Significance
Casitas B-cell lymphoma (Cbl) family proteins are RING finger-containing E3 ubiquitin ligases involved in degradation of activated tyrosine kinases. Previous studies in Cbl-deficient models focused primarily on the consequences of persistent tyrosine kinase signaling resulting in uncontrolled cell activation and proliferation. In the present study, we provide evidence that, in the complete absence of Cbl family proteins, failure to turn over active tyrosine kinases induces irreparable breakdown of the homeostasis of the protein milieu in primary mouse mammary epithelial cells and triggers stress-mediated cell death. Thus, our data reveal that well-regulated removal of active tyrosine kinases is essential for cell survival, an aspect of Cbl family protein functions that has not been previously fully appreciated.
Keywords: CBL, ubiquitin ligase, tyrosine kinase, protein degradation, stress response
Abstract
Casitas B-cell lymphoma (Cbl) family ubiquitin ligases negatively regulate tyrosine kinase-dependent signal transduction by promoting degradation of active kinases. We and others previously reported that loss of Cbl functions caused hyperproliferation in lymphoid and hematopoietic systems. Unexpectedly, Cbl deletion in Cbl-b–null, Cbl-c–null primary mouse mammary epithelial cells (MECs) (Cbl triple-deficiency) induced rapid cell death despite enhanced MAP kinase and AKT activation. Acute Cbl triple-deficiency elicited distinct transcriptional and biochemical responses with partial overlap with previously described cellular reactions to unfolded proteins and oxidative stress. Although the levels of reactive oxygen species were comparable, detergent-insoluble protein aggregates containing phosphorylated c-Src accumulated in Cbl triple-deficient MECs. Treatment with a broad-spectrum kinase inhibitor dasatinib blocked protein aggregate accumulation and restored in vitro organoid formation. This effect is most likely mediated through c-Src because Cbl triple-deficient MECs were able to form organoids upon shRNA-mediated c-Src knockdown. Taking these data together, the present study demonstrates that Cbl family proteins are required to protect MECs from proteotoxic stress-induced cell death by promoting turnover of active c-Src.
The Casitas B-cell lymphoma (Cbl) family proteins, Cbl, Cbl-b, and Cbl-c/Cbl-3 in mammals are evolutionarily conserved E3 ubiquitin ligases defined by a high degree of structural similarities, namely the presence of the N-terminal tyrosine kinase binding domain, a short linker region followed by the RING finger domain (1). By binding cognate phosphotyrosine motif-containing proteins in the tyrosine kinase binding domain (2, 3) and E2 ubiquitin-conjugating enzymes in the linker-RING finger domains (4–6), Cbl family proteins facilitate transfer of ubiquitin moieties from E2 to activated signaling molecules of the tyrosine kinase-regulated pathways (7–9). Because ubiquitination primarily serves as a signal for protein degradation (10), Cbl family proteins play essential roles in limiting the magnitude and duration of active signal transduction.
We and others previously demonstrated that Cbl functions are required for orderly activation of the immune and hematopoietic systems (reviewed in refs. 11 and 12). However, comparable studies in epithelial tissues have not been reported. Initial analyses of Cbl-deficient mice described increased mammary ductal density and branching in the absence of Cbl (13). A subsequent study suggested that Cbl functioned in the mammary stroma to support mammary organogenesis (14). On the other hand, despite its confirmed expression in the mammary gland, no significant defects were described in the mammary gland development of Cbl-c–deficient mice (15). In a separate study, transgenic overexpression of Cbl-c was shown to cause mild delay in mammary ductal elongation (16). However, considering high degrees of sequence similarities and overlapping expression, essential functions of Cbl family proteins may be obscured when only a single member of the family is manipulated. Definitive roles of Cbl family proteins may not become apparent unless all family members are deleted.
To address this issue, we generated Cbl, Cbl-b, and Cbl-c triple-deficient mouse mammary epithelial cells (MECs) by acutely deleting Cbl on the Cbl-b, Cbl-c double-deficient background. Using this system, we herein report that Cbl family proteins are indispensable for the survival and proliferation of normal adult MECs.
Results
MEC Survival Is Impaired in the Complete Absence of Cbl Family Proteins.
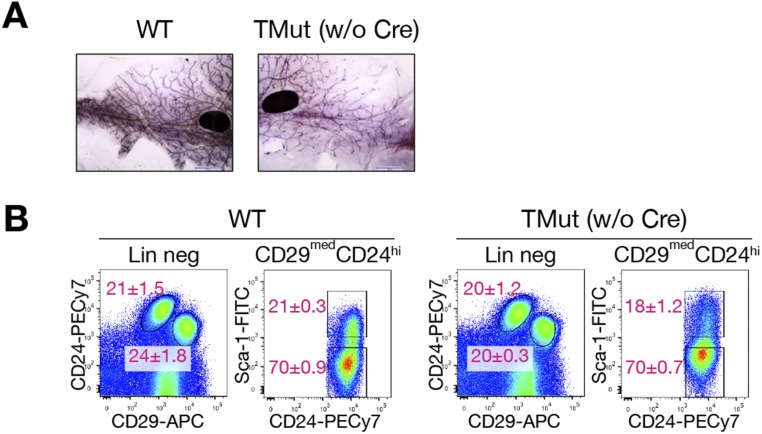
To investigate the roles of Cbl family proteins in the mammary gland, we previously attempted to delete Cbl in the MECs using the mammary gland-specific mouse mammary tumor virus (MMTV)-Cre on the Cbl-b–deficient background (MMTV-Cre;Cblflox;Cblbdel mice). In this model, however, mutant mice developed aggressive myeloproliferative disorders and most of them died by 8 wk of age (17). Early mortality hampered detailed analysis of the mammary gland phenotypes. We were also concerned that the poor overall health of the mice might cause secondary changes to the mammary gland development. To circumvent these problems, we elected to delete Cbl in vitro with adenovirus-expressing Cre DNA recombinase. Mice carrying Cblflox (conditional mutation) (18), Cblbdel (null mutation) (19), and Cblcdel (null mutation) (15) alleles were crossed to homozygosity to generate Cblflox;Cblbdel;Cblcdel (triple-mutant, TMut; without Cre expression, these mice are Cbl-b, Cbl-c double-deficient and express Cbl only) mice. We introduced the null allele of Cblc in this study because Cbl-c was reported to be expressed in epithelial tissues including the mammary gland (15). Thus, Cre-mediated deletion of Cbl from the TMut cells will render them deficient in all three Cbl family members. Both male and female TMut mice were apparently healthy up to 300 d of age and fertile. Mammary gland development of virgin female TMut mice was indistinguishable from that of WT controls (Fig. S1A) and dams successfully nursed multiple litters, indicating that normal mammary gland functions can be maintained in the absence of Cbl-b and Cbl-c. There was no significant difference in the ratio of CD29hiCD24+ (basal/myoepithelial cells; mammary epithelial stem cell activities also reside in this population) and CD29medCD24hi (luminal) populations or the ratio of Sca-1lo (luminal progenitors) and Sca-1hi (mature luminal) cells (20) within the luminal population between WT and TMut mammary glands (Fig. S1B). A mild increase in the levels of surface Sca-1 was detected reproducibly in TMut cells, but the reason for this observation is not clear.
Fig. S1.
Mammary glands develop normally in the absence of Cbl-b and Cbl-c. (A) Whole-mount staining of #4 mammary gland from 10-wk-old virgin WT and homozygous Cblflox;Cblbdel;Cblcdel (TMut) female mice. (Scale bars, 2 mm.) (B) Surface marker analysis of MECs from 10-wk-old WT and TMut female mice. After magnetic depletion of cells expressing nonepithelial lineage markers (CD45, CD31, TER119, and CD140α), MECs were stained for CD29, CD24, and Sca-1. Left panels of each genotype show CD29 and CD24 expression patterns of cells electronically gated for the absence of nonepithelial lineage markers (CD45, CD31, TER119, and CD140α). Right panels show CD24 and Sca-1 expression patterns of CD29medCD24hi population marked in the Left panels. Numbers represent mean ± SEM of the percentage of each population out of respective parent populations from three mice.
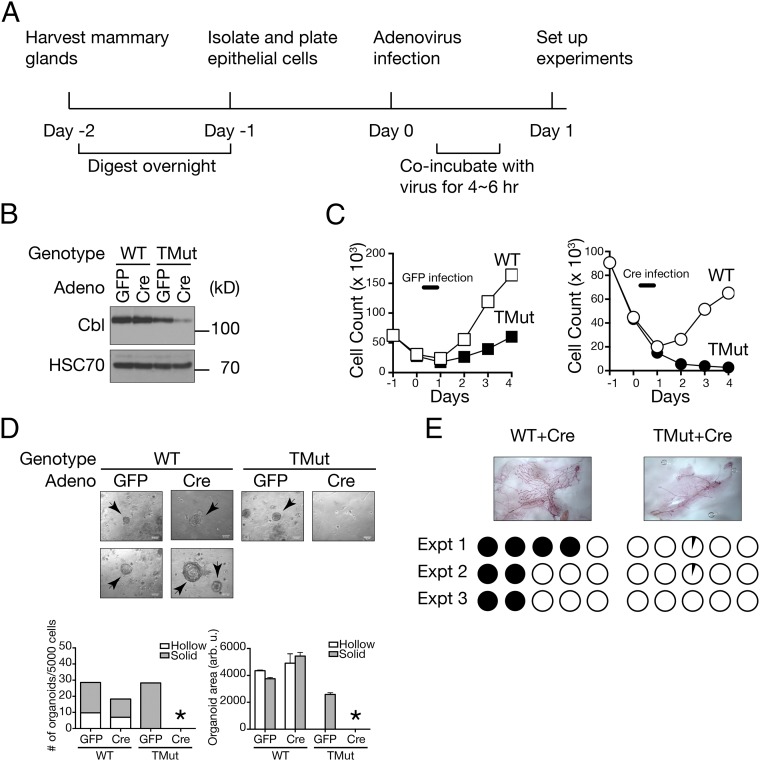
To delete Cbl from Cbl-b, Cbl-c double-deficient MECs, we isolated primary MECs from adult virgin TMut females and infected them with Ad5CMVCre-eGFP (Cre adenovirus; this virus expresses a fusion protein of Cre and GFP) in culture (Fig. 1A). To control for cytopathic effects of adenovirus, half of the cells were infected with Ad5CMVeGFP (GFP adenovirus). Cre-mediated Cbl deletion was confirmed by genomic DNA analysis, quantitative RT-PCR (qRT-PCR), and immunoblotting (Fig. 1B and Fig. S2). The low-level expression of Cbl protein in Cre-infected TMut MECs is likely a result of residual protein at the time of sample collection. We could not extend culture duration because of cell death, as described below. Although ∼10% of the cells did not score positive for GFP expression (Fig. S2), they never outgrew GFP (+) cells in multiple experiments we performed for this study, suggesting that essentially 100% of the cells underwent Cre-mediated DNA recombination.
Fig. 1.
Defective proliferation of Cbl triple-deficient MECs. (A) Experimental design for ex vivo generation of Cbl triple-deficient MECs. (B) Confirmation of Cbl deletion by immunoblotting. WT and homozygous Cblflox;Cblbdel;Cblcdel (TMut) MECs were infected with GFP or Cre-GFP adenovirus and total cell lysate was prepared 24 h later. HSC70 is shown as loading control. (C) Freshly isolated WT or TMut MECs were plated on day 1, infected with GFP (Left) or Cre-GFP (Right) adenovirus on day 0 and live cells were enumerated every day. A representative result from two independent experiments is shown. (D) MECs were prepared as above and cultured in Matrigel at 5,000 cells per well for 7 d. Organoids are indicated with arrows. (Scale bars, 0.14 mm.) Organoid areas were determined by measuring all organoids by ImageJ software. Data are from one representative experiment of more than 10 independent experiments. An asterisk indicates no organoid. (E) MECs were prepared as above and 1 × 105 cells were injected into the epithelium-cleared #4 mammary fat pads of 3-wk-old virgin female NSG mice. Each mouse received WT cells on one side and TMut cells on the contralateral side. Ductal outgrowth was evaluated 8 wk after transplantation. (Original magnification: 1×.) Each circle represents an individual recipient mouse fat pad. Filled circles indicate over 75% fat pad filled. Open circles indicate no ductal outgrowth. Partially filled circles indicate the presence of small outgrowth/cell clusters at the injection site.
Fig. S2.
Adenovirus infection efficiency and additional confirmation of Cbl deletion. (A) Five-hundred thousand (5 × 105) freshly-isolated WT MECs were infected with GFP or Cre-GFP adenovirus at 4 × 108 pfu/mL and cells were observed under a fluorescence microscope the next day. Of these, 90.2% of MECs infected with GFP adenovirus and 88.7% of MECs infected with Cre-GFP adenovirus were positive for GFP. More than 250 cells for each sample were scored. (Scale bars, 20 μm.) (B, Upper) A map of the WT and mutant Cbl genomic loci. Locations of the primers used for diagnostic PCR and expected product sizes are indicated. (Lower) Genomic DNA was isolated from MECs infected with indicated adenovirus and analyzed by PCR using indicated primers. Primer F4 will not bind to genomic DNA from Cre-infected TMut MECs. In the WT allele, the expected product size between F2 and R2 is 3.5 kb, which was not amplified routinely. (C) qRT-PCR analysis of Cbl family gene transcripts after Cre adenovirus infection. Expression levels of Cbl, Cblb, and Cblc relative to Gapdh was determined in WT and TMut MECs after Cre adenovirus infection. Expression levels in WT cells are set as 1. Data represent mean ± SEM of three independent experiments.
We first evaluated the effects of Cbl triple-deficiency on MEC growth in a monolayer culture on tissue culture-treated plastics. Without adenovirus infection, both WT and TMut MECs proliferated. Upon infection with the control GFP adenovirus, the cell viability reduced acutely, but both WT and TMut cells recovered and started to proliferate by day 2 (Fig. 1C, Left). When infected with the Cre virus, however, although its effects on WT cells were comparable to GFP virus infection, TMut cells never showed signs of proliferation, and by day 3 all cells detached from the plate (Fig. 1C, Right).
Considering that Cbl, Cbl-b double-deficient lymphocytes, and bone marrow cells proliferated significantly more robustly than WT cells (17, 18), death of Cbl triple-deficient MECs was rather unexpected. We next placed them in the laminin-rich extracellular matrix (Matrigel) for 3D culture. Cre-infected primary MECs from WT mice formed organoids in 7–10 d (Fig. 1D). Some organoids developed hollow lumens (Fig. 1D, Lower), whereas other did not (Fig. 1D, Upper). However, MECs from TMut mice infected with Cre virus failed to form organoids, indicating that Cbl family proteins are required for MECs to generate such structures in vitro. Of note, whereas MECs from TMut mice infected with the control GFP adenovirus were capable of forming organoids, they were generally smaller in size than those from WT MECs and they rarely formed hollow lumens. Combined with reduced proliferation in a monolayer culture after GFP virus infection (Fig. 1C), these observations may indicate that, although functionally competent in vivo (as judged by their ability to nurse their young), Cbl-b, Cbl-c double-deficient MECs are more vulnerable to certain stressful conditions, such as in vitro culture or adenovirus infection. Although organoid-forming efficiency and its morphology have been previously linked to the lineage and the developmental stages of the MECs (21), we did not pursue these aspects further in the present study. Of note, because Matrigel is a laminin-rich extracellular matrix that can engage integrins (22), it is unlikely that failed organoid formation by Cbl triple-deficient MECs in 3D culture was because of anoikis.
We also tested whether Cbl triple-deficient MECs could grow in vivo. Cre adenovirus-infected MECs were injected into the cleared mammary fat pads of immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Cells were injected 1 d after adenovirus infection, when both WT and TMut cells showed comparable viability (Fig. 1C). As expected, WT MECs formed ductal outgrowths. However, Cbl triple-deficient MECs were incapable of repopulating cleared mammary fat pads (Fig. 1E).
From these results, we concluded that Cbl family proteins are required for normal primary mouse MEC survival and growth.
Cbl Triple-Deficiency Blocks DNA Synthesis and Induces Cell Death in Primary Mouse MECs.
Defective cell survival and growth of Cbl triple-deficient MECs may be because of increased cell death or impairment of cell proliferation. To address these possibilities, cells were prepared as outlined in Fig. 1A and analyzed 1 d after Cre adenovirus infection, when both WT and Cbl triple-deficient MECs showed comparable cell numbers and viability.
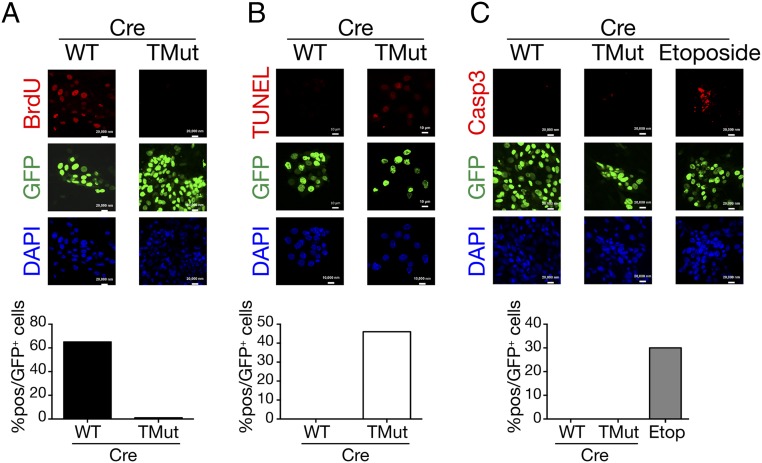
To evaluate cell proliferation, we measured DNA synthesis by pulsing the cultures with BrdU for the last 6 h of incubation. Whereas more than 60% of WT cells incorporated BrdU, Cbl triple-deficient cells showed essentially no BrdU incorporation (Fig. 2A), indicating that DNA synthesis is impaired when Cbl family proteins are acutely removed.
Fig. 2.
DNA synthesis defect and death of Cbl triple-deficient MECs. Cells were prepared as in Fig. 1A, stained, and scored in 10 randomly selected fields. Shown are representative results from more than two independent experiments with similar results. (A) Culture was pulsed with 10 μM BrdU for the last 6 h before fixation. BrdU incorporation was detected by anti-BrdU antibody. A total of 442 WT and 430 TMut cells were scored under the microscope. (Scale bars, 20 μm.) (B) DNA fragmentation was detected using the In Situ Cell Death Detection Kit. A total of 179 WT and 131 TMut cells were scored. (Scale bars, 10 μm.) (C) Cleaved caspase 3 was detected by specific antibody. WT MECs infected with Cre adenovirus and treated overnight with 100 μM etoposide are shown as control. A total of 298 WT, 338 TMut, and 229 etoposide-treated WT cells were scored. (Scale bars, 20 μm.)
We next assessed DNA fragmentation by TUNEL assay. As shown in Fig. 2B, nearly half of Cbl triple-deficient MECs stained positive, whereas no WT cells showed DNA fragmentation. Interestingly, cleaved caspase 3 was not detected in either WT or Cbl triple-deficient MECs (Fig. 2C), suggesting that cell death in Cbl triple-deficient MECs is mediated through caspase-independent mechanisms.
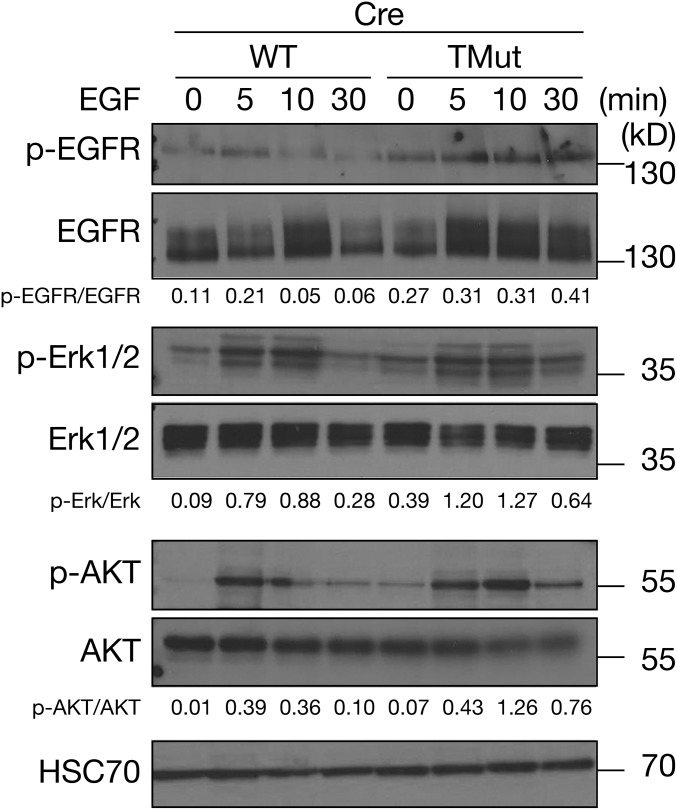
Multiple independent reports demonstrated that Cbl family proteins were required to facilitate degradation of active receptor tyrosine kinases (23–25). In line with these previous observations, we found that phosphorylated EGFR persisted up to 30 min after ligand stimulation in Cbl triple-deficient MECs, but peaked at 5 min after stimulation and declined swiftly in WT cells (Fig. 3). Furthermore, activation of downstream signaling molecules, such as Erk and AKT in Cbl triple-deficient MECs was mildly elevated, indicating that these biochemical events can be uncoupled from cell proliferation and survival (Fig. 3).
Fig. 3.
Biochemical pathways downstream of EGFR are intact in Cbl triple-deficient MECs. MECs were prepared as described in Fig. 1A, starved of EGF for 6 h, stimulated with 5 nM EGF, and lysed in RIPA buffer. Levels of phosphorylated and total EGFR, Erk, and AKT were determined by immunoblotting. HSC70 is shown as loading control. Signals were quantitated by densitometry and ratios of phosphorylated proteins to total proteins are indicated.
Altogether, we conclude that DNA synthesis is blocked and MECs cannot survive in the complete absence of Cbl family proteins although conventional EGFR-downstream signaling pathways are preserved.
Cbl Deficiency Triggers Stress Responses.
Loss of Cbl functions has been associated with hyperactivation of the Ras-Raf-MAPK pathway (1). In human, germ-line mutations in CBL manifest many clinical features shared with congenital Ras pathway gene mutations and they are collectively known as the RASopathies (26). At the cell biological level, activating mutations in the Ras pathway genes are known to induce oncogene-induced senescence in untransformed cells (27). However, unlike Cbl triple-deficient MECs, cells expressing active Ras undergo a few rounds of cell division before they cease to divide (28). Furthermore, senescent cells are still alive and metabolically active. Clearly, our observations in Cbl triple-deficient MECs do not conform to the definitions of senescence.
In light of the rapid onset of cell death, we considered that Cbl triple-deficient MECs might be undergoing stress instead. To test this, we analyzed the stress-related gene-expression profiles using a commercial PCR-based gene-expression array. This array covers multiple stress and cell death-related pathways including apoptosis, autophagy, cell cycle arrest, DNA damage and repair, hypoxia, inflammatory response, necrosis, osmotic stress, oxidative stress, and the unfolded protein response (UPR). Primary mouse MECs were prepared as above and RNA was isolated 24 h after Cre infection. As shown in Fig. 4A, we could not identify a single pathway that was systematically altered, but the genes that showed greater than 50% up-regulation in Cbl triple-deficient MECs fell into either the autophagy, hypoxia, inflammatory response, osmotic stress, oxidative stress, or UPR pathways.
Fig. 4.
Cbl triple-deficiency triggers stress response. (A) Stress and cell death-related gene expression was compared using the Mouse Stress & Toxicity PathwayFinder PCR Array between Cre adenovirus-infected WT and TMut MECs prepared as in Fig. 1A. RNA was isolated 24 h after infection. The solid line indicates equal expression. Dotted lines indicate 50% changes relative to WT. Raw data are available as Dataset S1. (B) qRT-PCR analysis of representative stress pathway transcripts in Cbl triple-deficient MECs. Expression levels of Ddit3, Hspa5, spliced and total Xbp1 and Hmox1 relative to Gapdh were determined. WT cells treated overnight with 200 nM thapsigargin (Thap) or 10 μM hydrogen peroxide (H2O2; Hmox1 only) were used as control. Levels in WT cells are set as 1. Data represent mean ± SEM of three independent experiments. (C) Immunoblot analysis of stress pathway molecules. For select molecules, lysates from L929 mouse cells treated with 1 μM thapsigargin for 6 h were used as a positive control (Thap). Phospho-IRE1α migrates faster than IRE1α (52).
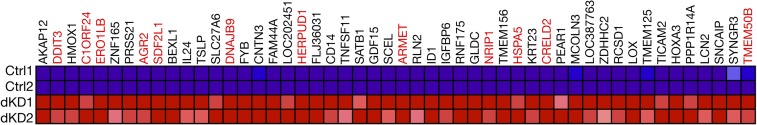
In parallel, we analyzed the gene-expression profile of MCF10A human untransformed MEC line expressing shRNA against Cbl and Cbl-b [Cbl, Cbl-b double knockdown (dKD); still expresses Cbl-c]. We previously reported that, although dKD MCF10A cells were viable and EGFR degradation was delayed, loss of Cbl and Cbl-b did not enhance cell growth in response to growth factors (29). Gene-set enrichment analysis on the genes up-regulated more than 50% in dKD MCF10A cells found a significant enrichment in UPR-associated genes (Fig. S3 and Table S1).
Fig. S3.
The top 50 genes up-regulated in Cbl, Cbl-b dKD MCF10A cells. Stress pathway genes are highlighted in red.
Table S1.
Top 10 Reactome pathways enriched among the genes up-regulated >50% in Cbl, Cbl-b dKD MCF10A cells over control cells
| Term | Overlap | P value | Adjusted P value | Z-score | Combined score |
| Unfolded protein response (UPR) | 15/79 | 1.83809E-11 | 9.20884E-09 | −1.946148267 | 36.0097789 |
| IRE1α activates chaperones | 10/48 | 2.36377E-08 | 5.92124E-06 | −1.930889588 | 23.24205058 |
| XBP1(S) activates chaperone genes | 9/46 | 1.97574E-07 | 3.29948E-05 | −1.888849525 | 19.49133983 |
| ATF6-α activates chaperones | 5/12 | 5.53459E-06 | 0.000554566 | −1.736535884 | 13.01937413 |
| ATF6-α activates chaperone genes | 5/10 | 2.75676E-06 | 0.000345284 | −1.531439766 | 12.20732616 |
| PERK regulates gene expression | 4/28 | 0.001683934 | 0.1406085 | −2.074476627 | 4.069658138 |
| Class I MHC mediated antigen processing and presentation | 10/255 | 0.010818693 | 0.494316978 | −2.151411285 | 1.515837731 |
| SHC1 events in ERBB4 signaling | 3/28 | 0.013553964 | 0.510670007 | −2.213166992 | 1.487318324 |
| GRB2 events in ERBB2 signaling | 3/30 | 0.016070847 | 0.510670007 | −2.124871487 | 1.427980947 |
| SHC1 events in ERBB2 signaling | 3/32 | 0.018832923 | 0.510670007 | −2.115029487 | 1.421366812 |
Because the UPR genes are highly represented in both Cbl triple-deficient MECs and dKD MCF10A cells, we decided to investigate this pathway in more detail. Additionally, we were also intrigued by the up-regulation of the heme oxygenase 1 (Hmox1) transcript in both datasets. Previously, Cbl was shown to be involved in responses to toxicants and oxidative stresses (30–32). Although we did not observe global up-regulation of oxidative stress-induced genes in the present datasets, because of these previous reports we decided to examine the oxidative stress pathway as well.
First, we repeated qRT-PCR on select stress-related genes to confirm findings from the commercial array. In the UPR pathway, we found Ddit3 (DNA-damage–inducible transcript 3; also known as Chop or Gadd153) and Hspa5 (heat shock protein 5; also known as Bip or Grp78) up-regulated in Cbl triple-deficient MECs, whereas there was no increase in the spliced form of Xbp1 (X-box binding protein 1) (Fig. 4B). In the oxidative stress pathway, we confirmed Hmox1 up-regulation.
These changes in gene expression were corroborated at the protein level. Among the three arms of the UPR, the protein kinase R-like endoplasmic reticulum (ER) kinase (PERK) and Inositol-requiring enzyme 1 (IRE1) pathways are commonly linked to stress-mediated cell death (33). As anticipated from Ddit3 up-regulation, phosphorylation of PERK as well as the level of CHOP (C/EBP homologous protein, the protein product of Ddit3) were elevated in Cbl triple-deficient MECs compared with WT cells. Despite an increase in PERK phosphorylation, we observed only a marginal increase in the phosphorylation of eukarotic initiation factor 2α (eIF2α) 24 h after Cre infection. Furthermore, there was no discernable increase in the levels of phosphorylated IRE1α (Fig. 4C). In the oxidative stress pathway, consistent with the elevated transcript level, heme oxygenase 1 protein level was increased in Cbl triple-deficient MECs.
Active c-Src Partitions into Detergent-Insoluble Fraction in Cbl Triple-Deficient MECs.
We then examined how these molecular/biochemical changes were reflected at the functional and morphological levels. Despite the up-regulation of the oxidative stress-inducible gene Hmox1, the reactive oxygen species (ROS) levels were comparable between WT and Cbl triple-deficient MECs (Fig. 5A).
Fig. 5.
Cbl triple-deficiency disrupts active tyrosine kinase turnover. MECs were prepared as in Fig. 1A. (A) Detection of ROS. MECs were incubated with CellROX Deep Red reagent and analyzed by flow cytometry. Only GFP+ (Cre-GFP–infected) cells were analyzed. (B) Transmission electron micrographs of MECs. (Right) The area indicated in the Left panel at a higher magnification. Yellow arrow, Golgi; yellow arrowhead, mitochondrion; magenta arrow, ER; magenta arrowhead, multivesicular body. [Scale bars, 2 μm (Left) and 500 nm (Right).] (C) Detection of protein aggregates. ProteoStat fluorescent dye was applied to MECs and cells were examined by fluorescence microscopy. (Scale bars, 5 μm.) (D) Colocalization of protein aggregates with organelle markers. Adeno-Cre infected TMut MECs were stained with ProteoStat and antibodies against EEA1 (Left), LAMP1 (Center), or LC3A/B (Right). Because Cre adenovirus is tagged with GFP, nuclei show green fluorescence. Insets show outlined areas in higher (100×) magnifications. Arrowheads indicate colocalization of LC3A/B and aggregates. (Scale bars, 5 μm.) (E) Detection of EGFR, Src, and HSP90 in RIPA-soluble and -insoluble fractions. MECs were lysed with RIPA buffer, and RIPA-insoluble materials were lysed in SDS sample buffer. Twenty micrograms of RIPA-soluble protein and a proportionate amount of RIPA-insoluble material was analyzed.
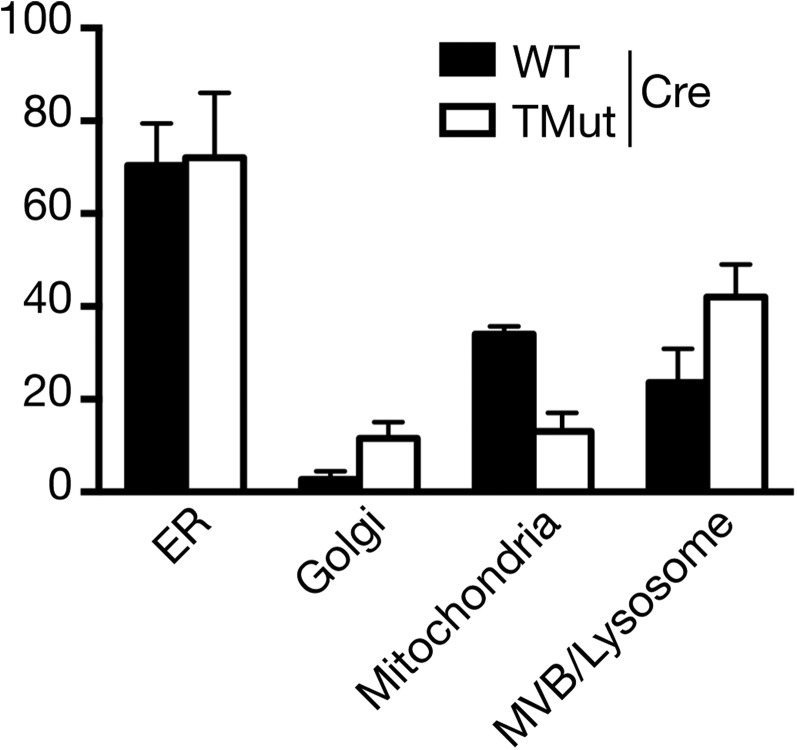
Certain types of cell death can be identified by characteristic morphological changes. Therefore, MEC ultrastructure was analyzed by transmission electron microscopy. More membrane-bound structures (morphologically identified as multivesicular bodies and lysosomes) with electron-dense deposits were found in Cbl triple-deficient MECs than in WT MECs, although the difference did not reach a statistically significant level (Fig. 5B and Fig. S4). We did not observe nuclear condensation or enlarged ER, features generally associated with apoptosis and ER stress, respectively.
Fig. S4.
Enumeration of organelles. Electron microscope images were taken at an original magnification of 40,000× and organelles were counted from three sets of 15, 7, and 7 cells for WT and two sets of 11 and 8 cells for TMut. The difference between WT and TMut for respective organelles was not statistically significant by multiple t test.
Because of the presence of electron-dense deposits and up-regulation of UPR genes, we investigated whether protein aggregates could be detected by additional means. ProteoStat is a molecular rotor-type fluorescent dye that exhibits a significant increase in fluorescence quantum yield in the presence of protein aggregates (34). When we applied this reagent to primary MECs, we found strong fluorescence in the cytoplasm of Cbl triple-deficient MECs but not in WT MECs, indicating protein aggregates formed in the absence of Cbl family proteins (Fig. 5C). To determine if these ProteoStat (+) aggregates are associated with specific intracellular organelles, we stained Cbl triple-deficient MECs with representative markers of the protein degradation pathway. We focused on this pathway because Cbl has been primarily associated with protein degradation. As shown in Fig. 5D, ProteoStat (+) aggregates did not colocalize with EEA1 or LAMP1. However, they showed considerable overlap with LC3A/B staining, suggesting an involvement of the autophagy pathway.
Next, we sought to define the composition of the aggregates. After extracting mild detergent-soluble proteins with RIPA buffer, lysates were centrifuged and pellets were dissolved directly in SDS sample buffer. As an initial analysis, we probed for EGFR and c-Src, both well-characterized Cbl targets (1). Fig. 5E demonstrates that more EGFR and c-Src were present in the RIPA-insoluble fraction of Cbl triple-deficient MECs than that of WT MECs. Furthermore, there was a noticeable decrease in the chaperone HSP90 in the RIPA-soluble fraction of Cbl triple-deficient MECs and concomitant increase in the RIPA-insoluble fraction.
It is well established that the primary function of Cbl family proteins is to ubiquitinate tyrosine phosphorylated signal transduction molecules and facilitate their degradation. Therefore, if the increase of c-Src and EGFR in the RIPA-insoluble compartment was a result of loss of Cbl-mediated ubiquitination, accumulated proteins should be tyrosine phosphorylated. When the phosphorylation status was investigated using specific antibodies, phosphorylated (Tyr416) c-Src was detectable in both RIPA-soluble and -insoluble fractions (Fig. 5E). However, phosphorylated EGFR was present only in the RIPA-soluble fraction and not in the RIPA-insoluble fraction.
PERK Inhibition Partially Restores Organoid Formation by Cbl Triple-Deficient MECs.
To confirm causative roles of the stress pathway activation in cell death, we treated Cbl triple-deficient MECs with chemical inhibitors of stress pathways and examined whether they could restore cell growth.
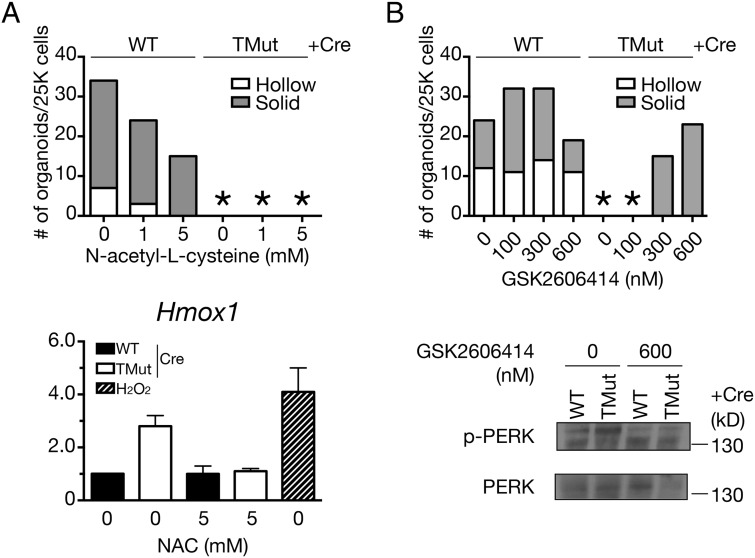
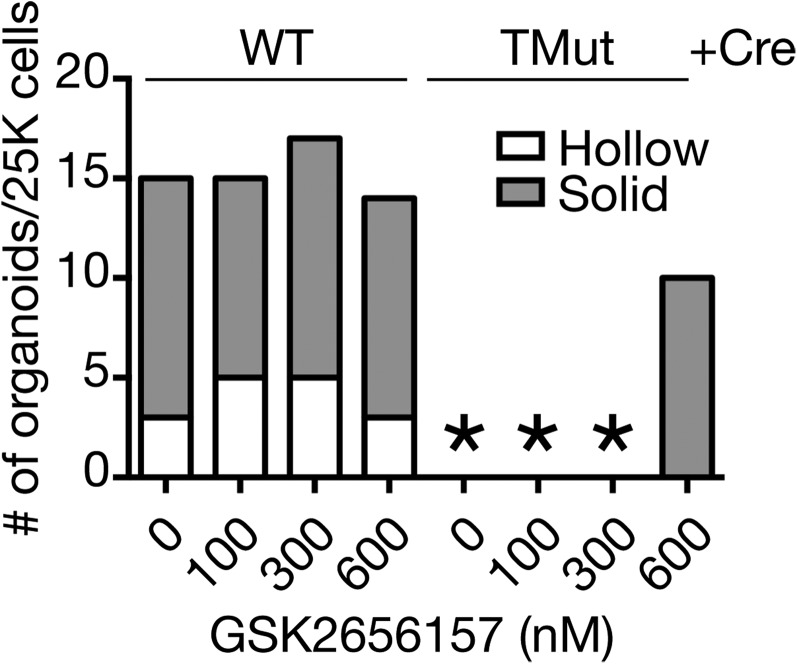
Treatment with a ROS scavenger N-acetyl-l-cysteine (NAC) had no effect on the inability of Cbl triple-deficient MECs to form organoids, even though the up-regulation of the oxidative stress-inducible gene Hmox1 was inhibited (Fig. 6A). In contrast, the PERK inhibitor GSK2606414 (35) partially restored the organoid formation of Cbl triple-deficient MECs in Matrigel (Fig. 6B). In the presence of GSK2606414, organoid formation of WT MECs was greatly accelerated so that, at the highest drug concentrations, some of the organoids started to collapse by the time they were enumerated on day 7. To verify the specificity of this compound, we tested another PERK inhibitor, GSK2656157. Although not as robust as GSK2606414, GSK2656157 also partially restored organoid formation by Cbl triple-deficient MECs (Fig. S5).
Fig. 6.
PERK inhibition partially restores organoid formation by Cbl triple-deficient MECs. (Upper) MECs were cultured in the presence of indicated concentrations of (A) the ROS scavenger NAC or (B) the PERK inhibitor GSK2606414 in Matrigel for 7 d and organoids were enumerated. A representative result from more than two independent experiments is shown. An asterisk indicates no organoid. (Lower) To confirm inhibition of stress pathways, MECs were prepared as in Fig. 1A and cultured in the presence of indicated inhibitors for 18 h before sample preparation. Oxidative stress pathway inhibition by NAC was verified by Hmox1 expression. H2O2 was used as positive control. Data for 0 mM NAC treatment are from the same experiments as in Fig. 4B.
Fig. S5.
Effect of a PERK inhibitor GSK2656157 on organoid formation in Matrigel. An asterisk indicates no organoid. A representative result from two independent experiments is shown.
Dasatinib Blocks Protein Aggregate Formation and Restores Organoid Formation by Cbl Triple-Deficient MECs.
Sandilands et al. previously reported that Cbl functions as a selective autophagy cargo receptor to protect FAK-deficient mouse squamous carcinoma cells from cell death caused by active Src turnover failure (36). Although their system differs from our present model in a number of aspects, colocalization of ProteoStat (+) aggregates with an autophagosome marker LC3, as well as the presence of active c-Src in detergent-insoluble fractions in our system, led us to ask whether the death of Cbl triple-deficient MECs was because of accumulation of active c-Src. Active c-Src is thermodynamically unstable and prone to aggregate (37).
To address this question, we first treated Cbl triple-deficient MECs with a Src/Abl family kinase inhibitor, dasatinib. We found dasatinib potently restored organoid formation by Cbl triple-deficient MECs (Fig. 7A). On the other hand, treatment with an EGFR tyrosine kinase inhibitor gefitinib showed no effect on organoid formation by Cbl triple-deficient MECs (Fig. 7B). Combined with the absence of phosphorylated EGFR in the detergent-insoluble fraction (Fig. 5E), these data support the idea that death of Cbl triple-deficient MECs is not the consequence of EGFR overactivation. Gefitinib inhibited organoid formation by WT MECs in a dose-dependent manner, underscoring the requirement for EGFR signaling in this process.
Fig. 7.
Dasatinib blocks protein aggregate formation and restores organoid formation by Cbl triple-deficient MECs. (A and B) MECs were cultured in the presence of indicated concentrations of dasatinib (broad spectrum Src/Abl family kinase inhibitor) or gefitinib (EGFR tyrosine kinase inhibitor) in Matrigel for 7 d and organoids were enumerated. A representative result from more than two independent experiments is shown. An asterisk indicates no organoid. (C) MECs were cultured in monolayer in the presence of indicated inhibitors. Protein aggregates were detected with ProteoStat reagent. (Scale bars, 5 μm.) The number of aggregates/GFP+ cells was enumerated in three randomly selected fields for each condition. An asterisk indicates no aggregate.
To further investigate the link between ProteoStat (+) aggregate formation and cell death, we treated Cbl triple-deficient MECs with stress pathway inhibitors (NAC and GSK2606414) and kinase inhibitors (dasatinib and gefitinib). As shown in Fig. 7C, treatment with dasatinib almost completely abolished aggregate formation in Cbl triple-deficient MECs, whereas cells treated with other chemicals still formed aggregates. Interestingly, the PERK inhibitor GSK2606414 reduced the amount of aggregates but did not completely eradicate them.
Loss of c-Src Restores Organoid Formation by Cbl Triple-Deficient MECs.
Dasatinib was originally developed as a Src/Abl family kinase inhibitor (38) and approved for the treatment of BCR-ABL [the fusion gene product of BCR (breakpoint cluster region) and the Abl1 kinase gene] (+) chronic myeloid leukemia. However, its target spectrum is much broader than other clinically approved BCR-ABL inhibitors. Potential dasatinib targets include receptor tyrosine kinases, such as c-Kit, PDGFR, the ErbB family and ephrin receptors, and nonreceptor tyrosine kinases, such as the TEC family, FAK and Syk, as well as serine/threonine kinases in the MAP kinase pathways (39).
Therefore, it is conceivable that, although we found c-Src in a detergent-insoluble fraction, cyto-protective effect of dasatinib may be mediated through any of the numerous kinases that interact with this compound. To decipher the precise mechanism of dasatinib action in the present system, we first asked whether the cyto-protective effects of dasatinib can be attributed to its broad range of targets. To this end, we performed organoid formation assays in the presence of a nonspecific kinase inhibitor, staurosporine. Unlike dasatinib, staurosporine treatment did not restore organoid formation by Cbl triple-deficient MECs, suggesting involvement of specific dasatinib targets in this process (Fig. S6).
Fig. S6.
Effect of a nonspecific kinase inhibitor staurosporine on organoid formation in Matrigel. An asterisk indicates no organoid. A representative result from two independent experiments is shown.
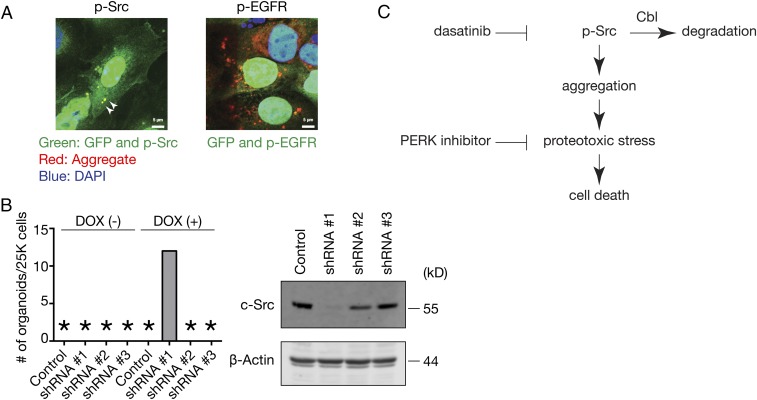
We next evaluated the expression levels of known and proposed dasatinib targets using the EMBL-EBI Expression Atlas (https://www.ebi.ac.uk/gxa/home) and concluded that c-Src is a likely candidate based on its abundant expression in the adult mammary gland. We first confirmed that phosphorylated c-Src colocalized with ProteoStat (+) aggregates in Cbl triple-deficient MECs, whereas phosphorylated EGFR failed to do so, which is anticipated from the inability of gefitinib to restore organoid formation (Fig. 8A).
Fig. 8.
c-Src knockdown restores organoid formation by Cbl triple-deficient MECs. (A) Adeno-Cre infected TMut MECs were stained with ProteoStat and antibodies against phosphorylated c-Src (Left) or phosphorylated EGFR (Right). Because Cre adenovirus is tagged with GFP, nuclei show green fluorescence. Arrowheads indicate colocalization of p-Src and aggregates. (Scale bars, 5 μm.) (B) TMut MECs were infected with lentivirus encoding tetracycline-inducible shRNA against c-Src (shRNA #1, #2, and #3) or nontargeting control followed by Cre adenovirus and subjected to organoid formation assay in Matrigel in the presence or absence of 1 μg/mL doxycycline. Organoids were enumerated 7 d later. An asterisk indicates no organoid. A representative result from two independent experiments is shown. (C) Proposed mechanism of cell death induced in the absence of Cbl family proteins.
To test the role of c-Src directly, we infected Cbl TMut MECs with lentivirus-encoding tetracycline-inducible shRNA against c-Src, deleted Cbl by Cre adenovirus infection, and subjected the cells to organoid formation assay in the presence and absence of doxycycline. One of three shRNAs restored organoid formation upon doxycycline treatment, and this was the only clone that reduced c-Src expression significantly (Fig. 8B).
Based on these observations, we conclude that Cbl family proteins are required to promote turnover of tyrosine kinases, most notably c-Src, and that failure of this process leads to aggregate formation and induces cell death through biochemical pathways involving PERK activation (Fig. 8C).
Discussion
In the present study, we uncovered an essential role of Cbl family proteins in active c-Src turnover in normal MECs, and that loss of this clearance mechanism results in stress-mediated cell death. Previous studies focused primarily on their roles in ubiquitinating active receptor tyrosine kinases and promoting trafficking through the endosomal pathway for degradation. Delayed clearance of phosphorylated EGFR in Cbl triple-deficient MECs (Fig. 3) is consistent with this well-established functions of Cbl family proteins, but our data demonstrate that enhanced MAPK and AKT activation cannot override proteostatic stress in primary MECs completely deficient in Cbl family proteins.
Because receptor tyrosine kinases and Src family kinases have been widely accepted as Cbl targets, we focused on EGFR and c-Src in the current study. Between these two tyrosine kinases, although active c-Src was detectable in the detergent-insoluble fraction as well as in the ProteoStat (+) aggregates, phosphorylated EGFR was conspicuously absent from these pools, and inhibition of EGFR tyrosine kinase activity had no effect on the Cbl triple-deficient MEC survival. The prevailing consensus is that EGFR binds to Cbl only upon ligand-induced phosphorylation (40) and becomes ubiquitinated (8, 9), which serves as a signal for degradation in the lysosome (41). Because nonubiquitinated EGFR is not efficiently sorted into the multivesicular bodies and recycles back to the plasma membrane (42, 43), ligand-engaged EGFR should enter the recycling pathway in the complete absence of Cbl family proteins. In light of this model, we consider that unphosphorylated EGFR in the detergent-insoluble fraction may have originated from the de novo synthesis pathway or, if they are derived from the cell surface pool, they must have been completely dephosphorylated after ligand engagement. Because Cbl is not known to interact with unphosphorylated EGFR, accumulation of unphosphorylated EGFR in the detergent-insoluble fraction may be secondary to the proteostasis disruption because of active c-Src accumulation. Alternatively, Cbl may possess previously undescribed functions in the quality control of select membrane proteins, including nascent EGFR. Taking these data together, we feel that future studies should clarify the molecular basis behind these observations.
Present findings are consistent with an earlier report by Sandilands et al. that Cbl is required for autophagic targeting of active c-Src in squamouse carcinoma cells (36), providing a confirmatory evidence that a similar molecular pathway is operational in normal epithelial cells. Because of technical challenges associated with manipulating primary cells, we have yet to investigate whether this function requires ubiquitin ligase activity of Cbl family proteins.
The phenotypes of Cbl triple-deficient MECs we described herein are in stark contrast to previous reports, including our own, in the immune and hematopoietic systems. Do Cbl family proteins have tissue-specific functions? Or does this reflect an intrinsic difference between Cbl, Cbl-b double-deficiency and Cbl, Cbl-b, and Cbl-c triple-deficiency? In this regard, we found UPR-associated genes up-regulated in Cbl, Cbl-b dKD MCF10A cells (Fig. S3), which indicates that both double- and triple-deficiencies induce proteostatic stress in MECs. As for tissue-specific functions, our preliminary experiments indicate that bone marrow cells proliferate poorly when all Cbl family proteins are removed acutely (Fig. S7). Therefore, it is conceivable that the published phenotypes of Cbl-deficient immune and hematopoietic cells are the consequences of in vivo adaptation. Presumably, a significant proportion of cells fail to proliferate from proteotoxicity of active tyrosine kinase accumulation in the absence of Cbl family proteins. However, if cells successfully up-regulate chaperones or activate alternative pathways to clear accumulated active tyrosine kinases, such cells may survive and gain growth advantage. This model is consistent with paradoxical observations that treatment with dasatinib or a BCR-ABL inhibitor imatinib aggravated the myeloproliferative disease phenotype of Cbl RING finger mutant (44) or MMTV-Cre;Cblflox;Cblbdel mice (45), respectively. Clearly, limiting the tyrosine kinase activation promotes proliferation of hematopoietic cells in the absence of Cbl activity.
Fig. S7.
Acute Cbl deletion on the Cbl-b, Cbl-c–deficient background does not confer proliferative advantage to bone marrow cells. WT control and TMut mice were crossed to mice carrying the Cre activity reporter Tg(CAG-cat,-EGFP)39Miya (53) and the tamoxifen-inducible Cre Gt(ROSA)26Sor <tm1(cre/ERT2)Tyj> (54). Bone marrow cells were isolated, cultured in the presence (solid lines) or absence (dotted lines) of 10 nM 4-hydroxytamoxifen, and GFP expression was analyzed by flow cytometry.
Based on the present results, we consider it is highly likely that some of the phenotypes of Cbl-deficient cells and animals may be secondary to the proteostasis disruption from the accumulation of active tyrosine kinases. Reassessment of functions associated with Cbl family proteins may be necessary to incorporate the extra layer of complexity provided through the pleiotropic nature of the proteotoxic stress response.
Materials and Methods
Mice.
The Cblflox;Cblbdel;Cblcdel mouse strain (Cbl TMut) was generated by crossing B6.Cg-Cbltm2Hua (conditional allele), B6.Cg-Cblbtm1Hua (null allele), and B6.Cg-Cblctm1Pngr (null allele) mice to homozygosity. WT mice were C57BL/6J or other C57BL/6-based WT mice from the M.N. laboratory breeding colony. The NSG mice were originally from The Jackson Laboratory and maintained in-house. All mice were housed in microisolators in the US Department of Agriculture/American Association of Laboratory Animal Science-accredited facility at the University of Nebraska Medical Center on a 12-h light, 12-h dark cycle. All mouse experiments were approved by the University of Nebraska Institutional Animal Care and Use Committee. Mice were used between 6 and 12 wk of age unless otherwise specified.
Chemicals, Antibodies, and Related Reagents.
The following antibodies and reagents were obtained from commercial sources.
Chemicals.
GSK2606414 (R&D Systems), GSK2656157 (Toronto Research Chemical), NAC (MP Biomedicals), Dasatinib (Selleck Chemicals).
MEC isolation.
Biotin–anti-CD31 (BD Pharmingen, 553371, clone MEC 13.3), biotin–anti-CD45 (BD Pharmingen, 553078, clone 30-F11), biotin–anti-TER119 (BD Pharmingen, 553672, clone TER-119), biotin–anti-CD140α (eBioscience, 13–1401, clone APA5), biotin–anti-CD24 (eBioscience, 13–0242, clone M1/69).
Flow cytometry.
Anti–CD24-PECy7 (eBioscience, 25–0242, clone M1/69), anti–CD29-APC (eBioscience, 17–0291, clone HMb1-1), anti–Sca-1-FITC (BD Pharmingen, 553335, clone E13-161.7), CellROX Deep Red (Molecular Probes).
Fluorescence microscopy.
Anti-BrdU (eBioscience, 14–5071, clone BU20A), TUNEL (Roche, 12156792910), anticleaved caspase-3 (Cell Signaling Technology, 9661, polyclonal), Proteostat (Enzo Life Science, ENZ-51035), anti-EEA1 (Cell Signaling Technology, 3288, clone C45B10), anti-LAMP1 (Developmental Studies Hybridoma Bank at the University of Iowa, 1D4B), anti-LC3A/B (Cell Singaling Technology, 12741, clone D3U4C).
Immunoblot.
Anti-Cbl (Cell Signaling Technology, 2747, polyclonal), anti-HSC70 (Santa Cruz Biotechnology, sc-7298, clone B6), antiphospho-Erk1/2 (Thr202/Tyr204) (Cell Signaling Technology, 9101, polyclonal), anti-Erk1/2 (Cell Signaling Technology, 9102, polyclonal), antiphospho-AKT (Ser473) (Cell Signaling Technology, 4060, clone D9E), anti-AKT1/2/3 (Santa Cruz Biotechnology, sc-8312, H-136, polyclonal), anti-HSP90 (Santa Cruz Biotechnology, sc-382636, H-41, polyclonal), anti-EGFR (Santa Cruz Biotechnology, sc-03, polyclonal), anti-phospho-EGFR (Tyr1068) (Cell Signaling, 3777, clone D7A5), anti-c-Src (Cell Signaling Technology, 2109, clone 36D10), antiphospho-Src family (Tyr416) (Cell Signaling Technology, 6943, clone D49G4), anti-Tubulin (Proteintech, 66031-1-Ig, clone 1E4C11), antiphospho-PERK (Cell Signaling Technology, 3179, clone 16F8), anti-PERK (Rockland, 100–401-962, polyclonal), antiphospho-eIF2α (Cell Signaling Technology, 3597, clone 119A11), anti-eIF2α (Santa Cruz Biotechnology, sc-11386, polyclonal), anti-CHOP (Pierce, MA1-250, clone 9C8), antiphospho-IRE1α (Novus, NB100-2323, polyclonal), anti-IRE1α (Cell Signaling Technology, 3294, clone 14C10), antiheme oxigenase-1 (Cell Signaling Technology, 5061, polyclonal), antiphospho-tyrosine (EMD Millipore, 05–321, clone 4G10), anti–β-actin (Proteintech, 20536–1-AP, polyclonal).
Secondary reagents.
HRP –anti-mouse IgG (H+L) (Pierce), HRP–anti-rabbit IgG (H+L) (Jackson ImmunoResearch), Alexa Fluor 488 donkey anti-rabbit IgG (H+L) (Invitrogen), Alexa Fluor 594 donkey anti-mouse IgG (H+L) (Invitrogen), Streptavidin Pacific Blue (BD Biosciences).
Isolation of Primary Mouse MECs.
Primary mouse MECs were prepared from 6- to 12-wk-old virgin female mice as previously described, with some modifications (46–48). Mammary gland numbers 3, 4 (without lymph node), and 5 were isolated and minced with a scalpel. Five-milliliter collagenase digestion buffer [RPMI-1640, 25 mM Hepes, 5% (vol/vol) FBS (HyClone), 100 units/mL penicillin 100 μg/mL streptomycin (Life Technologies), and 2 mg/mL collagenase IV (Sigma-Aldrich)] was added to each gram of tissue, and the mixture was incubated overnight at 37 °C with moderate agitation. After separating mammary organoids from fat by centrifugation, single-cell preparation was made by incubating organoids in 0.25% trypsin-EDTA for 20 min followed by 0.1 mg/mL DNase I (Sigma-Aldrich) treatment for 3 min and filtration through a 70-μm mesh. To remove the hematopoietic, endothelial, erythroid, and fibroblast lineage cells, the cell preparation was labeled with biotinylated anti-CD45, anti-CD31, anti-TER119, and anti-CD140α antibodies followed by MACS antibiotin MicroBeads (Miltenyi Biotec) and separated by AutoMACS or MiniMACS (Miltenyi). Lineage-depleted cells were further enriched for MECs by labeling with biotinylated anti-CD24 followed by MACS anti-biotin MicroBeads and positively selecting for the CD24+ population.
Primary Mouse MEC Culture and Adenovirus Infection.
Primary MECs were cultured in complete Epicult B media [Epicult B base media supplemented with proliferation supplement (both from Stemcell Technologies), 5% (vol/vol) FBS, 10 ng/mL FGF (BD Biosciences), 5 ng/mL EGF (Life Technologies), 4 μg/mL heparin (Sigma-Aldrich), 100 units/mL penicillin, 100 μg/mL streptomycin and 50 μg/mL gentamycin (Life Technologies)].
For adenovirus infection, primary MECs were plated in a monolayer at 5 × 105 cells per well in 12-well plates in complete Epicult B media. The next day, cells were exposed to 0.5 mL of serum-free Epicult B media containing either Ad5CMVeGFP or Ad5CMVCre-eGFP (University of Iowa Gene Transfer Vector Core, Iowa City, IA) at 4 × 108 pfu/mL for 6 h. After infection, cells were returned to complete Epicult B media and cultured for additional 16 h before experiments. Approximately 90% of the cells were routinely infected by the virus as assessed by GFP expression (Fig. S2A) and therefore used without sorting for GFP-expressing cells in all subsequent experiments to minimize damage to the cells from excessive handling.
For 3D culture, 5 × 103 adenovirus-infected MECs were suspended in 0.4 mL complete Epicult B media with 2% (vol/vol) growth factor-reduced Matrigel (BD Biosciences) and seeded in a well of an eight-well chamber slide (BD Biosciences) on top of a polymerized layer of 100% Matrigel. The cultures were fed with fresh complete Epicult B media every 2 d. For chemical inhibitor studies, cells were fed daily. Phase-contrast images were obtained under 10× magnification after 7 d.
All cells were cultured at 37 °C in 5% (vol/vol) CO2/20% O2 environment.
Lentivirus Infection.
For lentivirus infection, primary MECs were prepared as above and infected with lentivirus at 2.5 × 105 TU/mL for 6 h in the presence of 5 μg/mL polybrene. After adenovirus infection on the following day, cells were plated in Matrigel and cultured with or without 1 μg/mL doxycycline to induce shRNA. For immunoblot experiments, cells were maintained in the presence of 250 ng/mL puromycin to select for lentivirus-infected cells.
Packaged lentivirus clones were obtained from Dharmacon/GE Healthcare. The following tetracycline-inducible SMARTvector clones with mCMV/TurboRFP were used: nontargeting control (VSC6571), c-Src shRNA #1 (V3IMMMCR_12062595, ATATTTGCGGCTACCCCAG), c-Src shRNA #2 (V3IMMMCR_11382729, GGAGCGGCTGCAGATTGTC), c-Src shRNA #3 (V3IMMMCR_12220731, TCAACTCCTCGGACACCGT).
MEC Transplant.
Three-week-old female NSG mice were anesthetized and #4 mammary fat pads were exposed with an inverted Y incision in the abdomen. The developing endogenous mammary gland was excised and ∼1 × 105 adenovirus-infected MECs were injected into the cleared fat pad. Each mouse received WT MECs on one side and TMut MECs on the contralateral side. Mammary gland repopulation was assessed by carmine staining of the whole-mount preparation 8 wk after transplantation.
qRT-PCR.
Total RNA was isolated using RNAqueous-Micro Kit (Life Technologies). cDNA was synthesized from 1 μg of total RNA using iScript cDNA Synthesis Kit (Bio-Rad). Reaction mixture was incubated for 5 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, and then held at 4 °C. cDNA samples were diluted to 1:10 and mixed with 300 nM gene-specific primers and SYBER Green Supermix (Bio-Rad) in a final volume of 25 μL. qRT-PCR was performed in the CFX96 real time detection system (Bio-Rad) by incubating for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 40 s at 55 °C, and 30 s at 72 °C. A melt curve was generated at the end of each run to ensure product quality. From the amplification plot of each sample, a threshold cycle value (Ct) was calculated from the exponential phase. Reactions were performed in duplicate, and relative amounts of cDNAs were normalized to that of Gapdh.
The Mouse Stress & Toxicity PathwayFinder PCR Array was from Qiagen.
Flow Cytometry.
MECs were suspended in ice-cold FACS buffer (PBS/1% BSA) at 106 cells/200 μL, incubated for 20 min with anti–CD29-APC, anti–CD24-PECy7, and anti–Sca-1-FITC antibodies, washed, and resuspended in FACS buffer. Nonepithelial lineage cells were electronically gated out by labeling with biotinylated anti-CD45, anti-CD31, anti-TER119, and anti-CD140α antibodies followed by streptavidin-Pacific Blue. ROS was detected using CellROX Deep Red Reagent (Invitrogen) per the manufacturer’s instructions. Data were acquired on an LSRII (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Immunoblot Assays.
Immunoblot assays were performed as described previously (49). Cells were lysed with M-PER mammalian protein extraction reagent (Pierce) for PERK, eIF2α, CHOP, and IRE1α. For fractionation of soluble and insoluble compartments, cells were first lysed with RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0). RIPA-insoluble materials were lysed in SDS sample buffer.
Immunofluorescence Microscopy.
Immunofluorescence microscopy analysis was performed as previously described (49). Confocal images were acquired with an LSM710 fluorescence confocal microscope (Carl Zeiss). For enumeration, more than 10 fields were randomly selected and a total of 130∼450 cells were visually scored for each sample.
For BrdU incorporation analysis, cells were pulsed with 10 μM BrdU for the last 6 h of incubation. After fixation, permeablilization, and blocking, cells were incubated for 1 h with 10 Kunitz units/mL DNase I before incubation with mouse anti-BrdU antibody followed by detection with Alexa Fluor 594-labeled anti-mouse IgG.
DNA fragmentation was detected with an In Situ Cell Death Detection Kit TMR Red (Roche Diagnostics).
Protein aggregate was detected with Proteostat Aggresome Detection Kit (Enzo Life Sciences).
Electron Microscopy.
Twenty-four hours after Cre adenovirus infection, cells were trypsinized and centrifuged. Cell pellets were fixed overnight in 0.1 M phosphate buffer with 2% (wt/vol) paraformaldehyde and 2% (wt/vol) glutaraldehyde. Following postfixing in 1% osmium tetroxide, samples were dehydrated in graded ethanol with propylene oxide as a transition fluid for infiltration of Araldite resin. After overnight polymerization, samples were thin-sectioned at 100 nm and stained with 2% (wt/vol) uranyl acetate and Reynold’s lead citrate for 10 min each. Samples were examined under a Zeiss EM10A electron microscope or a FEI Technai G2 Spirit transmission electron microscope (FEI) operated at 80 keV.
Microarray Analysis.
Total RNA was isolated using TriReagent (Life Technologies) and hybridized with GeneChip Human Genome U133 Plus 2.0 array (Affymetrix). Raw data were processed using the GenePattern platform (50). Gene-set enrichment analysis was performed on the Enrichr platform (51). Microarray data have been deposited in the Gene Expression Omnibus Database with the accession number GSE67301.
Genomic DNA Preparation.
Genomic DNA was prepared using Gentra Puregene kit (Qiagen).
Oligonucleotides.
Genomic DNA PCR:
Cbl F2: CCAGCTTCTTTGTTTCCTCCTTACCTCCCC,
Cbl F4: CAATGGCCCTGTTCTCAGCTGTGGGCATCC,
Cbl R2: CACCACTTGCCTACCCCAAAGGGATAAACT.
qRT-PCR:
Cbl: AGCTGATGCTGCCGAATTT and TTGCAGGTCAGATCAATAGTGG,
Cblb: GGAGCTTTTTGCACGGACTA and TGCATCCTGAATAGCATCAA,
Cblc: CCACCTGCCTGCTTTGAC and GCTACTTGGAGAGGTGGCAAAG,
Ddit3: CCACCACACCTGAAAGCAGAA and AGGTGAAAGGCAGGGACTCA,
Hspa5: TTCAGCCAATTATCAGCAAACTCT and TTTTCTGATGTATCCTCTTCACCAGT,
spliced Xbp1: CTGAGTCCGAATCAGGTGCAG and GTCCATGGGAAGATGTTCTGG,
total Xbp1: TGGCCGGGTCTGCTGAGTCCG and GTCCATGGGAAGATGTTCTGG,
Hmox1: GACACCTGAGGTCAAGCACA and CTAGCAGGCCTCTGACGAAG,
Gapdh: CCTGGAGAAACCTGCCAAGTATG and GAGTGGGAGTTGCTGTTGAAGTC.
Supplementary Material
Acknowledgments
The authors thank the University of Nebraska Medical Center Comparative Medicine, Flow Cytometry, DNA Microarray Core, Advanced Microscope Core and Electron Microscopy Core Facilities for their expert assistance; Dr. Michael A. Hollingsworth and Mr. Thomas Caffrey for the MACS cell separation apparatus; Drs. Samikshan Dutta and Kaustubh Datta for help with electron micrographs; Dr. Melissa Teoh-Fitzgerald for reactive oxygen species assay; and Dr. Josef Penninger for Cbl-c mutant mice. Support for the core facilities are provided, in part, by the Nebraska Research Initiative, The University of Nebraska Foundation, the Nebraska Banker’s Fund, the Fred and Pamela Buffett Cancer Center Support Grant P30CA036727, and an Institutional Development Award from the National Institute of General Medical Sciences (P30GM106397). Additional funding for confocal microscopes were provided by NIH Grants S10RR027301 and S10RR019278. This work was supported by US Department of Defense Grant W81 XWH-10-1-0740 (to M.N.); Nebraska Department of Health and Human Services Grant LB606 Stem Cell 2011-06 (to M.N.); Nebraska Center for Cell Signaling pilot project Grant 5P30GM106397 (to M.N.); and the University of Nebraska Medical Center–Buffett Cancer Center (M.N.). C.M. was supported by a graduate fellowship from the University of Nebraska Medical Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE67301).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615677113/-/DCSupplemental.
References
- 1.Thien CB, Langdon WY. Cbl: Many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2(4):294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 2.Lupher ML, Jr, Reedquist KA, Miyake S, Langdon WY, Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J Biol Chem. 1996;271(39):24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 3.Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398(6722):84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 4.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102(4):533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 5.Kobashigawa Y, et al. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci USA. 2011;108(51):20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou H, et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19(2):184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joazeiro CA, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286(5438):309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 8.Levkowitz G, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4(6):1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 9.Yokouchi M, et al. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274(44):31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 10.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 11.Naramura M, Band V, Band H. Indispensable roles of mammalian Cbl family proteins as negative regulators of protein tyrosine kinase signaling: Insights from in vivo models. Commun Integr Biol. 2011;4(2):159–162. doi: 10.4161/cib.4.2.14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naramura M, et al. Mutant Cbl proteins as oncogenic drivers in myeloproliferative disorders. Oncotarget. 2011;2(3):245–250. doi: 10.18632/oncotarget.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy MA, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18(8):4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crowley MR, Bowtell D, Serra R. TGF-beta, c-Cbl, and PDGFR-alpha the in mammary stroma. Dev Biol. 2005;279(1):58–72. doi: 10.1016/j.ydbio.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths EK, et al. Cbl-3-deficient mice exhibit normal epithelial development. Mol Cell Biol. 2003;23(21):7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore F, et al. Abnormal mammary gland development in MMTV-CBLC transgenic mouse. In Vivo. 2009;23(2):225–228. [PubMed] [Google Scholar]
- 17.Naramura M, Nandwani N, Gu H, Band V, Band H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc Natl Acad Sci USA. 2010;107(37):16274–16279. doi: 10.1073/pnas.1007575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naramura M, et al. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol. 2002;3(12):1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 19.Chiang YJ, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403(6766):216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 20.Sleeman KE, et al. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reginato MJ, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5(8):733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 23.Miyake S, Lupher ML, Jr, Druker B, Band H. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc Natl Acad Sci USA. 1998;95(14):7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levkowitz G, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12(23):3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee PS, et al. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 1999;18(13):3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 29.Duan L, et al. Negative regulation of EGFR-Vav2 signaling axis by Cbl ubiquitin ligase controls EGF receptor-mediated epithelial cell adherens junction dynamics and cell migration. J Biol Chem. 2011;286(1):620–633. doi: 10.1074/jbc.M110.188086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Dong T, Pröschel C, Noble M. Chemically diverse toxicants converge on Fyn and c-Cbl to disrupt precursor cell function. PLoS Biol. 2007;5(2):e35. doi: 10.1371/journal.pbio.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, et al. Neuronal Cbl controls biosynthesis of insulin-like peptides in Drosophila melanogaster. Mol Cell Biol. 2012;32(18):3610–3623. doi: 10.1128/MCB.00592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakoub S, et al. The proto-oncoprotein c-Cbl protects cells against oxidative stress by down-regulating apoptosis and is highly expressed in several cancers. J Cancer Sci Ther. 2014;6:122–135. [Google Scholar]
- 33.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen D, et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys. 2011;60(3):173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Axten JM, et al. Discovery of 7-methyl-5-(1-[3-(trifluoromethyl)phenyl]acetyl-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) J Med Chem. 2012;55(16):7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 36.Sandilands E, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2011;14(1):51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- 37.Boczek EE, et al. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc Natl Acad Sci USA. 2015;112(25):E3189–E3198. doi: 10.1073/pnas.1424342112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das J, et al. 2-aminothiazole as a novel kinase inhibitor template. Structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1- piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J Med Chem. 2006;49(23):6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- 39.Hantschel O, Rix U, Superti-Furga G. Target spectrum of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib. Leuk Lymphoma. 2008;49(4):615–619. doi: 10.1080/10428190801896103. [DOI] [PubMed] [Google Scholar]
- 40.Galisteo ML, Dikic I, Batzer AG, Langdon WY, Schlessinger J. Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J Biol Chem. 1995;270(35):20242–20245. doi: 10.1074/jbc.270.35.20242. [DOI] [PubMed] [Google Scholar]
- 41.Haglund K, et al. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5(5):461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 42.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314(17):3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duyvestyn JM, et al. Dasatinib targets B-lineage cells but does not provide an effective therapy for myeloproliferative disease in c-Cbl RING finger mutant mice. PLoS One. 2014;9(4):e94717. doi: 10.1371/journal.pone.0094717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An W, et al. Loss of Cbl and Cbl-b ubiquitin ligases abrogates hematopoietic stem cell quiescence and sensitizes leukemic disease to chemotherapy. Oncotarget. 2015;6(12):10498–10509. doi: 10.18632/oncotarget.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 47.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 48.Welm BE, Dijkgraaf GJP, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2(1):90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhopadhyay C, Zhao X, Maroni D, Band V, Naramura M. Distinct effects of EGFR ligands on human mammary epithelial cell differentiation. PLoS One. 2013;8(10):e75907. doi: 10.1371/journal.pone.0075907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 51.Chen EY, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamoto S, et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470(3):263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 54.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.