Abstract
Folate metabolism has been associated with cancers via alterations in nucleotide synthesis, DNA methylation, and DNA repair. We hypothesized that genetic variants in methylenetetrahydrofolate reductase (MTHFR), a key enzyme of folate metabolism, would affect the prognosis of prostate cancer. Three haplotype-tagging single-nucleotide polymorphisms (SNPs) across the MTHFR gene region were genotyped in a cohort of 458 patients with clinically localized prostate cancer treated with radical prostatectomy. One SNP, rs9651118, was associated with disease recurrence, and the association persisted after multivariate analyses adjusting for known risk factors. Public dataset analyses suggested that rs9651118 affects MTHFR expression. Quantitative real-time polymerase chain reaction analysis revealed that MTHFR expression is significantly upregulated in prostate tumor tissues when compared with adjacent normal tissues. Furthermore, overexpression of MTHFR correlates with cancer recurrence and death in two independent publicly available prostate cancer datasets. In conclusion, our data provide rationale to further validate the clinical utility of MTHFR rs9651118 as a biomarker for prognosis in prostate cancer.
Keywords: prostate cancer, radical prostatectomy, recurrence, genetic variation, methylenetetrahydrofolate reductase
1. Introduction
Accumulating evidence has suggested the involvement of folate status in modulating the risk of multiple cancers [1]. Folate can donate a methyl group to deoxyuridine monophosphate, converting it to thymidine monophosphate, which is used for DNA replication and repair. Folate deficiency can result in the compromised production of thymidine and misincorporation of uracil during cell division, leading to chromosomal instability. The enzyme methylenetetrahydrofolate reductase (MTHFR) catalyzes the irreversible reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, the predominant form of folate in plasma, which in turn provides the methyl group for the remethylation of homocysteine to convert to S-adenosylmethionine, the universal methyl group donor for numerous cellular methylation reactions including DNA methylation [2]. Low levels of folate may induce DNA hypomethylation and potentially activate oncogene transcription, leading to carcinogenesis [3].
Many studies have linked genetic variants in MTHFR to the risk of various types of cancer [4,5,6], including prostate cancer. A functional MTHFR gene polymorphism, rs1801133 (C677T), codes for an alanine to valine substitution at the folate binding site, and the enzyme activity for heterozygotes and variant homozygotes is reduced to approximately 60% and 30%, respectively, of that seen in wild-type homozygotes [7]. Meta-analyses have revealed that rs1801133 T might confer a protective effect against prostate cancer [4], but the prognostic effects of these variants on disease progression remain undetermined. Thus, we performed a systematic evaluation of common MTHFR gene variants in relation to the biochemical recurrence (BCR) after radical prostatectomy for patients with clinically localized prostate cancer.
2. Results
The clinical features of all participants are summarized in Table S1. After a median follow-up of 54 months, 184 (40.2%) patients had disease recurrence. BCR was significantly related to prostate-specific antigen (PSA) varieties at diagnosis, pathologic Gleason score, stage, surgical margin, and lymph node metastasis (p < 0.001).
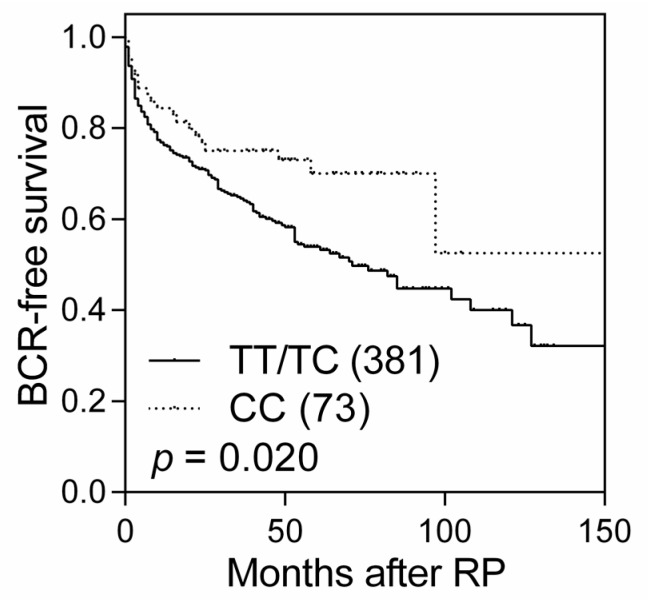
Kaplan-Meier plots and log-rank tests were first used to assess the associations of MTHFR single-nucleotide polymorphisms (SNPs) with BCR in different genetic models (Table 1). A significant association was found between rs9651118 and BCR in the recessive model (p = 0.020). Patients with MTHFR rs9651118 CC genotype exhibited a 42% lower risk of recurrence (hazard ratio (HR) 0.58, 95% confidence interval (CI) 0.37–0.93, p = 0.023; Table 2 and Figure 1) when compared to those with TT and TC genotypes. The association persisted after adjusting for age, PSA at diagnosis, pathologic Gleason score, pathologic stage, surgical margin, and lymph node metastasis (HR 0.51, 95% CI 0.27–0.98, p = 0.044; Table 2).
Table 1.
Association between haplotype tagging SNPs in MTHFR and BCR in clinically localized prostate cancer patients treated with RP.
| SNP ID | Location | Chromosome | Position | Alleles | MAF | p | ||
|---|---|---|---|---|---|---|---|---|
| Additive | Dominant | Recessive | ||||||
| rs3753582 | Intron 1 | 1 | 11805485 | T > G | 0.136 | 0.971 | 0.463 | - |
| rs9651118 | Intron 2 | 1 | 11802157 | T > C | 0.404 | 0.096 | 0.555 | 0.020 |
| rs1801133 | Exon 5 | 1 | 11796321 | C > T | 0.300 | 0.535 | 0.538 | 0.751 |
Abbreviations: SNP, single-nucleotide polymorphism; MTHFR, methylenetetrahydrofolate reductase; BCR, biochemical recurrence; RP, radical prostatectomy; MAF, minor allele frequency. p-values for log-rank test. p < 0.05 is in boldface.
Table 2.
Univariate and multivariate analyses of MTHFR rs9651118 and BCR after RP.
| SNP Genotype | Patients, n | BCR, n (%) | 5-Year BCR-Free Survival, % | HR (95% CI) | p | HR (95% CI) a | p a |
|---|---|---|---|---|---|---|---|
| rs9651118 | |||||||
| TT | 163 | 66 (40.5) | 54.6 | 1.00 | 1.00 | ||
| TC | 218 | 94 (43.1) | 53.3 | 1.03 (0.75–1.41) | 0.846 | 1.00 (0.68–1.47) | 0.997 |
| CC | 73 | 20 (27.4) | 70.1 | 0.59 (0.36–0.98) | 0.041 | 0.51 (0.26–1.02) | 0.057 |
| TC/CC vs. TT | 0.91 (0.67–1.24) | 0.559 | 0.89 (0.61–1.29) | 0.537 | |||
| CC vs. TT/TC | 0.58 (0.37–0.93) | 0.023 | 0.51 (0.27–0.98) | 0.044 | |||
| Trend | 0.84 (0.68–1.04) | 0.100 | 0.81 (0.62–1.07) | 0.132 |
Abbreviations: n, number; HR, hazard ratio; CI, confidence interval; p, p-value; PSA, prostate-specific antigen. a Adjusted by age, PSA at diagnosis, pathologic Gleason score, pathologic stage, surgical margin, and lymph node metastasis. p < 0.05 are in boldface.
Figure 1.
Kaplan-Meier curves comparing BCR-free survival by MTHFR rs9651118 genotypes. Numbers in parentheses indicate the number of patients. RP, radical prostatectomy.
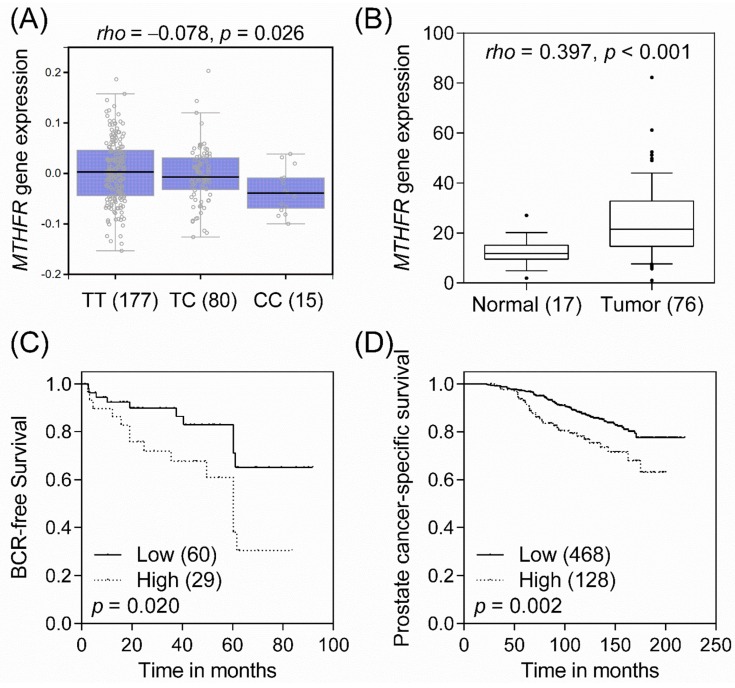
In order to provide biologically plausible support for the observed associations, we initially used Genotype-Tissue Expression (GTEx) datasets to evaluate the association of rs9651118 with MTHFR expression. We observed that the rs9651118 T to C transition was negatively associated with MTHFR expression (Spearman’s rank correlation coefficient rho = −0.078, p = 0.026; Figure 2A), indicating that rs9651118 might be an expression quantitative trait loci for MTHFR. Subsequently, we examined differences in MTHFR gene expression between normal and cancer tissues. MTHFR gene expression was significantly higher in prostate tumor tissues compared to adjacent normal tissues (rho = 0.397, p < 0.001; Figure 2B). We further evaluated the prognostic value of MTHFR expression in prostate cancer progression using publicly available prostate cancer microarray datasets. Patients were dichotomized by MTHFR gene expression using an optimization algorithm for the minimum p value. High expression of MTHFR was associated with shorter BCR-free survival (p = 0.020; Figure 2C) and shorter prostate cancer-specific survival (p = 0.002; Figure 2D) in two independent datasets [8,9]. Taken together, the minor allele C of rs9651118 might decrease MTHFR expression and thus protect patients from cancer recurrence and death.
Figure 2.
Functional analyses of MTHFR rs9651118 with prostate cancer progression: (A) Correlation of rs9651118 genotypes with MTHFR mRNA expression. Boxplot represents MTHFR mRNA expression according to the rs9651118 genotypes (GTEx dataset). There is a trend toward decreased MTHFR mRNA expression in the cells of rs9651118 C carriers; (B) MTHFR mRNA expression in 76 human prostate cancers and 17 adjacent normal tissue specimens, as determined by qRT-PCR, indicates that MTHFR is upregulated in the tumor tissues; (C) Kaplan-Meier analysis of BCR-free survival based on MTHFR mRNA expression in an independent set of prostate cancer microarray data [8]; (D) Kaplan-Meier analysis of prostate cancer-specific survival based on MTHFR mRNA expression in an independent set of public microarray data [9]. Numbers in parentheses indicate the number of patients.
3. Discussion
In the present study, we investigated the clinical relevance of genetic variants in MTHFR for prostate cancer recurrence after radical prostatectomy. We showed that inheritance of a MTHFR rs9651118 CC genotype was associated with increased BCR-free survival. These results might be biologically credible, since cells carrying the C allele tend to have decreased expression of MTHFR. Validation of our initial findings in two independent cohorts further confirms that higher levels of MTHFR are associated with prostate cancer development and poorer patient outcomes.
The SNP rs9651118 is located in the intron 2 of MTHFR, and has low linkage disequilibrium with rs1801133 and rs3753582 (r2 < 0.30). Functional annotation using HaploReg revealed that rs9651118 coincides with regions of open chromatin, which probably correspond to the promoters or enhancers of MTHFR (Table S2). Specifically, the protective allele C is predicted to destroy a putative binding site for E1A binding protein p300 (EP300), which might result in lower MTHFR expression. EP300 is a transcriptional coactivator and has histone acetyltransferase activity favoring transcription via chromatin remodeling [10]. It has been shown that EP300 is upregulated by androgen ablation, and its expression correlates with worse prognosis in prostate cancer [11]. Similar to our results, a study reported that rs9651118 C was associated with reduced risk of lung cancer [12] and better survival of breast cancer [13]. The protective effects of rs9651118 C could be due to the decreased expression of MTHFR, resulting in increased availability of 5,10-methylenetetrahydrofolate and prevention of uracil misincorporation by aiding thymidine biosynthesis. Additionally, Kang and colleagues demonstrated that individuals carrying low enzyme activity of MTHFR polymorphisms showed a reduced level of aberrant hypermethylation in the promoter region of O-6-methylguanine-DNA methyltransferase, a DNA repair gene protecting cells from the cytotoxicity from alkylating agents [14]. Although MTHFR rs1801133 (C677T) has been shown to be related to prostate cancer risk [4], it was not associated with disease recurrence in this study. The reasons for this need further investigation, but our results suggest that different pathways might be involved during prostate cancer development and progression. Thus, further functional characterizations are warranted to validate these genotype-phenotype correlations.
4. Materials and Methods
4.1. Patient Recruitment and Data Collection
A total of 458 patients with clinically localized prostate cancer following radical prostatectomy as initial therapy were recruited from National Taiwan University (Taipei, Taiwan), E-Da (Kaohsiung, Taiwan), Kaohsiung Medical University (Kaohsiung, Taiwan), and Kaohsiung Veterans General hospitals (Kaohsiung, Taiwan), as described previously [15,16,17,18]. Patient baseline characteristics and treatment outcomes were collected from medical records. BCR was defined as PSA values of 0.2 ng/mL or more after radical prostatectomy [19,20]. Written informed consent was obtained from all patients, and the study was approved by the Institutional Review Board of Kaohsiung Medical University Hospital (#KMUHIRB-2013132; 21 January 2014; Kaohsiung, Taiwan) in accordance with the approved procedures.
4.2. Single-Nucleotide Polymorphism (SNP) Selection and Genotyping
Genomic DNA was isolated from peripheral blood using the QIAamp DNA Blood Maxi Kit (Qiagen, Valencia, CA, USA), and was stored until use. We utilized a haplotype-tagging SNP approach to capture common genetic variations in the MTHFR gene region. Haplotype-tagging SNPs were selected from Han Chinese in Beijing and Southern Han Chinese 1000 genomes data [21] using the Haploview Tagger (Broad Institute, Cambridge, MA, USA)with pairwise tagging (minor-allele frequency ≥0.1 and r2 ≥ 0.8) [22]. We identified eight SNPs, which were genotyped using Agena Bioscience MassARRAY iPLEX system at the National Center for Genome Medicine, Taipei, Taiwan. Genotyping quality control was performed, and the concordance rate was 100% among 10 duplicated samples. Any SNP that failed the assay design, deviated from Hardy-Weinberg equilibrium (p < 0.05), or was below a genotyping call rate of 0.8, was removed (number, n = 5). Thus, three SNPs were included for further statistical analyses.
4.3. Human Tissue Complementary DNA (cDNA) Array and TaqMan Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
The expression levels of MTHFR and ACTB (β-actin) were measured using the TissueScan human prostate cancer cDNA array II and III, including 17 normal and 79 prostate cancer samples (OriGene Technologies, Rockville, MD, USA). qRT-PCR was performed using pre-validated TaqMan gene expression assays (Applied Biosystems, Foster city, CA, USA), MTHFR (Hs01114487_m1) and ACTB (Hs01060665_g1), on an Applied Biosystems 7500 according to the manufacturer's instructions. Quantification of each sample was normalized to the expression levels of the housekeeping gene ACTB.
4.4. Statistical Analysis
Kaplan-Meier plots and log-rank tests were used to evaluate the significance of patient characteristics, genotypes, and MTHFR gene expressions in relation to BCR. Crude or adjusted HR and 95% CI for BCR were estimated by univariate or multivariate Cox regression with adjustment of age, PSA at diagnosis, pathologic Gleason score, pathologic stage, surgical margin, and lymph node metastasis, as previously described [16]. The trend of MTHFR gene expression among normal and tumor tissues was analyzed by Spearman correlation. Statistical analysis was performed using the Statistical Package for the Social Sciences version 22.0.0 (IBM, Armonk, NY, USA), and two-tailed p < 0.05 was considered statistically significant.
4.5. Bioinformatics Analysis
HaploReg v4.1 [23] was used to identify the regulatory potential of the SNP. GTEx data were used to correlate the relationships between rs9651118 and MTHFR gene expression in transformed human fibroblasts [24]. Publicly available transcriptomic datasets [8,9] were used to evaluate the association of MTHFR expression and prostate cancer progression.
5. Conclusions
To the best of our knowledge, this is the first report to systematically study the prognostic impact of MTHFR genetic variants in patients with prostate cancer. We chose the BCR end point based on serum PSA due to its clinical relevance. In a population of patients treated with radical prostatectomy and did not receive additional therapies before disease progression, the median actuarial time from BCR to metastasis was five years. Once metastatic prostate cancer develops, the median time to prostate cancer-specific mortality was less than five years [25]. A rising PSA is often the first indication of the development of progressive disease and triggers a change in therapy. However, considering the limited number of patients and the homogeneous Taiwanese population in this study, these findings should be interpreted with caution and need to be validated in larger independent cohorts. Nevertheless, we performed multivariate analysis to reduce the false-positive findings, which revealed that our results remained significant even after adjusting for known clinical outcome predictors. Moreover, rs9651118 affects MTHFR expression and the expression is significantly correlated with prostate cancer development and progression in independent studies. Our study has provided further support for the prognostic value of MTHFR genetic variants, and has revealed the importance of folate metabolism in prostate cancer recurrence.
Acknowledgments
This work was supported by the Ministry of Science and Technology of Taiwan (grant number: 102-2628-B-039-005-MY3, 103-2314-B-037-060, 104-2314-B-650-006, 104-2314-B-037-052-MY3, and 105-2314-B-650-003-MY3), the Kaohsiung Medical University Hospital (grant number: KMUH103-3R43), the E-Da Hospital (grant number: EDPJ104059 and EDPJ105054), and the China Medical University (grant number: CMU105-S-42). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Chao-Shih Chen for data analysis and the National Center for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support. The results published here are based in part on data generated by the GTEx and HaploReg projects.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/12/1996/s1.
Author Contributions
Victor C. Lin, Chao-Yuan Huang, Chia-Cheng Yu, Shu-Pin Huang, and Bo-Ying Bao conceived and designed the experiments. Te-Ling Lu, Hsin-Ling Yin, Sheau-Fang Yang, Yung-Chin Lee, Chia-Chu Liu, and Ta-Yuan Chang performed the experiments and analyzed the data. All authors wrote, reviewed, and approved submission of the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kim Y.I. Folate and carcinogenesis: Evidence, mechanisms, and implications. J. Nutr. Biochem. 1999;10:66–88. doi: 10.1016/S0955-2863(98)00074-6. [DOI] [PubMed] [Google Scholar]
- 2.Duthie S.J., Narayanan S., Brand G.M., Pirie L., Grant G. Impact of folate deficiency on DNA stability. J. Nutr. 2002;132:2444S–2449S. doi: 10.1093/jn/132.8.2444S. [DOI] [PubMed] [Google Scholar]
- 3.Fang J.Y., Zhu S.S., Xiao S.D., Jiang S.J., Shi Y., Chen X.Y., Zhou X.M., Qian L.F. Studies on the hypomethylation of c-myc, c-ha-ras oncogenes and histopathological changes in human gastric carcinoma. J. Gastroenterol. Hepatol. 1996;11:1079–1082. doi: 10.1111/j.1440-1746.1996.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 4.Guo S., Jiang X., Chen X., Chen L., Li X., Jia Y. The protective effect of methylenetetrahydrofolate reductase C677T polymorphism against prostate cancer risk: Evidence from 23 case-control studies. Gene. 2015;565:90–95. doi: 10.1016/j.gene.2015.03.067. [DOI] [PubMed] [Google Scholar]
- 5.Kumar P., Yadav U., Rai V. Methylenetetrahydrofolate reductase gene C677T polymorphism and breast cancer risk: Evidence for genetic susceptibility. Meta Gene. 2015;6:72–84. doi: 10.1016/j.mgene.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Yang L.J., Deng M.Z., Luo Y.Y., Wu S., Xiong L., Wang D., Liu Y., Liu H. Mthfr C677T and A1298C polymorphisms and risk of lung cancer: A comprehensive evaluation. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15027615. [DOI] [PubMed] [Google Scholar]
- 7.Frosst P., Blom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G., Boers G.J., den Heijer M., Kluijtmans L.A., van den Heuvel L.P., et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 8.Gulzar Z.G., McKenney J.K., Brooks J.D. Increased expression of nusap in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa T., Kollmeyer T.M., Morlan B.W., Anderson S.K., Bergstralh E.J., Davis B.J., Asmann Y.W., Klee G.G., Ballman K.V., Jenkins R.B. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS ONE. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh A.K., Varga J. The transcriptional coactivator and acetyltransferase p300 in fibroblast biology and fibrosis. J. Cell. Physiol. 2007;213:663–671. doi: 10.1002/jcp.21162. [DOI] [PubMed] [Google Scholar]
- 11.Debes J.D., Comuzzi B., Schmidt L.J., Dehm S.M., Culig Z., Tindall D.J. P300 regulates androgen receptor-independent expression of prostate-specific antigen in prostate cancer cells treated chronically with interleukin-6. Cancer Res. 2005;65:5965–5973. doi: 10.1158/0008-5472.CAN-04-2837. [DOI] [PubMed] [Google Scholar]
- 12.Swartz M.D., Peterson C.B., Lupo P.J., Wu X., Forman M.R., Spitz M.R., Hernandez L.M., Vannucci M., Shete S. Investigating multiple candidate genes and nutrients in the folate metabolism pathway to detect genetic and nutritional risk factors for lung cancer. PLoS ONE. 2013;8:e53475. doi: 10.1371/journal.pone.0053475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q., Jiang K., Li Q., Ji Y.J., Chen W.L., Xue X.H. Polymorphisms in the mthfr gene are associated with breast cancer risk and prognosis in a Chinese population. Tumor Biol. 2015;36:3757–3762. doi: 10.1007/s13277-014-3016-4. [DOI] [PubMed] [Google Scholar]
- 14.Kang S., Kim J.W., Kang G.H., Park N.H., Song Y.S., Kang S.B., Lee H.P. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol. Oncol. 2005;96:173–180. doi: 10.1016/j.ygyno.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 15.Bao B.Y., Pao J.B., Lin V.C., Huang C.N., Chang T.Y., Lan Y.H., Lu T.L., Lee H.Z., Chen L.M., Ting W.C., et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin. Chim. Acta. 2010;411:1232–1237. doi: 10.1016/j.cca.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 16.Huang S.P., Huang L.C., Ting W.C., Chen L.M., Chang T.Y., Lu T.L., Lan Y.H., Liu C.C., Yang W.H., Lee H.Z., et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol. Biomark. Prev. 2009;18:3068–3074. doi: 10.1158/1055-9965.EPI-09-0665. [DOI] [PubMed] [Google Scholar]
- 17.Huang S.P., Lan Y.H., Lu T.L., Pao J.B., Chang T.Y., Lee H.Z., Yang W.H., Hsieh C.J., Chen L.M., Huang L.C., et al. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011;107:486–492. doi: 10.1111/j.1464-410X.2010.09512.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu C.C., Lin V.C., Huang C.Y., Liu C.C., Wang J.S., Wu T.T., Pu Y.S., Huang C.H., Huang C.N., Huang S.P., et al. Prognostic significance of cyclin d1 polymorphisms on prostate-specific antigen recurrence after radical prostatectomy. Ann. Surg. Oncol. 2013;20:S492–S499. doi: 10.1245/s10434-013-2869-x. [DOI] [PubMed] [Google Scholar]
- 19.Freedland S.J., Sutter M.E., Dorey F., Aronson W.J. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy: Prostate-specific antigen. Urology. 2003;61:365–369. doi: 10.1016/S0090-4295(02)02268-9. [DOI] [PubMed] [Google Scholar]
- 20.Huang S.P., Levesque E., Guillemette C., Yu C.C., Huang C.Y., Lin V.C., Chung I.C., Chen L.C., Laverdiere I., Lacombe L., et al. Genetic variants in micrornas and microrna target sites predict biochemical recurrence after radical prostatectomy in localized prostate cancer. Int. J. Cancer. 2014;135:2661–2667. doi: 10.1002/ijc.28904. [DOI] [PubMed] [Google Scholar]
- 21.Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of ld and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Ward L.D., Kellis M. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pound C.R., Partin A.W., Eisenberger M.A., Chan D.W., Pearson J.D., Walsh P.C. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.