Abstract
Ribosomes stalled during protein synthesis can be rescued by tmRNA, which acts first as a tRNA and then as an mRNA to direct addition of a C-terminal degradation tag to the nascent polypeptide. Ribosomal protein S1 binds tmRNA, but its functional role in tmRNA-mediated tagging is uncertain. To probe interactions between S1 and tmRNA, truncated variants missing one or more of the six contiguous S1 domains were studied. The third S1 domain (R1) plays a critical role in binding tmRNA and mRNA but requires additional N- or C-terminal S1 domains. The binding of S1 and its fragments to tmRNA and mRNA is positively cooperative, and the essential role of the R1 domain may be to mediate protein–protein interactions. Overproduction of N-terminal fragments of S1 in Escherichia coli displaces endogenous S1 from ribosomes, inhibits general protein synthesis, and slows growth but causes little if any disruption of tmRNA-mediated tagging. Moreover, tagging of proteins translated from model mRNAs with either no or an increased requirement for S1 is indistinguishable. These results raise the possibility that S1 plays little or no role in tmRNA-mediated tagging.
Keywords: ssrA tagging, trans-translation, ribosome rescue, protein–RNA interactions
SsrA tagging is a ubiquitous system for translational quality control in eubacteria (1). SsrA, often called tmRNA, acts both as a tRNA and mRNA to rescue stalled ribosomes and to mediate attachment of a degradation tag to the nascent polypeptide. Stalling occurs when ribosomes reach the 3′ end of an mRNA, at rare codons when the cognate tRNA is scarce, and at stop codons when translation termination is inefficient (2–5). tmRNA, charged with alanine, is recruited to stalled ribosomes and first performs a tRNA-like role in which the alanine becomes linked to the growing polypeptide chain. Then, through a poorly understood mechanism, the ribosome switches from translating the original mRNA to translating an ORF on tmRNA. This process results in attachment of a peptide tag that targets the incomplete protein for degradation and frees the ribosome to translate other cellular messages.
Several proteins are required for tmRNA-mediated tagging. SmpB, a dedicated tmRNA binding protein (6), mediates interactions with the ribosome, some of which are normally made by tRNA itself (7), whereas alanine tRNA synthetase and elongation factor Tu mediate aminoacylation and ribosome delivery, respectively (7, 8). Ribosomal protein S1 copurifies with tmRNA–SmpB complexes, crosslinks to tmRNA both in the presence and absence of ribosomes in vitro, and binds to tmRNA in vitro (9–11), but its role in tmRNA function is unclear.
Ribosomal protein S1 participates in translation initiation of many mRNAs (12–16) and may also play a role in translation elongation (13). During initiation, S1 binds to the mRNA leader upstream of the Shine–Dalgarno (SD) sequence (14). This interaction is of particular importance for initiation on messages with weak or very strong SD sequences (12, 15). Interestingly, translation of leaderless mRNAs does not require S1 (16). During normal translation, S1 interacts with the ribosome and with mRNA (12). Several roles for S1 in tmRNA function have been postulated, including assisting in determination of the tmRNA reading frame, delivery of tmRNA to stalled ribosomes, and protection of tmRNA from ribonuclease degradation (10). The gene for S1 (rpsA) is essential in Escherichia coli, precluding the generation of knockout strains that could be used to test for important roles in tmRNA function (12). S1 is absent in some species that contain tmRNA, suggesting that it is not essential for tmRNA function. However, it is difficult to exclude the possibility that another protein fulfills an S1-like role in these organisms.
E. coli S1 contains six contiguous S1 domains (refs. 12 and 17; Fig. 1a), which are thought to bind single-stranded RNA and RNA pseudoknots (18). The first two domains of S1 (jointly referred to here as the N-terminal fragment) are important for ribosome binding, the extreme C-terminal domain (R4) autogenously regulates S1 expression, and the central domains (R1, R2, and R3) appear to be involved in mRNA binding (12, 19). Here we investigate the role of S1 domains in tmRNA binding and function. Using a set of truncated variants, we show that the R1 domain is critical for tmRNA binding but also requires adjacent N- or C-terminal domains. To investigate S1's influence on tmRNA translation, overexpression of truncated variants was used to test for dominant-negative effects. As expected, overexpression of certain truncated S1 proteins severely reduced normal translation (20). Surprisingly, however, translation of tmRNA during tagging of proteins expressed from model mRNAs that stall ribosomes was not inhibited by the same truncated S1 variants. This result was observed irrespective of the requirement for S1 for translation of the tagging substrate. Taken together, these results suggest that S1 does not play a critical role in tmRNA-mediated tagging.
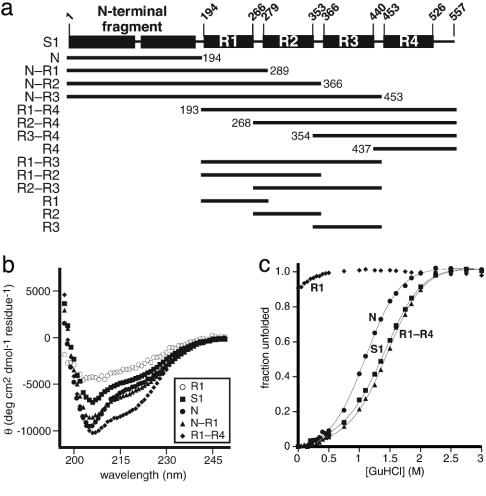
Fig. 1.
S1 domain structure and properties. (a) Schematic of ribosomal protein S1 highlighting the domains and fragments used for this study. (b) CD spectra for S1 (▪) and a representative group of fragments including N (•), N–R1 (▴), R1–R4 (♦), and R1 (○). (c) GuHCl denaturation monitored by tryptophan fluorescence of S1 (▪) and a representative group of fragments including N (•), R1–R4 (▴), and R1 (♦).
Materials and Methods
Bacterial Strains and Plasmids. E. coli strains were derivatives of X90 [F′ lacIq lac′ pro′/ara Δ(lac-pro) nalA argE(am) rif′ thi-1]. Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Plasmid constructs were verified by DNA sequencing (Massachusetts Institute of Technology Biopolymers Facility, Cambridge, MA).
For details of plasmid construction, see the supporting information, which is published on the PNAS web site. Coding sequences for ribosomal protein S1 and truncated variants were PCR amplified and cloned under control of the T7 RNA polymerase promoter between the NdeI and XhoI sites of pET21b (Novagen), resulting in a His-6 tag at the C terminus of each protein. pKW11 encodes wild-type tmRNA (2). pET800 encodes the arc-st11 gene (21). Plasmid pCH206, expressing tmRNA-FLAG encoding the nondegradable peptide tag ANDYKDDDDK in its ORF, was a gift from C. Hayes (Massachusetts Institute of Technology). Plasmid derivatives of pCH410 (22) encoded S1, N, N–R1, R1–R4, and R2–R4 under the control of the pBAD promoter. Plasmid derivatives of pCH405-FLAG-AraC encoded tmRNA-FLAG and S1, N, N–R1, N–R2, R1–R4, or R2–R4 under the control of the pBAD promoter. pPW500 encodes the N-terminal domain of λ cI repressor with a trpA terminator under the control of a pTrc promoter (2). pPW500-leaderless and pPW500-extended-SD encode the same protein but have altered mRNA leader sequences.
Protein Purification. BB101 cells [X90 (λDE3) slyD::kan] containing overexpression plasmids for S1 or S1 fragments were grown overnight in LB broth plus 100 μg/ml ampicillin, diluted 1,000-fold into fresh medium, grown to an OD600 of 0.6–0.8, and induced with isopropyl β-d-1-thiogalactopyranoside (1 mM final concentration) for 2 h. Cells were harvested by centrifugation and lysed in nickel lysis buffer [100 mM NaH2PO4/10 mM Tris·HCl/6 M guanidine hydrochloride (GuHCl), pH 8] for 30 min at 22°C. After centrifugation, His-6-tagged proteins in the supernatant were purified by Ni-nitrilotriacetic acid agarose (Qiagen) chromatography and exchanged into TE buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA) on a PD-10 column (Amersham Biosciences). The R1 fragment was exchanged into 50 mM MES (pH 6.5)/100 mM KCl. Some proteins were concentrated in a Millipore ultrafree centrifugal filter before buffer exchange. Protein concentrations were determined by UV absorbance at 280 nm by using extinction coefficients (M–1·cm–1) of 46730 (S1), 6970 (N or R4), 20910 (N–R1), 33690 (N–R2), 46350 (N–R3), 40660 (R1–R4), 28000 (R2–R4), 13940 (R3–R4), 39380 (R1–R3), 26720 (R1–R2 or R2–R3), and 14060 (R2).
Biophysical Studies. CD spectra were taken at protein concentrations from 5 to 10 μM in 10 mM sodium phosphate (pH 7.4), at 25°C, with an AVIV 60DS CD spectrometer. For denaturation studies, proteins (0.5 μM) were incubated at 22°C for at least 12 h in 120-μl reactions containing 0–4 M GuHCl, 50 mM Tris·HCl (pH 7.5), 100 mM ammonium chloride (NH4Cl), and 1 mM DTT. Fluorescence (excitation 270 nm; emission 300–425 nm) was measured by using a QM-2000–4SE spectrofluorometer (Photon Technology International, Lawrenceville, NJ). The fraction of folded protein at each GuHCl concentration was calculated from the center-of-mass fluorescence. Because the R1 and R1–R2 proteins were only partially folded, the folded and unfolded values for the R1–R3 protein were used to calculate the fraction folded for these proteins.
Binding Assays. The tmRNA and arc-st11 T7 transcription templates were amplified by PCR from pKW11 (2) and pET800 (21), respectively (see supplementary materials). RNAs were transcribed in vitro (23), treated with RNase-free-DNase I (New England Biolabs), purified with the Qiagen RNeasy mini kit, and dephosphorylated with calf intestinal phosphatase (New England Biolabs). RNA was purified in a 6% denaturing polyacrylamide gel, excised and eluted by soaking overnight in TE buffer plus 200 mM NaCl, precipitated with ethanol, and resuspended in TE buffer. Concentrations were determined by UV absorbance at 260 nm by using extinction coefficients (M–1·cm–1) of 3577000 (tmRNA) and 3328000 (arc-st11 mRNA). RNA was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) and purified with the Qiagen RNeasy mini kit.
Binding reactions (20 μl) contained S1 or S1 fragments, 20 pM radiolabeled RNA, 10 mM Tris·HCl (pH 7.5), 100 mM NH4Cl, 1 mM DTT, 100 μg/ml BSA (New England Biolabs), 100 μg/ml E. coli tRNA (Sigma), and 5% glycerol and were incubated at 22°C for 30–60 min (10). Inclusion of tRNA was required to observe discrete, shifted bands rather than a diffuse set of protein–RNA complexes. RNA was treated before the binding reaction by heating at 65°C for 3 min in TE buffer, incubating at 22°C for 2 min, and then incubating for 2 min in binding buffer. Samples were electrophoresed in a 5% native polyacrylamide gel (16 cm × 17 cm × 1.5 mm) prerun at 100 V for ≈1 h in 25 mM Tris/190 mM glycine/1 mM EDTA and then run for 3–6hat18 mA per gel at 4°C. Bound and unbound RNA was quantified by using a Molecular Dynamics Phosphorimager. Data from three or more independent experiments were fit to the following equation: fraction bound = 1/(1 + (Kapp/[protein])n).
Growth Curves, Translation, and Ribosome Binding. X90 cells transformed with derivatives of pCH410 expressing S1 or fragments were grown at 37°C in LB broth plus 20 μg/ml tetracycline. Overnight cultures were diluted 40-fold into 4 ml cultures, grown to OD600 ≈ 0.2, and used to seed 20- to 30-ml cultures at a 1:10 dilution. After growth at 37°C to OD600 ≈ 0.4, protein overexpression was induced by the addition of arabinose to 0.2%, and growth was continued for 3 h. OD600 values were plotted versus time by using data from two or three independent experiments. The pre- and postinduction portions of the curve were fit to exponential functions to determine doubling times. Before and 60 min after arabinose addition, 1 ml of each culture was transferred to a separate tube, [35S]methionine was added to 50 μCi/ml (1 Ci = 37 GBq), and growth at 37°C was continued for 5 min. Cells were harvested, resuspended in UL buffer [10 mM Tris·HCl, pH 8/100 mM NaCl/8 M urea], and frozen at –80°C for 20 min or more. Pellets were thawed, allowed to lyse at 22°C for at least 20 min, and cellular debris was removed by centrifugation. Supernatants were transferred to a new tube, and total protein was determined by Bradford analysis (Bio-Rad protein assay). Equal amounts of protein from each sample were electrophoresed in a 10% SDS/PAGE, and radiolabeled proteins were visualized and quantified by using a Molecular Dynamics Phosphorimager.
For ribosome purification, cultures were grown for 1 h after arabinose addition, and rifampicin was added to 200 μg/ml. After 30 min, cultures were chilled on ice and cells were harvested by centrifugation, resuspended in 0.5 ml of B-PER (Pierce), and frozen overnight at –80°C. Resuspended cells were thawed, lysozyme was added to 0.2 mg/ml, and lysis was allowed to proceed on ice for 10 min. Ribosome buffer [50 mM Trisacetate, pH 7.5/100 mM KCl/14 mM magnesium acetate] was added to bring the volume to 1.5 ml, DNase I was added to 1.33 units/ml, and debris was removed by centrifugation. Supernatants were layered onto 1 ml of 10% sucrose in ribosome buffer and centrifuged at 53,000 rpm for 3–4 h in a Beckman TLS-55 rotor. The supernatant was saved and the pellet was resuspended by shaking three times at 4°C with 150 μl of ribosome buffer for 20 min. Equal quantities of total protein from the supernatant and pellet fractions, respectively, were electrophoresed in two 10% SDS polyacrylamide gels. One gel was stained with Coomassie blue, and the other was transferred to an Immobilon-P polyvinylidine fluoride membrane (Millipore). His-6-tagged proteins were detected by using His-probe H15 (Santa Cruz Biotechnology) with ECL Plus Western blotting detection reagent (Amersham Biosciences) and x-ray film.
Tagging Assays. Plasmid derivatives of pCH405-FLAG-AraC encoding S1, N, N–R1, N–R2, R1–R4, and R2–R4 were transformed into X90 ssrA::cat cells (24) containing pPW500. Cultures were grown at 37°C in LB broth plus 20 μg/ml tetracycline and 100 μg/ml ampicillin. Overnight cultures were diluted 40-fold into 4-ml cultures, grown to OD600 ≈ 0.2, and used to seed 20- to 30-ml cultures at a 1:10 dilution. Cultures were grown to an OD600 of 0.2–0.4, and S1 protein expression was induced by addition of arabinose to 0.2%. After 30–45 min, expression of the λ-cI-N-trpAt protein encoded by pPW500 was induced with isopropyl β-d-1-thiogalactopyranoside (1 mM). After 2–2.5 h, cells were harvested and lysed, and His-6-tagged proteins were purified by Ni-nitrilotriacetic acid chromatography, electrophoresed in a 15% SDS-polyacrylamide gel, and stained with Coomassie blue. Gels were scanned with a SNAPSCAN 1212 scanner (AGFA) and quantified by using imagequant (Molecular Dynamics). The fraction of total λ-cI-N-trpAt protein that was tagged was determined from the average of three independent experiments. Experiments examining S1 effects on tagging of proteins from the leaderless or extended SD λ-cI-N-trpAt mRNAs were performed in a similar manner with the appropriate pPW500 derivatives and pCH405-FLAG-AraC, except that arabinose was not added and isopropyl β-d-1-thiogalactopyranoside induction was for 1.5 h.
Results
Folding of S1 and Truncated Variants. Intact S1 protein and the S1 fragments shown in Fig. 1a were expressed with C-terminal His-6 tags and purified. Using previously defined domains of S1 as a guide (12), each truncated protein contained one or more contiguous S1 domains. The portion of S1 designated as the “N-terminal fragment” contains two S1 motifs that are less homologous than the four C-terminal S1 domains, which are designated R1, R2, R3, and R4. All of the S1 variants shown in Fig. 1a were overexpressed and purified, except for the R3 domain, which had low solubility.
The structural integrity of the S1 fragments was assessed by CD and, in some cases, by GuHCl unfolding (Fig. 1 b and c show selected data). The truncated variants fell into two classes. The R1, R2, and R1–R2 proteins appeared to be largely unfolded, with relatively featureless CD spectra and no or noncooperative GuHCl transitions. By contrast, full-length S1 and the remaining variants, including N, N–R1, N–R2, N–R3, R4, R3–R4, R2–R4, R2–R3, R1–R3, and R1–R4, had characteristics expected for stably folded structures. The CD spectra of these proteins were consistent with a mixture of α-helix and β-sheet, as anticipated from the structures of homologous S1 domains (17, 25). Full-length S1 and the N, N–R2, N–R3, R1–R3, and R1–R4 fragments displayed cooperative unfolding transitions, with half-maximal denaturation between 1.1 and 1.7 M GuHCl (Fig. 1c).
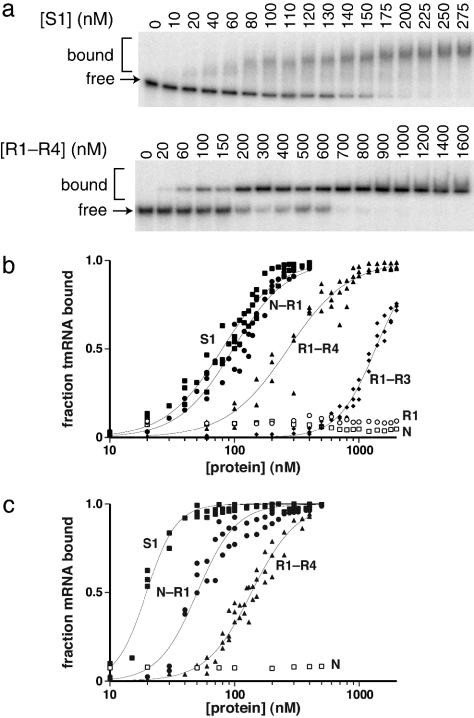
RNA Binding. Specific binding of S1 or its fragments to tmRNA or mRNA was assayed by changes in gel mobility in the presence of excess tRNA, with data fit to obtain the half-maximal binding concentration (Kapp) and a Hill coefficient (n). Full-length S1 bound tmRNA with a Kapp of 80 nM, but binding was not observed for the N, R1, R2, R4, R2–R4, R2–R3, or R3–R4 fragments (Kapp > 40 μM) (Fig. 2 a and b; Table 1; data not shown). By contrast, the N–R1, N–R2, and N–R3 fragments bound tmRNA with apparent affinities (80–100 nM) close to that of intact S1, the R1–R4 fragment (Kapp = 270 nM) bound 3- to 4-fold more weakly, and the R1–R3 fragment (Kapp = 1.4 μM) bound ≈18-fold less well (Fig. 2b; Table 1). R1 was present in all constructs that bound tmRNA but did not bind to tmRNA by itself. Therefore, R1 must work in concert with other S1 domains to bind tmRNA or stabilize binding.
Fig. 2.
Binding of S1 and fragments to RNA. (a) Mobility-shift assay gels for the binding of tmRNA to S1 and R1–R4 are shown. (b) Curves for tmRNA binding by full-length S1 (▪), N–R1 (•), R1–R4 (▴), R1–R3 (♦), N(□), and R1 (○). (c) Curves for Arc-st11 mRNA binding. Binding data were fit as described in Materials and Methods.
Table 1. Binding of S1 and fragments to RNA.
| Protein | Kapp, tmRNA, nM | Hill coefficient (tmRNA) | Kapp, mRNA, nM | Hill coefficient (mRNA) |
|---|---|---|---|---|
| S1 | 80 ± 2 | 1.9 ± 0.1 | 20 ± 1 | 3.6 ± 0.3 |
| N—R1 | 98 ± 3 | 2.0 ± 0.1 | 50 ± 2 | 2.8 ± 0.2 |
| N—R2 | 96 ± 2 | 1.7 ± 0.1 | nd | nd |
| N—R3 | 80 ± 2 | 1.8 ± 0.1 | nd | nd |
| R1—R4 | 270 ± 11 | 1.8 ± 0.1 | 136 ± 3 | 2.4 ± 0.1 |
| R1—R3 | 1,400 | 2.9 ± 0.1 | nd | nd |
nd, not determined.
We also assayed binding of intact S1 and some fragments to a model mRNA encoding Arc-st11 repressor (21). S1 bound this mRNA with an apparent affinity of 20 nM, the N1–R1 protein (50 nM) and R1–R4 protein (136 nM) bound less tightly, and the N and R2–R4 fragment showed no binding (Fig. 2c; Table 1; data not shown). These results are consistent with an important role for the R1 domain in mRNA as well as tmRNA binding.
The tmRNA-binding isotherms for intact S1 and the N–R1, N–R2, N–R3, and R1–R4 fragments showed positive cooperativity, with Hill coefficients of ≈2 (Table 1). This result indicates that binding requires two or more proteins, with relatively weak binding of the first molecule and stronger binding of subsequent molecules. A simple explanation for this behavior is that binding of subsequent proteins is stabilized by protein–protein as well as protein–RNA interactions (see Discussion). The participation of multiple S1 proteins or fragments in tmRNA binding is also consistent with the observed supershifting of the bound complex as the protein concentration is raised (Fig. 2a). S1 and fragment binding to our model mRNA was also positively cooperative with Hill coefficients ranging from 2.4 to 3.6 (Table 1). Hence, multisite binding and positive cooperativity appear to be a general property of the binding of S1 and its fragments to RNA.
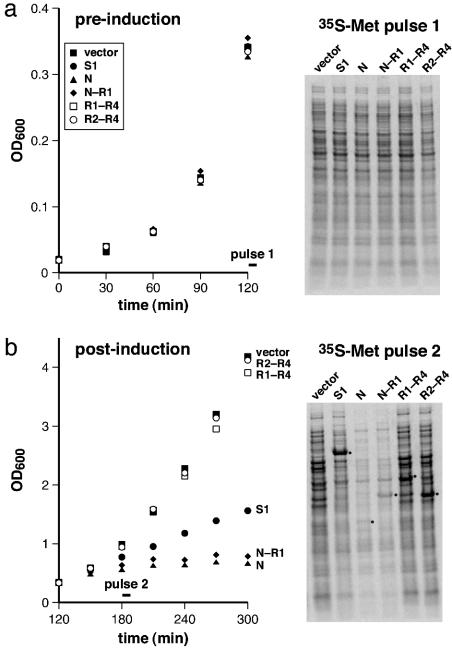
Overexpression Assays. Expression of S1 fragments has been shown to affect translation efficiency and cell growth adversely (20). To test the effects of full-length S1 and its N, N–R1, R1–R4, and R2–R4 fragments, we cloned these proteins into plasmids under the control of an arabinose-inducible promoter. Cells containing these plasmids grew at rates similar to a vector control before arabinose induction (Fig. 3a). After induction, cells containing the N or N–R1 fragments grew very slowly, and cells overexpressing S1 grew at about half the rate of the vector control and the R1–R4 and R2–R4 cultures (Fig. 3b). To test whether these growth defects correlated with a reduction in overall translation, portions of the cultures were pulse-labeled with [35S]methionine immediately before and 60 min after induction, and protein synthesis was assayed by SDS/PAGE and autoradiography. Preinduction synthesis of 35S proteins was essentially identical in all cultures (Fig. 3a), but synthesis was reduced about 10-fold after induction of the N or N–R1 fragments (Fig. 3b). Overexpression of full-length S1 reduced overall protein synthesis by roughly 2-fold, with the major synthesis product being S1 itself, whereas overexpression of the R1–R4 and R2–R4 fragments caused little change in protein synthesis beyond the fragments themselves (Fig. 3b). These results suggest that the slow growth in cells overexpressing N, N–R1, or S1 is caused by inhibition of protein synthesis.
Fig. 3.
Effects of overexpression of S1 variants on cell growth and translation. Growth curves before (a) and after (b) arabinose induction at 120 min. Strains contained the vector plasmid (▪) or plasmids encoding the full-length S1 (•), N(▴), N–R1 (♦), R1–R4 (□), and R2–R4 (○) proteins. A portion of each culture was pulse-labeled with [35S]methionine at 120 or 180 min and analyzed by SDS/PAGE and autoradiography. The bands corresponding to the overexpressed S1 variants are highlighted with dots.
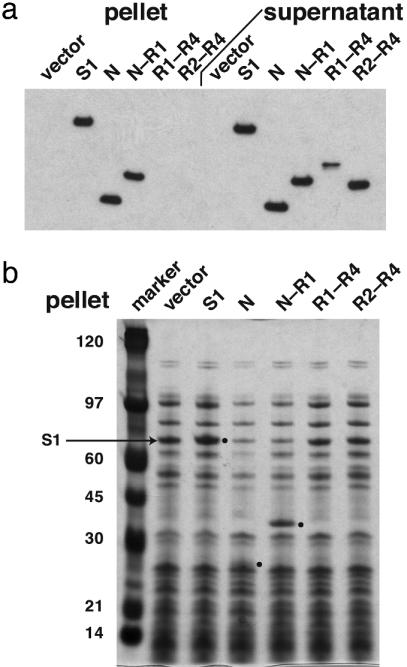
To test for ribosome binding by S1 or its fragments, ribosomes from different overexpression cultures were isolated by centrifugation, and portions of the ribosome-rich pellet and ribosome-free supernatant fractions were analyzed by SDS/PAGE and Western blots (Fig. 4a). Full-length S1 and the N and N–R1 fragments were recovered in both fractions, whereas the R1–R4 and R2–R4 fragments remained exclusively in the supernatant. The N and N–R1 fragments also appeared to displace S1 from ribosomes, because less endogenous S1 was observed in the N and N–R1 pellets compared to the vector control (Fig. 4b). Interestingly, when full-length S1 was overexpressed, there was a modest increase in the amount of S1 that copurified with ribosomes, suggesting that wild-type levels of S1 do not saturate ribosomes.
Fig. 4.
Association of S1 proteins with ribosomes in vivo. Ribosomes were isolated by centrifugation from cultures expressing S1 or its fragments. The same amount of total protein from the pellet and supernatant fractions, respectively, was run on SDS gels and detected by Western blot analysis with anti-His antibody (a) or by staining with Coomassie blue (b). The positions of the overexpressed S1 proteins are marked with dots.
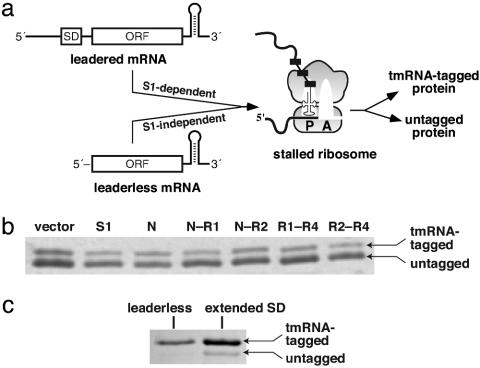
Truncated S1 Variants and tmRNA Function. The effects of S1 overexpression on tmRNA translation and tagging were assayed by using λ-cI-N-trpAt (2), a protein encoded by an mRNA without an in-frame stop codon (Fig. 5a). Ribosomes translate to the 3′ end of this message, and λ-cI-N-trpAt protein (which contains His-6 and FLAG epitopes) is either tagged by the tmRNA system or released in an untagged state (2). The tmRNA variant used for this experiment directs addition of a nondegradable peptide tag (Materials and Methods). Hence, the ratio of tagged to untagged λ-cI-N-trpAt protein provides an assay for the efficiency of tmRNA translation.
Fig. 5.
tmRNA-mediated tagging. (a) Cartoon of tagging of the λ-cI-N-trpAt protein. Leadered λ-cI-N-trpAt mRNA is translated in an S1-dependent manner, whereas the leaderless mRNA is translated in an S1-independent manner. Translation of both types of mRNA leads to a stalled ribosome complex, which is resolved by tmRNA-mediated tagging or ribosomal fall-off, resulting in tagged and untagged λ-cI-N-trpAt protein. (b) Coomassie-stained SDS gel of λ-cI-N-trpAt proteins isolated by nickel affinity after a tagging assay with leadered λ-cI-N-trpAt mRNA in which S1 variants were overexpressed. Tagged and untagged proteins are indicated by arrows. (c) λ-cI-N-trpAt proteins from tagging experiments employing leaderless and extended SD λ-cI-N-trpAt mRNA.
To determine whether overexpression of S1 variants that bind tmRNA reduces tmRNA-mediated tagging, S1 fragments were allowed to accumulate in the cell, expression of the λ-cI-N-trpAt protein was induced, and tagged and untagged λ-cI-N-trpAt were isolated by nickel affinity chromatography and separated by SDS/PAGE (Fig. 5b). When S1 or its N, N–R1, N–R2, R1–R4, or R2–R4 fragments were overexpressed, the amount of tagged λ-cI-N-trpAt varied from 25–35% of the total λ-cI-N-trpAt protein, with only small differences caused by the expression of different proteins. Even overproduction of N or N–R1, which have strong dominant-negative effects on normal translation, did not prevent tmRNA from recognizing stalled ribosomes in vivo or from directing translation of tag sequences. Additionally, tmRNA function was not affected by overexpression of the R1–R4 fragment, which binds tmRNA but not ribosomes.
To test whether tmRNA-mediated tagging is affected by the presence of S1 during translation initiation, we investigated tagging of proteins synthesized from a leaderless λ-cI-N-trpAt mRNA, which does not require S1 for translation (16), and from an extended-SD mRNA, which has an increased dependence on S1 (15). Although total expression of λ-cI-N-trpAt was significantly lower, ≈80% of the λ-cI-N-trpAt was tagged with both the leaderless and extended-SD mRNAs (Fig. 5c). Moreover, overexpression of the S1 fragments caused no change in tagging for the leaderless mRNA (data not shown). These experiments show that tagging of λ-cI-N-trpAt encoded by different mRNAs is independent of the need for S1 to initiate translation from these messages. Inefficient translation of the leaderless and extended-SD mRNAs appears to be the cause of the increased efficiency of tagging seen in these experiments (see Discussion).
Discussion
S1 adopts an elongated structure on the ribosome and in solution, suggesting that its domains are relatively independent (12, 26). Linker regions, which are poorly conserved among different S1 homologues, also appear to separate and structurally isolate individual domains. Together, these observations suggest a “beads on a string” model for S1. Our studies support this model. The isolated N-fragment and R3–R4 and R4 fragments had native CD spectra. The isolated R1 and R2 domains, by contrast, appeared to be largely unfolded. These domains may require interactions with other S1 domains or with RNA to fold stably. Nevertheless, the modular structure of S1 justifies functional studies with individual domains or groups of domains, as previous studies have shown (12, 20, 27).
In accord with previous work, our results support a model in which S1 interacts with ribosomes by using its N-terminal region (12). The N-terminal fragment, which consists of two S1 motifs, binds ribosomes in vitro, probably via interactions with ribosomal proteins (12). We find that the N-terminal fragment copurifies with ribosomes in the cell, displaces endogenous S1 from ribosomes, and inhibits protein synthesis and cell growth. Interestingly, overexpression of full-length S1 was also found to inhibit growth and protein synthesis modestly but to increase the amount of S1 associated with ribosomes. These results would be expected if S1 must bind simultaneously to ribosomes and mRNA to support translation. By this model, too much S1 would saturate ribosomes and compete with docking of S1 molecules already bound to mRNA. S1 associates relatively weakly with ribosomes in comparison with other ribosomal proteins (12). Our finding that too much S1 is detrimental to cells suggests that “weak” ribosome binding may be important for S1 function.
We find that the R1 domain of S1 is required both for strong tmRNA and mRNA binding. These results are consistent with prior studies using poly(U) RNA (12). By itself, however, R1 shows little or no binding and needs other S1 domains for high-affinity RNA interactions. Interestingly, domains N- or C-terminal to R1 are able to fulfill this role. These other domains may help the R1 domain to fold and to contact RNA and/or may make weak RNA contacts, which are strengthened by cooperative contacts mediated by R1. We favor the latter possibility for several reasons. First, S1 motifs are generic RNA-binding modules (17), and the R2, R3, and R4 domains of S1 have been implicated in RNA binding in other systems (12, 19, 27). For example, R4 binds rpsA mRNA (19) and, in our experiments, causes a substantial increase in tmRNA binding (compare R1–R3 with R1–R4 in Fig. 2a). Second, the binding of S1 and its fragments to tmRNA and mRNA is positively cooperative (Hill coefficient ≥2). Binding of the first molecule could make binding of the next more favorable through protein–protein interactions. At RNA-binding concentrations, full-length S1 is monomeric (ref. 12; data not shown), but high local concentrations on RNA could drive formation of protein–protein interactions. Indeed, a fragment similar to R1–R4 dimerizes in an R1-dependent and an RNA-independent fashion at high concentrations at pH 6.0 but not pH 8.0 (27). Thus, dimerization seems to be promoted by an acidic environment, which RNA binding would mimic. Taken together, these results suggest that R1 may function to strengthen RNA contacts made by other S1 domains through cooperative interactions. Other models of cooperativity are possible but seem less likely. For instance, binding of one protein could expose a second, stronger binding site in the RNA (28), but this same event would have to occur on tmRNA and our model mRNA, which differ dramatically in sequence and structure.
Simultaneous binding of S1 to tmRNA and ribosomes does not seem to be required for tmRNA function. If it were required, overexpression of R1–R4, which binds tightly to tmRNA but does not interact with ribosomes, should reduce tmRNA binding to stalled ribosomes and decrease tagging. Moreover, blocking the S1-binding site on ribosomes by overexpression of the N or N–R1 fragments should also reduce tmRNA tagging by this model. Neither result was observed. Overexpression of R1–R4 or the N fragment had no significant effect on tmRNA tagging. If S1 does help tmRNA interact with stalled ribosomes, it must do so by using sites distinct from those used in its normal ribosome interactions. Another model for S1 participation in tmRNA function is protection from cellular ribonucleases (10). If this model is correct, then overexpression of S1 or its fragments must not increase or decrease this protection significantly because tagging efficiency, and presumably tmRNA levels, was essentially unperturbed.
S1 could potentially function to set the ORF on tmRNA and/or to support reinitiation of translation on this ORF (10). The central role of S1 in normal translation appears to be assisting initiation by binding to mRNA sequences upstream of the SD sequence (14). This role seems especially important for mRNAs with weak or very strong SD sequences, but S1 inhibits translation of leaderless mRNAs without SD sequences (12, 15–16). We observed similar levels of tmRNA tagging by using a model mRNA with an extended SD sequence, which has an increased S1 requirement for translation (15), and by using a leaderless model mRNA, whose translation should be inhibited by S1 on the ribosome (16). These results indicate that tmRNA tagging is not affected by the initial S1 requirement for initiation of translation. Because tmRNA does not have a SD sequence, it may behave like a leaderless mRNA and not need S1 for translation initiation. Alternatively, S1 may affect the reinitiation of translation on tmRNA independently of initiation of translation on mRNA. The level of tmRNA-mediated tagging did increase significantly when leaderless or extended-SD mRNAs were used instead of an mRNA with a canonical SD sequence, but much lower translation levels of the former mRNAs probably cause this difference. The tmRNA system can be saturated when efficient translation of defective mRNAs leads to high intracellular concentrations of stalled ribosomes (S. Moore, personal communication), a possible explanation for the lower levels of tagging observed with higher protein expression levels.
Although S1 binds to both mRNA and tmRNA, translation of these RNAs has different S1 requirements. Translation of most mRNAs requires S1 and is inhibited by overexpression of proteins that contain the N-terminal fragment of S1. Translation of the tmRNA ORF, by contrast, does not appear to require S1's presence on the ribosome and is unaffected by overexpression of S1 fragments that inhibit normal translation or that bind tightly to tmRNA. S1, when it is not associated with the ribosome, may play a role in unwinding the tmRNA ORF (7). Nonetheless, at present, we find no compelling evidence that S1 is required for tmRNA tagging or ribosome rescue. S1, which is a highly abundant intracellular protein, probably binds to almost any RNA molecule, and its association with tmRNA may be functionally irrelevant.
Supplementary Material
Acknowledgments
We thank Chris Hayes, Dan Bolon, Randy Burton, Sean Moore, and Tom RajBhandary for advice and materials. This work was supported by National Institutes of Health Grant AI-16892 and a National Institutes of Health postdoctoral fellowship (to K.E.M.).
Abbreviations: GuHCl, guanidine hydrochloride; SD, Shine–Delgarno.
References
- 1.Karzai, A. W., Roche, E. D. & Sauer, R. T. (2000) Nat. Struct. Biol. 7, 449–455. [DOI] [PubMed] [Google Scholar]
- 2.Keiler, K. C., Waller, P. R. H. & Sauer, R. T. (1996) Science 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 3.Abo, T., Inada, T., Ogawa, K. & Aiba, H. (2000) EMBO J. 19, 3762–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roche, E. D. & Sauer, R. T. (1999) EMBO J. 18, 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes, C. S., Bose, B. & Sauer, R. T. (2002) J. Biol. Chem. 277, 33825–33832. [DOI] [PubMed] [Google Scholar]
- 6.Karzai, A. W., Susskind, M. M. & Sauer, R. T. (1999) EMBO J. 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle, M., Gillet, R., Kaur, S., Henne, A., Ramakrishnan, V. & Frank, J. (2003) Science 300, 127–130. [DOI] [PubMed] [Google Scholar]
- 8.Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K. & Inokuchi, H. (1994) Proc. Natl. Acad. Sci. USA 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karzai, A. W. & Sauer, R. T. (2001) Proc. Natl. Acad. Sci. USA 98, 3040–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wower, I. K., Zweib, C. W., Guven, S. A. & Wower, J. (2000) EMBO J. 19, 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordeau, V. & Felden, B. (2002) Biochemie 84, 723–729. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian, A.-P. (1983) Prog. Nucleic Acid Res. Mol. Biol. 28, 101–142. [DOI] [PubMed] [Google Scholar]
- 13.Potapov, A. P. & Subramanian, A. R. (1992) Biochem. Int. 27, 745–753. [PubMed] [Google Scholar]
- 14.Boni, I. V., Isaeva, D. M., Musychenko, M. L. & Tzareva, N. V. (1991) Nucleic Acids Res. 19, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komarova, A. V., Tchufistova, L. S., Supina, E. V. & Boni, I. V. (2002) RNA 8, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moll, I., Grill, S., Gualerzi, C. O. & Bläsi, U. (2002) Mol. Microbiol. 43, 239–246. [DOI] [PubMed] [Google Scholar]
- 17.Bycroft, M., Hubbard, T. J. P., Proctor, M., Freund, S. M. V. & Murzin, A. G. (1997) Cell 88, 234–242. [DOI] [PubMed] [Google Scholar]
- 18.Ringquist, S., Jones, T., Snyder, E. E., Gibson, T., Boni, I. & Gold, L. (1995) Biochemistry 34, 3640–3648. [DOI] [PubMed] [Google Scholar]
- 19.Boni, I. V., Artamonova, V. S. & Dreyfus, M. (2000) J. Bacteriol. 182, 5872–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnier, J., Stöffler, G. & Nishi, K. (1986) J. Biol. Chem. 261, 11866–11871. [PubMed] [Google Scholar]
- 21.Milla, M. E., Brown, B. M. & Sauer, R. T. (1994) Struct. Biol. 1, 518–523. [DOI] [PubMed] [Google Scholar]
- 22.Hayes, C. S. & Sauer, R. T. (2003) Mol. Cell 12, 903–911. [DOI] [PubMed] [Google Scholar]
- 23.Milligan, J. F. & Uhlenbeck, O. C. (1989) Methods Enzymol. 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 24.Roche, E. D. & Sauer, R. T. (2001) J. Biol. Chem. 276, 28509–28515. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, Y., Mah, T.-F., Greenblatt, J. & Friedman, D. I. (2002) J. Mol. Biol. 318, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 26.Sengupta, J., Agrawal, R. K. & Frank, J. (2001) Proc. Natl. Acad. Sci. USA 98, 11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisaglia, M., Laalami, S., Uzan, M. & Bontems, F. (2003) J. Biol. Chem. 278, 15261–15271. [DOI] [PubMed] [Google Scholar]
- 28.Draper, D. E., Pratt, C. W. & Von Hippel, P. H. (1977) Proc. Natl. Acad. Sci. USA 74, 4786–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.