Significance
Bacterial natural products (NPs) have served as inspiration for many therapeutics. The hunt for new bioactive NPs has led to a global search for natural ecosystems from which bacteria can be cultured. Here, we used NP-focused metagenome sequencing to explore biosynthetic diversity in urban park soil of New York City. Our analyses reveal rich biosynthetic diversity in these microbiomes and predict that gene clusters encoding many clinically approved NPs families discovered using bacteria cultured from around the world are actually present in the soil microbiomes of a single city. Contrary to traditional NP discovery efforts that involve shallow explorations of diverse environments, our data suggest that a deeper exploration of local microbiomes may prove equally, if not, more productive.
Keywords: natural products, metagenomics, biosynthesis
Abstract
Numerous therapeutically relevant small molecules have been identified from the screening of natural products (NPs) produced by environmental bacteria. These discovery efforts have principally focused on culturing bacteria from natural environments rich in biodiversity. We sought to assess the biosynthetic capacity of urban soil environments using a phylogenetic analysis of conserved NP biosynthetic genes amplified directly from DNA isolated from New York City park soils. By sequencing genes involved in the biosynthesis of nonribosomal peptides and polyketides, we found that urban park soil microbiomes are both rich in biosynthetic diversity and distinct from nonurban samples in their biosynthetic gene composition. A comparison of sequences derived from New York City parks to genes involved in the biosynthesis of biomedically important NPs produced by bacteria originally collected from natural environments around the world suggests that bacteria producing these same families of clinically important antibiotics, antifungals, and anticancer agents are actually present in the soils of New York City. The identification of new bacterial NPs often centers on the systematic exploration of bacteria present in natural environments. Here, we find that the soil microbiomes found in large cities likely hold similar promise as rich unexplored sources of clinically relevant NPs.
Bacterial natural products (NPs) have a rich history of serving as the inspiration for the development of diverse small-molecule therapeutics (1). The discovery of new bioactive bacterial metabolites conventionally begins with the culturing of bacteria from the environment and the subsequent examination of the molecules they produce in pure culture. The search for new bacteria to introduce into NP discovery pipelines has led to a global search for natural ecosystems from which diverse bacteria can be cultured (2, 3). In some instances, molecules isolated from bacteria originally obtained in remote environments have subsequently been found to be produced by organisms acquired closer to home. For example, the macrolactins are a series of antiviral compounds originally isolated from marine microorganisms (4) but later found to be encoded by common root-associated Bacillus species (5). We wondered whether urban park soils might represent rich reservoirs of NP biosynthetic diversity. Here, we used targeted metagenome sequencing to explore the biosynthetic diversity of the urban park soil microbiomes of New York City (NYC). In these studies, phylogenetic analysis of individual next-generation sequencing reads, derived from the conserved biosynthesis genes amplified from environmental DNA (eDNA), was used to predict the gene cluster families present in an environment. Our analysis revealed the presence of rich NP biosynthetic diversity throughout NYC park soils. A comparison of eDNA sequences to gene clusters that encode biomedically important NP families revealed that park soils likely contain bacteria that encode congeners of many biomedically important NPs, including clinically used antibiotics, antifungals, and anticancer agents. Although previous efforts to identify metabolites have focused on the global-scale culturing of bacteria from natural environments, NPs capable of improving human health may lie hidden much closer to home, in the urban park soil microbiomes of our large cities.
Results and Discussion
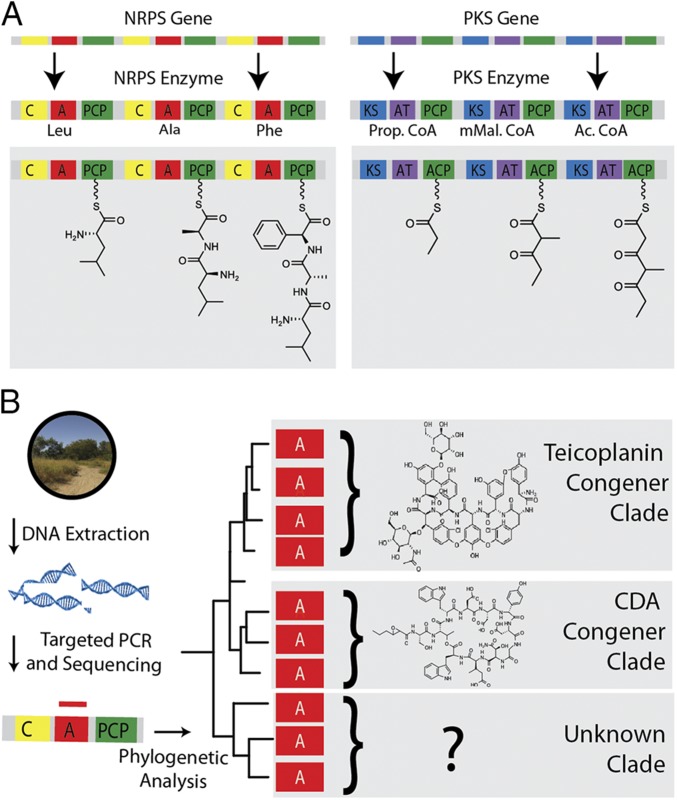
Despite supporting almost 9 million people at one of the highest population densities in the United States (6), the diverse ecosystems found in NYC parks support a large collection of flora and fauna, including 2,000 species of plants and 350 species of birds (7, 8). Microbial diversity surveys indicate that NYC is also home to diverse bacterial and fungal communities (9, 10). Although bacteria produce a tremendous diversity of small molecules, nonribosomal peptide (NRP) and polyketide (PK) biosynthesis is responsible for producing many of the most biomedically important NPs characterized from bacteria (1, 11). Both biosynthetic systems follow the same paradigm wherein molecules are generated in an iterative building process, using enzymes that are composed of conserved domains organized into modules (Fig. 1). A prototypical module contains three domains: one for selecting building blocks, one for connecting building blocks, and one for carrying the growing NP. We used nonribosomal peptide synthetase (NRPS) adenylation (AD) and polyketide synthase (PKS) ketosynthase (KS) domain-specific degenerate primers (12, 13) to amplify biosynthetic domains from eDNA isolated from 275 top soils collected across the five boroughs of NYC. (Fig. 2 A and B and Dataset S1) Phylogenetic analysis of these amplicons was used to assess the NP biosynthesis in urban park soils.
Fig. 1.
Surveying NP biosynthesis by phylogenetic analysis of PCR amplicons. (A) In NRP and PK biosynthesis, NPs are produced by megasynth(et)ases in thio-templated assembly line fashion. Each module in a megasynth(et)ase is composed of a highly conserved set of domains and is responsible for the incorporation of one amino acid or coenzyme A (CoA)-based building block into the growing NP. (B) Phylogenetic analysis of domain fragments PCR-amplified from eDNA can be used to study biosynthetic diversity hidden in soil microbiomes.
Fig. 2.
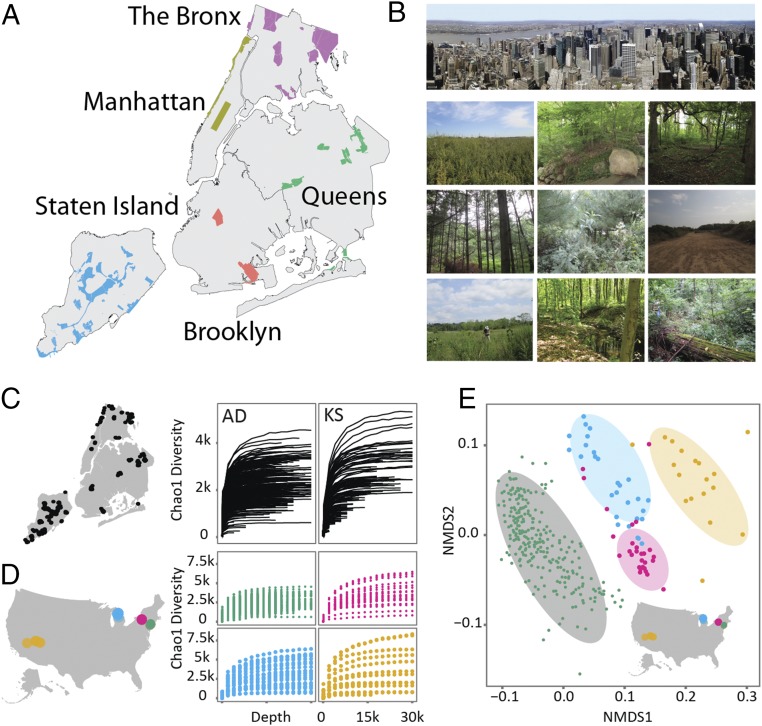
AD and KS domain analysis in soils from diverse NYC parks ecotypes. The five boroughs of NYC contain a large collection of parks, many of which contain both heavily groomed and “natural” environments. (A) Soil samples were collected from each of the parks marked on the NYC map. (B) Natural environments found in these parks represent diverse ecotypes. (C) AD and KS domain richness was determined for 275 different NYC park soils using Chao1-based rarefaction analysis of domain fragment PCR-amplified directly for eDNA. AD domains are responsible for selecting amino acid building blocks used in NRP biosynthesis, whereas KS domains are responsible for the formation of carbon−carbon bonds between CoA building blocks in PKS biosynthesis. (D) NYC AD domain richness (green) is compared with soils collected in nonurban areas of upstate New York (pink), Michigan (blue), and the western United States (yellow). (E) NMDS analysis of AD domains sequenced from soils collected in NYC parks and nonurban areas of upstate New York, Michigan, and the western United States.
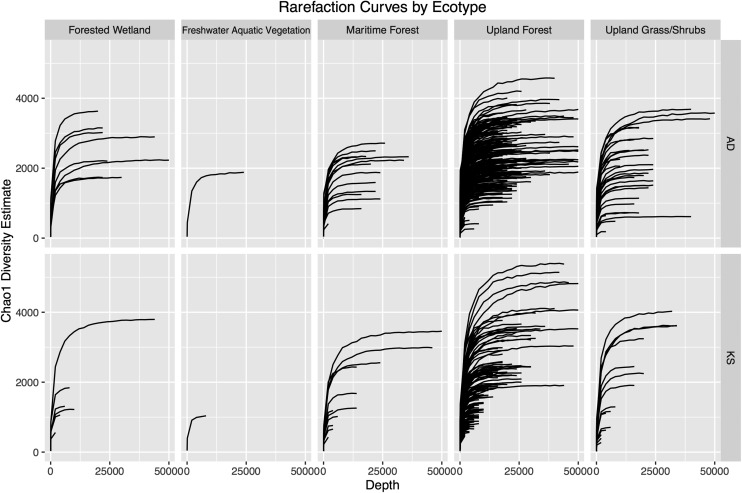
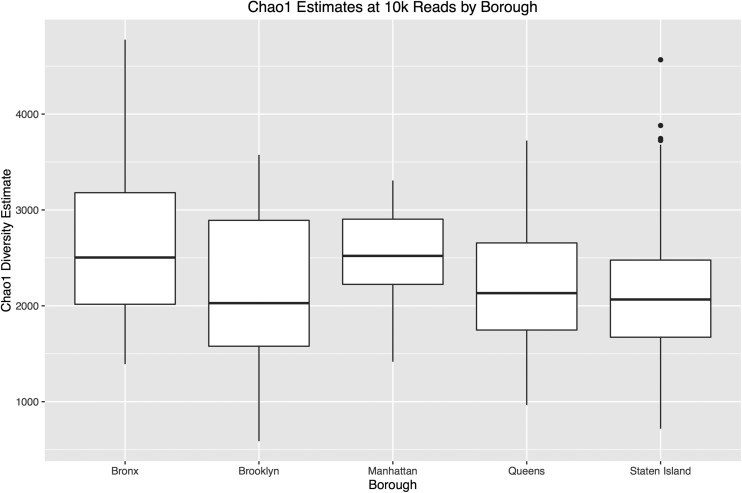
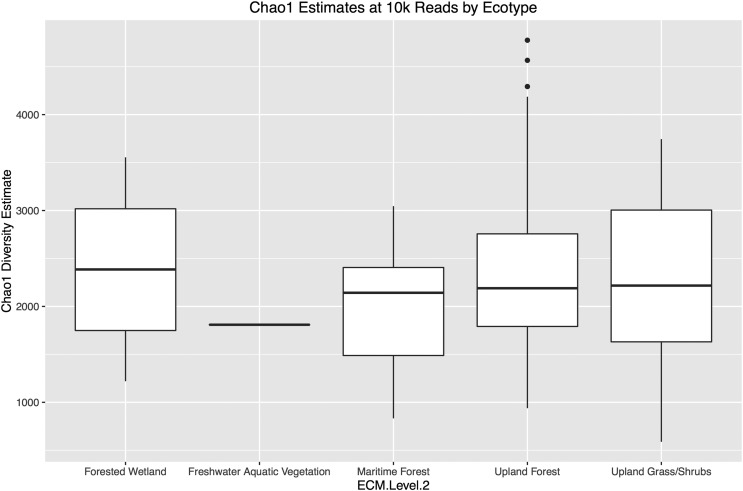
AD and KS domain operational taxonomic unit (OTU) diversity estimates were calculated for each site using Chao1 rarefaction methods. This analysis suggests the presence of thousands of unique AD and KS domain sequences in each park soil. Although diversity estimates varied from site to site (Fig. 2C), we did not observe significant differences between ecotypes or boroughs (Figs. S1–S3). Maritime Forest samples had the fewest predicted OTUs, and Bronx samples had the most; however, these differences were small and, in the case of ecotype-based differences, were strongly influenced by a few outliers.
Fig. S1.
Rarefaction curves shown in Fig. 2C broken down by ecotype.
Fig. S3.
Chao1 diversity estimates for samples sorted by borough using a sampling depth of 10,000 reads. The average estimated diversity obtained from 10 independent samplings is shown.
Fig. S2.
Chao1 diversity estimates for samples sorted by ecotype using a sampling depth of 10,000 reads. The average estimated diversity obtained from 10 independent samplings is shown.
For comparison purposes, we amplified and sequenced AD domains from eDNA isolated from 96 nonurban US soils, which were processed in the same manner as described above. Rarefaction analysis indicates slightly lower, but comparable, domain diversity in urban versus nonurban soil environments (Fig. 2D). To compare the NYC-derived AD domain sequences with those from other regions of the United States, we performed a nonmetric multidimensional scaling (NMDS) ordination analysis of intersample distances. In the NMDS ordination plot, AD domain sequences from samples collected in the four different areas (NYC, upstate New York, Midwest, and West) cluster into distinct groups (Fig. 2E). Similar geographic distance-dependent differences in species beta diversity as well as NP biosynthetic diversity have been observed in other environmental surveys of bacterial species (14, 15) (16, 17). It is still unclear whether these differences arise from selection in the environment or if they indicate limits to the natural dispersion of microorganisms over long distances. Taken together, the alpha and beta diversity analyses suggest that the NYC park soils are rich in biosynthetic diversity and that the collections of biosynthetic domains in these samples are distinct from nonurban environments.
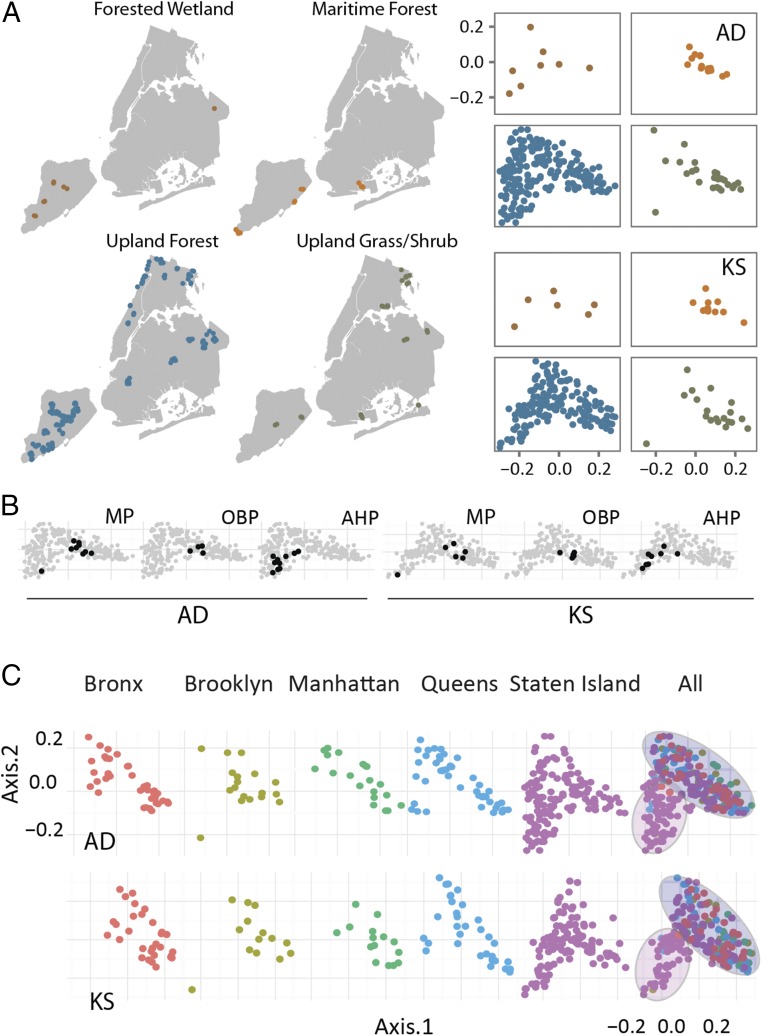
To look for relationships among populations of biosynthetic domain sequences derived from different urban park soils, we performed a principal component analysis (PCA) of AD and KS OTUs. Both ecotype and geography-dependent sample clustering patterns were present in a 2D PCA plot where the axes represent the first two principal components (Fig. 3). We also observed a correlation between AD and KS domain relationship patterns in the PCA plot. We believe this correlation can be explained by the fact that a particular sequence variant of a gene cluster is often specific to one strain and, therefore, specific AD and KS domain sequences would be expected to cosegregate with a specific strain or species in the environment.
Fig. 3.
PCA plots of New York Soil AD and KS domain intersample distances. Sample distances derived from KS and AD OTU tables were used to perform a PCA to look for both ecotype- and borough-based relationships. (A) Sample collection sites for each ecotype are marked on maps of NYC. Ecotype-specific PCA plots for AD and KS domain sequences reveal clustering of samples from some ecotypes. (B) Similar clustering is observed for samples from some parks, especially in the case of Marine Park (MP), Ocean Breeze Park (OBP), and Arden Heights Preserve (AHP). (C) Borough-specific PCA plots show a cluster of samples from all five boroughs as well as a cluster of samples that is almost exclusively derived from Staten Island.
When annotated according to ecotype (Fig. 3A), samples derived from the Maritime Forest and Upland Grass environments cluster together on the PCA plot, whereas the Upland Forest ecotype, which contains the majority of samples in NYC, is found throughout the plot. This finding suggests that the grouping defined by the Maritime Forest and Upland Grass delineates environments with largely similar microbiomes, whereas the Forest ecotype grouping does not. At the park level, we observed clustering of samples collected within a number of parks (Fig. 3B). Although this is not the case for all parks, it suggests that, as seen in nonurban environments, geography might be a factor in domain population differences. We therefore expanded our geographic analysis to look for borough-based clustering of domain populations. This analysis revealed a distinct cluster consisting almost exclusively of samples collected in Staten Island, seen at the bottom left hand corner of both the AD and the KS domain PCA plots in Fig. 3C. The remainder of the PCA plot shows an intermingling of samples collected throughout the five boroughs. These results indicate that Staten Island appears to contain not only soils that resemble those seen in the other four boroughs but also a set of soils that is biosynthetically distinct from most samples collected throughout the rest of the city (Fig. 3C). This is perhaps not surprising, as it is the least populated and most suburban of the boroughs and is separated from the rest of the city by the New York Bay.
Although the PCA analysis was able to detect patterns of OTU populations, the first two principle components, which were used to construct the PCA plots, account for less than 10% of the variance between samples, indicating significant differences in populations of AD and KS domain amplicons even from physically close samples. This finding suggests that soil microbiomes may encode geographically distinct core secondary metabolomes (Figs. 2 and 3). Furthermore, the large sample-to-sample variation we observed suggests that urban park environments will be rewarding starting points for NP discovery. Based on the domain richness and diversity, our data suggest that, in NYC, future urban NP discovery efforts should focus particular attention on collection sites in Staten Island.
To facilitate the assignment of environmental sequences to natural gene cluster families of interest, we previously developed the web-based analysis platform eSNaPD (environmental Surveyor of Natural Product Diversity) (18, 19). In brief, eSNaPD classifies amplicon sequences by using BLAST to compare them to a curated dataset of known gene clusters, as well as to all other AD and KS domain sequences found in GenBank. This BLAST analysis identifies environmental domains that are more closely related to a curated gene cluster of interest than to any other biosynthetic sequence in GenBank and is experimentally similar to cataloging bacterial species in a metagenome using 16S rRNA sequences (19). We have shown eSNaPD classification to be a robust indicator of a functional relationship between a microbiome-derived gene cluster yielding an AD or KS domain sequence tag and a curated known gene cluster (20–22). Based on empirical data from previous metagenomic analyses (20–23), the homology cutoffs used in this study (e value < 10−100) will identify gene clusters with a high likelihood of encoding either the same metabolite or a novel derivative (congener) of the metabolite encoded by the matching curated gene cluster.
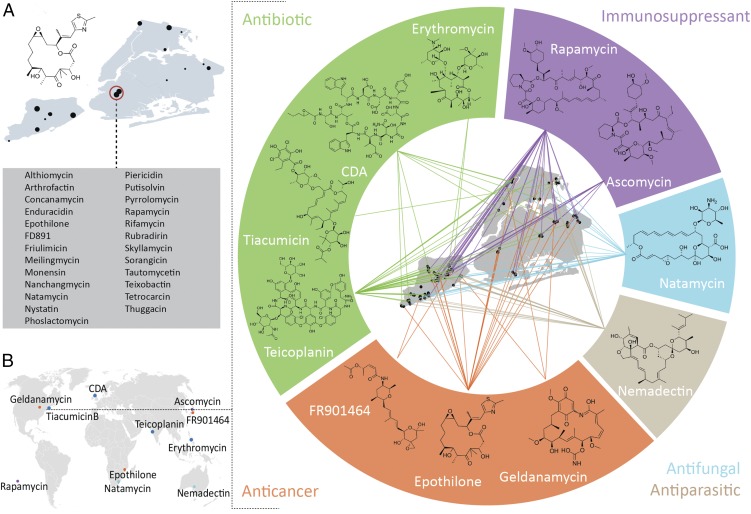
eSNaPD profiling of biosynthetic domain sequences from New York park soil microbiomes revealed AD and KS domain sequences that map to a diverse collection of gene cluster families that encode bioactive NPs. These data can be used to track the distribution of individual, biomedically relevant biosynthetic families across NYC park soil microbiomes. For example, Fig. 4A shows the distribution of metagenome-derived amplicon sequences assigned by eSNaPD to the epothilone gene cluster. Epothilone is a PKS-derived anticancer agent that interferes with microtubule formation. The epothilone analog, ixabepilone, is approved for use as a treatment for metastatic breast cancer (24). Alternatively, the NP-encoding capacity of a metagenome at a specific collection site can be profiled using domain sequence data. As an example of this, Fig. 4A lists a subset of biosynthetically and biomedically interesting gene cluster families detected by eSNaPD in one urban park, Prospect Park in Brooklyn.
Fig. 4.
Distribution of biomedically relevant NP gene cluster families in New York Park soils. (A) Park sites with domain sequences related to the biosynthetic gene cluster for the anticancer agent epothilone are shown as a representative chemical biographic map of NYC. Metagenomic AD and KS domain sequences related to gene clusters that encode the biosynthesis of the bioactive NPs listed here were found in a single soil collected from Prospect Park in Brooklyn. (B) Original collection sites for the bacteria that produce 11 therapeutically relevant NPs are shown on the world map. Metagenomic sequencing of biosynthetic domains found in NYC park soils indicates that bacteria containing gene clusters capable of encoding either these NPs or close congeners of these NPs are likely widely distributed in park soils from a single city. Original discovery sites for molecules included on the world map in B were obtained from the following references: epothilone (32), rapamycin (33), teicoplanin (34, 35), erythromycin (36), CDA (calcium-dependent antibiotic) (37), tiacumicin B (38), FR910464 (39), ascomycin (40), natamycin (41), geldanamycin (42), nemadectin (43, 44).
In an effort to better understand how NYC’s metagenomic biosynthetic diversity might compare with historical fermentation-based efforts to identify therapeutically relevant NPs, we mapped AD and KS domain sequences to gene clusters that encode 11 therapeutically relevant NPs. These 11 NPs include clinically approved anticancer, antibacterial, immunosuppressive, antifungal, and antiparasitic agents that were originally discovered using bacteria cultured from natural environments found all over the world (Fig. 4B, Inset). The distribution of eDNA-derived domain sequences that map to gene clusters that encode these NPs is shown in Fig. 4B. Remarkably, our domain sequence data suggest biosynthetic gene clusters with the potential to encode either these specific NPs or their close congeners likely lie hidden in the natural areas of a single city. Less than 1% of reads are assigned to a gene cluster using our existing eSNaPD database. Although this number will undoubtedly increase as more gene clusters are annotated and more annotated gene clusters are added to the eSNaPD database, it suggests the presence of a large reservoir of unknown gene clusters in these environments. Previous efforts to identify NPs with therapeutically relevant activities have largely focused on the worldwide collection and screening of bacteria cultured from natural environments. Our analyses of urban microbiomes show the existence of tremendous biosynthetic diversity throughout urban park soils, suggesting an equal effort should be applied to studying and cataloging urban microbiomes, as they appear to encode diverse, potentially biomedically relevant NPs. Because of its high population density and its status as an important point of entry for foreign visitors, NYC is an epicenter of infectious diseases in the United States. Interestingly, our metagenomic sequence tag analysis suggests that natural cures to many of these diseases may lie hidden in the NYC park soil microbiome.
Conclusions
The sequencing of DNA extracted directly from environmental isolates allows for the NP-encoding capacity of soil microbiomes to be explored in greater detail than with culture-based methods. Our data suggest that even environments such as small urban parks harbor extensive and largely unexplored chemical diversity. The identification of specific soils that are particularly rich in biosynthetic diversity should help guide the identification of productive starting points for future novel NP discovery efforts. We show that many gene cluster families that were first found in samples collected in disparate environments around the world are predicted to be present in the collective soil microbiome of a single city. Although we examined urban soil environments in this study, it is likely that similar observations would hold true for many complex environmental microbiomes. Our results suggest that it may prove more productive to focus on a deeper exploration of individual environmental samples in the search for new therapeutically relevant NPs, instead of scratching the surface of samples obtained from a large number of environments.
Materials and Methods
Soil Collection and DNA Isolation.
In collaboration with the Natural Areas Conservancy (NAC), the NYC Department of Parks and Recreation, and the Rockefeller University Summer Science Outreach Program, we collected 275 topsoil samples from parks located throughout NYC’s five boroughs (Fig. 2A). Collection sites were divided into five ecotypes based on the NAC’s preestablished Ecological Covertype Map of NYC, a map derived primarily from satellite imagery of the vegetation at each park site (25, 26). The five ecotypes used in this study—Upland Grass/Shrubs, Maritime Forest, Upland Forest, Forested Wetland, and Freshwater Aquatic Vegetation—represent high-level classifications that reflect the broad ecological patterns found in NYC parks (Fig. 2B). Soils were collected from existing forest assessment plots in each park, prioritizing parks where at least three distinct plots existed per ecotype. A soil core was used to collect 30 g of soil from within 15 cm of the soil surface at each site. DNA was extracted from 0.25 g of soil using the 96-well PowerSoil-htp soil DNA isolation kit (MoBio) according to the manufacture’s instructions. Sample collection information appears in Dataset S1. Soils in this study are largely collected from areas with parks that had been designated as natural areas by the NYC Department of Parks and Recreation under their Forever Wild initiative (New York Parks Forever Wild Initiative, https://www.nycgovparks.org/greening/nature-preserves).
Degenerate Primer PCR.
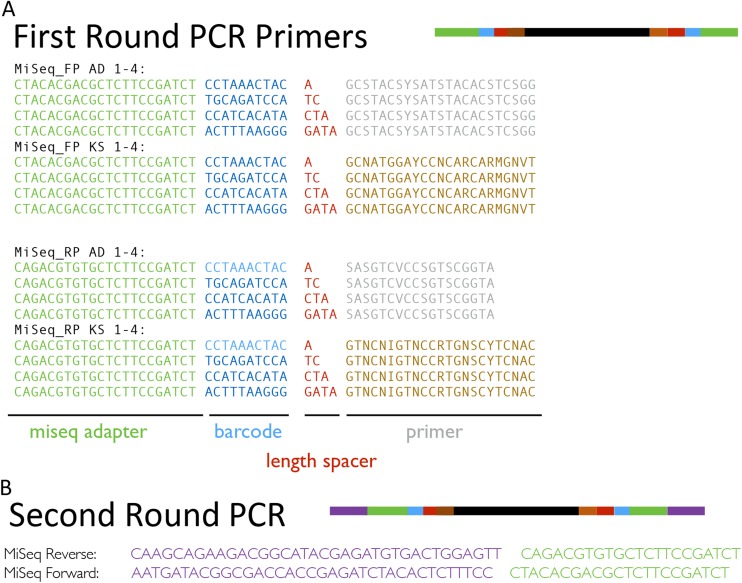
Degenerate primer pairs targeting NRP AD domains [A3F (5′-GCSTACSYSATSTACACSTCSGG) and A7R (5′-SASGTCVCCSGTSCGGTA)] (12) and PK KS domains [degKS2F (5′-GCNATGGAYCCNCARCARMGNVT) and degKS2R (5′-GTNCCNGTNCCRTGNSCYTCNAC)] (27) were used to PCR-amplify biosynthetic domains from each eDNA sample. To permit parallel sequencing of amplicons from each sample, we adopted a primer design strategy in which both forward and reverse primers contained barcodes that, together, can be used to uniquely identify a sample. Primers contained the invariant Illumina p5 or p7 sequence, an 8-bp barcode sequence, a spacer sequence needed to minimize the “strobe” effect of sequencing amplicons by phasing the amplicon sequences and the degenerate primer (Fig. S4) (28). After the first round of PCR, each amplicon contained two 8-bp barcodes that were used to uniquely identify the source of the amplicon. The 40-µL PCR reactions were set up as follows: 20 µL of FailSafe PCR Buffer G (AD) or Buffer E (KS) (Epicentre), 1 µL of Taq Polymerase (Bulldog Bio), 1.25 µL of each primer (100 μM), 14.5 µL of water and 2 µL of purified eDNA. Amplification conditions for AD domain primers were as follows: 95 °C for 4 min followed by 40 cycles of 94 °C for 30 s, 67.5 °C for 30 s, 72 °C for 1 min, and, finally, 72 °C for 5 min. Amplification conditions for KS domain primers were as follows: 95 °C for 4 min followed by 40 cycles of 94 °C for 40 s, 56.3 °C for 40 s, 72 °C for 75 s, and, finally, 72 °C for 5 min.
Fig. S4.
Example of PCR primer design strategy. The primer design strategy we used to produce AD and KS domain amplicons is shown pictorially. Each first-round amplicon incorporates a portion of the Illumina adaptor sequence and a unique 8-bp barcode on each end (Forward primer, MiSeq_FP; Reverse primer, MiSeq_RP) and a variable length spacer. The second round of amplification provides the remainder of the Illumina sequence. This amplicon barcoding approach is modeled after a strategy outlined by Fadrosh et al. (28).
Second-Round PCR for Sequencing.
First-round amplicons contained incomplete Illumina adaptors and therefore required a second round of PCR to append the remainder of the adaptor sequence. First-round amplicons were pooled as collections of 96 samples and cleaned using Agencourt Ampure XP magnetic beads (Beckman Coulter). Cleaned, pooled amplicons were used as template in a second 20-µL PCR using the following reaction conditions: 10 µL of FailSafe Buffer G (Epicenter), 5.8 µL of water, 0.4 µL of each primer (100 µM) (MiSeqForward, CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT; MiSeq Reverse AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT), 0.4 µL of Taq, and 3 µL of cleaned amplicon (50 ng to 100 ng). Amplification proceeded as follows: 95 °C for 5 min, six cycles of 95 °C for 30 s, 70 °C for 30 s, and 72 °C for 45 s, and, finally, 72 °C for 5 min.
Processing of KS and AD Domain Sequences.
Second-round PCR amplicons were cleaned twice with Agencourt Ampure XP magnetic beads (0.6:1 bead volume to DNA solution) and sequenced using Illumina MiSeq 2 × 300 technology (602 cycles: 301 × 301). Separate sequencing runs were performed for pooled AD and KS domain amplicons yielding 28 × 106 or 33 × 106 clusters, respectively. Reads were demultiplexed using a publicly available python package for debarcoding paired-end reads (https://github.com/zachcp/paired-end-debarcoder). The fastq files corresponding to the forward and reverse reads were split into sample specific fastq files based on the unique 16-bp barcode that was created by concatenating the first 8 bp of the forward read and the first 8 bp of the reverse read. Using the trimfq command in seqtk (29), quality bases were removed each read. Forward reads shorter than 240 bp and reverse reads shorter than 175 bp were removed. The remaining forward and reverse reads were trimmed to 240 bp and 175 bp, respectively. Paired end reads were concatenated using a single “N” spacer between the forward read and the reverse complement of the second read. A single fasta file of debarcoded, quality-filtered sequences of uniform length (416 bp) was created for each sample.
Clustering of Concatenated Sequences.
Concatenated reads were clustered using a variation of the UPARSE clustering pipeline (30). For each sample, reads were dereplicated and clustered at 97% identify using UPARSE. After the first round of clustering, all remaining singletons were discarded. In a second round of clustering, 97% centroid sequences from all samples were pooled and clustered at 95% identity using Usearch cluster_fast (30). Membership in these 95% identity clusters was used to construct a combined OTU table in Phyloseq (31).
Richness and Relationship Analyses.
Rarefaction.
AD and KS rarefaction curves were generating by subsampling the OTU table at regular intervals: 1 × 101, 1 × 102, 1 × 103, and every multiple of 1 × 103 until 30 × 103. The Chao1 diversity metric was used to predict the diversity at each depth (31). At each depth, we independently subsampled and calculated the Chao1 diversity metric 10 times and reported the mean of these 10 iterations.
NMDS ordination.
To compare geographically distant samples (i.e., NYC and non-NYC samples), we performed an ordination analysis of the OTU table using Bray−Curtis distances (31). For this analysis, we removed any OTUs that were not present in at least three samples and any samples that did not have at least 1,000 reads after the rare OTUs were removed. Read counts in a sample were normalized to be a fraction of total reads in the sample. NMDS ordination analysis of the normalized table was carried out using phyloseq’s ordinate command (31).
MDS ordination.
We used PCA to assess patterns within NYC samples. For this analysis, OTUs not present in two or more samples were discarded, and samples were normalized by read count. Principle component analysis was performed on the intersample distance matrix calculated with the Bray−Curtis distance metric. PCA plots show the first two principal components that account for 6.6% (Axis 1, 3.9%; Axis 2, 2.7%) and 7.7% (Axis 1, 4.4%; Axis 2, 3.3%) of the AD and KS domain data, respectively.
Assignment of AD and KS Domains to Known Gene Clusters.
Gene clusters predicted to encode medically important families of NPs are common in the urban park soil microbiome. Only a small fraction of sequences derived from most soil metagenomic sequencing efforts are closely related to functionally characterized genes; this is also true of our NYC AD and KS sequence data, which suggests not only that urban microbiomes encode structurally and functionally diverse NPs but that many of these NPs likely differ from known NPs. When using PCR amplicon data to study NP biosynthesis, the small numbers of sequences that are closely related to well-characterized clusters are informative, as they allow for detailed predictions about the types of metabolites that are encoded in soil microbiomes. AD and KS amplicons were assigned to known biosynthetic gene clusters using the eSNaPD program (18, 19). Empirical exploration with the eSNaPD algorithm has shown that e values as high as 10−40 to −60 return reliable gene cluster predictions (20–22). We used an expectation value to 10−100 for this analysis. The eSNaPD 2.0, populated with sequence data deposited in GenBank as of 2014, was used in this study. All hits displayed in Fig. 4 were manually validated by BLAST analysis against GenBank.
Supplementary Material
Acknowledgments
We thank the students of Rockefeller University's Summer Science Research Program and Barnard College’s Summer Research Institute for collecting soil samples. This work was supported by National Institutes of Health Grants U01 GM110714 (to S.F.B.) and Al110029 (to Z.C.-P.); National Science Foundation Coastal Science, Engineering, and Education for Sustainability (SEES) Grant 1325185 (to K.L.M.); and grants to the Natural Areas Conservancy (C.C.P., H.M.F., and S.C-P.) from the Doris Duke Charitable Foundation, The Mayor's Fund to Advance New York City, and Tiffany & Co.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the BioProject database (accession no. PRJNA338196).
See Commentary on page 14477.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615581113/-/DCSupplemental.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 3.Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot (Tokyo) 2012;65(8):441. doi: 10.1038/ja.2012.54. [DOI] [PubMed] [Google Scholar]
- 4.Gustafson K, Roman M, Fenical W. The macrolactins, a novel class of antiviral and cytotoxic macrolides from a deep-sea marine bacterium. J Am Chem Soc. 1989;111:7519–7524. [Google Scholar]
- 5.Chen XH, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25(9):1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 6.United States Census Bureau 2016 Quick Facts: New York City, New York, (US Census Bureau, Washington, DC). Available at www.census.gov/quickfacts/table/PST045215/3651000. Accessed August 1, 2016.
- 7.Kiviat E, Johnson EA. Biodiversity Assessment Handbook for New York City. Hudsonia Ltd.; Annandale-on-Hudson, NY: 2013. [Google Scholar]
- 8.Johnson EA. Legacy: Conserving New York State’s Biodiversity. New York State Biodiversity Project; New York: 2006. [Google Scholar]
- 9.Ramirez KS, et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc Biol Sci. 2014;281(1795):20141988. doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire KL, et al. Digging the New York City Skyline: Soil fungal communities in green roofs and city parks. PLoS One. 2013;8(3):e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cragg GM, Newman DJ. Natural products: A continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: Detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49(1):10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- 13.Schirmer A, et al. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol. 2005;71(8):4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA. 2011;108(19):7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chust G, et al. Dispersal similarly shapes both population genetics and community patterns in the marine realm. Sci Rep. 2016;6:28730. doi: 10.1038/srep28730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlop-Powers Z, Owen JG, Reddy BV, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Proc Natl Acad Sci USA. 2014;111(10):3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charlop-Powers Z, et al. Global biogeographic sampling of bacterial secondary metabolism. eLife. 2015;4:e05048. doi: 10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy BV, Milshteyn A, Charlop-Powers Z, Brady SF. eSNaPD: A versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem Biol. 2014;21(8):1023–1033. doi: 10.1016/j.chembiol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen JG, et al. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc Natl Acad Sci USA. 2013;110(29):11797–11802. doi: 10.1073/pnas.1222159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang HS, Brady SF. Arixanthomycins A−C: Phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem Biol. 2014;9(6):1267–1272. doi: 10.1021/cb500141b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen JG, et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc Natl Acad Sci USA. 2015;112(14):4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang FY, Ternei MA, Calle PY, Brady SF. Targeted metagenomics: Finding rare tryptophan dimer natural products in the environment. J Am Chem Soc. 2015;137(18):6044–6052. doi: 10.1021/jacs.5b01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallifidas D, Kang HS, Brady SF. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc. 2012;134(48):19552–19555. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Center for Biotechnology Information . PubChem Compound Summary for CID 6445540. Nat Cent Biotechnol Info; Bethesda, MD: 2016. [Google Scholar]
- 25.O’Neil-Dunne JPM, MacFaden SW, Forgione HM, Lu JWT. Urban Ecological Land Cover Mapping for New York City. Spatial Informatics Group; Burlington, VT: 2014. [Google Scholar]
- 26.Forgione HM, Pregitzer CC, Charlop-Powers S, Gunthier B. Advancing urban ecosystem governance in New York City: Shifting towards a unified perspective for conservation management. Environ Sci Policy. 2016;62:127–132. [Google Scholar]
- 27.Schirmer A, et al. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol. 2005;71(8):4840–4849. doi: 10.1128/AEM.71.8.4840-4849.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fadrosh DW, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H. seqtk: A Fast and Lightweight Tool for Processing Sequences. Broad Inst; Cambridge, MA: 2016. [Google Scholar]
- 30.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 31.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerth K, Bedorf N, Höfle G, Irschik H, Reichenbach H. Epothilons A and B: Antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). Production, physico-chemical and biological properties. J Antibiot (Tokyo) 1996;49(6):560–563. doi: 10.7164/antibiotics.49.560. [DOI] [PubMed] [Google Scholar]
- 33.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28(10):721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 34.Bardone MR, Paternoster M, Coronelli C. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J Antibiot (Tokyo) 1978;31(3):170–177. doi: 10.7164/antibiotics.31.170. [DOI] [PubMed] [Google Scholar]
- 35.McCormick MH, McGuire JM, Pittenger GE, Pittenger RC, Stark WM. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot Annu. 1955-1956;3:606–611. [PubMed] [Google Scholar]
- 36.Bunch RL, Mcguire JM. 1953. US Patent US 2653899A.
- 37.Hopwood DA. Forty years of genetics with Streptomyces: From in vivo through in vitro to in silico. Microbiology. 1999;145(Pt 9):2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- 38.Theriault RJ, et al. Tiacumicins, a novel complex of 18-membered macrolide antibiotics. I. Taxonomy, fermentation and antibacterial activity. J Antibiot (Tokyo) 1987;40(5):567–574. doi: 10.7164/antibiotics.40.567. [DOI] [PubMed] [Google Scholar]
- 39.Nakajima H, et al. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 1996;49(12):1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 40.Hatanaka H, Iwami M, Kino T, Goto T, Okuhara M. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. I. Taxonomy of the producing strain. J Antibiot (Tokyo) 1988;41(11):1586–1591. doi: 10.7164/antibiotics.41.1586. [DOI] [PubMed] [Google Scholar]
- 41.Struyk AP, et al. Pimaricin, a new antifungal antibiotic. Antibiot Annu. 1957−1958;5:878–885. [PubMed] [Google Scholar]
- 42.BeBoer C, Dietz A. The description and antibiotic production of Streptomyces hygroscopicus var. Geldanus. J Antibiot (Tokyo) 1976;29(11):1182–1188. doi: 10.7164/antibiotics.29.1182. [DOI] [PubMed] [Google Scholar]
- 43.Carter GT, et al. LL-F28249 antibiotic complex: A new family of antiparasitic macrocyclic lactones. Isolation, characterization and structures of LL-F28249 alpha, beta, gamma, lambda. J Antibiot (Tokyo) 1988;41(4):519–529. doi: 10.7164/antibiotics.41.519. [DOI] [PubMed] [Google Scholar]
- 44.Carter GT, Torrey MJ, Greenstein M. 1995. US Patent 5,418,168.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.