–30), we found that MUC2 from WT cells migrates over multiple crypts to cover and protect KO cells while maintaining bacterial-epithelial separation. Thus, cohesive/adhesive forces may be more important than mucus-goblet cell linkages in attaching the inner mucus layer to the epithelia, and covalent linkage may not be required to maintain a sterile inner mucus layer in female mosaic mice.
We also made the surprising discovery that Cosmc spatially regulates the gut microbiota, but in a regio-specific manner. Deletion of Cosmc throughout the intestine led to a loss of diversity in the colonic mucosa, but not in the overlying lumen or small intestine mucosa. Similarly, loss of Bacteroides, reduced mucosal diversity, and a dysbiotic mucosal but relatively preserved luminal microbiota are seen in human IBD (4, 31).
Mechanistically, our results indicate that nutrient selection, host genes, and bacterial adhesion function to establish the microbial gradient via Cosmc. In the mucosal-luminal axis, we found that the host glycocalyx, dependent on Cosmc, selects commensals in the mucosa despite a diet rich in polysaccharides, but not in the lumen (32). This finding contrasts with the conventional view that bacteria only rely on host glycans following dietary polysaccharide depletion and suggests that dietary polysaccharides may prevent migration of a dysbiotic mucosal microbiota to the lumen and subsequent fecal-oral transmission (32, 33). In the cephalo-caudal axis, we found that Cosmc differentially controls host genes in the small versus large bowel by indirect mechanisms downstream of altered glycoproteins to spatially regulate the microbiota and facilitates expression of host ligands for bacteria by directly regulating glycocalyx synthesis. These ligands select commensals in the slow transit times of the colon but not in the small intestine (34). Our data functionally validate Cosmc as an IBD risk gene that spatially regulates the gut microbiota and contributes to sex bias in pathology and sets the stage to evaluate the interaction of Cosmc, the glycocalyx, and the gut microbiota in human IBD.
Methods
Mice.
Cosmcf/+ females (7) were crossed with B6.SJL-Tg(Vil-cre)997Gum/J transgenic males from Jackson Laboratory. All mice were backcrossed to C57BL/6J for at least 10 generations. All studies adhered to approved Institutional Animal Care and Use Committee (IACUC) protocols (Emory University). Mouse genotypes were determined by PCR with primer for Vil-Cre (F: 5′GTGTGGGAC AGAGAACAAACC, R: 5′ACATCTTCAGGTTCTGCGGG) and Cosmcflox (F: 5′GCAACA CAAAGAAACCCTGGG, R: 5′TCGTCTTTGTTAGGGGCTTGC).
Immunoblot.
Epithelia were purified (35) and immunoblots performed (36) with antibodies against Cosmc (C4; Santa Cruz) and actin (H-10; Santa Cruz).
Enzyme Assays.
Enzyme activity assays for T-synthase and mannosidase were performed as described (36).
Tissue Staining.
Intestine was dissected and prepared by swiss-roll [formalin-fixed, paraffin-embedded (FFPE), distal portion of segment in center] for staining with monoclonal antibodies against Tn (CA3638, a kind gift from the late Georg Springer, University of Health Sciences/The Chicago Medical School, North Chicago, IL) and STn (TAG72, B72.3; Santa Cruz) antigens by immunohistochemistry (7). For mucus analysis, Carnoy’s was used to preserve the mucus layer and tissues were stained with PAS/Alcian Blue; MUC2 (sc-15334, H-300), STn (TAG72, B72.3; Santa Cruz), UEA-I (B-1065; Vector Laboratories), and DAPI; or with a universal fluorescent in situ hybridization probe against bacteria (37). Crypt length in colon or crypt-villus length in small intestine was measured on H&E swiss-rolled, FFPE sections as described in the legends for Fig. 4 and Fig. S4.
ELISA.
ELISAs for lipocalin-2 were performed as instructed (R&D, DuoSet) and for fecal IgA as described (38).
DSS Colitis.
DSS (2.5%; Affymetrix) was administered in drinking water to mice for 4 d before switching to autoclaved drinking water. Body weight, stool softness, and blood in the stool (Hemoccult SENSA; Beckman Coulter) was assessed daily. DAI was determined by averaging score for %body weight change [no loss (0), 1–5% (1), 5–10% (2), 10–20% (3), >20% (4)], stool consistency (hard (0), soft (2), diarrhea (4)], and blood [negative (0), positive (2), macroscopic (4)]. Histology was assessed on H&E-stained swiss-roll of colorectum from FFPE sections. Aperio digital microscope was used to measure CR length and length of ulcerated (complete loss of crypt architecture with inflammatory infiltrate) and eroded (partial loss of crypt architecture with inflammatory infiltrate) epithelia. Mortality was determined in mice whose body weight dropped >25%, necessitating euthanasia per Emory IACUC guidelines.
Microbiota.
Intestines were removed and dissected longitudinally. Fecal pellet was collected from distal colon for analysis. Otherwise, feces were gently removed and tissue was collected from distal colon and ileum for analysis of mucosal-associated bacteria. V4 region of rRNA gene was amplified by PCR and sequenced by MiSeq (Illumina). Taxonomic divisions were determined by MacQIIME (39). Antibiotics (NMVA) were administered as described (40). Feces were cultured on LB in aerobic conditions to confirm elimination of microbiota.
Gene Expression.
Intestinal epithelia were purified (35) and total RNA isolated by RNeasy (Qiagen) with RNA quality and purity assessed by Agilent BioAnalyzer and nanodrop, respectively. University of California, Los Angeles (UCLA) genomics core performed RNA amplification and hybridization on Illumina mouse ref 8 v2.0 expression chips. Individual genes with fold change > 1.5 and FDR < 0.05 were determined by significance analysis of microarrays. GSEA for disease pathways and Gene Ontology (GO) processes (Metacore, only relevant findings included) and gene networks (Ingenuity Pathway Analysis) were determined. Data were deposited in Gene Expression Omnibus under accession no. GSE84416.
Statistics.
Statistics were calculated with Prism GraphPad software as described in the figure legends.
Acknowledgments
We thank J. Heimburg-Molinaro for help in editing; W. Gu, X. Sun, D. Rios, and I. Williams for helpful discussions; R. Xavier for reviewing the manuscript; the Semel Institute UCLA Neurosciences Genomics Core; and the Emory Winship Pathology Core Laboratory. This work was supported by NIH Grants U01CA168930 (to R.D.C. and T.J.) and P41GM103694 (to R.D.C.), Emory Winship Cancer Center Support Grant P30CA138292 (to C.S.M.), 2RO1-1DK59888 and 2R01-DK055679 (to A.N.), the Burroughs Wellcome Trust Career Award for Medical Scientists and the NIH Early Independence Grants DP5OD019892 (to S.R.S.), 5R01-CA179424 and DK89763 (to R.M.J. and A.S.N.), and AI64462 (to A.S.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.C.M. is a Guest Editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE84416).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612158114/-/DCSupplemental.
References
- 1.Sauer CG, Kugathasan S. Pediatric inflammatory bowel disease: Highlighting pediatric differences in IBD. Med Clin North Am. 2010;94(1):35–52. doi: 10.1016/j.mcna.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14(1):20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steentoft C, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99(26):16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Cosmc is an essential chaperone for correct protein O-glycosylation. Proc Natl Acad Sci USA. 2010;107(20):9228–9233. doi: 10.1073/pnas.0914004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D, et al. Accounting for eXentricities: Analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS One. 2014;9(12):e113684. doi: 10.1371/journal.pone.0113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashyap PC, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA. 2013;110(42):17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Platelet biogenesis and functions require correct protein O-glycosylation. Proc Natl Acad Sci USA. 2012;109(40):16143–16148. doi: 10.1073/pnas.1208253109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chassaing B, et al. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 2012;7(9):e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Axelsson MA, Asker N, Hansson GC. O-glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J Biol Chem. 1998;273(30):18864–18870. doi: 10.1074/jbc.273.30.18864. [DOI] [PubMed] [Google Scholar]
- 14.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131(1):117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18(4):478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalili H. Risk of inflammatory bowel disease with oral contraceptives and menopausal hormone therapy: Current evidence and future directions. Drug Saf. 2016;39(3):193–197. doi: 10.1007/s40264-015-0372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lécuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40(4):608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Siegmund B. Interleukin-18 in intestinal inflammation: Friend and foe? Immunity. 2010;32(3):300–302. doi: 10.1016/j.immuni.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Pemberton AD, et al. Innate BALB/c enteric epithelial responses to Trichinella spiralis: Inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J Immunol. 2004;173(3):1894–1901. doi: 10.4049/jimmunol.173.3.1894. [DOI] [PubMed] [Google Scholar]
- 21.Karner M, et al. First multicenter study of modified release phosphatidylcholine “LT-02” in ulcerative colitis: A randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol. 2014;109(7):1041–1051. doi: 10.1038/ajg.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 23.Hogan SP, et al. Resistin-like molecule beta regulates innate colonic function: Barrier integrity and inflammation susceptibility. J Allergy Clin Immunol. 2006;118(1):257–268. doi: 10.1016/j.jaci.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrom KS, et al. Goblet cell derived RELM-β recruits CD4+ T cells during infectious colitis to promote protective intestinal epithelial cell proliferation. PLoS Pathog. 2015;11(8):e1005108. doi: 10.1371/journal.ppat.1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee S, et al. MEP1A allele for meprin A metalloprotease is a susceptibility gene for inflammatory bowel disease. Mucosal Immunol. 2009;2(3):220–231. doi: 10.1038/mi.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wesener DA, et al. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol. 2015;22(8):603–610. doi: 10.1038/nsmb.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schütte A, et al. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci USA. 2014;111(34):12396–12401. doi: 10.1073/pnas.1407597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelaseyed T, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260(1):8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 34.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe. 2016;19(4):550–559. doi: 10.1016/j.chom.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Neufert C, et al. Tumor fibroblast-derived epiregulin promotes growth of colitis-associated neoplasms through ERK. J Clin Invest. 2013;123(4):1428–1443. doi: 10.1172/JCI63748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudelka MR, et al. Cellular O-glycome reporter/amplification to explore O-glycans of living cells. Nat Methods. 2016;13(1):81–86. doi: 10.1038/nmeth.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 38.Knoop KA, et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J Immunol. 2009;183(9):5738–5747. doi: 10.4049/jimmunol.0901563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]