Abstract
Eukaryotic RNA polymerase (Pol) III is recruited to target promoters by a stable preinitiation complex containing transcription factors TFIIIC and TFIIIB. After the first transcription cycle, reinitiation proceeds through facilitated recycling, a process by which the terminating Pol III rapidly reloads onto the same transcription unit. Here, we show that Pol III is repeatedly recaptured in vitro by the first transcribed gene, even in the presence of a juxtaposed competitor promoter complex, thus suggesting that facilitated recycling is not merely due to a stochastic reassociation process favored by the small size of class III genes. The transcription factor requirements for facilitated reinitiation were investigated by taking advantage of Pol III templates that support both TFIIIC-dependent and TFIIIC-independent transcription. A TFIIIC-less transcription system, in which TFIIIB was reconstituted from recombinant TATA box-binding protein and Brf1 proteins and a crude fraction containing the Bdp1 component, was sufficient to direct efficient Pol III recycling on short (≈100 bp) class III genes. Unexpectedly, however, on longer (>300 bp) transcription units, reinitiation in the presence of TFIIIB alone was compromised, and TFIIIC was further required to reestablish a high reinitiation rate. Transcription reinitiation was also severely impaired when recombinant Bdp1 protein replaced the corresponding crude fraction in reconstituted TFIIIB. The data reveal an unexpected complexity in the Pol III reinitiation mechanism and suggest the existence of a handing-back network between Pol III, TFIIIC, and TFIIIB on actively transcribed class III genes.
Transcription reinitiation accounts for the bulk of RNA synthesis in living cells, and it appears to involve dedicated strategies in most transcription systems (1). In eukaryotes, the highest known reinitiation efficiency is attained by the RNA polymerase (Pol) III transcription system, devoted to the production of very abundant, noncoding RNAs, such as the tRNAs, the 5S rRNA, the U6 small nuclear RNA, and the 7SL RNA. Such a property can be reproduced in vitro, thus rendering the Pol III system the most suitable for the study of basic questions relating to transcription reinitiation and Pol recycling (1). In yeast, tRNA gene activation starts with the assembly of a preinitiation complex (PIC), comprising the two multiprotein transcription factors, TFIIIB and TFIIIC, that is stable enough to direct multiple rounds of transcription by Pol III (2). In addition, as shown by template competition experiments, the first transcription cycle on a given gene commits Pol III to reinitiate more rapidly on that gene, with the exclusion of a competitor template primed to recruit free Pol III (3). The molecular mechanism of Pol III-facilitated recycling has not been investigated in detail. This process is likely to involve a direct coupling between termination and reinitiation, as shown by the fact that run-off termination on truncated class III genes does not allow efficient recycling (3), and by the recent observation that a peptide nucleic acid roadblock downstream of the terminator interferes with reinitiation (4). Because class III genes are generally very small (≈100 bp), facilitated recycling might result mainly from the fact that the terminating polymerase is still in the vicinity of the PIC, and thus has a high probability of reattaching to the same transcription unit in a purely stochastic process. Alternatively, a specific reloading mechanism could maintain Pol III in contact with promoter-bound factors during consecutive transcription cycles (2). The transcription factor requirement for facilitated Pol III recycling has also not been investigated in detail. Previous work in yeast (3, 5, 6) has shown that TFIIIB alone, assembled upstream of the transcription start site, possess the ability to direct multiple cycles of Pol III transcription on the tRNA and U6 small nuclear RNA genes. These observations support the notion of TFIIIC as an assembly factor whose main role is to promote the assembly of TFIIIB, and that becomes thus dispensable for transcription once a TFIIIB–DNA complex has been formed. Other evidence in the human system, however, suggests that activities other than TFIIIB, such as the La autoantigen (7) or the TFIIIC-interacting NF1 proteins (8), have the potential to influence Pol III termination and reinitiation.
In this study, a reconstituted Pol III transcription system from Saccharomyces cerevisiae, capable of highly efficient reinitiation in vitro, has been used to address the molecular mechanism and transcription factor requirements of Pol III transcription reinitiation. The results suggest that the high efficiency of Pol III reinitiation rests on a complex, transcription factor-dependent polymerase recapture pathway.
Materials and Methods
DNA Templates. The construction of the DNA templates used in this study is described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Transcription Proteins and in Vitro Transcription Assays. In vitro transcription reaction conditions and procedures for RNA purification and analysis were essentially as described (5). In all transcription experiments, both TFIIIC-containing and TFIIIC-less PICs were assembled by incubating the template DNA (20 fmol of supercoiled plasmid) with transcription proteins at 20°C for 20 min. The following amounts of transcription proteins were used: 150 ng of TFIIIC purified up to the DEAE-Sephadex A-25 step (or 50 ng of tDNA-affinity-purified TFIIIC) (9); 40 ng of pure, recombinant TATA box-binding protein (rTBP) and 80 ng of rBrf1, which were both purified from overexpressing Escherichia coli cells (9); and 0.5 μg of B′′ fraction, which was partially purified, up to the Bio-Rex 70 chromatography step, from chromatin pellets generated during yeast nuclear extract preparation (10). As estimated by SDS/PAGE analysis followed by Coomassie blue staining, this amount of crude fraction provided no more than 20 ng of Bdp1 polypeptide. With these amounts of transcription proteins, ≈5 fmol of active PICs were assembled on plasmid-borne tRNA genes, as evaluated from accurate quantification of the output of single-round transcription reactions (11). rBdp1 was purified from a crude extract of High Five cells (Invitrogen) infected with a baculovirus encoding 8his-Bdp1 (12). S. cerevisiae Pol III was purified according to published procedures (9). The details of both rBdp1 and Pol III purification are reported in Supporting Materials and Methods. Stable ternary complexes were formed by incubating PICs with Pol III and the appropriate sets of three NTPs for 10 min at 20°C. In most experiments, 10 ng of purified Pol III was used, corresponding to a limiting concentration of the enzyme (based on titration experiments, conducted under multiple round transcription conditions, in which Pol III concentration was varied in the presence of fixed amounts of the other components). In the competition experiments with physically linked U6 and tDNA genes, the plasmid was first incubated 20 min at 20°C with the above reported amounts of rTBP, rBrf1, and B′′ fraction. Limiting Pol III was then added together with an NTP mixture lacking ATP; after that, PICs were allowed to assemble on tDNAGly by adding TFIIIC and TFIIIB components in the above reported amounts, and the competition was started 20 min later by the addition of ATP. All of the transcription assays were performed at 500 μM ATP, CTP, GTP, and 100 μM UTP, except for tDNAIle(TAT) transcription, that was carried out in the presence of 25 μMUTP. Heparin at a 200 μg/ml concentration was used in single-round transcription reactions.
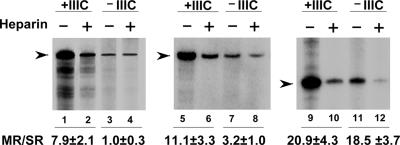
Results
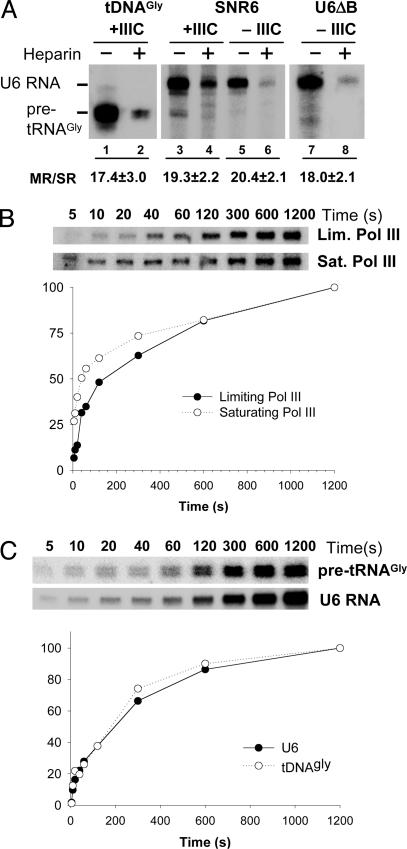
Facilitated Pol III Reinitiation on Different Class III Genes. On most yeast tRNA genes (tDNAs), TFIIIB can only be recruited through a TFIIIC-mediated pathway, whereas the SNR6 gene (coding for the U6 small nuclear RNA), both in its wild type (wt) form and in a mutant version lacking the extragenic B block promoter element (U6ΔB), can be efficiently transcribed in the absence of TFIIIC in purified in vitro systems, due to the presence of a TATA box capable to recruit TFIIIB through interaction with its TBP component (13, 14). In Fig. 1, the transcription reinitiation frequency was determined for a tDNAGly(TTC) and for the U6 RNA gene by comparing the output of multiple-versus single-round in vitro transcription reactions (11) performed with a limiting Pol III concentration. Stalled ternary complexes were formed upon incubation of DNA templates with either TFIIIC plus TFIIIB (tDNAGly, lanes 1 and 2; SNR6, lanes 3 and 4) or TFIIIB alone (SNR6, lanes 5 and 6; U6ΔB, lanes 7 and 8), followed by the addition of Pol III and the appropriate subsets of three NTPs. The missing NTP was then added, alone (lanes 1, 3, 5, and 7) or in association with heparin at a concentration that completely inhibit reinitiation (lanes 2, 4, 6, and 8), and transcription proceeded for 5 min. Approximately 18–20 transcription cycles took place on both tDNAGly and SNR6, corresponding to ≈16 s per cycle. Reinitiation frequency on wt SNR6 was unaffected by TFIIIC (compare lanes 3 and 4 with lanes 5 and 6 in Fig. 1 A), and it was very similar to the reinitiation frequency on U6ΔB (lanes 7 and 8). A distinguishing feature of facilitated reinitiation is the slower rate of the first transcription cycle with respect to subsequent cycles (3). We thus analyzed (Fig. 1B) the kinetics of first-round initiation on the U6 RNA gene. In particular, we determined the time course of formation of stalled, heparin-resistant ternary complexes by freshly added Pol III at two different concentrations: the same, limiting concentration used in the experiment in Fig. 1 A (0.2 ng/μl), and a Pol III concentration of 2 ng/μl, determined to be saturating by titration experiments conducted under multiple-round transcription conditions. These two polymerase concentrations were tested in the absence of TFIIIC. The time course of initiation by limiting Pol III was also analyzed in the presence of TFIIIC for both tDNAGly and SNR6 templates (Fig. 1C). The plots of these time course experiments evidence two interesting features of first round initiation by Pol III. First, as shown in Fig. 1B, initiation is considerably faster at saturating than at limiting Pol III concentrations (≈40 s versus ≈160 s half-times). Second, the presence of TFIIIC in the PIC does not influence first-round initiation significantly: initiation half-time by limiting Pol III in the presence of TFIIIC was ≈160 s on both tDNAGly and SNR6 (Fig. 1C). Most importantly, there is a general discrepancy between the rate of initiation of the first transcription cycle and the higher rate at which new transcription cycles are performed (Fig. 1 A, 16 s per complete cycle). The observation that the first-round initiation rate can be increased considerably by increasing the polymerase concentration suggests that PIC finding by Pol III is a rate-limiting step in the first transcription cycle, and that this step is facilitated in the subsequent cycles. In principle, the recycling mechanism could facilitate PIC finding either by simply increasing the local concentration of Pol III or by modifying the polymerase–promoter complex interaction. According to the first hypothesis, it should be possible to reduce the time for the first cycle to values close to 16 s (the recycling time observed in Fig. 1) by further increasing Pol III concentration. A time course analysis of first-round initiation was carried out at a Pol III concentration of 6 ng/μl (3-fold higher than the saturating concentration of Fig. 1B). No further reduction of the initiation half-time was observed, instead, transcription was significantly inhibited at this high Pol III concentration, where a dominant-negative effect of inactive Pol III molecules could become more pronounced (data not shown).
Fig. 1.
Facilitated transcription reinitiation on tRNA and U6 RNA genes. (A) Stable PICs, containing both TFIIIB (reconstituted from rTBP, rBrf1, and crude B′′) and TFIIIC (+IIIC) or TFIIIB alone (–IIIC) were formed on tDNAGly(TCC) (lanes 1 and 2), SNR6 (lanes 3–6), or U6ΔB (lanes 7 and 8) templates. Pol III (10 ng) was then added together with NTP mixtures lacking CTP (lanes 1 and 2) or ATP (lanes 3–8), and the incubation continued for 10 min. Transcription was resumed by the addition of the missing nucleotide, with (lanes 2, 4, 6, and 8) or without (lanes 1, 3, 5, and 7) heparin, and was run for 5 min. The migration positions of U6 RNA and pre-tRNAGly are indicated. The ratios between the amounts of single- and multiple-round transcription products, derived from PhosphorImager counting, are reported below the lanes with the SE of three independent experiments. (B) Stable PICs were assembled on the U6ΔB template for 20 min in the absence of TFIIIC. Limiting (10 ng, upper gel) or saturating (100 ng, lower gel) amounts of Pol III were then added together with an NTP mixture lacking ATP. After the indicated time periods, the missing nucleotide was added in association with heparin, to allow for completion of the transcription cycle by heparin-resistant complexes. The plot shown is derived from quantification of the shown gels. (C) Stable PICs containing both TFIIIB and TFIIIC were assembled for 20 min on either tDNAGly(TCC) (upper gel) or on the wt SNR6 template (lower gel). Limiting (10 ng) amounts of Pol III were then added together with NTP mixtures lacking either CTP (for the tDNA) or ATP (for SNR6). After the indicated time periods, the missing nucleotides were added in association with heparin, to allow for completion of the transcription cycle by heparin-resistant complexes. The plot shown is derived from quantification of the shown gels.
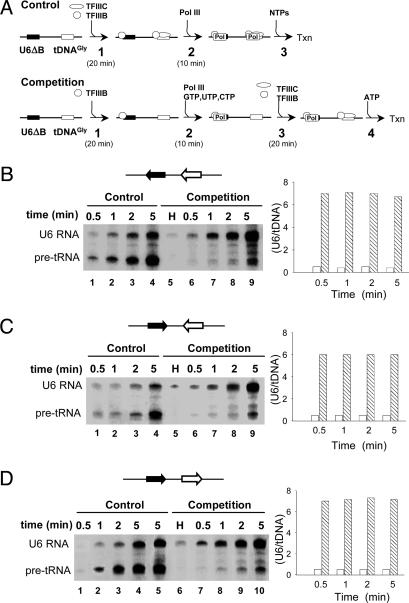
Pol III Gene Commitment in the Framework of Physically Linked Transcription Units. Template exclusion experiments with tDNAs placed on distinct plasmid molecules have previously revealed the commitment of recycling Pol III to the first transcribed gene (3). Because of the small size of tDNAs, in those experiments the local concentration of the test gene, with respect to the terminating polymerase, was much higher than the concentration of competitor DNA, and gene commitment could have been a consequence of the higher probability, for the terminating polymerase, to find the test promoter relative to the competitor promoter. To allow for competition between promoters at a comparable local concentration, both the test and the competitor genes were inserted into the same plasmid molecule in three different relative orientations, with a 400- to 600-bp spacing (Fig. 2 B–D). The U6ΔB template, supporting TFIIIC-independent Pol III recycling, and tDNAGly(TCC), requiring TFIIIC for transcription, were chosen as a test gene and competitor gene, respectively. In the competition reaction protocol (Fig. 2 A Lower), the plasmid carrying the two genes is preincubated with TFIIIB (step 1), which can load onto U6ΔB but not onto the tDNA. When a limiting amount of Pol III is added, together with a U6-specific subset of three NTPs (step 2), Pol III is sequestered in stable, arrested ternary complexes on the U6 gene. PICs are then assembled onto the competitor gene (tDNAGly) by adding a large excess of TFIIIC and fresh TFIIIB (step 3). Finally, U6 transcription is resumed by the addition of ATP (step 4). Upon U6 transcription termination, Pol III could in principle reinitiate on either of two similarly distant, initiation-competent templates: the just-transcribed U6ΔB test gene or the tDNAGly competitor gene. In the “control” experiment (Fig. 2 A Upper), TFIIIC is added to the preincubation mixture before Pol III (steps 1 and 2), to allow for polymerase recruitment by both promoters. The results of transcription competition experiments for each of the three constructs are shown in Fig. 2 B–D. In each image, lanes 1–4 refer to the control reaction, and lanes 5–9 (lanes 6–10 in Fig. 2D) to the corresponding competition experiment. Lane 5 in Fig. 2 B and C and lane 6 in Fig. 2D show the results of competition reactions in which transcription was limited to a single round. When Pol III was made to perform the first transcription cycle on the U6 gene, it predominantly reinitiated on the same gene, at least during the first 5 min, independent of the orientation of the two genes within the plasmid. The transcription output, calculated by comparing U6 transcript amounts in Fig. 2D, lanes 6 and 10, and those in Fig. 2 B and C, lanes 5 and 9, corresponded to ≈20 transcription cycles. Conversely, when Pol III was initially assembled on both promoters, it transcribed both genes with a constant, two-fold preference for tDNAGly over the U6 RNA gene (Fig. 2 B–D, lanes 1–4). As more precisely shown by the bar plots in Fig. 2 B–D, the U6 versus tDNAGly transcription ratio is close to 0.5 in the control experiment, regardless of reinitiation time, whereas it becomes 7 (6.5 after 10 min of reinitiation; data not shown) when Pol III is made to transcribe the U6 gene first. Fig. 2D, lane 5 shows the products of a control reaction in which Pol III and the four NTPs were added to PICs (preassembled on both genes) simultaneously, followed by a 5-min incubation. In this reaction, the tDNAGly–TFIIIC–TFIIIB complex was exposed for 5 min to the same amount of free, unbound Pol III one would expect to be available to the tDNAGly PIC during a 5-min competition (i.e., the reaction conditions of lane 10 of the same image) if Pol III completely releases after each cycle. In this control reaction, the tDNAGly was 5-fold more actively transcribed than in the lane 10 reaction, thus indicating that transcription of the competitor tDNAGly is actually excluded. What distinguishes the above experiments from those reported in our previous study of Pol III reinitiation (3) is the fact that the challenge gene is now provided on the same plasmid as the test gene. However, the present competition experiments can only be considered in cis if functional PICs are effectively assembled on the competitor (tDNAGly) gene. Accurate determination of tDNA occupancy by functional PICs revealed a 25% occupancy under the experimental conditions of Fig. 2. As detailed in Supporting Materials and Methods, and Fig. 6, which is published as supporting information on the PNAS web site, a stochastic redistribution model for reinitiation predicts that, at a 25% occupancy of the competitor tDNA, the U6/tDNA transcript ratio would be 3.4 after 4 transcription cycles, and 2.9 after 20 transcription cycles, whereas we observe a constant ratio of 7, even after 20 cycles. We thus conclude that the first transcription cycle specifically commits Pol III to reinitiate on the U6–TFIIIB complex, either by virtue of a polymerase retention mechanism avoiding polymerase release after each cycle, and/or as a consequence of a slowly acquired, Pol III-induced conformational change of the TFIIIB–DNA complex to an activated state that rapidly allows binding and initiation by Pol III in the subsequent cycles. In the latter hypothesis, Pol III could in principle be released during each round of transcription; however, the observed exclusion of tDNAGly during a 5-min competition (Fig. 2) can only be accounted for by assuming that the time required for the Pol III-dependent tDNA PIC activation is in the order of minutes.
Fig. 2.
Template exclusion experiments with physically linked transcription units. (A) The schemes illustrate the control (Upper) and competition (Lower) reaction protocols, respectively (see text for details). (B–D) In vitro transcription was performed after the control (lanes 1–4) or the competition (lanes 5–9 in B and C and lanes 6–10 in D) reaction schemes using the templates illustrated above each gel. Filled and empty arrows indicate the direction of transcription and represent the U6ΔB and the tDNAGly templates, respectively. Samples of the reaction mixtures were stopped at the indicated times. Lane 5 in D refers to a control reaction in which Pol III and the four NTPs were added simultaneously to PICs preassembled on both templates, and transcription was then run for 5 min. Lane 5 in B and C and lane 6 in D show the products of reactions (competition protocol) in which transcription was limited to a single round by the addition of heparin (H). The positions of U6 RNA and pre-tRNAGly are indicated on the left. The bar plots report the ratios between the amount of U6ΔB and tDNAGly transcription products accumulated at given times with the control (white bars) or the competition (hatched bars) reaction protocol.
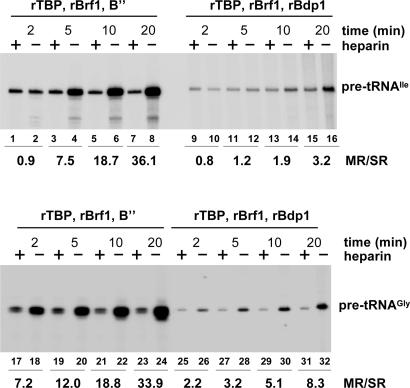
Reinitiation Properties of rTFIIIB. The experiments in Figs. 1 and 2 show that a PIC lacking TFIIIC is sufficient to direct facilitated Pol III recycling on the U6 RNA gene, thereby leading to attribute the recycling properties to TFIIIB components. However, in the in vitro transcription system used for those experiments, only two of the three known TFIIIB components, namely TBP and Brf1, were provided in a pure, recombinant form, whereas the third component, Bdp1, was provided as a crude fraction. The reason for using crude instead of rBdp1 is that the former generally allows much higher levels of transcription in vitro (15). Fig. 7, which is published as supporting information on the PNAS web site shows such an effect for the tDNAGly(TCC) and a tDNAIle(TAT) template (5). In principle, the low transcriptional output in the presence of all-rTFIIIB might be due to either inefficient PIC formation, or decreased reinitiation competence, or both. This point was directly addressed by the experiment in Fig. 3. The tDNAIle(TAT) (Fig. 3 Upper) and the tDNAGly(TCC) (Fig. 3 Lower) templates were transcribed in vitro in the presence of TFIIIC and either all-rTFIIIB reconstituted with 100 ng of rBdp1 from insect cells (lanes 9–16 and 25–32) or TFIIIB reconstituted with the crude B′′ fraction (≤ 20 ng of Bdp1 protein, lanes 1–8 and 17–24). The comparison of lanes 1, 3, 5, and 7 with lanes 9, 11, 13, and 15, and of lanes 17, 19, 21, and 23 with lanes 25, 27, 29, and 31, showing the single-round transcription outputs, revealed that ≈3-fold less functional transcription complexes were formed with rBdp1 than with the crude fraction in the case of tDNAIle, which was 5-fold less in the case of tDNAGly [the failure to attain with rBdp1 single-round transcription outputs similar to those produced by B′′, even in the presence of an 8-fold higher rBdp1 concentration (data not shown), might be explained by a lesser stability of rBdp1-containing PICs to heparin challenge]. Strikingly, the PICs formed on tDNAIle with all-rTFIIIB supported only two transcription cycles in 10 min, three in 20 min, whereas the PICs formed with crude B′′ supported an ≈10-fold faster reinitiation. In the case of tDNAGly, the PICs formed with crude B′′ supported an ≈4-fold faster reinitiation than PICs assembled with all-rTFIIIB. Faster reinitiation with crude B′′ with respect to all recombinant TFIIIB was also observed for the U6 RNA gene (data not shown). All-rTFIIIB is thus significantly less effective than TFIIIB containing crude B′′ in promoting transcription reinitiation by Pol III (we return to the implications of this finding below). The higher reinitiation rate observed with rBdp1 on tDNAGly compared with tDNAIle (compare lanes 9–16 with lanes 25–32) further suggests that different tDNAs may be endowed with different reinitiation properties.
Fig. 3.
Reinitiation properties of all-rTFIIIB. PICs were assembled for 30 min on tDNAIle(TAT) (Upper) or tDNAGly(TCC) (Lower) by using TFIIIC and different types of reconstituted TFIIIB. In lanes 1–8 and 17–24, TFIIIB was reconstituted from rTBP and rBrf1 proteins plus crude B′′. In lanes 9–16 and 25–32, pure rBdp1 protein from insect cells (100 ng) was used instead of B′′. Pol III (10 ng) was then added together with an NTP mixture lacking CTP, and the incubation was continued for 15 min. Transcription was resumed by the addition of CTP, either in the presence or in the absence of heparin, and the incubation continued for the indicated times. The ratios between the amounts of single- and multiple-round transcription products (MR/SR) are reported below the lanes.
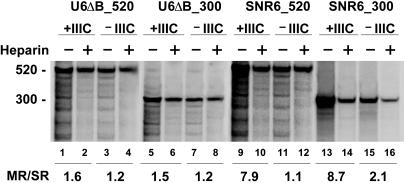
Reinitiation on Long Class III Genes. The small size of class III genes might favor Pol III retention for consecutive transcription cycles. We thus constructed variants of both SNR6 and U6ΔB templates in which the length of the U6 transcription unit was artificially increased to 300 bp (U6ΔB_300 and SNR6_300) or 520 bp (U6ΔB_520 and SNR6_520). As shown in Fig. 4, lanes 1–8, the reinitiation frequency on both the U6ΔB long-size derivatives was dramatically reduced with respect to the short (100 bp) U6ΔB (see Fig. 1), with only one to two cycles completed in 5 min. As expected, transcription of these B block-less templates was not significantly affected by TFIIIC (compare Fig. 4, lanes 3 and 4, with lanes 1 and 2, and lanes 7 and 8 with lanes 5 and 6). The TFIIIC-independent Pol III recycling on the U6 gene is thus compromised if the length of the transcription unit is increased. Surprisingly, however, TFIIIC could restore transcription reinitiation on both the SNR6 long-size derivatives, in which a TFIIIC-binding B block is present. In the presence of TFIIIC, eight to nine transcription rounds were completed in 5 min on both SNR6_520 and SNR6_300 (Fig. 4, lanes 9 and 10 and lanes 13 and 14), whereas reinitiation was much reduced in the absence of TFIIIC (Fig. 4, lanes 11, 12, 15, and 16), as observed for the U6ΔB derivatives. (In Fig. 4, lanes 1 and 9, several bands are present below the 520 nt U6ΔB_520 transcript, that tend to disappear in the presence of heparin (Fig. 4, lanes 2 and 10). These bands are TFIIIB-dependent and might represent paused elongation complexes). To further assess the role of TFIIIC in Pol III recycling, we analyzed the reinitiation properties of SCR1, a naturally long (520 bp) yeast class III gene that can be transcribed in vitro through both TFIIIC-dependent and TFIIIC-independent pathways, due to the presence of a TATA element at position –31 (16). As shown in Fig. 5, lanes 1 and 2, approximately eight transcription cycles took place in 5 min on SCR1 in the presence of TFIIIC, indicating efficient reinitiation in agreement with a previous analysis (16). Strikingly, however, reinitiation on SCR1 was abolished in the absence of TFIIIC (Fig. 5, lanes 3 and 4). The requirement for TFIIIC in SCR1 transcription reinitiation was also observed with a U6-SCR1 chimeric template in which the 5′-flanking region of SNR6 was fused to the transcribed portion of SCR1 (Fig. 5, lanes 5–8). At variance with SCR1, some TFIIIC-independent reinitiation took place on U6-SCR1, possibly due to better reloading of Pol III on the TFIIIB–DNA complex assembled on the U6 promoter region. Importantly, Fig. 5, lanes 9–12, show that reducing the length of the U6-SCR1 transcription unit from 522 to 90 bp (by insertion of a terminator sequence at +90) restores Pol III-facilitated recycling in the absence of TFIIIC to levels (≈20 cycles in 5 min) comparable with those observed with U6ΔB. As further shown in Fig. 8, which is published as supporting information on the PNAS web site, TFIIIC increased the rate of initial recruitment of Pol III by SCR1 ≈2-fold (a result reproduced in two independent experiments), and reproducibly increased the elongation rate by ≈30%, the combination of which would not account for the observed 8-fold difference in reinitiation rate.
Fig. 4.
Reinitiation properties of Pol III on long-size derivatives of the U6 RNA gene. Stable PICs containing TFIIIB alone, reconstituted from rTBP, rBrf1, and crude B′′ (–IIIC), or both TFIIIB and TFIIIC (+IIIC), were formed for 20 min on the U6ΔB_520 (lanes 1–4), U6ΔB_300 (lanes 5–8), SNR6_520 (lanes 9–12), or SNR6_300 (lanes 13–16) templates. Pol III (10 ng) was then added together with GTP, CTP, and UTP, and the incubation was continued for 10 min. Transcription was resumed by the addition of ATP, either with or without heparin, and was run for 5 min. The ratios between the amounts of multiple- and single-round transcription products (MR/SR) are reported below the gel.
Fig. 5.
Reinitiation on the SCR1 gene. Stable PICs, containing TFIIIB alone, reconstituted from rTBP, rBrf1, and crude B′′ (–IIIC) or both TFIIIB and TFIIIC (+IIIC) were formed for 20 min on the SCR1-C4T template (ref. 16; lanes 1–4), the U6-SCR1 template, which consisted of the 5′-flanking region of SNR6 fused to SCR1 (lanes 5–8), or the U6-SCR1 mini template, and was generated by inserting a terminator at position +90 of U6-SCR1. After PIC assembly, Pol III (10 ng) was added together with an NTP mixture lacking CTP. Transcription was resumed 10 min later by adding CTP, either with (lanes 2, 4, 6, 8, 10, and 12) or without (lanes 1, 3, 5, 7, 9, and 11) heparin, and was run for 5 min. The positions of the SCR1-derived transcription products are indicated by arrow-heads. The ratios between the amounts of multiple- and single-round transcription products (MR/SR) are reported below with the SE of three independent experiments.
Discussion
This work provides insight into the Pol III reinitiation mechanism by showing that facilitated recycling relies on a specific polymerase recapture pathway involving promoter-bound transcription factors. Recapture might simply take place through physical retention of the polymerase on the same transcription unit, without enzyme dissociation after each cycle. Alternatively (or in addition), a Pol III-induced conformational change in the PIC after the first transcription cycle might shift it to an active state, allowing rapid binding and initiation by Pol III. The finding that Pol III, when loaded on the U6 promoter by TFIIIB only, is refractory to rebinding to a TFIIIB–TFIIIC-directed tDNA promoter located on the same plasmid strongly suggests, by itself, the existence of a of Pol III-reloading mechanism only relying on TFIIIB–Pol III interactions. Two lines of evidence, however, argue in favor of a more complex mechanism of Pol III reinitiation. First, all-rTFIIIB (reconstituted from pure rTBP, rBrf1, and rBdp1 components) supported reinitiation much less efficiently than TFIIIB containing a crude B′′ fraction. Second, TFIIIC strongly stimulated reinitiation on long (>300 bp) transcription units. The reinitiation defect observed with all-rTFIIIB might be due either to the lack of a reinitiation-stimulatory component(s) present in the crude Bdp1 fraction, or to the absence, in the recombinant proteins, of a particular covalent modification profile required for reinitiation. The stimulatory component may be one of the components found in an equivalent crude B′′ fraction that confers stringent start site selection (17). Also, we have shown (15) that a fraction called TFIIIE can stimulate in vitro transcription by all-rTFIIIB. Such a stimulatory effect has been reproducibly observed with several preparations of both TFIIIE and recombinant TFIIIB components (G.D., unpublished observations). The extent of TFIIIE stimulation, however, generally does not reach the transcription levels observed with the crude B′′ fraction, thus rendering a possible reinitiation role of TFIIIE difficult to evaluate. Moreover, the failure to secure a purification of the TFIIIE fraction that retains the reinitiation function, leaves these findings in limbo. The same limitation applies to the Nhp6 protein, which has been shown to stimulate U6 RNA gene transcription with all-rTFIIIB, but only to a limited extent (18).
The gene-length-dependent involvement of TFIIIC in Pol III reinitiation further points to the complexity of this process. We propose that Pol III gene commitment depends on Pol III–TF interactions that are either maintained during the entire transcription cycle or are specifically put in place upon termination. On short (≈100 bp) class III genes, Pol III is retained on the transcribed gene even in the absence of TFIIIC, through interactions with the TFIIIB complex that require additional components and/or a specific covalent modification profile of TFIIIB. On longer genes, such as SCR1, the terminating Pol III is too far away from the transcription initiation region to interact with promoter-bound TFIIIB, and gene commitment is only made possible by interactions with TFIIIC, which contacts the DNA template further downstream. TFIIIB bends the DNA significantly (19), and a recent atomic force microscopy study (20) has shown that stalled elongation complexes of yeast Pol III compact the DNA by ≈30 nm by wrapping it around the surface of the polymerase. With short class III genes, the compactness produced by DNA bending and wrapping is probably sufficient to favor the macromolecular contacts responsible for facilitated reinitiation, whereas on long transcription units, TFIIIC–polymerase contacts would be further required. In keeping with this view, the Tfc4 subunit of TFIIIC has been reported to specifically interact with both the ABC10α and the C53 subunits of yeast Pol III (21, 22), and human Pol III–TFIIIC interactions have also been observed (23, 24). The fact that TFIIIC can stimulate reinitiation, even when the B block is placed at 600 bp downstream of the transcription start site is surprising, and further points to the ability of this factor to cause higher-order rearrangements of the DNA [such as looping (25)], thus contributing to transcription unit compaction (a more detailed discussion of TFIIIC versatility in promoting reinitiation can be found as a comment to Fig. 9, which is published as supporting information on the PNAS web site). The stimulation of Pol III reinitiation brought about by TFIIIC might in principle be due to polypeptides, other than the six known yeast TFIIIC subunits, that might be present in our TFIIIC preparations. Even if such a possibility cannot be formally excluded, we tend to disfavor it for two reasons. First, the reinitiation activity was fully present in an affinity-purified TFIIIC preparation that did not contain any major polypeptide apart from the known TFIIIC subunits (data not shown). Second, the effect of TFIIIC on reinitiation strictly depended on the presence of a B block (see Fig. 4), implying that, if the reinitiation activity of TFIIIC is due to a previously unknown component, the action of such a component should depend on TFIIIC–B block interaction. Finally, it should be noted that an in cis competition analysis as the one reported in Fig. 2 could not be applied to the long genes, because of their TFIIIC requirement. Therefore, mechanisms other than Pol III recapture, such as facilitated disentanglement and release at the terminator by TFIIIC, might in principle account for improved reinitiation.
Supplementary Material
Acknowledgments
We thank André Sentenac for valuable suggestions and continuous encouragement, Simone Ottonello, Stefania Petrucco, and Jean-Louis Sikorav for discussions and critical reading of the manuscript, Matteo Forloni (University of Parma, Parma, Italy) for the experiment in Fig. 7, and the reviewers for their insightful criticism. This work was supported by Human Frontier Science Program Organization Grant RGY0011/2002-C (to G.D.) and by the Italian Ministry of Education, University, and Research (2003 Fondo pergli Investimenti della Ricerca di Base and COFIN Programs).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TBP, TATA box-binding protein; Pol, RNA polymerase; PIC, preinitiation complex; r, recombinant.
References
- 1.Dieci, G. & Sentenac, A. (2003) Trends. Biochem. Sci. 28, 202–209. [DOI] [PubMed] [Google Scholar]
- 2.Geiduschek, E. P. & Kassavetis, G. A. (2001) J. Mol. Biol. 310, 1–26. [DOI] [PubMed] [Google Scholar]
- 3.Dieci, G. & Sentenac, A. (1996) Cell 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 4.Guffanti, E., Corradini, R., Ottonello, S. & Dieci, G. (2004) J. Biol. Chem. 279, 20708–20716. [DOI] [PubMed] [Google Scholar]
- 5.Dieci, G., Percudani, R., Giuliodori, S., Bottarelli, L. & Ottonello, S. (2000) J. Mol. Biol. 299, 601–613. [DOI] [PubMed] [Google Scholar]
- 6.Kassavetis, G. A., Braun, B. R., Nguyen, L. H. & Geiduschek, P. E. (1990) Cell 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 7.Fan, H., Sakulich, A. L., Goodier, J. L., Zhang, X., Qin, J. & Maraia, R. J. (1997) Cell 88, 707–715. [DOI] [PubMed] [Google Scholar]
- 8.Wang, Z., Bai, L., Hsieh, Y. J. & Roeder, R. G. (2000) EMBO J. 19, 6823–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huet, J., Manaud, N., Dieci, G., Peyroche, G., Conesa, C., Lefebvre, O., Ruet, A., Riva, M. & Sentenac, A. (1996) Methods Enzymol. 273, 249–267. [DOI] [PubMed] [Google Scholar]
- 10.Kassavetis, G. A., Joazeiro, C. A. P., Pisano, M., Geiduschek, E. P., Colbert, T., Hahn, S. & Blanco, J. A. (1992) Cell 71, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 11.Kassavetis, G. A., Riggs, D. L., Negri, R., Nguyen, L. H. & Geiduschek, E. P. (1989) Mol. Cell. Biol. 9, 2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumay-Odelot, H., Acker, J., Arrebola, R., Sentenac, A. & Marck, C. (2002) Mol. Cell. Biol. 22, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moenne, A., Camier, S., Anderson, G., Margottin, F., Beggs, J. & Sentenac, A. (1990) EMBO J. 9, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joazeiro, C. A., Kassavetis, G. A. & Geiduschek, E. P. (1994) Mol. Cell. Biol. 14, 2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rüth, J., Conesa, C., Dieci, G., Lefebvre, O., Düsterhoft, A., Ottonello, S. & Sentenac, A. (1996) EMBO J. 15, 1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 16.Dieci, G., Giuliodori, S., Catellani, M., Percudani, R. & Ottonello, S. (2002) J. Biol. Chem. 277, 6903–6914. [DOI] [PubMed] [Google Scholar]
- 17.Andrau, J. C. & Werner, M. (2001) Eur. J. Biochem. 268, 5167–5175. [DOI] [PubMed] [Google Scholar]
- 18.Lopez, S., Livingstone-Zatchej, M., Jourdain, S., Thoma, F., Sentenac, A. & Marsolier, M. C. (2001) Mol. Cell. Biol. 21, 3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leveillard, T., Kassavetis, G. A. & Geiduschek, E. P. (1991) J. Biol. Chem. 266, 5162–5168. [PubMed] [Google Scholar]
- 20.Rivetti, C., Codeluppi, S., Dieci, G. & Bustamante, C. (2003) J. Mol. Biol. 326, 1413–1426. [DOI] [PubMed] [Google Scholar]
- 21.Dumay, H., Rubbi, L., Sentenac, A. & Marck, C. (1999) J. Biol. Chem. 274, 33462–33468. [DOI] [PubMed] [Google Scholar]
- 22.Flores, A., Briand, J. F., Gadal, O., Andrau, J. C., Rubbi, L., Van Mullem, V., Boschiero, C., Goussot, M., Marck, C., Carles, C., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh, Y. J., Kundu, T. K., Wang, Z., Kovelman, R. & Roeder, R. G. (1999) Mol. Cell. Biol. 19, 7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, Y. J., Wang, Z., Kovelman, R. & Roeder, R. G. (1999) Mol. Cell. Biol. 19, 4944–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnol, A. F., Margottin, F., Schultz, P., Marsolier, M. C., Oudet, P. & Sentenac, A. (1993) J. Mol. Biol. 233, 644–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.